rDNA Clusters Make Contact with Genes that Are Involved in Differentiation and Cancer and Change Contacts after Heat Shock Treatment
Abstract
1. Introduction
2. Materials and methods
2.1. 4C Procedure
2.2. Computer Treatments
2.3. Control of Reproducibility of 4C Sequencing
2.4. Filtering for Contact Detection and Search for Gene-Contact Coordinates
2.5. Viewpoint and Estimation of Proximity Effects for Cis-Interactions
2.6. Gene Quantification and Differential 4C Analysis
2.7. Genome-Wide Profiles
2.8. RNA-Seq Analysis
2.9. Statistical Calculations
3. Results
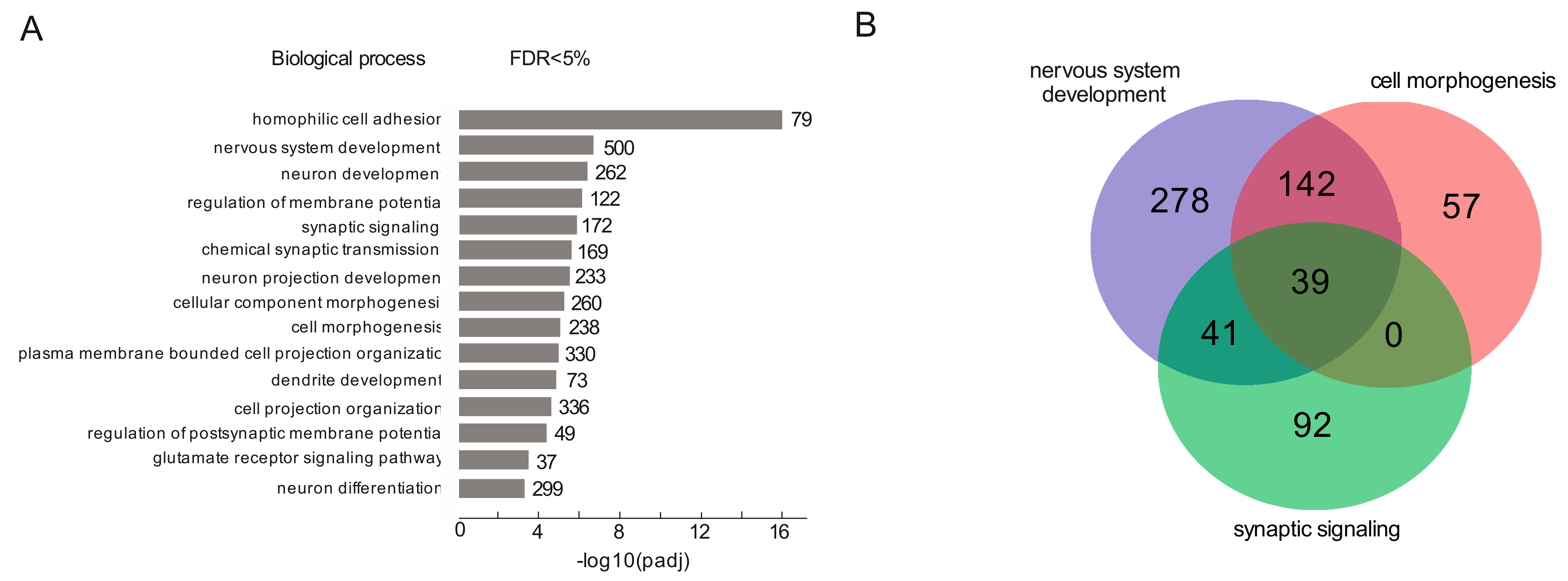
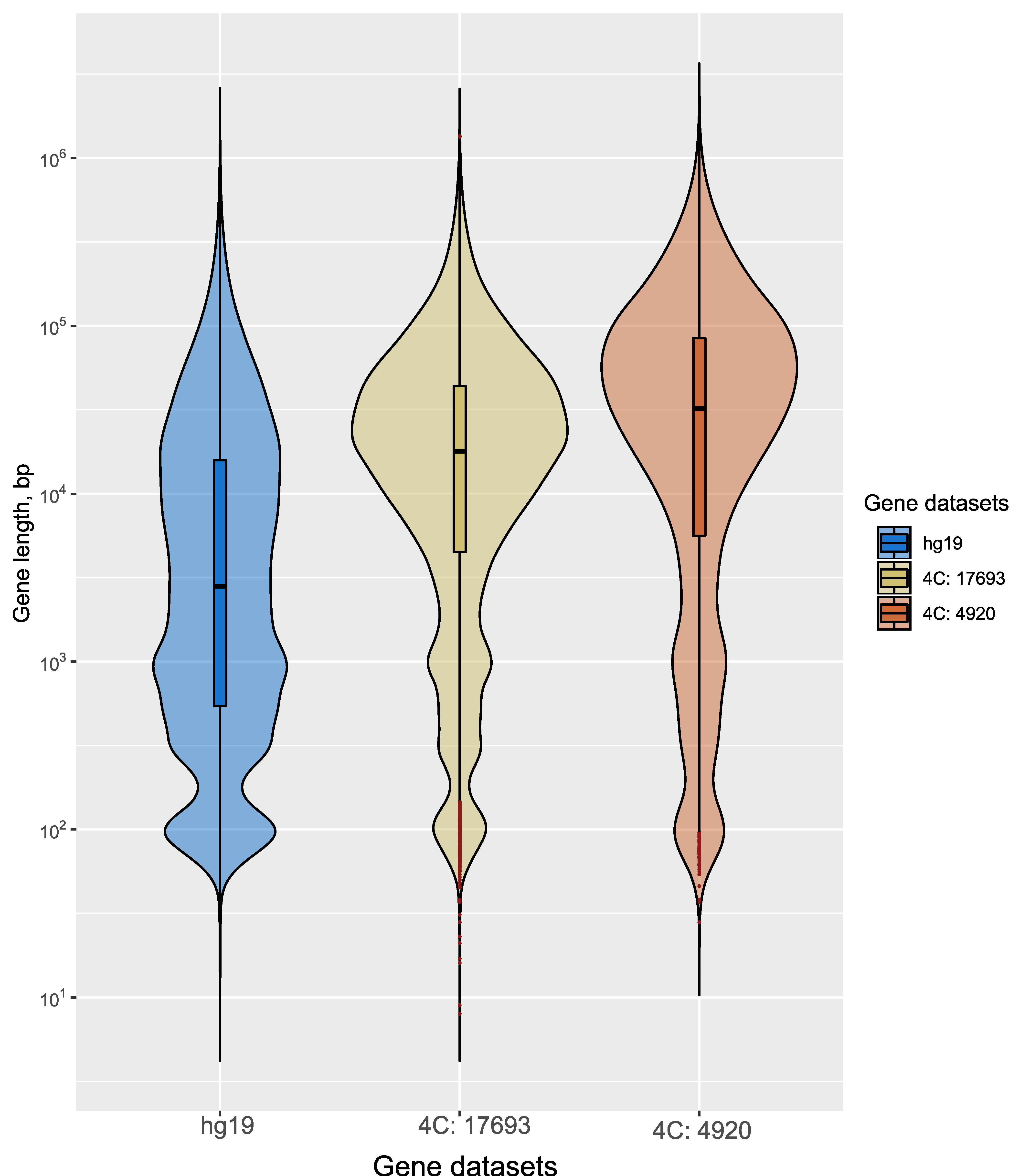
3.1. rDNA Clusters Contact the Genes Involved in Differentiation of Neurons in HEK293T Cells
3.2. rDNA-Contacting Genes Are Associated with Different Cancers
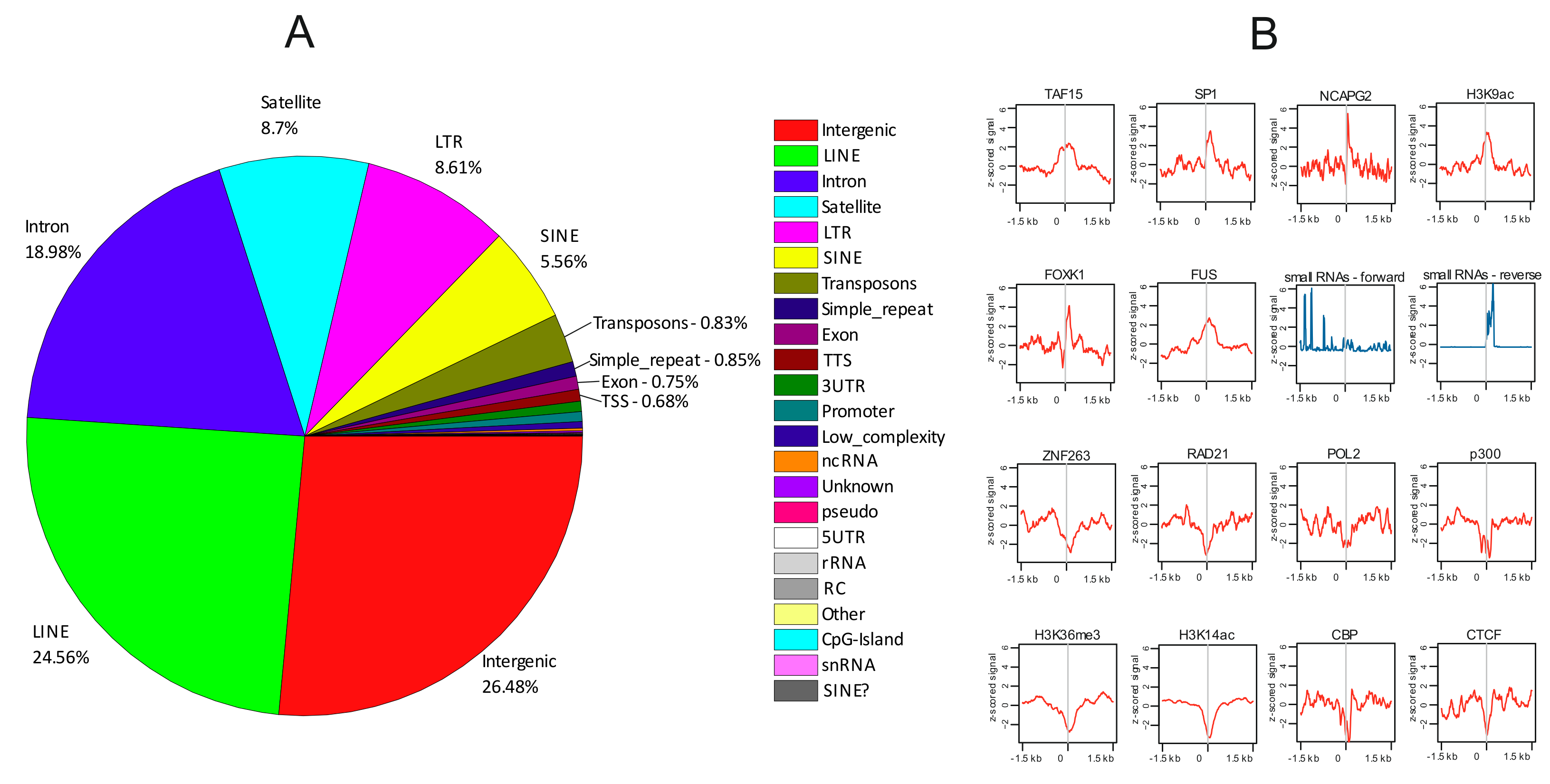
3.3. rDNA-Contacting Sites at Genes Possess a Specific Set of Epigenetic Marks
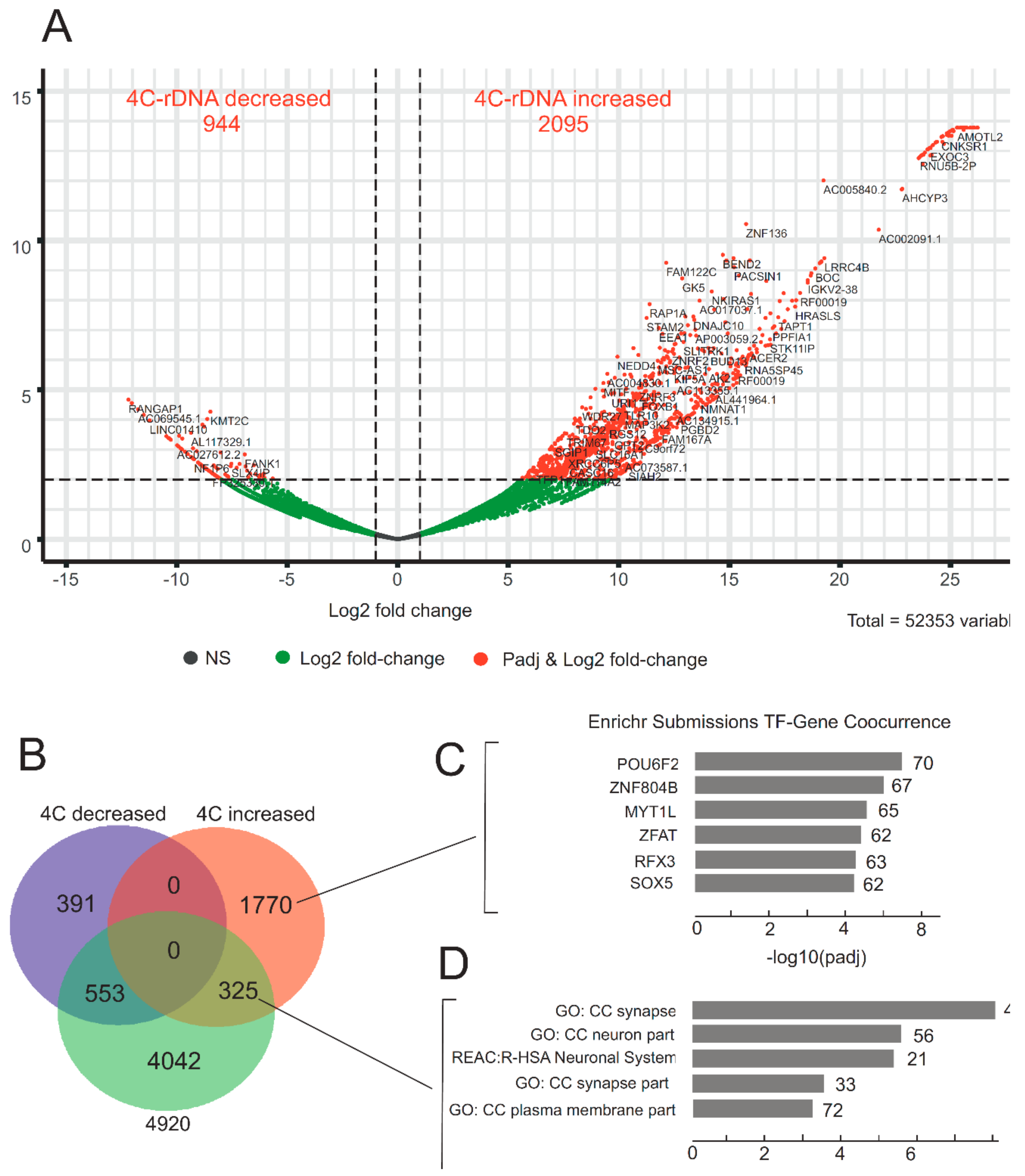
3.4. Heat Shock Treatment Dramatically Changes the Pattern of rDNA-Contacting Sites
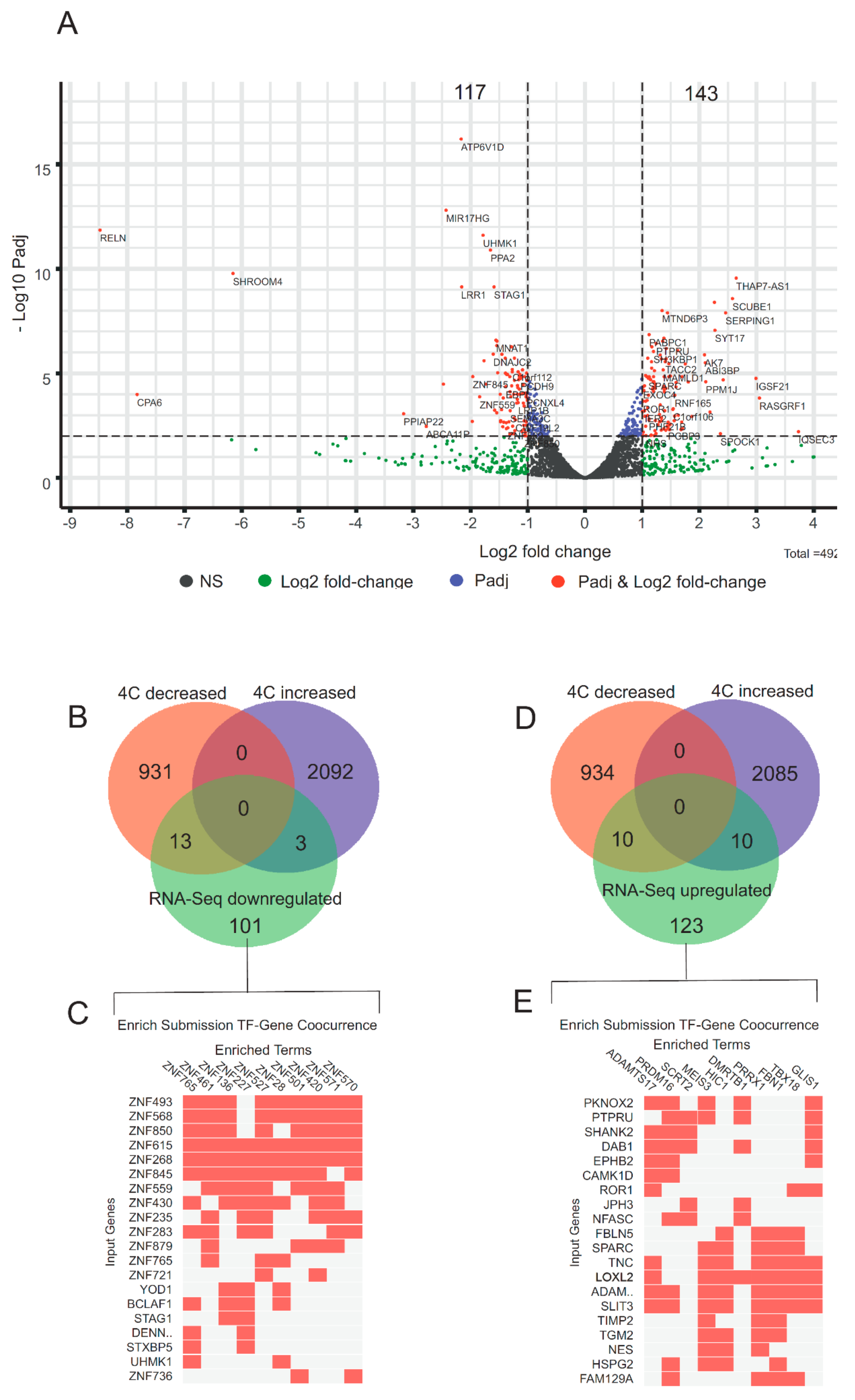
3.5. rDNA-Contacting Genes Specifying Transcription Factors Change Their Expression after Heat Shock Treatment and Induce an Epigenetic Switch
3.6. Genomic Browser Analysis Indicates Changes in rDNA Contacts after Heat Shock Treatment
