The Influence of (5′R)- and (5′S)-5′,8-Cyclo-2′-Deoxyadenosine on UDG and hAPE1 Activity. Tandem Lesions are the Base Excision Repair System’s Nightmare
Abstract
1. Introduction
2. Materials and Methods
2.1. Oligonucleotide Synthesis and Purification
2.2. Oligonucleotide Concentration
2.3. Mass Spectroscopy of Oligonucleotides
2.4. Thermal ds-Oligonucleotide Stability—Melting Temperature Measurement
2.5. Circular Dichroism Analysis of Double-Stranded Oligonucleotides
2.6. Preparation of 5′-End-Labeled Oligonucleotides
2.7. Oligonucleotide Hybridisation and UDG and hAPE1 Digestion Assay with Subsequent Piperidine Treatment
2.8. Theoretical Computation Methodology of ONIOM Studies
3. Results and Discussion
3.1. The Influence of 5′R-cdA and 5′S-cdA on UDG and hAPE1 Activity
3.2. Theoretical Study
4. Conclusions
- -
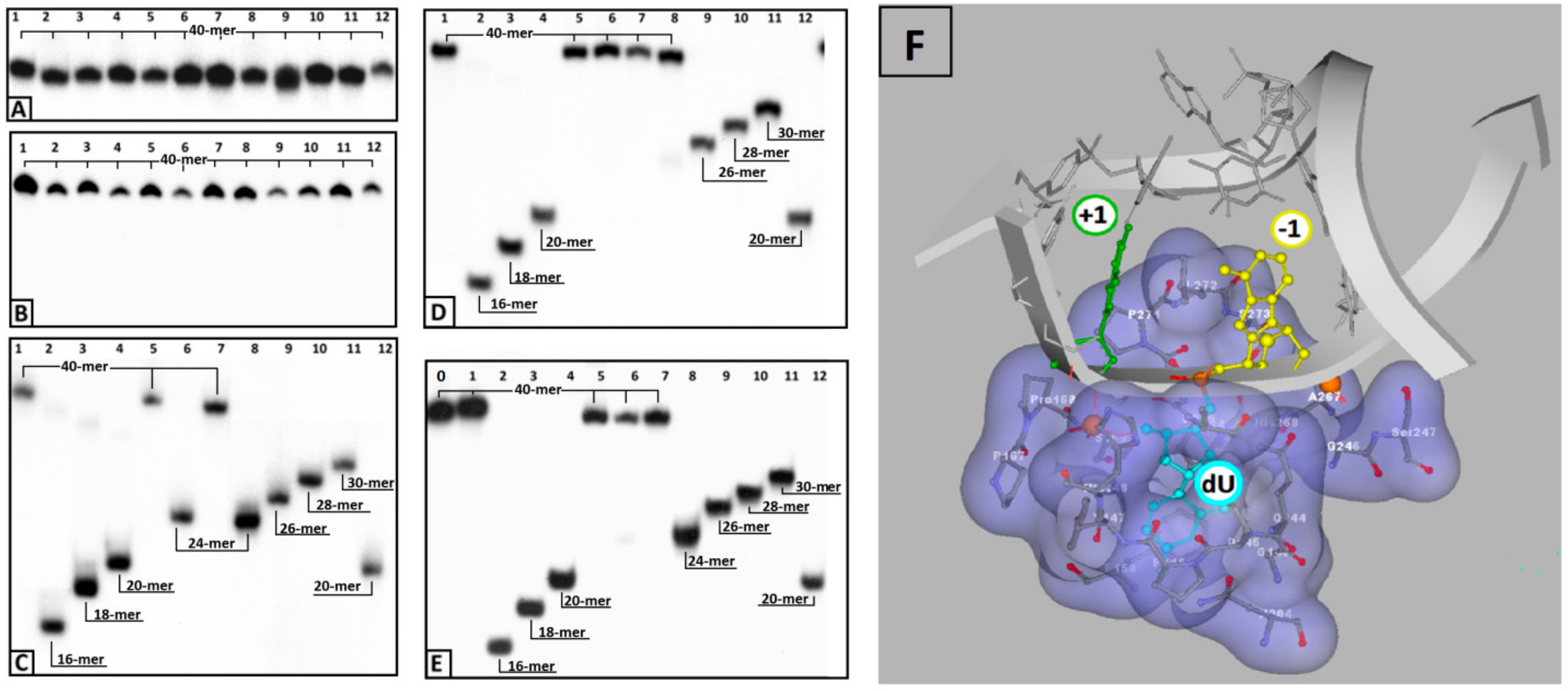
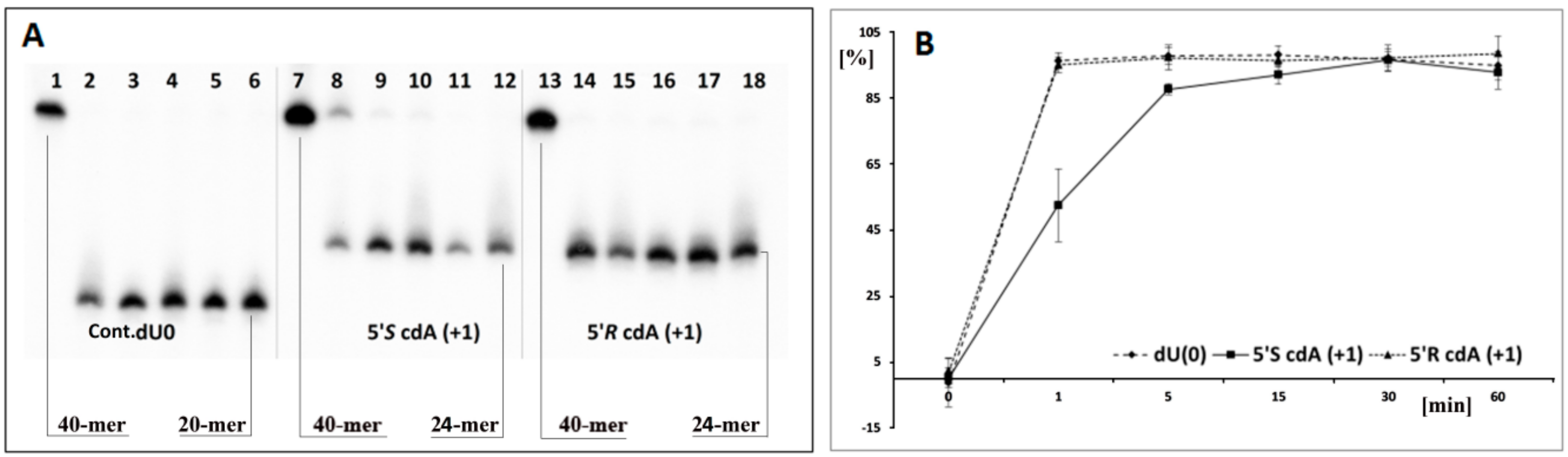
