Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition
Abstract
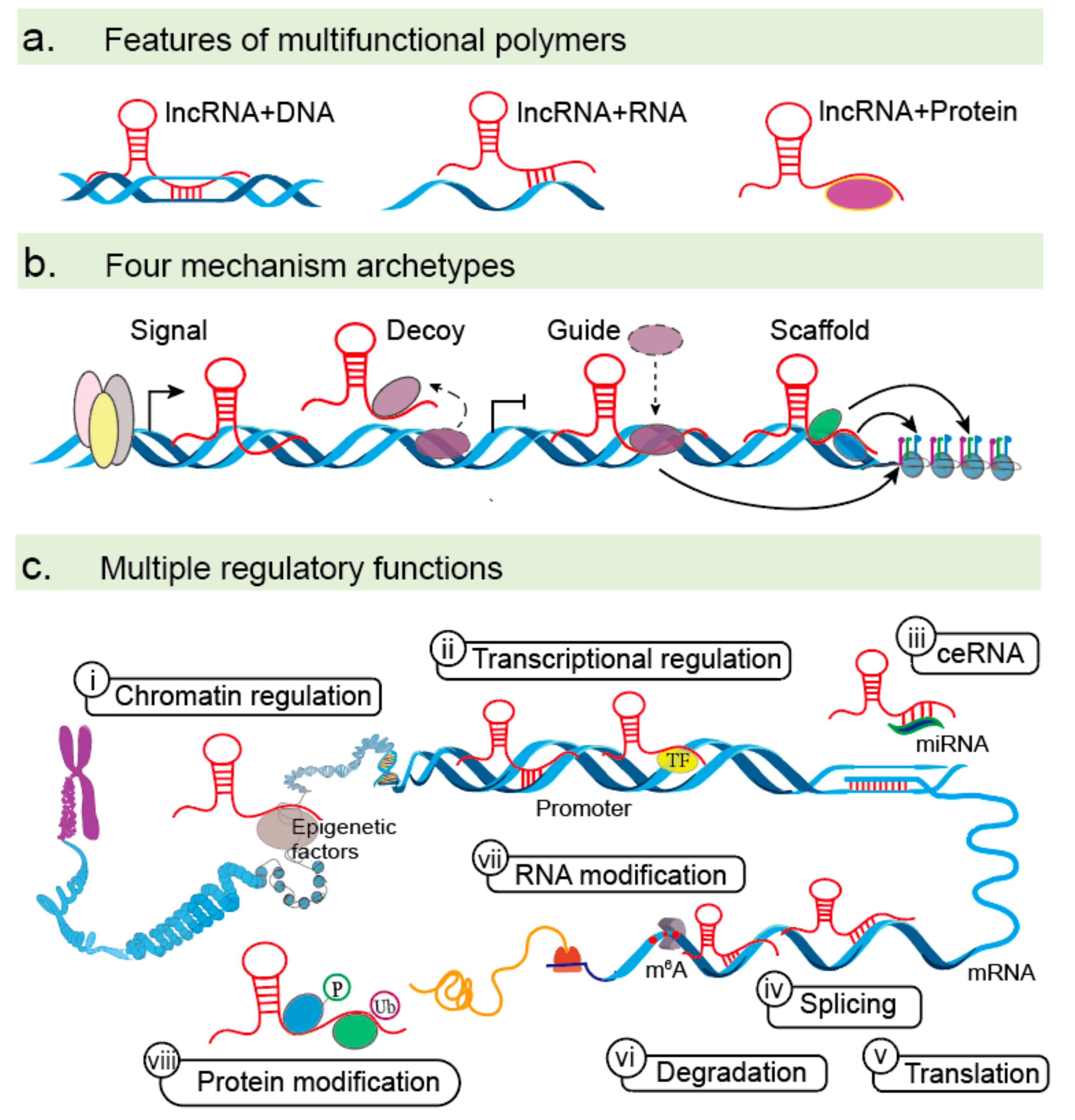
:1. Introduction
2. LncRNAs as Emerging EMT Regulators
3. The Molecular Mechanisms of lncRNAs in EMTs
3.1. LncRNAs Acting on EMTs at the Epigenetic and Transcriptional Levels
3.1.1. Chromatin Modification and Regulation
3.1.2. Transcriptional Activators
3.1.3. Transcriptional Repressors
3.2. LncRNAs in EMTs at the Post-Transcriptional Level
3.2.1. Interactions with miRNAs
3.2.2. Regulation of mRNA Stability and Splicing
3.2.3. Protein and mRNA Modifications
3.2.3.1. Protein Modification
3.2.3.2. mRNA Modification
4. EMT Pathways Control by lncRNAs
5. LncRNAs as Biomarkers and Therapeutic Targets
6. Conclusions and Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Ransohoff, J.D.; Wei, Y.; Khavari, P.A. The functions and unique features of long intergenic non-coding RNA. Nat. Rev. Mol. Cell Biol. 2018, 19, 143–157. [Google Scholar] [CrossRef] [PubMed]
- Morris, K.V.; Mattick, J.S. The rise of regulatory RNA. Nat. Rev. Genet. 2014, 15, 423–437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chew, C.L.; Conos, S.A.; Unal, B.; Tergaonkar, V. Noncoding RNAs: Master Regulators of Inflammatory Signaling. Trends Mol. Med. 2018, 24, 66–84. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Egranov, S.D.; Yang, L.; Lin, C. Molecular mechanisms of long noncoding RNAs-mediated cancer metastasis. Genes Chromosomes Cancer 2019, 58, 200–207. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Luo, J.; Luan, S.; He, C.; Li, Z. Long non-coding RNAs involved in cancer metabolic reprogramming. Cell. Mol. Life Sci. 2019, 76, 495–504. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Wang, Y.Y.; Xin, H.W.; Wang, L.; Arfuso, F.; Dharmarajan, A.; Kumar, A.P.; Wang, H.; Tang, F.R.; Warrier, S.; et al. The expanding roles of long non-coding RNAs in the regulation of cancer stem cells. Int. J. Biochem. Cell Biol. 2019, 108, 17–20. [Google Scholar] [CrossRef] [PubMed]
- Kalluri, R.; Weinberg, R.A. The basics of epithelial-mesenchymal transition. J. Clin. Investig. 2009, 119, 1420–1428. [Google Scholar] [CrossRef] [Green Version]
- Nieto, M.A.; Huang, R.Y.; Jackson, R.A.; Thiery, J.P. EMT: 2016. Cell 2016, 166, 21–45. [Google Scholar] [CrossRef] [Green Version]
- Pastushenko, I.; Brisebarre, A.; Sifrim, A.; Fioramonti, M.; Revenco, T.; Boumahdi, S.; Van Keymeulen, A.; Brown, D.; Moers, V.; Lemaire, S.; et al. Identification of the tumour transition states occurring during EMT. Nature 2018, 556, 463–468. [Google Scholar] [CrossRef]
- Clemson, C.M.; Hutchinson, J.N.; Sara, S.A.; Ensminger, A.W.; Fox, A.H.; Chess, A.; Lawrence, J.B. An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol. Cell 2009, 33, 717–726. [Google Scholar] [CrossRef]
- Battistelli, C.; Sabarese, G.; Santangelo, L.; Montaldo, C.; Gonzalez, F.J.; Tripodi, M.; Cicchini, C. The lncRNA HOTAIR transcription is controlled by HNF4alpha-induced chromatin topology modulation. Cell Death Differ. 2018, 26, 890–901. [Google Scholar] [CrossRef] [PubMed]
- Battistelli, C.; Cicchini, C.; Santangelo, L.; Tramontano, A.; Grassi, L.; Gonzalez, F.J.; de Nonno, V.; Grassi, G.; Amicone, L.; Tripodi, M. The Snail repressor recruits EZH2 to specific genomic sites through the enrollment of the lncRNA HOTAIR in epithelial-to-mesenchymal transition. Oncogene 2017, 36, 942–955. [Google Scholar] [CrossRef] [PubMed]
- Hirata, H.; Hinoda, Y.; Shahryari, V.; Deng, G.; Nakajima, K.; Tabatabai, Z.L.; Ishii, N.; Dahiya, R. Long Noncoding RNA MALAT1 Promotes Aggressive Renal Cell Carcinoma through Ezh2 and Interacts with miR-205. Cancer Res. 2015, 75, 1322–1331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wu, C.; Zhang, C.; Li, Z.; Zhu, T.; Chen, J.; Ren, Y.; Wang, X.; Zhang, L.; Zhou, X. TGF-beta-induced STAT3 overexpression promotes human head and neck squamous cell carcinoma invasion and metastasis through malat1/miR-30a interactions. Cancer Lett. 2018, 436, 52–62. [Google Scholar] [CrossRef] [PubMed]
- Terashima, M.; Ishimura, A.; Wanna-Udom, S.; Suzuki, T. MEG8 long noncoding RNA contributes to epigenetic progression of the epithelial-mesenchymal transition of lung and pancreatic cancer cells. J. Biol. Chem. 2018, 293, 18016–18030. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Q.; Deng, F.; Qin, Y.; Zhao, Z.; Wu, Z.; Xing, Z.; Ji, A.; Wang, Q.J. Long non-coding RNA regulation of epithelial-mesenchymal transition in cancer metastasis. Cell Death Dis. 2016, 7, e2254. [Google Scholar] [CrossRef]
- Tang, Y.; Cheung, B.B.; Atmadibrata, B.; Marshall, G.M.; Dinger, M.E.; Liu, P.Y.; Liu, T. The regulatory role of long noncoding RNAs in cancer. Cancer Lett. 2017, 391, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Gugnoni, M.; Ciarrocchi, A. Long Noncoding RNA and Epithelial Mesenchymal Transition in Cancer. Int. J. Mol. Sci. 2019, 20, 1924. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Yang, F.; Jia, L.-T.; Yang, A.-G. Missing Links in Epithelial-Mesenchymal Transition: Long Non-Coding RNAs Enter the Arena. Cell. Physiol. Biochem. 2017, 44, 1665–1680. [Google Scholar] [CrossRef] [Green Version]
- Guttman, M.; Rinn, J.L. Modular regulatory principles of large non-coding RNAs. Nature 2012, 482, 339–346. [Google Scholar] [CrossRef] [Green Version]