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Moss, T.; Langlois, F.; Gagnon-Kugler, T.; Stefanovsky, V. A housekeeper with power of attorney: The rRNA genes in ribosome biogenesis. Cell. Mol. Life Sci. 2017, 64, 29–49. [Google Scholar] [CrossRef] [PubMed]
- Ananiev, E.V.; Barsky, V.E.; Ilyin, Y.V.; Churikov, N.A. Localization of nucleoli in Drosophila melanogaster polytene chromosomes. Chromosoma 1981, 81, 619–628. [Google Scholar] [CrossRef] [PubMed]
- Worton, R.G.; Sutherland, J.; Sylvester, J.E.; Willard, H.F.; Bodrug, S.; Dube, I.; Duff, C.; Kean, V.; Ray, P.N.; Schmickel, R.D.; et al. Human ribosomal RNA genes: Orientation of the tandem array and conservation of the 5′end. Science 1988, 239, 64–68. [Google Scholar] [CrossRef] [PubMed]
- Santoro, R.; Grummt, I. Epigenetic mechanism of rRNA gene silencing: Temporal order of NoRC-mediated histone modification, chromatin remodeling, and DNA methylation. Mol. Cell. Biol. 2005, 25, 2539–2546. [Google Scholar] [CrossRef] [PubMed]
- Guetg, C.; Lienemann, P.; Sirri, V.; Grummt, I.; Hernandez-Verdun, D.; Hottiger, M.O.; Fussenegger, M.; Santoro, R. The NoRC complex mediates the heterochromatin formation and stability of silent rRNA genes and centromeric repeats. EMBO J. 2010, 29, 2135–2146. [Google Scholar] [CrossRef]
- Guetg, C.; Scheifele, F.; Rosenthal, F.; Hottiger, M.O.; Santoro, R. Inheritance of silent rDNA chromatin is mediated by PARP1 via noncoding RNA. Mol. Cell 2012, 45, 790–800. [Google Scholar] [CrossRef]
