Off-Target Deletion of Conditional Dbc1 Allele in the Foxp3YFP-Cre Mouse Line under Specific Setting
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mice
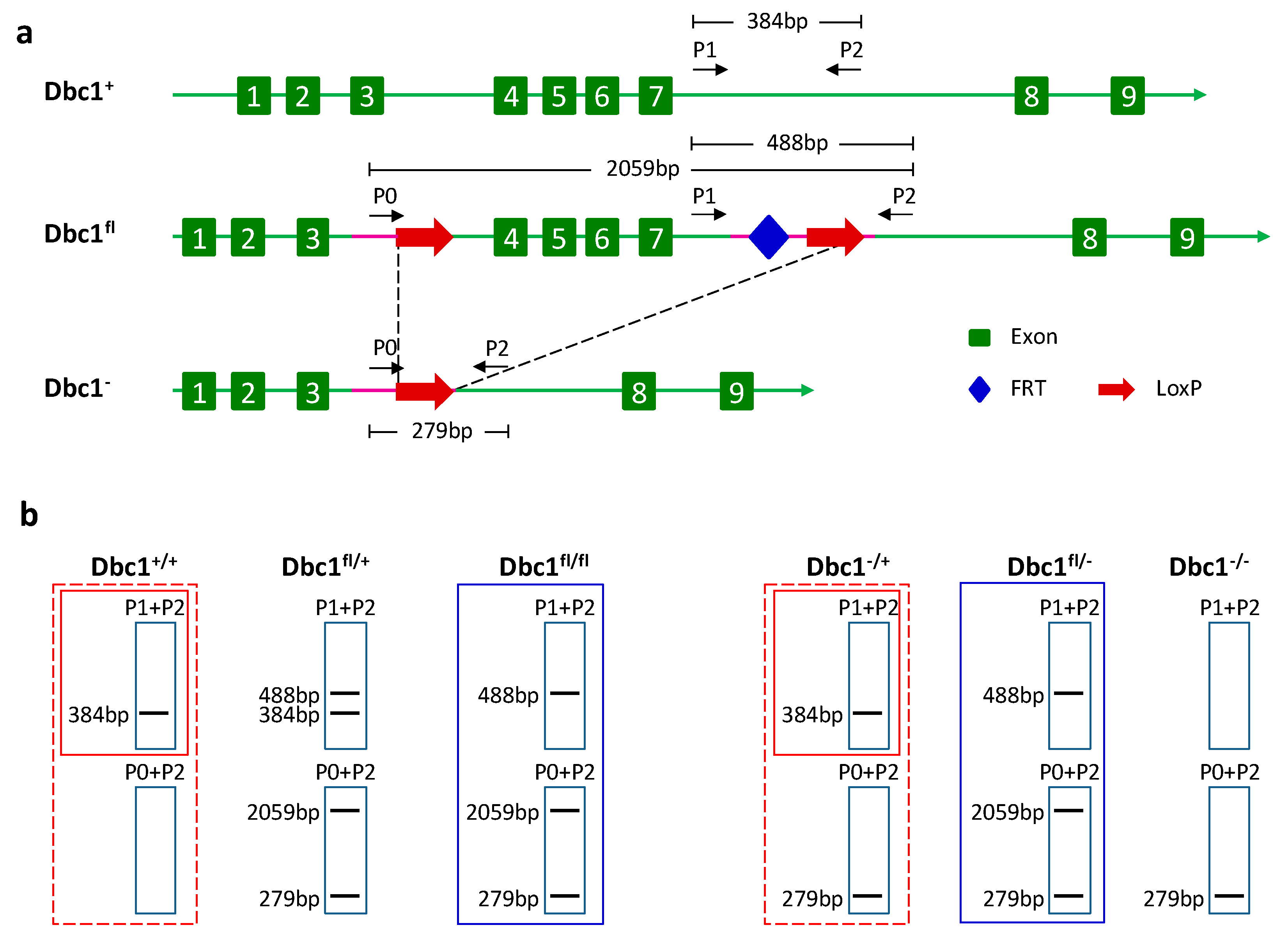
2.2. Genomic PCR
2.3. Quantitative PCR (qPCR)
2.4. Immunoblot Analysis
3. Results
3.1. The Conditional Dbc1 Allele Was Completely Knocked Out in Some Progeny of Foxp3Cre/Y Dbc1fl/+ × Foxp3Cre/Cre Dbc1fl/Fl Mice
3.2. Germline Recombination of Floxed Dbc1 Allele Occurs in the Male Foxp3Cre/Y Dbc1fl/+ Mice
3.3. The Floxed Dbc1 Allele Was Recombined in non-T Cells of Some Foxp3Cre Dbc1fl Mice
3.4. Expression of Cre Recombinase in the Fetus Results in Germline Recombination and Dbc1 Knock out Mice
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Sauer, B. Functional expression of the cre-lox site-specific recombination system in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 1987, 7, 2087–2096. [Google Scholar] [CrossRef] [PubMed]
- Hou, L.; Yau, Y.Y.; Wei, J.; Han, Z.; Dong, Z.; Ow, D.W. An open-source system for in planta gene stacking by Bxb1 and Cre recombinases. Mol. Plant 2014, 7, 1756–1765. [Google Scholar] [CrossRef] [PubMed]
- Sauer, B.; Henderson, N. Site-specific DNA recombination in mammalian cells by the Cre recombinase of bacteriophage P1. Proc. Natl. Acad. Sci. USA 1988, 85, 5166–5170. [Google Scholar] [CrossRef] [PubMed]
- Gu, H.; Marth, J.D.; Orban, P.C.; Mossmann, H.; Rajewsky, K. Deletion of a DNA polymerase beta gene segment in T cells using cell type-specific gene targeting. Science 1994, 265, 103–106. [Google Scholar] [CrossRef] [PubMed]
- Tsien, J.Z.; Chen, D.F.; Gerber, D.; Tom, C.; Mercer, E.H.; Anderson, D.J.; Mayford, M.; Kandel, E.R.; Tonegawa, S. Subregion- and cell type-restricted gene knockout in mouse brain. Cell 1996, 87, 1317–1326. [Google Scholar] [CrossRef]
- Wu, J.; Zhang, H.; Shi, X.; Xiao, X.; Fan, Y.; Minze, L.J.; Wang, J.; Ghobrial, R.M.; Xia, J.; Sciammas, R.; et al. Ablation of Transcription Factor IRF4 Promotes Transplant Acceptance by Driving Allogenic CD4(+) T Cell Dysfunction. Immunity 2017, 47, 1114–1128.e6. [Google Scholar] [CrossRef] [PubMed]
- Ramalingam, R.; Larmonier, C.B.; Thurston, R.D.; Midura-Kiela, M.T.; Zheng, S.G.; Ghishan, F.K.; Kiela, P.R. Dendritic cell-specific disruption of TGF-beta receptor II leads to altered regulatory T cell phenotype and spontaneous multiorgan autoimmunity. J. Immunol. 2012, 189, 3878–3893. [Google Scholar] [CrossRef]
- Liput, D.J. Cre-Recombinase Dependent Germline Deletion of a Conditional Allele in the Rgs9cre Mouse Line. Front. Neural. Circuits 2018, 12, 68. [Google Scholar] [CrossRef]