- Marchese, F.P.; Raimondi, I.; Huarte, M. The multidimensional mechanisms of long noncoding RNA function. Genome Biol. 2017, 18, 206. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.J.; Gao, C.; Ma, Z.; Cong, R.; Zhang, Q.; Guo, A.Y. lncRInter: A database of experimentally validated long non-coding RNA interaction. J. Genet. Genom./Yi Chuan Xue Bao 2017, 44, 265–268. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.C.; Chang, H.Y. Molecular mechanisms of long noncoding RNAs. Mol. Cell 2011, 43, 904–914. [Google Scholar] [CrossRef] [PubMed]
- Bohmdorfer, G.; Wierzbicki, A.T. Control of Chromatin Structure by Long Noncoding RNA. Trends Cell Biol. 2015, 25, 623–632. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.Q.; Jiang, D.M.; Hu, S.S.; Zhao, L.; Wang, L.; Yang, M.H.; Ai, M.L.; Jiang, H.J.; Han, Y.; Ding, Y.Q.; et al. SATB2-AS1 suppresses colorectal carcinoma aggressiveness by inhibiting SATB2-dependent Snail transcription and epithelial-mesenchymal transition. Cancer Res. 2019, 79, 3542–3556. [Google Scholar] [CrossRef] [PubMed]
- Li, W.; Zhang, Z.; Liu, X.; Cheng, X.; Zhang, Y.; Han, X.; Zhang, Y.; Liu, S.; Yang, J.; Xu, B.; et al. The FOXN3-NEAT1-SIN3A repressor complex promotes progression of hormonally responsive breast cancer. J. Clin. Investig. 2017, 127, 3421–3440. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.H.; Yang, F.; Wang, F.; Ma, J.Z.; Guo, Y.J.; Tao, Q.F.; Liu, F.; Pan, W.; Wang, T.T.; Zhou, C.C.; et al. A long noncoding RNA activated by TGF-beta promotes the invasion-metastasis cascade in hepatocellular carcinoma. Cancer Cell 2014, 25, 666–681. [Google Scholar] [CrossRef]
- Wu, Y.; Yang, X.; Chen, Z.; Tian, L.; Jiang, G.; Chen, F.; Li, J.; An, P.; Lu, L.; Luo, N.; et al. m(6)A-induced lncRNA RP11 triggers the dissemination of colorectal cancer cells via upregulation of Zeb1. Mol. Cancer 2019, 18, 87. [Google Scholar] [CrossRef]
- Hu, Q.; Li, C.; Wang, S.; Li, Y.; Wen, B.; Zhang, Y.; Liang, K.; Yao, J.; Ye, Y.; Hsiao, H.; et al. LncRNAs-directed PTEN enzymatic switch governs epithelial-mesenchymal transition. Cell Res. 2019, 29, 286–304. [Google Scholar] [CrossRef]
- Cebria-Costa, J.P.; Millanes-Romero, A.; de Herreros, A.G.; Peiro, S. The Epithelial-to-Mesenchymal Transition (EMT), a Particular Case. Mol. Cell. Oncol. 2014, 1, e960770. [Google Scholar] [CrossRef]
- Yu, C.Y.; Kuo, H.C. The Trans-Spliced Long Noncoding RNA tsRMST Impedes Human Embryonic Stem Cell Differentiation Through WNT5A-Mediated Inhibition of the Epithelial-to-Mesenchymal Transition. Stem Cells 2016, 34, 2052–2062. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.P.; Duan, F.F.; Zhao, Y.T.; Gu, K.L.; Liao, L.Q.; Su, H.B.; Hao, J.; Zhang, K.; Yang, N.; Wang, Y. A TRIM71 binding long noncoding RNA Trincr1 represses FGF/ERK signaling in embryonic stem cells. Nat. Commun. 2019, 10, 1368. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Li, Y.; Xu, Q.; Yao, W.; Wu, Q.; Yuan, J.; Yan, W.; Xu, T.; Ji, X.; Ni, C. Long non-coding RNA-ATB promotes EMT during silica-induced pulmonary fibrosis by competitively binding miR-200c. Biochim. Biophys. Acta. Mol. Basis Dis. 2018, 1864, 420–431. [Google Scholar] [CrossRef] [PubMed]
- Yan, W.; Wu, Q.; Yao, W.; Li, Y.; Liu, Y.; Yuan, J.; Han, R.; Yang, J.; Ji, X.; Ni, C. MiR-503 modulates epithelial-mesenchymal transition in silica-induced pulmonary fibrosis by targeting PI3K p85 and is sponged by lncRNA MALAT1. Sci. Rep. 2017, 7, 11313. [Google Scholar] [CrossRef] [PubMed]
- Wang, S.; Zuo, H.; Jin, J.; Lv, W.; Xu, Z.; Fan, Y.; Zhang, J.; Zuo, B. Long noncoding RNA Neat1 modulates myogenesis by recruiting Ezh2. Cell Death Dis. 2019, 10, 505. [Google Scholar] [CrossRef]
- Hou, P.; Zhao, Y.; Li, Z.; Yao, R.; Ma, M.; Gao, Y.; Zhao, L.; Zhang, Y.; Huang, B.; Lu, J. LincRNA-ROR induces epithelial-to-mesenchymal transition and contributes to breast cancer tumorigenesis and metastasis. Cell Death Dis. 2014, 5, e1287. [Google Scholar] [CrossRef]
- Chen, Y.; Peng, Y.; Xu, Z.; Ge, B.; Xiang, X.; Zhang, T.; Gao, L.; Shi, H.; Wang, C.; Huang, J. LncROR Promotes Bladder Cancer Cell Proliferation, Migration, and Epithelial-Mesenchymal Transition. Cell. Physiol. Biochem. 2017, 41, 2399–2410. [Google Scholar] [CrossRef]
- Li, L.; Gu, M.; You, B.; Shi, S.; Shan, Y.; Bao, L.; You, Y. Long non-coding RNA ROR promotes proliferation, migration and chemoresistance of nasopharyngeal carcinoma. Cancer Sci. 2016, 107, 1215–1222. [Google Scholar] [CrossRef]
- Terashima, M.; Tange, S.; Ishimura, A.; Suzuki, T. MEG3 Long Noncoding RNA Contributes to the Epigenetic Regulation of Epithelial-Mesenchymal Transition in Lung Cancer Cell Lines. J. Biol. Chem. 2017, 292, 82–99. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.B.; Wei, H.; Wang, J.S.; Li, L.; Chen, A.Y.; Li, Z.G. Down-regulated expression of LINC00518 prevents epithelial cell growth and metastasis in breast cancer through the inhibition of CDX2 methylation and the Wnt signaling pathway. Biochim. Biophys. Acta. Mol. Basis Dis. 2019, 1865, 708–723. [Google Scholar] [CrossRef]
- Li, Z.; Hou, P.; Fan, D.; Dong, M.; Ma, M.; Li, H.; Yao, R.; Li, Y.; Wang, G.; Geng, P.; et al. The degradation of EZH2 mediated by lncRNA ANCR attenuated the invasion and metastasis of breast cancer. Cell Death Differ. 2016, 24, 59–71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Peng, L.; Jiang, B.; Yuan, X.; Qiu, Y.; Peng, J.; Huang, Y.; Zhang, C.; Zhang, Y.; Lin, Z.; Li, J.; et al. Super-Enhancer-Associated Long Noncoding RNA HCCL5 Is Activated by ZEB1 and Promotes the Malignancy of Hepatocellular Carcinoma. Cancer Res. 2019, 79, 572–584. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Ma, C.; Zhang, L.; Li, N.; Zhang, X.; He, J.; He, R.; Shao, M.; Wang, J.; Kang, L.; et al. LncRNAAC132217.4, a KLF8-regulated long non-coding RNA, facilitates oral squamous cell carcinoma metastasis by upregulating IGF2 expression. Cancer Lett. 2017, 407, 45–56. [Google Scholar] [CrossRef] [PubMed]