- Savić, N.; Bär, D.; Leone, S.; Frommel, S.C.; Weber, F.A.; Vollenweider, E.; Ferrari, E.; Ziegler, U.; Kaech, A.; Shakhova, O.; et al. lncRNA maturation to initiate heterochromatin formation in the nucleolus is required for exit from pluripotency in ESCs. Cell Stem Cell 2014, 15, 720–734. [Google Scholar] [CrossRef]
- Van Koningsbruggen, S.; Gierliński, M.; Schofield, P.; Martin, D.; Barton, G.J.; Ariyurek, Y.; den Dunnen, J.T.; Lamond, A.I. High-resolution whole genome sequencing reveals that specific chromatin domains from most human chromosomes associate with nucleoli. Mol. Biol. Cell. 2010, 21, 3735–3748. [Google Scholar] [CrossRef]
- Németh, A.; Conesa, A.; Santoyo-Lopez, J.; Medina, I.; Montaner, D.; Péterfia, B.; Solovei, I.; Cremer, T.; Dopazo, J.; Längst, G. Initial genomics of the human nucleolus. PLoS Genet. 2010, 6, e1000889. [Google Scholar] [CrossRef]
- Pontvianne, F.; Carpentier, M.C.; Durut, N.; Pavlištová, V.; Jaške, K.; Schořová, Š.; Parrinello, H.; Rohmer, M.; Pikaard, C.S.; Fojtová, M.; et al. Identification of Nucleolus-Associated Chromatin Domains Reveals a Role for the Nucleolus in 3D Organization of the A. thaliana Genome. Cell Rep. 2016, 16, 1574–1587. [Google Scholar] [CrossRef]