- the attaching of 5′S or 5′R cdA to the dU 3′ hydroxyl group in a single-stranded oligo completely blocks UDG activity opposite to the cdA adjacent to the dU 5′-end, allowing UDG glycosidic bond hydrolysis.
- -
- probably due to steric hindrance caused by 5′S-cdA in ss-ScdA(+1), dU glycosidic bond hydrolysis is slower than that found for ss-RcdA(+1) and the reference ss-DNA (Cont.dU(0)).
- -
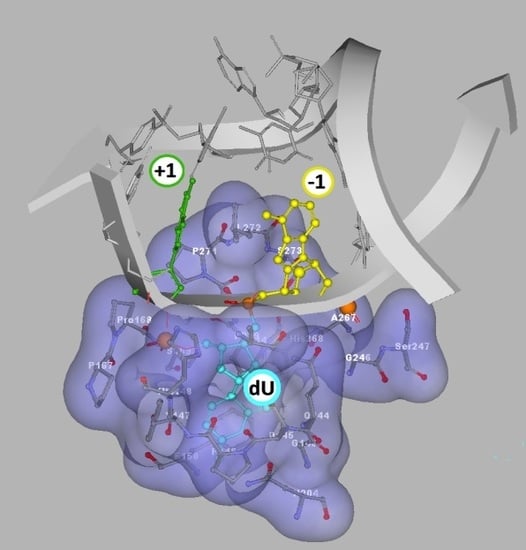
- as shown in Figure 2C,E, both the 5′S and 5′R diastereomers of cdA inhibit uracil release from the ds-RcdA(−1) and ds-ScdA(−1) oligos. Surprisingly, only ds-RcdA(+1) is a substrate for 2′-deoxyuridine glycosylase and no digestion was observed for ds-ScdA(+1). The distance extent between 5′S-cdA and dU ranges from −/+3 to −/+7 and probably further base pairs abolished the inhibition effect on glycosylase. Moreover, these tandem lesions were recognised as a good substrate for further AP-site lysis by hAPE1.
- -
- the AP-site formed during ds-RcdA(+1) digestion by UDG was identified as an unsuitable substrate for hAPE1.