- Sakai, S.; Ohhata, T.; Kitagawa, K.; Uchida, C.; Aoshima, T.; Niida, H.; Suzuki, T.; Inoue, Y.; Miyazawa, K.; Kitagawa, M. Long noncoding RNA ELIT-1 acts as a Smad3 cofactor to facilitate TGF-beta/Smad signaling and promote epithelial-mesenchymal transition. Cancer Res. 2019, 79, 2821–2838. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M.; Bucci, G.; Rizzotto, D.; Bordo, D.; Marzi, M.J.; Puppo, M.; Flinois, A.; Spadaro, D.; Citi, S.; Emionite, L.; et al. LncRNA EPR controls epithelial proliferation by coordinating Cdkn1a transcription and mRNA decay response to TGF-beta. Nat. Commun. 2019, 10, 1969. [Google Scholar] [CrossRef] [PubMed]
- Deng, S.J.; Chen, H.Y.; Ye, Z.; Deng, S.C.; Zhu, S.; Zeng, Z.; He, C.; Liu, M.L.; Huang, K.; Zhong, J.X.; et al. Hypoxia-induced LncRNA-BX111 promotes metastasis and progression of pancreatic cancer through regulating ZEB1 transcription. Oncogene 2018, 37, 5811–5828. [Google Scholar] [CrossRef]
- Yan, X.; Zhang, D.; Wu, W.; Wu, S.; Qian, J.; Hao, Y.; Yan, F.; Zhu, P.; Wu, J.; Huang, G.; et al. Mesenchymal Stem Cells Promote Hepatocarcinogenesis via lncRNA-MUF Interaction with ANXA2 and miR-34a. Cancer Res. 2017, 77, 6704–6716. [Google Scholar] [CrossRef] [PubMed]
- Jiang, H.; Li, T.; Qu, Y.; Wang, X.; Li, B.; Song, J.; Sun, X.; Tang, Y.; Wan, J.; Yu, Y.; et al. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor Slug and promotes colon cancer progression. Cancer Lett. 2018, 425, 78–87. [Google Scholar] [CrossRef]
- Lu, Z.; Li, Y.; Che, Y.; Huang, J.; Sun, S.; Mao, S.; Lei, Y.; Li, N.; Sun, N.; He, J. The TGFbeta-induced lncRNA TBILA promotes non-small cell lung cancer progression in vitro and in vivo via cis-regulating HGAL and activating S100A7/JAB1 signaling. Cancer Lett. 2018, 432, 156–168. [Google Scholar] [CrossRef]
- Luo, L.; Tang, H.; Ling, L.; Li, N.; Jia, X.; Zhang, Z.; Wang, X.; Shi, L.; Yin, J.; Qiu, N.; et al. LINC01638 lncRNA activates MTDH-Twist1 signaling by preventing SPOP-mediated c-Myc degradation in triple-negative breast cancer. Oncogene 2018, 37, 6166–6179. [Google Scholar] [CrossRef]
- Wang, X.; Yu, H.; Sun, W.; Kong, J.; Zhang, L.; Tang, J.; Wang, J.; Xu, E.; Lai, M.; Zhang, H. The long non-coding RNA CYTOR drives colorectal cancer progression by interacting with NCL and Sam68. Mol. Cancer 2018, 17, 110. [Google Scholar] [CrossRef] [PubMed]
- Cai, W.; Wu, B.; Li, Z.; He, P.; Wang, B.; Cai, A.; Zhang, X. LncRNA NBR2 inhibits epithelial-mesenchymal transition by regulating Notch1 signaling in osteosarcoma cells. J. Cell. Biochem. 2018, 120, 2015–2027. [Google Scholar] [CrossRef]
- Liang, W.C.; Ren, J.L.; Wong, C.W.; Chan, S.O.; Waye, M.M.; Fu, W.M.; Zhang, J.F. LncRNA-NEF antagonized epithelial to mesenchymal transition and cancer metastasis via cis-regulating FOXA2 and inactivating Wnt/beta-catenin signaling. Oncogene 2018, 37, 1445–1456. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Han, Z.; Sun, Z.; Wang, Y.; Zheng, M.; Song, C. LncRNA SLCO4A1-AS1 facilitates growth and metastasis of colorectal cancer through beta-catenin-dependent Wnt pathway. J. Exp. Clin. Cancer Res. 2018, 37, 222. [Google Scholar] [CrossRef]
- Kong, J.; Sun, W.; Li, C.; Wan, L.; Wang, S.; Wu, Y.; Xu, E.; Zhang, H.; Lai, M. Long non-coding RNA LINC01133 inhibits epithelial-mesenchymal transition and metastasis in colorectal cancer by interacting with SRSF6. Cancer Lett. 2016, 380, 476–484. [Google Scholar] [CrossRef] [PubMed]
- Han, Q.; Xu, L.; Lin, W.; Yao, X.; Jiang, M.; Zhou, R.; Sun, X.; Zhao, L. Long noncoding RNA CRCMSL suppresses tumor invasive and metastasis in colorectal carcinoma through nucleocytoplasmic shuttling of HMGB2. Oncogene 2018, 38, 3019–3032. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wei, Z.; Wu, K.; Dai, W.; Zhang, C.; Peng, J.; He, Y. Long noncoding RNA B3GALT5-AS1 suppresses colon cancer liver metastasis via repressing microRNA-203. Aging 2018, 10, 3662–3682. [Google Scholar] [CrossRef]
- Li, G.Y.; Wang, W.; Sun, J.Y.; Xin, B.; Zhang, X.; Wang, T.; Zhang, Q.F.; Yao, L.B.; Han, H.; Fan, D.M.; et al. Long non-coding RNAs AC026904.1 and UCA1: A “one-two punch” for TGF-beta-induced SNAI2 activation and epithelial-mesenchymal transition in breast cancer. Theranostics 2018, 8, 2846–2861. [Google Scholar] [CrossRef]
- Ge, X.; Li, G.Y.; Jiang, L.; Jia, L.; Zhang, Z.; Li, X.; Wang, R.; Zhou, M.; Zhou, Y.; Zeng, Z.; et al. Long noncoding RNA CAR10 promotes lung adenocarcinoma metastasis via miR-203/30/SNAI axis. Oncogene 2019, 38, 3061–3076. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Yuan, J.H.; Huang, J.F.; Yang, F.; Wang, T.T.; Ma, J.Z.; Zhang, L.; Zhou, C.C.; Wang, F.; Yu, J.; et al. Long noncoding RNA FTX inhibits hepatocellular carcinoma proliferation and metastasis by binding MCM2 and miR-374a. Oncogene 2016, 35, 5422–5434. [Google Scholar] [CrossRef]
- Yan, T.T.; Ren, L.L.; Shen, C.Q.; Wang, Z.H.; Yu, Y.N.; Liang, Q.; Tang, J.Y.; Chen, Y.X.; Sun, D.F.; Zgodzinski, W.; et al. miR-508 Defines the Stem-like/Mesenchymal Subtype in Colorectal Cancer. Cancer Res. 2018, 78, 1751–1765. [Google Scholar] [CrossRef] [Green Version]
- Li, T.; Xie, J.; Shen, C.; Cheng, D.; Shi, Y.; Wu, Z.; Deng, X.; Chen, H.; Shen, B.; Peng, C.; et al. Amplification of Long Noncoding RNA ZFAS1 Promotes Metastasis in Hepatocellular Carcinoma. Cancer Res. 2015, 75, 3181–3191. [Google Scholar] [CrossRef]