- Dillinger, S.; Straub, T.; Németh, A. Nucleolus association of chromosomal domains is largely maintained in cellular senescence despite massive nuclear reorganisation. PLoS ONE 2017, 12, e0178821. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Fedoseeva, D.M.; Sosin, D.V.; Snezhkina, A.V.; Melnikova, N.V.; Kudryavtseva, A.V.; Kravatsky, Y.V.; Kretova, O.V. Hot spots of DNA double-strand breaks and genomic contacts of human rDNA units are involved in epigenetic regulation. J. Mol. Cell. Biol. 2015, 7, 366–382. [Google Scholar] [CrossRef] [PubMed]
- Tchurikov, N.A.; Kretova, O.V.; Fedoseeva, D.M.; Sosin, D.V.; Grachev, S.A.; Serebraykova, M.V.; Romanenko, S.A.; Vorobieva, N.V.; Kravatsky, Y.V. DNA double strand breaks coupled with PARP1 and HNRNPA2B1 binding sites flank coordinately expressed domains in human chromosomes. PLoS Genet. 2013, 9, e1003429. [Google Scholar] [CrossRef] [PubMed]
- Dekker, J.; Rippe, K.; Dekker, M.; Kleckner, N. Capturing chromosome conformation. Science 2002, 95, 1306–1311. [Google Scholar] [CrossRef]
- Osborne, C.S.; Chakalova, L.; Brown, K.E.; Carter, D.; Horton, A.; Debrand, E.; Goyenechea, B.; Mitchell, J.A.; Lopes, S.; Reik, W.; et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004, 36, 1065–1071. [Google Scholar] [CrossRef]
- FastQC. Available online: https://www.bioinformatics.babraham.ac.uk/projects/fastqc (accessed on 4 November 2019).