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Travers, A.; Muskhelishvili, G. DNA structure and function. FEBS J. 2015, 282, 2279–2295. [Google Scholar] [CrossRef] [PubMed]
- Tubbs, A.; Nussenzweig, A. Endogenous DNA damage as a source of genomic instability in Cancer. Cell. 2017, 168, 644–656. [Google Scholar] [CrossRef] [PubMed]
- Lindahl, T. Instability and decay of the primary structure of DNA. Nature 1993, 362, 709–715. [Google Scholar] [CrossRef] [PubMed]
- Shrinivas, S.A.; Shanta, S.H.; Prajakta, B.B. DNA: Damage and repair mechanisms in humans. Glob. J. Pharm. Sci. 2017, 3, 1–8. [Google Scholar]
- Chatterjee, N.; Walker, G.C. Mechanisms of DNA damage, repair and mutagenesis. Environ. Mol. Mutagen. 2017, 58, 235–263. [Google Scholar] [PubMed]
- Barnes, J.L.; Zubair, M.; Kaarthik, J.; Poirier, M.C.; Martin, F.L. Carcinogens and DNA damage. Biochem. Soc. Trans. 2018, 46, 1213–1224. [Google Scholar] [CrossRef] [PubMed]
- Livnat, A. Interaction-based evolution: how natural selection and nonrandom mutation work together. Biol. Direct. 2013, 8, 1–53. [Google Scholar] [CrossRef]
- Bernatchez, L. On the maintenance of genetic variation and adaptation to environmental change: considerations from population genomics in fishes. J. Fish. Biol. 2016, 89, 2519–2556. [Google Scholar] [CrossRef]
- Nickoloff, J.A.; Boss, M.-K.; Allen, C.P.; LaRue, S.M. Translational research in radiation-induced DNA damage signaling and repair. Transl. Cancer Res. 2017, 6, 875–891. [Google Scholar] [CrossRef]
- De Bont, R.; van Larebeke, N. Endogenous DNA damage in humans: a review of quantitative data. Mutagenesis 2004, 19, 169–185. [Google Scholar] [CrossRef]
- Goodhead, D.T. The initial physical damage produced by ionizing radiations. Int. J. Radiat Biol. 1989, 56, 623–634. [Google Scholar] [CrossRef] [PubMed]
- Eccles, L.J.; O’Neill, P.; Lomax, M.E. Delayed repair of radiation induced clustered DNA damage: Friend or foe? Mutat Res. 2011, 711, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G.; O’Neill, P.; Stewart, R.D. Induction and repair of clustered DNA lesions: what do we know so far? Radiat. Res. 2013, 180, 100–109. [Google Scholar] [CrossRef] [PubMed]
- Cunniff, S.; O’Neill, P.; Greenberg, M.M.; Lomax, M.E. Reduced repair capacity of a DNA clustered damage site comprised of 8-oxo-7,8-dihydro-2′-deoxyguanosine and 2-deoxyribonolactone results in an increased mutagenic potential of these lesions. Mutation Res. 2014, 762, 32–39. [Google Scholar] [CrossRef]
- Pearson, C.G.; Shikazono, N.; Thacker, J.; O’Neill, P. Enhanced mutagenic potential of 8-oxo-7,8-dihydroguanine when present within a clustered DNA damage site. Nucleic. Acids Res. 2004, 32, 263–270. [Google Scholar] [CrossRef]
- Susan, S.; Wallace, S.S.; Murphy, D.L.; Sweasy, J.B. Base excision repair and cancer. Cancer Lett. 2012, 327, 73–89. [Google Scholar]
- Jacobs, A.L.; Schär, P. DNA glycosylases: in DNA repair and beyond. Chromosome 2012, 121, 1–20. [Google Scholar] [CrossRef]
- Abbotts, R.; Madhusudan, S. Human AP endonuclease 1 (APE1): from mechanistic insights to druggable target in cancer. Cancer Treat. Rev. 2010, 36, 425–435. [Google Scholar] [CrossRef]
- Bailly, V.; Verly, W.G. The roles of beta-and delta-eliminations in the repair of AP sites in DNA. In DNA Repair Mechanisms and Their Biological Implications in Mammalian Cells; Lambert, M.W., Laval, J., Eds.; NATO ASI Series; Springer: Boston, MA, USA, 1989; Volume 182, pp. 19–23. [Google Scholar]
- Osakabe, A.; Arimura, Y.; Matsumoto, S.; Horikoshi, N.; Sugasawa, K.; Kurumizaka, H. Polymorphism of apyrimidinic DNA structures in the nucleosome. Sci. Rep. 2017, 7, 41783. [Google Scholar] [CrossRef]