- Marchese, F.P.; Huarte, M. Long non-coding RNAs and chromatin modifiers: Their place in the epigenetic code. Epigenetics 2014, 9, 21–26. [Google Scholar] [CrossRef] [PubMed]
- Rinn, J.L.; Kertesz, M.; Wang, J.K.; Squazzo, S.L.; Xu, X.; Brugmann, S.A.; Goodnough, L.H.; Helms, J.A.; Farnham, P.J.; Segal, E.; et al. Functional demarcation of active and silent chromatin domains in human HOX loci by noncoding RNAs. Cell 2007, 129, 1311–1323. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.W.; Sun, M.; Xia, R.; Zhang, E.B.; Liu, X.H.; Zhang, Z.H.; Xu, T.P.; De, W.; Liu, B.R.; Wang, Z.X. LincHOTAIR epigenetically silences miR34a by binding to PRC2 to promote the epithelial-to-mesenchymal transition in human gastric cancer. Cell Death Dis. 2015, 6, e1802. [Google Scholar] [CrossRef] [PubMed]
- Margueron, R.; Reinberg, D. The Polycomb complex PRC2 and its mark in life. Nature 2011, 469, 343–349. [Google Scholar] [CrossRef] [Green Version]
- Di Croce, L.; Helin, K. Transcriptional regulation by Polycomb group proteins. Nat. Struct. Mol. Biol. 2013, 20, 1147–1155. [Google Scholar] [CrossRef]
- Wu, Q.; Xiang, S.; Ma, J.; Hui, P.; Wang, T.; Meng, W.; Shi, M.; Wang, Y. Long non-coding RNA CASC15 regulates gastric cancer cell proliferation, migration and epithelial mesenchymal transition by targeting CDKN1A and ZEB1. Mol. Oncol. 2018, 12, 799–813. [Google Scholar] [CrossRef]
- Fang, C.; He, W.; Xu, T.; Dai, J.; Xu, L.; Sun, F. Upregulation of lncRNA DGCR5 correlates with better prognosis and inhibits bladder cancer progression via transcriptionally facilitating P21 expression. J. Cell. Physiol. 2019, 234, 6254–6262. [Google Scholar] [CrossRef]
- Neumann, P.; Jae, N.; Knau, A.; Glaser, S.F.; Fouani, Y.; Rossbach, O.; Kruger, M.; John, D.; Bindereif, A.; Grote, P.; et al. The lncRNA GATA6-AS epigenetically regulates endothelial gene expression via interaction with LOXL2. Nat. Commun. 2018, 9, 237. [Google Scholar] [CrossRef]
- Zhao, J.; Ohsumi, T.K.; Kung, J.T.; Ogawa, Y.; Grau, D.J.; Sarma, K.; Song, J.J.; Kingston, R.E.; Borowsky, M.; Lee, J.T. Genome-wide identification of polycomb-associated RNAs by RIP-seq. Mol. Cell 2010, 40, 939–953. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Wang, Y.; Wang, Y.; Zhang, S.; Yu, L.; Guo, C.; Xu, H. Transcriptional and posttranscriptional regulation of HOXA13 by lncRNA HOTTIP facilitates tumorigenesis and metastasis in esophageal squamous carcinoma cells. Oncogene 2017, 36, 5392–5406. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cai, K.; Wang, J.; Wang, X.; Cheng, K.; Shi, F.; Jiang, L.; Zhang, Y.; Dou, J. MiR-7, inhibited indirectly by lincRNA HOTAIR, directly inhibits SETDB1 and reverses the EMT of breast cancer stem cells by downregulating the STAT3 pathway. Stem Cells 2014, 32, 2858–2868. [Google Scholar] [CrossRef] [PubMed]
- Ding, Q.; Miyazaki, Y.; Tsukasa, K.; Matsubara, S.; Yoshimitsu, M.; Takao, S. CD133 facilitates epithelial-mesenchymal transition through interaction with the ERK pathway in pancreatic cancer metastasis. Mol. Cancer 2014, 13, 15. [Google Scholar] [CrossRef] [PubMed]
- Latorre, E.; Carelli, S.; Raimondi, I.; D’Agostino, V.; Castiglioni, I.; Zucal, C.; Moro, G.; Luciani, A.; Ghilardi, G.; Monti, E.; et al. The Ribonucleic Complex HuR-MALAT1 Represses CD133 Expression and Suppresses Epithelial-Mesenchymal Transition in Breast Cancer. Cancer Res. 2016, 76, 2626–2636. [Google Scholar] [CrossRef] [PubMed]
- Bosson, A.D.; Zamudio, J.R.; Sharp, P.A. Endogenous miRNA and target concentrations determine susceptibility to potential ceRNA competition. Mol. Cell 2014, 56, 347–359. [Google Scholar] [CrossRef] [PubMed]
- Barrallo-Gimeno, A.; Nieto, M.A. The Snail genes as inducers of cell movement and survival: Implications in development and cancer. Development 2005, 132, 3151–3161. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wang, R.; Fang, L.; Ge, X.; Chen, L.; Zhou, M.; Zhou, Y.; Xiong, W.; Hu, Y.; Tang, X.; et al. HCP5 is a SMAD3-responsive long non-coding RNA that promotes lung adenocarcinoma metastasis via miR-203/SNAI axis. Theranostics 2019, 9, 2460–2474. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.; Zhang, S.; Wang, Y.; Wang, Y.; Nice, E.; Guo, C.; Zhang, E.; Yu, L.; Li, M.; Liu, C.; et al. Functional Role of a Novel Long Noncoding RNA TTN-AS1 in Esophageal Squamous Cell Carcinoma Progression and Metastasis. Clin. Cancer Res. 2018, 24, 486–498. [Google Scholar] [CrossRef]
- Dong, H.; Hu, J.; Zou, K.; Ye, M.; Chen, Y.; Wu, C.; Chen, X.; Han, M. Activation of LncRNA TINCR by H3K27 acetylation promotes Trastuzumab resistance and epithelial-mesenchymal transition by targeting MicroRNA-125b in breast Cancer. Mol. Cancer 2019, 18, 3. [Google Scholar] [CrossRef]
- Grelet, S.; Link, L.A.; Howley, B.; Obellianne, C.; Palanisamy, V.; Gangaraju, V.K.; Diehl, J.A.; Howe, P.H. A regulated PNUTS mRNA to lncRNA splice switch mediates EMT and tumour progression. Nat. Cell Biol. 2017, 19, 1105–1115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zeisberg, M.; Neilson, E.G. Biomarkers for epithelial-mesenchymal transitions. J. Clin. Investig. 2009, 119, 1429–1437. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, H.Y.; Yang, J. Unraveling the TWIST between EMT and cancer stemness. Cell Stem Cell 2015, 16, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Li, R.H.; Chen, M.; Liu, J.; Shao, C.C.; Guo, C.P.; Wei, X.L.; Li, Y.C.; Huang, W.H.; Zhang, G.J. Long noncoding RNA ATB promotes the epithelial-mesenchymal transition by upregulating the miR-200c/Twist1 axe and predicts poor prognosis in breast cancer. Cell Death Dis. 2018, 9, 1171. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Acloque, H.; Huang, R.Y.; Nieto, M.A. Epithelial-mesenchymal transitions in development and disease. Cell 2009, 139, 871–890. [Google Scholar] [CrossRef] [PubMed]