- Cutadapt. Available online: https://cutadapt.readthedocs.io (accessed on 4 November 2019).
- Langmead, B.; Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 2012, 9, 259–357. [Google Scholar] [CrossRef]
- Li, H.; Durbin, R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics 2010, 26, 589–595. [Google Scholar] [CrossRef]
- BWA 0.7.12 method mem. Available online: http://bio-bwa.sourceforge.net (accessed on 4 November 2019).
- Li, H.; Handsaker, B.; Wysoker, A.; Fennell, T.; Ruan, J.; Homer, N. 1000 Genome Project Data Processing Subgroup. The Sequence Alignment/Map format and SAMtools. Bioinformatics 2009, 25, 2078–2079. [Google Scholar] [CrossRef]
- Samtools 1.6. Available online: http://www.htslib.org (accessed on 4 November 2019).
- Ramírez, F.; Ryan, D.P.; Grüning, B.; Bhardwaj, V.; Kilpert, F.; Richter, A.S.; Heyne, S.; Friederike Dündar, F.; Manke, T. DeepTools2: A next generation web server for deep-sequencing data analysis. Nucleic Acids Res. 2016, 44, W160–W165. [Google Scholar] [CrossRef]
- Quinlan, A.R. BEDTools: The Swiss-Army Tool for Genome Feature Analysis. Curr. Protoc. Bioinformatics 2014, 47, 1–34. [Google Scholar] [CrossRef]
- Klein, F.A.; Pakozdi, T.; Anders, S.; Ghavi-Helm, Y.; Furlong, E.E.; Huber, W. FourCSeq: Analysis of 4C sequencing data. Bioinformatics 2015, 31, 3085–3091. [Google Scholar] [CrossRef] [PubMed]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [PubMed]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef] [PubMed]
- EnhancedVolcano R package. Available online: https://github.com/kevinblighe (accessed on 4 November 2019).
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, T.; Meyer, C.A.; Eeckhoute, J.; Johnson, D.S.; Bernstein, B.E.; Nusbaum, C.; Myers, R.M.; Brown, M.; Li, W.; et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008, 9, R137. [Google Scholar] [CrossRef]
- Stempor, P.; Ahringer, J. SeqPlots-Interactive software for exploratory data analyses: Pattern discovery and visualization in genomics. Wellcome Open Res. 2016, 1, 14. [Google Scholar] [CrossRef]
- Bioinformatics. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn (accessed on 4 November 2019).