- Keck, K. Bildung von cyclonudleotiden bei bestrahlung wassiriger losungen von purinenucleotiden. Z. Nat. 1968, 23, 1034–1043. [Google Scholar]
- Dizdaroglu, M. Free-radical-induced formation of an 8,5′-cyclo-2′-deoxyguanosine moiety in deoxyribonucleic acid. Biochem. J. 1986, 238, 247–254. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Dirksen, M.L.; Jiang, H.X.; Robbins, J.H. Ionizing-radiation-induced damage in the DNA of cultured human cells. Identification of 8,50-cyclo-2-deoxyguanosine. Biochem. J. 1987, 241, 929–932. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T.; Grand, A.; Cadet, J. 5′,8-Cyclo-2′-deoxyadenosine (cdA) formation by γ-radiation. Theoretical quantum mechanics study. Acta Biochim. Pol. 2009, 56, 655–662. [Google Scholar] [CrossRef] [PubMed]
- Kuraoka, I.; Robins, P.; Masutani, C.; Hanaoka, F.; Gasparutto, D.; Cadet, J.; Wood, D.R.; Lindhal, T. Oxygen free radical damage to DNA. Translesion synthesis by human DNA polymerase and resistance to exonuclease action at cyclopurine deoxynucleoside residues. J. Biol. Chem. 2001, 52, 49283–49288. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.; Xu, M.; Lai, Y.; Laverde, E.E.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Liu, Y. Bypass of a 5′,8-cyclopurine-2′-deoxynucleoside by DNA polymerase β during DNA replication and base excision repair leads to nucleotide misinsertions and DNA strand breaks. DNA Repair 2015, 33, 24–34. [Google Scholar] [CrossRef] [PubMed]
- Xu, M.; Lai, Y.; Jiang, Z.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Liu, Y. 5′,8-cyclo-2′-deoxypurine lesion induces trinucleotide repeat deletion via a unique lesion bypass by DNA polymerase β. Nucleic Acids Res. 2014, 42, 13749–13763. [Google Scholar] [CrossRef]
- You, C.; Swanson, A.L.; Dai, X.; Yuan, B.; Wang, J.; Wang, Y. Translesion Synthesis of 8,5-Cyclopurine-2-deoxynucleosides by DNA Polymerases η, ι, and ζ. J. Biol. Chem. 2013, 288, 28548–28556. [Google Scholar] [CrossRef]
- Walmacq, C.; Wang, L.; Chong, J.; Scibelli, K.; Lubkowska, L.; Gnatt, A.; Brooks, P.J.; Wangb, D.; Kashleva, M. Mechanism of RNA polymerase II bypass of oxidative cyclopurine DNA lesions. Proc. Natl. Acad. Sci. USA 2015, 112, 410–419. [Google Scholar] [CrossRef]
- Brooks, P.J.; Wise, D.S.; Berry, D.A.; Kosmoski, J.V.; Smerdon, M.J.; Somers, R.L.; Mackie, H.; Spoonde, A.Y.; Ackerman, E.J.; Coleman, K.; et al. The Oxidative DNA lesion 8,5′-(S)-cyclo-2′-deoxyadenosine Is repaired by the nucleotide excision repair pathway and blocks gene expression in mammalian cells. J. Biol. Chem. 2000, 275, 22355–22362. [Google Scholar] [CrossRef]
- Kuraoka, I.; Bender, C.; Romieu, A.; Cadet, J.; Wood, R.D.; Lindahl, T. Removal of oxygen free-radical-induced 5′,8-purine cyclodeoxynucleosides from DNA by the nucleotide excision-repair pathway in human cells. Proc. Natl. Acad. Sci. USA 2000, 97, 3832–3837. [Google Scholar] [CrossRef]
- Mir, O.; Ropert, S.; Goldwasser, F. Cisplatin as a cornerstone of modern chemotherapy. Lancet Oncol. 2009, 10, 304. [Google Scholar] [CrossRef]
- Kropachev, K.; Ding, S.; Terzidis, M.A.; Masi, A.; Liu, Z.; Cai, Y.; Kolbanovskiy, M.; Chatgilialoglu, C.; Broyde, S.; Geacintov, N.E.; et al. Structural basis for the recognition of diastereomeric 5′,8-cyclo-2′-deoxypurine lesions by the human nucleotide excision repair system. Nucleic Acids Res. 2014, 42, 5020–5032. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T.; Bellon, S.; O’Neill, P.; Lomax, M.E.; Cadet, J. Effects of (5′S)-5′,8-cyclo-2′-deoxyadenosine on the base excision repair of oxidatively generated clustered DNA damage. A biochemical and theoretical study. Org. Biomol. Chem. 2014, 43, 8671–8682. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Geacintov, N.E.; Krokidis, M.G.; Liu, Y.; Masi, A.; Shafirovich, V.; Terzidis, M.A.; Tsegay, P.S. 5′,8-Cyclopurine lesions in DNA damage: chemical, analytical, biological, and diagnostic significance. Cells 2019, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Terzidis, M.A. Purine 5′,8-cyclonucleoside lesions: chemistry and biology. Chem. Soc. Rev. 2011, 40, 1368–1382. [Google Scholar] [CrossRef] [PubMed]