- Thiery, J.P.; Sleeman, J.P. Complex networks orchestrate epithelial-mesenchymal transitions. Nat. Rev. Mol. Cell Biol. 2006, 7, 131–142. [Google Scholar] [CrossRef] [PubMed]
- Zhang, P.F.; Wang, F.; Wu, J.; Wu, Y.; Huang, W.; Liu, D.; Huang, X.Y.; Zhang, X.M.; Ke, A.W. LncRNA SNHG3 induces EMT and sorafenib resistance by modulating the miR-128/CD151 pathway in hepatocellular carcinoma. J. Cell. Physiol. 2019, 234, 2788–2794. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Guo, S.; Zhao, R.; Liu, Y.; Yang, G. STAT3-activated lncRNA LUCAT1 drives cell proliferation, migration and invasion in hepatoblastoma through regulating miR-301b/STAT3 axis. Hum. Gene Ther. 2018, 30, 702–713. [Google Scholar] [CrossRef]
- Liu, Y.; Xue, M.; Du, S.; Feng, W.; Zhang, K.; Zhang, L.; Liu, H.; Jia, G.; Wu, L.; Hu, X.; et al. Competitive endogenous RNA is an intrinsic component of EMT regulatory circuits and modulates EMT. Nat. Commun. 2019, 10, 1637. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Rai, A.; Chen, M.; Suwakulsiri, W.; Greening, D.W.; Simpson, R.J. Extracellular vesicles in cancer - implications for future improvements in cancer care. Nat. Rev. Clin. Oncol. 2018, 15, 617–638. [Google Scholar] [CrossRef]
- Li, Z.; Jiang, P.; Li, J.; Peng, M.; Zhao, X.; Zhang, X.; Chen, K.; Zhang, Y.; Liu, H.; Gan, L.; et al. Tumor-derived exosomal lnc-Sox2ot promotes EMT and stemness by acting as a ceRNA in pancreatic ductal adenocarcinoma. Oncogene 2018, 37, 3822–3838. [Google Scholar] [CrossRef] [PubMed]
- Wu, D.M.; Deng, S.H.; Liu, T.; Han, R.; Zhang, T.; Xu, Y. TGF-beta-mediated exosomal lnc-MMP2-2 regulates migration and invasion of lung cancer cells to the vasculature by promoting MMP2 expression. Cancer Med. 2018, 7, 5118–5129. [Google Scholar] [CrossRef] [PubMed]
- Xue, M.; Chen, W.; Xiang, A.; Wang, R.; Chen, H.; Pan, J.; Pang, H.; An, H.; Wang, X.; Hou, H.; et al. Hypoxic exosomes facilitate bladder tumor growth and development through transferring long non-coding RNA-UCA1. Mol. Cancer 2017, 16, 143. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Liang, W.; Fu, M.; Huang, Z.H.; Li, X.; Zhang, W.; Zhang, P.; Qian, H.; Jiang, P.C.; Xu, W.R.; et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J. Cancer Res. Clin. Oncol. 2017, 143, 991–1004. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Li, H.; Lu, X.; Wen, C.; Huo, Z.; Shi, M.; Tang, X.; Chen, H.; Peng, C.; Fang, Y.; et al. Melittin-induced long non-coding RNA NONHSAT105177 inhibits proliferation and migration of pancreatic ductal adenocarcinoma. Cell Death Dis. 2018, 9, 940. [Google Scholar] [CrossRef] [PubMed]
- Song, K.; Yuan, X.; Li, G.; Ma, M.; Sun, J. Long noncoding RNA CASC11 promotes osteosarcoma metastasis by suppressing degradation of snail mRNA. Am. J. Cancer Res. 2019, 9, 300–311. [Google Scholar] [PubMed]
- Hinman, M.N.; Lou, H. Diverse molecular functions of Hu proteins. Cell. Mol. Life Sci. 2008, 65, 3168–3181. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdelmohsen, K.; Gorospe, M. Posttranscriptional regulation of cancer traits by HuR. Wiley Interdiscip. Rev. RNA 2010, 1, 214–229. [Google Scholar] [CrossRef]
- Wu, X.; Yan, T.; Wang, Z.; Wu, X.; Cao, G.; Zhang, C. LncRNA ZEB2-AS1 promotes bladder cancer cell proliferation and inhibits apoptosis by regulating miR-27b. Biomed. Pharmacother. 2017, 96, 299–304. [Google Scholar] [CrossRef] [PubMed]
- Wu, F.; Gao, H.; Liu, K.; Gao, B.; Ren, H.; Li, Z.; Liu, F. The lncRNA ZEB2-AS1 is upregulated in gastric cancer and affects cell proliferation and invasion via miR-143-5p/HIF-1alpha axis. Onco Targets Ther. 2019, 12, 657–667. [Google Scholar] [CrossRef]
- Beltran, M.; Puig, I.; Pena, C.; Garcia, J.M.; Alvarez, A.B.; Pena, R.; Bonilla, F.; de Herreros, A.G. A natural antisense transcript regulates Zeb2/Sip1 gene expression during Snail1-induced epithelial-mesenchymal transition. Genes Dev. 2008, 22, 756–769. [Google Scholar] [CrossRef] [PubMed]
- Wapinski, O.; Chang, H.Y. Long noncoding RNAs and human disease. Trends Cell Biol 2011, 21, 354–361. [Google Scholar] [CrossRef] [PubMed]
- Zhao, B.S.; Roundtree, I.A.; He, C. Post-transcriptional gene regulation by mRNA modifications. Nat. Rev. Mol. Cell Biol. 2017, 18, 31–42. [Google Scholar] [CrossRef] [PubMed]
- Roundtree, I.A.; Evans, M.E.; Pan, T.; He, C. Dynamic RNA Modifications in Gene Expression Regulation. Cell 2017, 169, 1187–1200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dominissini, D.; Moshitch-Moshkovitz, S.; Schwartz, S.; Salmon-Divon, M.; Ungar, L.; Osenberg, S.; Cesarkas, K.; Jacob-Hirsch, J.; Amariglio, N.; Kupiec, M.; et al. Topology of the human and mouse m6A RNA methylomes revealed by m6A-seq. Nature 2012, 485, 201–206. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhao, B.S.; Zhou, A.; Lin, K.; Zheng, S.; Lu, Z.; Chen, Y.; Sulman, E.P.; Xie, K.; Bogler, O.; et al. m(6)A Demethylase ALKBH5 Maintains Tumorigenicity of Glioblastoma Stem-like Cells by Sustaining FOXM1 Expression and Cell Proliferation Program. Cancer Cell 2017, 31, 591–606.e6. [Google Scholar] [CrossRef]
- Lin, X.; Chai, G.; Wu, Y.; Li, J.; Chen, F.; Liu, J.; Luo, G.; Tauler, J.; Du, J.; Lin, S.; et al. RNA m(6)A methylation regulates the epithelial mesenchymal transition of cancer cells and translation of Snail. Nat. Commun. 2019, 10, 2065. [Google Scholar] [CrossRef] [PubMed]