- Dobin, A.; Davis, C.A.; Schlesinger, F.; Drenkow, J.; Zaleski, C.; Jha, S.; Batut, P.; Chaisson, M.; Gingeras, T.R. STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 2013, 29, 15–21. [Google Scholar] [CrossRef]
- Reimand, J.; Kull, M.; Peterson, H.; Hansen, J.; Vilog, J.G. Profiler—A web-based toolset for functional profiling of gene lists from large-scale experiments. Nucleic Acids Res. 2007, 35, W193–W200. [Google Scholar] [CrossRef]
- Shaw, G.; Morse, S.; Ararat, M.; Graham, F.L. Preferential transformation of human neuronal cells by human adenoviruses and the origin of HEK 293 cells. FASEB J. 2002, 16, 869–971. [Google Scholar] [CrossRef]
- Wu, Q.; Maniatis, T. A striking organization of a large family of human neural cadherin-like cell adhesion genes. Cell 1999, 97, 779–790. [Google Scholar] [CrossRef]
- Kuleshov, M.V.; Jones, M.R.; Rouillard, A.D.; Fernandez, N.F.; Duan, Q.; Wang, Z.; Koplev, S.; Jenkins, S.L.; Jagodnik, K.M.; Lachmann, A.; et al. Enrichr: A comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016, 44, W90–W97. [Google Scholar] [CrossRef] [PubMed]
- Graham, F.L.; Smiley, J.; Russell, W.C.; Nairn, R. Characteristics of a human cell line transformed by DNA from human adenovirus type 5. J. Gen. Virol. 1977, 36, 59–74. [Google Scholar] [CrossRef] [PubMed]
- Karmodiya, K.; Krebs, A.R.; Oulad-Abdelghani, M.; Kimura, H.; Tora, L. H3K9 and H3K14 acetylation co-occur at many gene regulatory elements, while H3K14ac marks a subset of inactive inducible promoters in mouse embryonic stem cells. BMC Genom. 2012, 13, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Macias, S.; Plass, M.; Stajuda, A.; Michlewski, G.; Eyras, E.; Cáceres, J.F. DGCR8 HITS-CLIP reveals novel functions for the Microprocessor. Nat. Struct. Mol. Biol. 2012, 19, 760–766. [Google Scholar] [CrossRef] [PubMed]
- Ono, T.; Losada, A.; Hirano, M.; Myers, M.P.; Neuwald, A.F.; Hirano, T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell 2003, 115, 109–121. [Google Scholar] [CrossRef]
- Rao, S.S.; Huang, S.C.; St Hilaire, B.G.; Engreitz, J.M.; Perez, E.M.; Kieffer-Kwon, K.R.; Sanborn, A.L.; Johnstone, S.E.; Bascom, G.D.; Bochkov, I.D.; et al. Cohesin Loss Eliminates All Loop Domains. Cell 2017, 171, 305–320. [Google Scholar] [CrossRef]
- Zhao, Z.; Dammert, M.A.; Hoppe, S.; Bierhoff, H.; Grummt, I. Heat shock represses rRNA synthesis by inactivation of TIF-IA and lncRNA-dependent changes in nucleosome positioning. Nucleic Acids Res. 2016, 44, 8144–8152. [Google Scholar] [CrossRef]
- Zhao, Z.; Sentürk, N.; Song, C.; Grummt, I. lncRNA PAPAS tethered to the rDNA enhancer recruits hypophosphorylated CHD4/NuRD to repress rRNA synthesis at elevated temperatures. Genes Dev. 2018, 32, 836–848. [Google Scholar] [CrossRef]
- Humburg, P.; Maugeri, N.; Lee, W.; Mohr, B.; Knight, J.C. Characterisation of the global transcriptional response to heat shock and the impact of individual genetic variation. Genome Med. 2016, 8, 87. [Google Scholar] [CrossRef]
- Kraushar, M.L.; Viljetic, B.; Wijeratne, H.R.; Thompson, K.; Jiao, X.; Pike, J.W.; Medvedeva, V.; Groszer, M.; Kiledjian, M.; Hart, R.P.; et al. Thalamic WNT3 Secretion Spatiotemporally Regulates the Neocortical Ribosome Signature and mRNA Translation to Specify Neocortical Cell Subtypes. J. Neurosci. 2015, 35, 10911–10926. [Google Scholar] [CrossRef]
- Kraushar, M.L.; Popovitchenko, T.; Volk, N.L.; Rasin, M.R. The frontier of RNA metamorphosis and ribosome signature in neocortical development. Int. J. Dev. Neurosci. 2016, 55, 131–139. [Google Scholar] [CrossRef] [PubMed]