- Brooks, P.J. The 8,5′-cyclopurine-2′-deoxynucleosides: candidate neurodegenerative DNA lesions in xeroderma pigmentosum, and unique probes of transcription and nucleotide excision repair. DNA Repair 2008, 7, 1168–1179. [Google Scholar] [CrossRef]
- Romieu, A.; Gasparutto, D.; Cadet, J. Synthesis and characterization of oligonucleotides containing 5′,8-cyclopurine-2′-Deoxyribonucleosides: (5′R)-5′,8-cyclo-2′-deoxyadenosine (5′S)-5′,8-cyclo-2′-deoxyguanosine, and (5′R)-5′,8-cyclo-2′-deoxyguanosine. Chem. Res. Toxicol. 1999, 12, 412–421. [Google Scholar] [CrossRef]
- Kibbe, W.A. OligoCalc: An online oligonucleotide properties calculator. Nucleic Acids Res. 2007, 35. [Google Scholar] [CrossRef]
- Parikh, S.S.; Walcher, G.; Jones, G.D.; Slupphaug, G.; Krokan, H.E.; Blackburn, G.M.; Tainer, J.A. Uracil-DNA glycosylase-DNA substrate and product structures: conformational strain promotes catalytic efficiency by coupled stereoelectronic effects. Proc. Natl. Acad. Sci. USA 2000, 97, 5083–5088. [Google Scholar] [CrossRef]
- Dinner, A.R.; Blackburn, G.M.; Karplus, M. Uracil-DNA glycosylase acts by substrate autocatalysis. Nature 2001, 413, 752–755. [Google Scholar] [CrossRef]
- Gu, J.; Xie, Y.; Schaefer, H.F. Electron attachment to nucleotides in aqueous solution. Chem. Phys. Chem. 2006, 7, 1885–1887. [Google Scholar] [CrossRef] [PubMed]
- Gu, J.; Xie, Y.; Schaefer, H.F. Electron attachment to DNA single strands: gas phase and aqueous solution. Nucleic Acids Res. 2007, 35, 5165–5172. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Dapprich, S.; Komáromi, I.; Byun, K.S.; Morokuma, K.; Frisch, M.J. A new ONIOM implementation in Gaussian98. Part, I. The calculation of energies, gradients, vibrational frequencies and electric field derivatives. J. Mol. Struct. 1990, 462, 1–21. [Google Scholar] [CrossRef]
- Lin, H.; Truhlar, D.G. Redistributed charge and dipole schemes for combined quantum mechanical and molecular mechanical calculations. J. Phys. Chem. A 2005, 109, 3991–4004. [Google Scholar] [CrossRef] [PubMed]
- Hehre, W.J. Ab Initio Molecular Orbital Theory. Acc. Chem. Res. 1976, 9, 63–101. [Google Scholar] [CrossRef]
- Rugne, E.; Gross, E.K.U. Density-Functional Theory for Time-Dependent Systems. Phys. Rev. Lett. 1984, 52, 997–1000. [Google Scholar]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09, Revision, A.02; Gaussian Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- BIOVIA. Discovery Studio Visualizer; v16.1.0.15350; BIOVIA: San Diego, CA, USA, 2015. [Google Scholar]
- Tokuyama, Y.; Furusawa, Y.; Ide, H.; Yasui, A.; Terato, H. Role of isolated and clustered DNA damage and the post-irradiating repair process in the effects of heavy ion beam irradiation. J. Rad. Res. 2015, 56, 446–455. [Google Scholar] [CrossRef]
- Mouret, S.; Baudouin, C.; Charveron, M.; Favier, A.; Cadet, J.; Douki, T. Cyclobutane pyrimidine dimers are predominant DNA lesions in whole human skin exposed to UVA radiation. Proc. Natl. Acad. Sci. USA 2006, 103, 13765–13770. [Google Scholar] [CrossRef]
- Barak, Y.; Cohen-Fix, O.; Livneh, Z. Deamination of cytosine-containing pyrimidine photodimers in UV-irradiated DNA. Significance for UV light mutagenesis. J. Biol. Chem. 1995, 270, 24174–24179. [Google Scholar] [CrossRef]