- Dongre, A.; Weinberg, R.A. New insights into the mechanisms of epithelial-mesenchymal transition and implications for cancer. Nat. Rev. Mol. Cell Biol. 2019, 20, 69–84. [Google Scholar] [CrossRef]
- Moustakas, A.; de Herreros, A.G. Epithelial-mesenchymal transition in cancer. Mol. Oncol. 2017, 11, 715–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ong, M.S.; Cai, W.; Yuan, Y.; Leong, H.C.; Tan, T.Z.; Mohammad, A.; You, M.L.; Arfuso, F.; Goh, B.C.; Warrier, S.; et al. ‘Lnc’-ing Wnt in female reproductive cancers: Therapeutic potential of long non-coding RNAs in Wnt signalling. Br. J. Pharmacol. 2017, 174, 4684–4700. [Google Scholar] [CrossRef]
- Sethi, G.; Chatterjee, S.; Rajendran, P.; Li, F.; Shanmugam, M.K.; Wong, K.F.; Kumar, A.P.; Senapati, P.; Behera, A.K.; Hui, K.M.; et al. Inhibition of STAT3 dimerization and acetylation by garcinol suppresses the growth of human hepatocellular carcinoma in vitro and in vivo. Mol. Cancer 2014, 13, 66. [Google Scholar] [CrossRef] [PubMed]
- Subramaniam, A.; Shanmugam, M.K.; Ong, T.H.; Li, F.; Perumal, E.; Chen, L.; Vali, S.; Abbasi, T.; Kapoor, S.; Ahn, K.S.; et al. Emodin inhibits growth and induces apoptosis in an orthotopic hepatocellular carcinoma model by blocking activation of STAT3. Br. J. Pharmacol. 2013, 170, 807–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abdulghani, J.; Gu, L.; Dagvadorj, A.; Lutz, J.; Leiby, B.; Bonuccelli, G.; Lisanti, M.P.; Zellweger, T.; Alanen, K.; Mirtti, T.; et al. Stat3 promotes metastatic progression of prostate cancer. Am. J. Pathol. 2008, 172, 1717–1728. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, R.B.; Contino, G.; Deshpande, V.; Tzatsos, A.; Conrad, C.; Benes, C.H.; Levy, D.E.; Settleman, J.; Engelman, J.A.; Bardeesy, N. STAT3 plays a critical role in KRAS-induced pancreatic tumorigenesis. Cancer Res. 2011, 71, 5020–5029. [Google Scholar] [CrossRef] [PubMed]
- Ko, J.H.; Nam, D.; Um, J.Y.; Jung, S.H.; Sethi, G.; Ahn, K.S. Bergamottin Suppresses Metastasis of Lung Cancer Cells through Abrogation of Diverse Oncogenic Signaling Cascades and Epithelial-to-Mesenchymal Transition. Molecules 2018, 23, 1601. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.H.; Lee, J.H.; Ko, J.H.; Jung, S.H.; Sethi, G.; Ahn, K.S. Brassinin Represses Invasive Potential of Lung Carcinoma Cells through Deactivation of PI3K/Akt/mTOR Signaling Cascade. Molecules 2019, 24, 1584. [Google Scholar] [CrossRef] [PubMed]
- Baek, S.H.; Ko, J.H.; Lee, J.H.; Kim, C.; Lee, H.; Nam, D.; Lee, J.; Lee, S.G.; Yang, W.M.; Um, J.Y.; et al. Ginkgolic Acid Inhibits Invasion and Migration and TGF-beta-Induced EMT of Lung Cancer Cells Through PI3K/Akt/mTOR Inactivation. J. Cell. Physiol. 2017, 232, 346–354. [Google Scholar] [CrossRef]
- Zhang, L.; Yang, F.; Yuan, J.H.; Yuan, S.X.; Zhou, W.P.; Huo, X.S.; Xu, D.; Bi, H.S.; Wang, F.; Sun, S.H. Epigenetic activation of the MiR-200 family contributes to H19-mediated metastasis suppression in hepatocellular carcinoma. Carcinogenesis 2013, 34, 577–586. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.J.; Deng, J.; Chen, C.; Hu, W.; Yuan, Y.C.; Xia, Z.K. LncRNA H19 promotes epithelial mesenchymal transition and metastasis of esophageal cancer via STAT3/EZH2 axis. Int. J. Biochem. Cell Biol. 2019, 113, 27–36. [Google Scholar] [CrossRef]
- Zhou, Z.; Wu, Q.; Yan, Z. Extracellular RNA in a single droplet of human serum reflects physiologic and disease states. Proc. Natl. Acad. Sci. USA 2019, 116, 19200–19208. [Google Scholar] [CrossRef] [Green Version]
- Fujita, K.; Nonomura, N. Urinary biomarkers of prostate cancer. Int. J. Urol. 2018, 25, 770–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, P.; Zhou, X.Y.; Du, X. Circulating long non-coding RNAs in cancer: Current status and future perspectives. Mol. Cancer 2016, 15, 39. [Google Scholar] [CrossRef] [PubMed]
- Hong, D.; Kurzrock, R.; Kim, Y.; Woessner, R.; Younes, A.; Nemunaitis, J.; Fowler, N.; Zhou, T.; Schmidt, J.; Jo, M.; et al. AZD9150, a next-generation antisense oligonucleotide inhibitor of STAT3 with early evidence of clinical activity in lymphoma and lung cancer. Sci. Transl. Med. 2015, 7, 314ra185. [Google Scholar] [CrossRef] [PubMed]
- Eide, P.W.; Eilertsen, I.A. Long noncoding RNA MIR31HG is a bona fide prognostic marker with colorectal cancer cell-intrinsic properties. Int. J. Cancer 2019, 144, 2843–2853. [Google Scholar] [CrossRef] [PubMed]
- Azmi, A.S.; Bao, B.; Sarkar, F.H. Exosomes in cancer development, metastasis, and drug resistance: A comprehensive review. Cancer Metastasis Rev. 2013, 32, 623–642. [Google Scholar] [CrossRef] [PubMed]
- Peinado, H.; Olmeda, D.; Cano, A. Snail, Zeb and bHLH factors in tumour progression: An alliance against the epithelial phenotype? Nat. Rev. Cancer 2007, 7, 415–428. [Google Scholar] [CrossRef] [PubMed]
- Brabletz, T.; Kalluri, R.; Nieto, M.A.; Weinberg, R.A. EMT in cancer. Nat. Rev. Cancer 2018, 18, 128–134. [Google Scholar] [CrossRef]
- Chen, D.L.; Chen, L.Z.; Lu, Y.X.; Zhang, D.S.; Zeng, Z.L.; Pan, Z.Z.; Huang, P.; Wang, F.H.; Li, Y.H.; Ju, H.Q.; et al. Long noncoding RNA XIST expedites metastasis and modulates epithelial-mesenchymal transition in colorectal cancer. Cell Death Dis. 2017, 8, e3011. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Lin, S.; Guan, L.; Yuan, H.; Liu, K.; Liu, C.; Ye, W.; Liao, Y.; Jia, J.; Zhang, R. Long non-coding RNA XIST inhibited breast cancer cell growth, migration, and invasion via miR-155/CDX1 axis. Biochem. Biophys. Res. Commun. 2018, 498, 1002–1008. [Google Scholar] [CrossRef]