- Song, A.J.; Palmiter, R.D. Detecting and Avoiding Problems When Using the Cre-lox System. Trends Genet. 2018, 34, 333–340. [Google Scholar] [CrossRef]
- Hu, H.; Cavendish, J.Z.; Agmon, A. Not all that glitters is gold: Off-target recombination in the somatostatin-IRES-Cre mouse line labels a subset of fast-spiking interneurons. Front. Neural Circuits 2013, 7, 195. [Google Scholar] [CrossRef]
- Rempe, D.; Vangeison, G.; Hamilton, J.; Li, Y.; Jepson, M.; Federoff, H.J. Synapsin I Cre transgene expression in male mice produces germline recombination in progeny. Genesis 2006, 44, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Kobayashi, Y.; Hensch, T.K. Germline recombination by conditional gene targeting with Parvalbumin-Cre lines. Front. Neural Circuits 2013, 7, 168. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, J.; Dublin, P.; Griemsmann, S.; Klein, A.; Brehm, R.; Bedner, P.; Fleischmann, B.K.; Steinhauser, C.; Theis, M. Germ-line recombination activity of the widely used hGFAP-Cre and nestin-Cre transgenes. PLoS ONE 2013, 8, e82818. [Google Scholar] [CrossRef] [PubMed]
- He, Y.; Sun, X.; Wang, L.; Mishina, Y.; Guan, J.L.; Liu, F. Male germline recombination of a conditional allele by the widely used Dermo1-cre (Twist2-cre) transgene. Genesis 2017, 55. [Google Scholar] [CrossRef]
- Horwitz, D.A.; Zheng, S.G.; Gray, J.D.; Wang, J.H.; Ohtsuka, K.; Yamagiwa, S. Regulatory T cells generated ex vivo as an approach for the therapy of autoimmune disease. Semin. Immunol. 2004, 16, 135–143. [Google Scholar] [CrossRef]
- Hu, G.; Liu, Z.; Zheng, C.; Zheng, S.G. Antigen-non-specific regulation centered on CD25+Foxp3+ Treg cells. Cell Mol. Immunol. 2010, 7, 414–418. [Google Scholar] [CrossRef] [Green Version]
- Rubtsov, Y.P.; Rasmussen, J.P.; Chi, E.Y.; Fontenot, J.; Castelli, L.; Ye, X.; Treuting, P.; Siewe, L.; Roers, A.; Henderson, W.R., Jr.; et al. Regulatory T cell-derived interleukin-10 limits inflammation at environmental interfaces. Immunity 2008, 28, 546–558. [Google Scholar] [CrossRef]
- Weinberg, S.E.; Singer, B.D.; Steinert, E.M.; Martinez, C.A.; Mehta, M.M.; Martinez-Reyes, I.; Gao, P.; Helmin, K.A.; Abdala-Valencia, H.; Sena, L.A.; et al. Mitochondrial complex III is essential for suppressive function of regulatory T cells. Nature 2019, 565, 495–499. [Google Scholar] [CrossRef]
- Hwang, S.M.; Sharma, G.; Verma, R.; Byun, S.; Rudra, D.; Im, S.H. Inflammation-induced Id2 promotes plasticity in regulatory T cells. Nat. Commun. 2018, 9, 4736. [Google Scholar] [CrossRef]