- Chau, K.F.; Shannon, M.L.; Fame, R.M.; Fonseca, E.; Mullan, H.; Johnson, M.B.; Sendamarai, A.K.; Springel, M.W.; Laurent, B.; Lehtinen, M.K. Downregulation of ribosome biogenesis during early forebrain development. ELife 2018, 10, 7. [Google Scholar] [CrossRef] [PubMed]
- Ensembl FTP server. Available online: http://ftp.ensembl.org/pub/grch37/current/gtf (accessed on 4 November 2019).
- Hnisz, D.; Abraham, B.J.; Lee, T.I.; Lau, A.; Saint-André, V.; Sigova, A.A.; Hoke, H.A.; Young, R.A. Super-enhancers in the control of cell identity and disease. Cell 2013, 155, 934–947. [Google Scholar] [CrossRef] [PubMed]
- Spellman, P.T.; Rubin, G.M. Evidence for large domains of similarly expressed genes in the Drosophila genome. J. Biol. 2002, 1, 5. [Google Scholar] [CrossRef]
- Tolhuis, B.; Muijrers, I.; de Wit, E.; Teunissen, H.; Talhout, W.; van Steensel, B.; van Lohuizen, M. Genome-wide profiling of PRC1 and PRC2 Polycomb chromatin binding in Drosophila melanogaster. Nat. Genet. 2006, 38, 649–694. [Google Scholar] [CrossRef]
- Le Dily, F.; Baù, D.; Pohl, A.; Vicent, G.P.; Serra, F.; Soronellas, D.; Castellano, G.; Wright, R.H.; Ballare, C.; Filion, G.; et al. Distinct structural transitions of chromatin topological domains correlate with coordinated hormone-induced gene regulation. Genes Dev. 2014, 28, 2151–2162. [Google Scholar] [CrossRef]
- Tchurikov, N.A. Molecular mechanisms of epigenetics. Biochemistry 2005, 70, 406–423. [Google Scholar] [CrossRef]
- Lewis, E.B. The theory and application of a new method of detecting chromosomal rearrangements in Drosophila melanogaster. Am. Nat. 1954, 88, 225–239. [Google Scholar] [CrossRef]
- Feinberg, A.P. The Nucleolus Gets the Silent Treatment. Cell Stem Cell 2014, 15, 675–676. [Google Scholar] [CrossRef][Green Version]
- Richter, K.; Haslbeck, M.; Buchner, J. The heat shock response: Life on the verge of death. Mol. Cell 2010, 40, 253–266. [Google Scholar] [CrossRef]
- King, I.F.; Yandava, C.N.; Mabb, A.M.; Hsiao, J.S.; Huang, H.S.; Pearson, B.L.; Calabrese, J.M.; Starmer, J.; Parker, J.S.; Magnuson, T.; et al. Topoisomerases facilitate transcription of long genes linked to autism. Nature 2013, 501, 58–62. [Google Scholar] [CrossRef] [PubMed]
- Sugino, K.; Hempel, C.M.; Okaty, B.W.; Arnson, H.A.; Kato, S.; Dani, V.S.; Nelson, S.B. Cell-type-specific repression by methyl-CpG-binding protein 2 is biased toward long genes. J. Neurosci. 2014, 34, 12877–12883. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Lemos, B. The long-range interaction map of ribosomal DNA arrays. PLoS Genet. 2018, 14, e1007258. [Google Scholar] [CrossRef] [PubMed]
- Percharde, M.; Lin, C.J.; Yin, Y.; Guan, J.; Peixoto, G.A.; Bulut-Karslioglu, A.; Biechele, S.; Huang, B.; Shen, X.; Ramalho-Santos, M. A LINE1-nucleolin partnership regulates early development and ESC identity. Cell 2018, 174, 391–405. [Google Scholar] [CrossRef]
- Kretova, O.V.; Fedoseeva, D.M.; Kravatsky, Y.V.; Alembekov, I.R.; Slovohotov, I.Y.; Tchurikov, N.A. Homeotic DUX4 Genes that Control Human Embryonic Development at the Two-Cell Stage Are Surrounded by Regions Contacting with rDNA Gene Clusters. Mol. Biol. 2019, 53, 268–273. [Google Scholar] [CrossRef]
- Pelkmans, L. Cell Biology. Using cell-to-cell variability–A new era in molecular biology. Science 2012, 336, 425–426. [Google Scholar] [CrossRef]
- Boija, A.; Klein, I.A.; Sabari, B.R.; Dall’Agnese, A.; Coffey, E.L.; Zamudio, A.V.; Li, C.H.; Shrinivas, K.; Manteiga, J.C.; Hannett, N.M.; et al. Transcription Factors Activate Genes through the Phase-Separation Capacity of Their Activation Domains. Cell 2018, 175, 1842–1855.e16. [Google Scholar] [CrossRef]