- Fan, S.; Fan, C.; Liu, N.; Huang, K.; Fang, X.; Wang, K. Downregulation of the long non-coding RNA ZFAS1 is associated with cell proliferation, migration and invasion in breast cancer. Mol. Med. Rep. 2018, 17, 6405–6412. [Google Scholar] [CrossRef]
- Xie, S.; Ge, Q.; Wang, X.; Sun, X.; Kang, Y. Long non-coding RNA ZFAS1 sponges miR-484 to promote cell proliferation and invasion in colorectal cancer. Cell Cycle 2018, 17, 154–161. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Z.; Liu, F.; Yang, F.; Liu, Y. Kockdown of OIP5-AS1 expression inhibits proliferation, metastasis and EMT progress in hepatoblastoma cells through up-regulating miR-186a-5p and down-regulating ZEB1. Biomed. Pharmacother. 2018, 101, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Yang, N.; Chen, J.; Zhang, H.; Wang, X.; Yao, H.; Peng, Y.; Zhang, W. LncRNA OIP5-AS1 loss-induced microRNA-410 accumulation regulates cell proliferation and apoptosis by targeting KLF10 via activating PTEN/PI3K/AKT pathway in multiple myeloma. Cell Death Dis. 2017, 8, e2975. [Google Scholar] [CrossRef] [PubMed]
- Krebs, A.M.; Mitschke, J.; Lasierra Losada, M.; Schmalhofer, O.; Boerries, M.; Busch, H.; Boettcher, M.; Mougiakakos, D.; Reichardt, W.; Bronsert, P.; et al. The EMT-activator Zeb1 is a key factor for cell plasticity and promotes metastasis in pancreatic cancer. Nat. Cell Biol. 2017, 19, 518–529. [Google Scholar] [CrossRef] [Green Version]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Zheng, X.; Carstens, J.L.; Kim, J.; Scheible, M.; Kaye, J.; Sugimoto, H.; Wu, C.C.; LeBleu, V.S.; Kalluri, R. Epithelial-to-mesenchymal transition is dispensable for metastasis but induces chemoresistance in pancreatic cancer. Nature 2015, 527, 525–530. [Google Scholar] [CrossRef] [Green Version]
- Hur, K.; Toiyama, Y.; Takahashi, M.; Balaguer, F.; Nagasaka, T.; Koike, J.; Hemmi, H.; Koi, M.; Boland, C.R.; Goel, A. MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT) in human colorectal cancer metastasis. Gut 2013, 62, 1315–1326. [Google Scholar] [CrossRef]
- Welch, D.R.; Hurst, D.R. Defining the Hallmarks of Metastasis. Cancer Res. 2019, 79, 3011–3027. [Google Scholar] [CrossRef]
- Liu, X.; Li, J.; Cadilha, B.L. Epithelial-type systemic breast carcinoma cells with a restricted mesenchymal transition are a major source of metastasis. Sci. Adv. 2019, 5, eaav4275. [Google Scholar] [CrossRef] [Green Version]



| Category | LncRNA | Partners | Expression | EMT Markers | Pathways | Function | Mechanism | Tumor Types | Reference |
|---|---|---|---|---|---|---|---|---|---|
| lncRNA: chromatin regulators | MEG8 | EZH2 | ↑ | SNAI1/2 | TGF-β | Promote EMT | Interacts with EZH2 and represses miR-34a and miR-203, resulting in up-regulated of SNAI1/2. | Lung and pancreatic cancer | [15] |
| MEG3 | EZH2 | ↑ | ZEB family | TGF-β | Promote EMT | Interacts with JARID2 and EZH2 and represses CDH1 and miRNA-200 family, resulting in up-regulated of ZEB. | Lung cancer | [39] | |
| LINC00518 | CDX2 | ↑ | ZEB1/2 Twist1 | Wnt | Inhibit EMT | Binds to the promoter region of CDX2 gene and promotes CDX2 methylation by recruiting DNA methyltransferase through activating the Wnt signaling pathway. | Breast cancer | [40] | |
| MALAT1 | Ezh2 | ↑ | E-cadherin | Wnt/β-catenin | Promote EMT and metastasis. | Activated by c-Fos and interacts with Ezh2, resulting in E-cadherin expression was decreased. | RCC | [13] | |
| ANCR | EZH2 | ↓ | E-cadherin | Wnt/β-catenin | Inhibit EMT and metastasis. | Interacts with EZH2 to increase the binding of CDK1 with EZH2 and to promote the degradation of EZH2, resulting in the up-regulated of E-cadherin. | Breast cancer | [41] | |
| lncRNA promoter: TFs | MALAT1 | STAT3 | ↑ | Snail | TGF-β/STAT3 | Promote EMT | STAT3 binds to the MALAT1 promoter region and transcriptionally activate MALAT1 expression. | HNSCC | [14] |
| HCCL5 | ZEB1 | ↑ | ZEB1 E-cadherin | TGF-β1 | Promote EMT and metastasis. | ZEB1 can bind to both the identified super-enhancer and promoter of HCCL5. HCCL5 was significantly and frequently overexpressed. | HCC | [42] | |
| AC132217.4 | KLF8 | ↑ | E-cadherin | AKT | Promote EMT | KLF8 binds to the upstream sequence of AC132217.4, activating its expression at the transcriptional level. | OSCC | [43] | |
| lncRNA: TFs | ELIT-1 | Smad3 | ↑ | Snail | TGF-β | Promote EMT | Binds to Smad3 and enhances Smad -responsive promoter activities by recruiting Smad3 to the promoters of its target genes including Snail, other TGF-β-target genes, and ELIT-1 itself. | lung adenocarcinoma gastric cancer | [44] |
| EPR | SMAD3 | ↓ | SNAI1 | TGF-β | Inhibit EMT | Interacts with SMAD3 and promotes Cdkn1a gene expression, resulting in the down-regulation of SNAI1. | Breast cancer | [45] | |
| tsRMST | NANOG, SUZ12 | ↓ | SNAI2 TWIST1 | Wnt | Inhibit EMT | Binds to NANOG and SUZ12 to repress the expression of WNT5A, resulting in the down-regulation of SNAI2 and TWIST1. | hESCs | [31] | |
| BX111 | YB1 | ↑ | ZEB1 MMP2 E-cadherin | — | Promote EMT and metastasis. | Activates transcription of ZEB1 via recruiting YB1 to its promoter region, resulting in the up-regulation of ZEB1. | Pancreatic cancer | [46] | |
| NEAT1 | FOXN3 | ↓ | E-cadherin | — | Inhibit EMT | Interacts with FOXN3 and SIN3A and represses the target genes including GATA3 and TJP1, resulting in the up-regulated of E-cadherin. | Breast cancer | [26] | |