- Lomax, M.E.; Cunniffe, S.; O’Neill, P. 8-OxoG retards the activity of the ligase III/XRCC1 complex during the repair of a single-strand break, when present within a clustered DNA damage site. DNA Repair 2004, 3, 289–299. [Google Scholar] [CrossRef]
- Bellon, S.; Shikazono, N.; Cunniffe, S.; Lomax, M.; O’Neill, P. Processing of thymine glycol in a clustered DNA damage site: mutagenic or cytotoxic. Nucleic Acid. Res. 2009, 37, 4430–4440. [Google Scholar] [CrossRef] [PubMed]
- Gollapalle, E.; Wang, R.; Adetolu, R.; Tsao, D.; Francisco, D.; Sigounas, G.; Georgakilas, A.G. Detection of oxidative clustered DNA lesions in X-irradiated mouse skin tissues and human MCF-7 breast cancer cells. Radiat. Res. 2007, 167, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Georgakilas, A.G.; Bennett, P.V.; Wilson, D.M. Sutherland BM. Processing of bistranded abasic DNA clusters in γ-irradiated human hematopoietic cells. Nucleic Acids Res. 2004, 32, 5609–5620. [Google Scholar] [CrossRef] [PubMed]
- Moreira, P.I.; Nunomura, A.; Nakamura, M.; Takeda, A.; Shenk, J.C.; Aliev, G.; Smith, M.A.; Perry, G. Nucleic acid oxidation in Alzheimer disease. Free Rad. Biol. Med. 2008, 44, 1493–1505. [Google Scholar] [CrossRef]
- Leandro, G.S.; Sykora, P.; Bohra, V.A. The Impact of Base Excision DNA Repair in Age-Related Neurodegenerative Diseases. Mutat Res. 2015, 776, 31–39. [Google Scholar] [CrossRef]
- Muller, F.L.; Lustgarten, M.S.; Jang, Y.; Richardson, A.; Van Remmen, H. Trends on oxidative angina theories. Free Radic. Biol. Med. 2007, 43, 477–503. [Google Scholar] [CrossRef]
- Brooks, P.J. The cyclopurine deoxynucleosides: DNA repair, biological effects, mechanistic insights, and unanswered questions. Free Radic. Biol. Med. 2017, 107, 90–100. [Google Scholar] [CrossRef]
- Schormann, N.; Ricciardi, R.; Chattopadhyay, D. Uracil-DNA glycosylases-structural and functional perspectives on an essential family of DNA repair enzymes. Protein Sci. 2014, 23, 1667–1685. [Google Scholar] [CrossRef]
- Slupphaug, G.; Eftedal, I.; Kavli, B.; Bharati, S.; Helle, N.M.; Haug, T.; Levine, D.W.; Krokan, H.E. Properties of a recombinant human uracil-DNA glycosylase from the UNG gene and evidence that UNG encodes the major uracil-DNA glycosylase. Biochemistry 1995, 34, 128–138. [Google Scholar] [CrossRef]
- Li, M.; Wilson, D.M. Human Apurinic/Apyrimidinic Endonuclease 1. Antioxid. Redox Signal. 2014, 20, 678–707. [Google Scholar] [CrossRef]
- Gros, L.; Ishchenko, A.A.; Ide, H.; Elder, R.H.; Saparbaev, M.K. The major human AP endonuclease (Ape1) is involved in the nucleotide incision repair pathway. Nucleic Acids Res. 2004, 32, 73–81. [Google Scholar] [CrossRef] [PubMed]
- Margolin, Y.; Dedon, P.C. A general method for quantifying sequence effects on nucleobase oxidation in DNA. Methods Mol. Biol. 2010, 610, 325–340. [Google Scholar] [PubMed]
- Ashton, N.W.; Bolderson, E.; Cubeddu, L.; O’Byrne, K.J.; Richard, D.J. Human single-stranded DNA binding proteins are essential for maintaining genomic stability. BCM Mol. Biol. 2013, 14, 1–20. [Google Scholar] [CrossRef] [PubMed]
- Breslauer, K.J.; Frank, R.; Blocker, H.; Marky, L.A. Predicting DNA duplex stability from the base sequence. Proc. Natl. Acad. Sci. USA 1986, 83, 3746–3750. [Google Scholar] [CrossRef] [PubMed]
- Karwowski, B.T. The role of (5′R) and (5′S) 5′,8-cyclo-2′-deoxyadenosine in ds-DNA structure: A comparative QM/MM theoretical study. Comput. Chem. 2013, 1010, 38–44. [Google Scholar]
- Zaliznyak, T.; Lukin, M.; de los Santos, C. Structure and stability of duplex DNA containing (5′S)-5′,8-cyclo-2′-deoxyadenosine: an oxidatively generated lesion repaired by NER. Chem. Res. Toxicol. 2012, 25, 2103–2111. [Google Scholar] [CrossRef]