- Ghosh, S.; Roy-Chowdhuri, S.; Kang, K.; Im, S.H.; Rudra, D. The transcription factor Foxp1 preserves integrity of an active Foxp3 locus in extrathymic Treg cells. Nat. Commun. 2018, 9, 4473. [Google Scholar] [CrossRef]
- Xu, M.; Pokrovskii, M.; Ding, Y.; Yi, R.; Au, C.; Harrison, O.J.; Galan, C.; Belkaid, Y.; Bonneau, R.; Littman, D.R. c-MAF-dependent regulatory T cells mediate immunological tolerance to a gut pathobiont. Nature 2018, 554, 373–377. [Google Scholar] [CrossRef] [PubMed]
- Franckaert, D.; Dooley, J.; Roos, E.; Floess, S.; Huehn, J.; Luche, H.; Fehling, H.J.; Liston, A.; Linterman, M.A.; Schlenner, S.M. Promiscuous Foxp3-cre activity reveals a differential requirement for CD28 in Foxp3(+) and Foxp3(−) T cells. Immunol. Cell Biol. 2015, 93, 417–423. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Tang, J.; Chen, W.; Li, Q.; Nie, J.; Lin, F.; Wu, Q.; Chen, Z.; Gao, Z.; Fan, H.; et al. Inflammation negatively regulates FOXP3 and regulatory T-cell function via DBC1. Proc. Natl. Acad. Sci. USA 2015, 112, E3246–E3254. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, P.P.; Fitzpatrick, D.R.; Beard, C.; Jessup, H.K.; Lehar, S.; Makar, K.W.; Perez-Melgosa, M.; Sweetser, M.T.; Schlissel, M.S.; Nguyen, S.; et al. A critical role for Dnmt1 and DNA methylation in T cell development, function, and survival. Immunity 2001, 15, 763–774. [Google Scholar] [CrossRef]
- Israelsen, W.J.; Dayton, T.L.; Davidson, S.M.; Fiske, B.P.; Hosios, A.M.; Bellinger, G.; Li, J.; Yu, Y.; Sasaki, M.; Horner, J.W.; et al. PKM2 isoform-specific deletion reveals a differential requirement for pyruvate kinase in tumor cells. Cell 2013, 155, 397–409. [Google Scholar] [CrossRef] [PubMed]
- Westendorf, K.; Durek, P.; Ayew, S.; Mashreghi, M.F.; Radbruch, A. Chromosomal localisation of the CD4cre transgene in B6. Cg-Tg(Cd4-cre)1Cwi mice. J. Immunol. Methods 2016, 436, 54–57. [Google Scholar] [CrossRef] [PubMed]
- Lu, L.; Lan, Q.; Li, Z.; Zhou, X.; Gu, J.; Li, Q.; Wang, J.; Chen, M.; Liu, Y.; Shen, Y.; et al. Critical role of all-trans retinoic acid in stabilizing human natural regulatory T cells under inflammatory conditions. Proc. Natl. Acad. Sci. USA 2014, 111, E3432–E3440. [Google Scholar] [CrossRef]
- Singh, A.K.; Khare, P.; Obaid, A.; Conlon, K.P.; Basrur, V.; DePinho, R.A.; Venuprasad, K. SUMOylation of ROR-gammat inhibits IL-17 expression and inflammation via HDAC2. Nat. Commun. 2018, 9, 4515. [Google Scholar] [CrossRef]
- Peng, W.X.; Zhu, S.L.; Zhang, B.Y.; Shi, Y.M.; Feng, X.X.; Liu, F.; Huang, J.L.; Zheng, S.G. Smoothened Regulates Migration of Fibroblast-Like Synoviocytes in Rheumatoid Arthritis via Activation of Rho GTPase Signaling. Front. Immunol. 2017, 8, 159. [Google Scholar] [CrossRef] [Green Version]