| lncRNA:Protein | MUF | ANXA2 | ↑ | Snail | Wnt | Promote EMT | Binds to the protein ANXA2 and ANXA2 can alter the subcellular localization of β-catenin to activate the Wnt cascade. | HCC | [47] |
| SNHG15 | Slug | ↑ | Slug | — | Promote EMT | Interacts with protein Slug via its C-terminal domain containing five zinc finger motifs, and promote Slug expression. | colon cancer | [48] | |
| TBILA | S100A7 | ↑ | SNAI1 ZEB1, | S100A7/JAB1 | Promote EMT | Binds to the S100A7 protein and promotes S100A7/JAB1 pathway activation, resulting in the up-regulation of SNAI1 and ZEB1. | NSCLC | [49] | |
| RP11 | hnRNPA2B1 | ↑ | Zeb1 | — | Promote EMT | Interacts with the protein hnRNPA2B1 and accelerates the mRNA degradation of Siah1 and Fbxo45, and subsequently prevented the proteasomal degradation of Zeb1. | CRC | [28] | |
| GAEA | MEX3C | ↑ | SNAI1 TWIST1, | AKT | Promote EMT | Binds to the MEX3C and catalyze K27-linked polyUb of PTEN. PTENK27-PolyUb removed phospho-groups from serine/threonine residues in substrates including TWIST1, SNAI1, and YAP1. | Human and mouse breast epithelial cells | [29] | |
| LINC01638 | c-Myc | ↑ | Twist1 | MTDH-Twist1 | Promote EMT | Interacts with protein c-Myc to prevent SPOP-induced c-Myc ubiquitination and degradation and then activate MTDH-Twist1 signaling to maintain mesenchymal traits with EMT and CSC-like features. | TNBC | [50] | |
| CYTOR | NCL Sam68 | ↑ | Twist E-cadherin | NF-ΚB | Promote EMT | NCL and Sam68 could recognize their specific motifs and directly bind to Exon1 of CYTOR, then activating the NF-κB pathway and promoting the expression of Twist. | CRC | [51] | |
| NBR2 | Notch1 | ↓ | E-cadherin | Notch | Inhibit EMT | Binds to the Notch1 protein and promotes Notch1 expression, resulting in the up-regulation of E-cadherin. | osteosarcoma | [52] | |
| NEF | β-catenin | ↓ | E-cadherin | Wnt | Inhibit EMT and metastasis. | Activated by FOXA2 and can interact with β-catenin, leading to the suppression on Wnt/β-catenin signaling and activation of FOXA2 expression. | HCC | [53] | |
| SLCO4A1-AS1 | β-catenin | ↑ | E-cadherin | Wnt | Promote EMT and metastasis. | Activates Wnt signaling through enhancing the stability of β-catenin by attenuating the interaction of β-catenin with GSKβ. | CRC | [54] | |
| LINC01133 | SRSF6 | ↓ | E-cadherin | TGF-β | Inhibit EMT and metastasis. | Binds to SRSF6 and blocking its critical domain, resulting in inhibition of EMT. | CRC | [55] | |
| CRCMSL | HMGB2 | ↓ | OCT4 | — | Inhibit EMT and metastasis. | Binds to protein HMGB2 and stabilizes the localization in the cytoplasm, attenuating the interaction between HMGB2 and OCT4 and inhibiting EMT. | CRC | [56] | |
| lncRNA: miRNA | B3GALT5-AS1 | miR-203 | ↓ | ZEB2, SNAI2 | — | Promote EMT and metastasis. | Directly binds to the promoter of miRNA-203 and represses miR-203 expression, resulting in the up-regulated of ZEB2 and SNAI2. | Colon cancer | [57] |
| UCA1 | miR-1, miR-203a | ↑ | Slug | TGF-β | Promote EMT | Promotes Slug expression at the post- transcriptional level, by directly titrating miR-1 and miR-203a. | Breast cancer | [58] | |
| CAR10 | miR-30, miR-203 | ↑ | SNAI1/2 | — | Promote EMT and metastasis. | Induces EMT by directly binding with miR-30 and miR-203 and then regulating the expression of Snail1 and Slug. | LUAD | [59] | |
| FTX | miR-374a | ↑ | Snail ZEB1 | Wnt/β-catenin | Promote EMT | Competitively binding miR-374a, thus resulting in the up-regulated of Snail and ZEB1 | HCC | [60] | |
| lnc-ATB | miR-200s | ↑ | ZEB1/2 | TGF-β | Promote EMT | lnc-ATB upregulated ZEB1 and ZEB2 by competitively binding the miR-200 family and then induced EMT and invasion. | HCC | [27] | |
| AK000053 | miR-508 | ↑ | ZEB1 | TGF-β | Promote EMT | Competitively interact with miR-508 and negatively regulated, resulting in the up-regulated of ZEB1. | CRC | [61] | |
| ZFAS1 | miR-150 | ↑ | ZEB1, MMP14/16 | — | Promote EMT and metastasis. | Competitively binding miR-150, resulting in the up-regulated of ZEB1, MMP14/16 | HCC | [62] | |
| lncRNA: mRNA | RP11 | Fbxo45, Siah1 | ↑ | Zeb1 | — | Promote EMT | Interacted with the 3’UTR of Fbxo45 mRNA and CDS of Siah1 mRNA, and subsequently prevented the proteasomal degradation of Zeb1. | CRC | [28] |
| AC132217.4 | IGF2 | ↑ | E-cadherin | AKT | Promote EMT and metastasis. | Interacted with the 3’UTR of IGF2 mRNA and activated AKT signalling by increasing IGF2 mRNA stability, remarkably down-regulated of E-cadherin. | OSCC | [43] | |
| lnc-ATB | IL-11 | ↑ | E-cadherin | IL-11/STAT3 | Promote EMT | Binds to IL-11 mRNA, thus increasing IL-11 mRNA stability, causing autocrine induction of IL-11, and then activating STAT3 signaling. | HCC | [27] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cheng, J.-T.; Wang, L.; Wang, H.; Tang, F.-R.; Cai, W.-Q.; Sethi, G.; Xin, H.-W.; Ma, Z. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells 2019, 8, 1178. https://doi.org/10.3390/cells8101178
Cheng J-T, Wang L, Wang H, Tang F-R, Cai W-Q, Sethi G, Xin H-W, Ma Z. Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells. 2019; 8(10):1178. https://doi.org/10.3390/cells8101178
Chicago/Turabian StyleCheng, Jun-Ting, Lingzhi Wang, Hong Wang, Feng-Ru Tang, Wen-Qi Cai, Gautam Sethi, Hong-Wu Xin, and Zhaowu Ma. 2019. "Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition" Cells 8, no. 10: 1178. https://doi.org/10.3390/cells8101178
APA StyleCheng, J.-T., Wang, L., Wang, H., Tang, F.-R., Cai, W.-Q., Sethi, G., Xin, H.-W., & Ma, Z. (2019). Insights into Biological Role of LncRNAs in Epithelial-Mesenchymal Transition. Cells, 8(10), 1178. https://doi.org/10.3390/cells8101178