- Huang, H.; Das, R.S.; Basu, A.K.; Stone, M.P. Structure of (5′S)-8,5′-cyclo-2′-deoxyguanosine in DNA. J. Am. Chem. Soc. 2011, 133, 20357–20368. [Google Scholar] [CrossRef]
- Huang, H.; Das, R.S.; Basu, A.K.; Stone, M.P. Structures of (5′S)-8,5′-cyclo-2′-deoxyguanosine mismatched with dA or dT. Chem. Res. Toxicol. 2012, 25, 478–490. [Google Scholar] [CrossRef]
- Haromy, T.P.; Raleigh, J.; Sundaralingam, M. Enzyme-bound conformations of nucleotide substrates. X-ray structure and absolute configuration of 8,5′-cycloadenosine monohydrate. Biochemistry 1980, 19, 1718–1722. [Google Scholar] [CrossRef]
- Takahara, P.M.; Rosenzweig, A.C.; Frederick, C.A.; Lippard, S.J. Crystal structure of double-stranded DNA containing the major adduct of the anticancer drug cisplatin. Nature 1995, 377, 649–652. [Google Scholar] [CrossRef]
- Gray, D.M.; Ratliff, R.L.; Vaughan, M.R. Circular dichroism spectroscopy of DNA. Methods Enzymol. 1992, 211, 389–406. [Google Scholar] [PubMed]
- Miyahara, T.; Nakatsuji, H.; Sugiyama, H. Helical structure and circular dichroism spectra of DNA: A theoretical study. J. Phys. Chem. 2013, 117, 42–55. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Price, N.E.; Johnson, K.M.; Wang, Y.; Gates, K.S. Interstrand cross-links arising from strand breaks at true abasic sites in duplex DNA. Nucleic Acids Res. 2017, 45, 6275–6283. [Google Scholar] [CrossRef][Green Version]
- Barrett, T.E.; Sawa, R.; Panayotou, G.; Barlow, T.; Brown, T.; Jiricny, J.; Pearl, L.H. Crystal structure of a G:T/U mismatch-specific DNA glycosylase: mismatch recognition by complementary-strand interactions. Cell 1998, 92, 117–129. [Google Scholar] [CrossRef]
- Zharkov, D.O.; Mechetin, G.V.; Nevinsky, G.A. Uracil DNA glycosylase: structural, thermodynamic and kinetic aspects of lesion search and recognition. Mutat. Res. 2010, 685, 11–20. [Google Scholar] [CrossRef]
- Gokey, T.; Hang, B.; Guliaev, A.B. Cadmium(II) inhibition of human uracil-DNA glycosylase by catalytic water supplantation. Sci. Rep. 2016, 6, 1–11. [Google Scholar] [CrossRef]
- Dong, H.; Hua, W.; Li, S. Estimation on the Individual Hydrogen-Bond Strength in Molecules with Multiple Hydrogen Bonds. J. Phys. Chem. A. 2007, 111, 2941–2945. [Google Scholar] [CrossRef]
- Gil, A.; Branchadell, V.; Bertran, J.; Oliva, A. An analysis of the different behavior of DNA and RNA through the study of the mutual relationship between stacking and hydrogen bonding. J. Phys. Chem. B 2009, 113, 4907–4914. [Google Scholar] [CrossRef]
- Altona, C.; Sundaralingam, M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J. Am. Chem. Soc. 1972, 94, 8205–8212. [Google Scholar] [CrossRef]
- Karwowski, B.T.; Gaillard, J.; Granda, A.; Cadet, J. Effect of (5′S)-5′,8-cyclo-2′-deoxyadenosine on the conformation of di and trinucleotides. A NMR and DFT study. Org. Biomol. Chem. 2008, 6, 3408–3413. [Google Scholar] [CrossRef]
- Venkhataraman, R.; Donald, C.D.; Roy, R.; You, H.J.; Doetsch, P.W.; Kow, Y.W. Enzymatic processing of DNA containing tandem dihydrouracil by endonucleases III and VIII. Nucleic Acids Res. 2001, 29, 407–414. [Google Scholar] [CrossRef] [PubMed]




| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 20 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 30 | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 40 | Tm [oC] | ||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Matrix | 3′ | T | A | G | A | A | C | A | G | T | C | C | T | T | A | T | A | A | C | A | G | A | G | A | T | A | C | G | A | G | G | G | T | G | G | T | T | T | C | C | G | |