- Jasurda, J.S.; Jung, D.O.; Froeter, E.D.; Schwartz, D.B.; Hopkins, T.D.; Farris, C.L.; McGee, S.; Narayan, P.; Ellsworth, B.S. The forkhead transcription factor, FOXP3: A critical role in male fertility in mice. Biol. Reprod. 2014, 90, 4. [Google Scholar] [CrossRef]
- Gray, P.A.; Fu, H.; Luo, P.; Zhao, Q.; Yu, J.; Ferrari, A.; Tenzen, T.; Yuk, D.I.; Tsung, E.F.; Cai, Z.; et al. Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 2004, 306, 2255–2257. [Google Scholar] [CrossRef] [PubMed]
- Diez-Roux, G.; Banfi, S.; Sultan, M.; Geffers, L.; Anand, S.; Rozado, D.; Magen, A.; Canidio, E.; Pagani, M.; Peluso, I.; et al. A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 2011, 9, e1000582. [Google Scholar] [CrossRef] [PubMed]
- Schmidt-Supprian, M.; Rajewsky, K. Vagaries of conditional gene targeting. Nat. Immunol. 2007, 8, 665–668. [Google Scholar] [CrossRef] [PubMed]




| Breeding Mice | Mouse No. | Frequency of Recombined Dbc1 Fragment | Litter Size | Number of Dbc1−/+ Mice in Progeny |
|---|---|---|---|---|
| Foxp3Cre/YDbc1fl/+ × Wt | YN-3, 4, 5 | low | 89 | 0 |
| Foxp3Cre/YDbc1fl/+ × Wt | YN-1, 2 | high | 67 | 2 |
| Wt × Foxp3Cre/+ Dbc1fl/+ | VN-3, 4 | low | 55 | 0 |
| Wt × Foxp3Cre/+ Dbc1fl/+ | VN-1, 2, 5, 6 | high | 64 | 0 |
| Foxp3Cre/YDbc1fl/fl × Wt | YM-1, 2 | low | 75 | 0 |
| Wt × Foxp3Cre/Cre Dbc1fl/fl | XM-1, 2 | low | 79 | 0 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, C.; Zhu, F.; Wang, J.; Zhang, W.; Bellanti, J.A.; Li, B.; Brand, D.; Olsen, N.; Zheng, S.G. Off-Target Deletion of Conditional Dbc1 Allele in the Foxp3YFP-Cre Mouse Line under Specific Setting. Cells 2019, 8, 1309. https://doi.org/10.3390/cells8111309
Xie C, Zhu F, Wang J, Zhang W, Bellanti JA, Li B, Brand D, Olsen N, Zheng SG. Off-Target Deletion of Conditional Dbc1 Allele in the Foxp3YFP-Cre Mouse Line under Specific Setting. Cells. 2019; 8(11):1309. https://doi.org/10.3390/cells8111309
Chicago/Turabian StyleXie, Chichu, Fangming Zhu, Julie Wang, Weizhou Zhang, Joseph A. Bellanti, Bin Li, David Brand, Nancy Olsen, and Song Guo Zheng. 2019. "Off-Target Deletion of Conditional Dbc1 Allele in the Foxp3YFP-Cre Mouse Line under Specific Setting" Cells 8, no. 11: 1309. https://doi.org/10.3390/cells8111309
APA StyleXie, C., Zhu, F., Wang, J., Zhang, W., Bellanti, J. A., Li, B., Brand, D., Olsen, N., & Zheng, S. G. (2019). Off-Target Deletion of Conditional Dbc1 Allele in the Foxp3YFP-Cre Mouse Line under Specific Setting. Cells, 8(11), 1309. https://doi.org/10.3390/cells8111309