| ScdA(-7) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | U | G | T | C | T | C | T | cdA | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C | 79.02 |
| ScdA(-5) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | U | C | T | C | T | cdA | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C | 79.02 |
| ScdA(-3) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | U | C | T | cdA | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C | 79.02 |
| ScdA(-1) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | U | cdA | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C | 79.02 |
| RcdA(-1) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | U | cdA | T | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C | 79.02 |
| ScdA(+1) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | cdA | U | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C | 80.00 |
| RcdA(+1) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | cdA | U | G | C | T | C | C | C | A | C | C | A | A | A | G | G | C | 79.02 |
| ScdA(+3) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | cdA | T | G | U | T | C | C | C | A | C | C | A | A | A | G | G | C | 77.00 |
| ScdA(+5) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | cdA | T | G | C | T | U | C | C | A | C | C | A | A | A | G | G | C | 76.02 |
| ScdA(+7) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | cdA | T | G | C | T | C | C | U | A | C | C | A | A | A | G | G | C | 76.02 |
| Cont.dU(0) | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | U | C | T | A | T | G | C | T | C | C | T | A | C | C | A | A | A | G | G | C | 76.02 |
| Native | 5′ | C | T | C | T | T | G | T | C | A | G | G | A | A | T | A | T | T | G | T | C | T | C | T | A | T | G | C | T | C | C | T | A | C | C | A | A | A | G | G | C | 82.00 |
| Nucleoside | υ2 * | Phase | Amplitude | Conformation |
|---|---|---|---|---|
| nucleoside relaxed | ||||
| dA | −32.67 | 165.11 | 33.80 | C2′-endo |
| 5′R-cdA | 10.98 | 283.23 | 47.99 | O4′-exo |
| 5′S-cdA | 10.92 | 283.35 | 47.31 | O4′-exo |
| nucleoside stressed | ||||
| 5′R-cdA(+1) | −1.07 | 268.75 | 48.78 | O4′-exo |
| 5′R-cdA(−1) | −28.68 | 236.65 | 52.18 | C3′-exo |
| 5′S-cdA(+1) | −2.01 | 267.58 | 47.67 | O4′-exo |
| 5′S-cdA(−1) | −29.63 | 235.62 | 52.47 | C3′-exo |
| dA(+1) | −27.20 | 193.68 | 27.99 | C2′-endo |
| dA(-1) | −28.79 | 178.13 | 28.81 | C2′-endo |

| Nucleoside | pdU Position +1 | pdU Position −1 |
|---|---|---|
| 5′S-cdA | −0.61 | −15.91 |
| 5′R-cdA | −0.37 | −18.16 |
| dA | −4.62 | −4.74 |
| cdA Position to pdU | ds-DNA | ||
|---|---|---|---|
| Cont.pdU | ScdA | RcdA | |
| +1 | 15.23 | 5.05 | 14.88 |
| −1 | 12.48 | 5.23 | 5.23 |
© 2019 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Karwowski, B.T. The Influence of (5′R)- and (5′S)-5′,8-Cyclo-2′-Deoxyadenosine on UDG and hAPE1 Activity. Tandem Lesions are the Base Excision Repair System’s Nightmare. Cells 2019, 8, 1303. https://doi.org/10.3390/cells8111303
Karwowski BT. The Influence of (5′R)- and (5′S)-5′,8-Cyclo-2′-Deoxyadenosine on UDG and hAPE1 Activity. Tandem Lesions are the Base Excision Repair System’s Nightmare. Cells. 2019; 8(11):1303. https://doi.org/10.3390/cells8111303
Chicago/Turabian StyleKarwowski, Bolesław T. 2019. "The Influence of (5′R)- and (5′S)-5′,8-Cyclo-2′-Deoxyadenosine on UDG and hAPE1 Activity. Tandem Lesions are the Base Excision Repair System’s Nightmare" Cells 8, no. 11: 1303. https://doi.org/10.3390/cells8111303
APA StyleKarwowski, B. T. (2019). The Influence of (5′R)- and (5′S)-5′,8-Cyclo-2′-Deoxyadenosine on UDG and hAPE1 Activity. Tandem Lesions are the Base Excision Repair System’s Nightmare. Cells, 8(11), 1303. https://doi.org/10.3390/cells8111303