- Sabari, B.R.; Dall’Agnese, A.; Boija, A.; Klein, I.A.; Coffey, E.L.; Shrinivas, K.; Abraham, B.J.; Hannett, N.M.; Zamudio, A.V.; Manteiga, J.C.; et al. Coactivator condensation at super-enhancers links phase separation and gene control. Science 2018, 361. [Google Scholar] [CrossRef]
- Latonen, L. Phase-to-Phase with Nucleoli-Stress Responses, Protein Aggregation and Novel Roles of RNA. Front. Cell Neurosci. 2019, 13, 151. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Tycko, B. The history of cancer epigenetics. Nat. Rev. Cancer 2004, 4, 143–153. [Google Scholar] [CrossRef]
- Feinberg, A.P.; Koldobskiy, M.A.; Göndör, A. Epigenetic modulators, modifiers and mediators in cancer aetiology and progression.Nat Rev Genet. Nat. Rev. Genet. 2016, 17, 284–299. [Google Scholar] [CrossRef] [PubMed]
- Diesch, J.; Bywater, M.J.; Sanij, E.; Cameron, D.P.; Schierding, W.; Brajanovski, N.; Son, J.; Sornkom, J.; Hein, N.; Evers, M.; et al. Changes in long-range rDNA-genomic interactions associate with altered RNA polymerase II gene programs during malignant transformation. Commun. Biol. 2019, 2, 39. [Google Scholar] [CrossRef] [PubMed]
- Weeks, S.E.; Metge, B.J.; Samant, R.S. The nucleolus: A central response hub for the stressors that drive cancer progression. Cell. Mol. Life Sci. 2019, 1–14. [Google Scholar] [CrossRef] [PubMed]

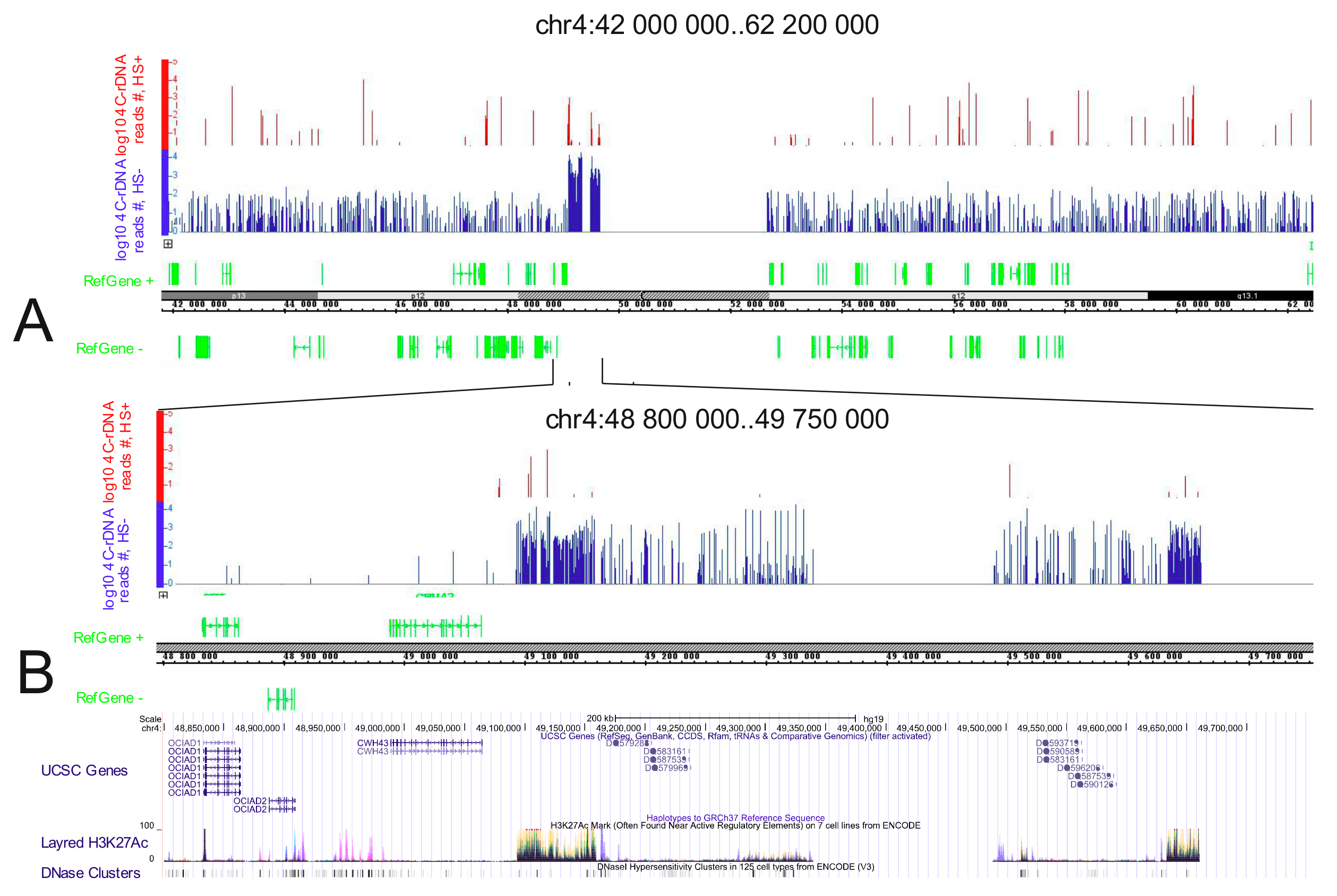
| GO.ID | Description | padj | Genes |
|---|---|---|---|
| GO:MF | |||
| GO:0000981 | DNA-binding transcription factor activity, RNA polymerase II-specific | 3.0020889186981556e-16 | PRDM16,SCRT2,MEIS3,HIC1,DMRTB1,PRRX1,TBX18,GLIS1,LHX4,GLI2,HIVEP3,NFATC4,EBF4,ALX4,ELF4,HES3,ZBTB7C,PAX7,HEYL,ZNF536,GLIS2 |
| GO:0043565 | sequence-specific DNA binding | 6.766317740279593e-16 | PRDM16,SCRT2,MEIS3,HIC1,DMRTB1,PRRX1,TBX18,GLIS1,LHX4,GLI2,NFATC4,EBF4,ALX4,ELF4,HES3,PAX7,HEYL,ZNF536,GLIS2 |
| GO:0003700 | DNA-binding transcription factor activity | 1.6218692524044966e-15 | PRDM16,SCRT2,MEIS3,HIC1,DMRTB1,PRRX1,TBX18,GLIS1,LHX4,GLI2,HIVEP3,NFATC4,EBF4,ALX4,ELF4,HES3,ZBTB7, PAX7,HEYL,ZNF536,GLIS2 |
| GO:0140110 | transcription regulator activity | 5.730692260911566e-14 | PRDM16,SCRT2,MEIS3,HIC1,DMRTB1,PRRX1,TBX18,GLIS1,LHX4,GLI2,HIVEP3,NFATC4,EBF4,ALX4,ELF4,HES3,ZBTB7C,PAX7,HEYL,ZNF536,GLIS2 |
| GO:0003677 | DNA binding | 1.078772693704691e-10 | PRDM16,SCRT2,MEIS3,HIC1,DMRTB1,PRRX1,TBX18,GLIS1,LHX4,GLI2,HIVEP3,NFATC4,EBF4,ALX4,ELF4,HES3,PAX7,HEYL,ZNF536,GLIS2 |
| GO:0003676 | nucleic acid binding | 5.094046931162048e-10 | ADAMTS17,PRDM16,SCRT2,MEIS3,HIC1,DMRTB1,PRRX1,TBX18,GLIS1,LHX4,GLI2,HIVEP3,NFATC4,EBF4,ALX4,ELF4,HES3,ZBTB7C,PAX7,HEYL,ZNF536,DPF3,GLIS2 |
| GO:0000976 | transcription regulatory region sequence-specific DNA binding | 3.1980974069268857e-9 | SCRT2,PRRX1,TBX18,GLIS1,GLI2,NFATC4,EBF4,ALX4,ELF4,HES3,HEYL,ZNF536,GLIS2 |
| GO:1990837 | sequence-specific double-stranded DNA binding | 6.201300596296805e-9 | SCRT2,PRRX1,TBX18,GLIS1,GLI2,NFATC4,EBF4,ALX4,ELF4,HES3,HEYL,ZNF536,GLIS2 |
| GO:0044212 | transcription regulatory region DNA binding | 1.717308909160837e-8 | SCRT2,PRRX1,TBX18,GLIS1,GLI2,NFATC4,EBF4,ALX4,ELF4,HES3,HEYL,ZNF536,GLIS2 |
| GO:0001067 | regulatory region nucleic acid binding | 1.7651201530658695e-8 | SCRT2,PRRX1,TBX18,GLIS1,GLI2,NFATC4,EBF4,ALX4,ELF4,HES3,HEYL,ZNF536,GLIS2 |
| GO:0003690 | double-stranded DNA binding | 2.223504167721871e-8 | SCRT2,PRRX1,TBX18,GLIS1,GLI2,NFATC4,EBF4,ALX4,ELF4,HES3,HEYL,ZNF536,GLIS2 |
| GO:0000977 | RNA polymerase II regulatory region sequence-specific DNA binding | 3.2487899485312494e-8 | SCRT2,PRRX1,GLIS1,GLI2,NFATC4,EBF4,ALX4,ELF4,HES3,HEYL,ZNF536,GLIS2 |
| GO:0001012 | RNA polymerase II regulatory region DNA binding | 3.510295518605051e-8 | SCRT2,PRRX1,GLIS1,GLI2,NFATC4,EBF4,ALX4,ELF4,HES3,HEYL,ZNF536,GLIS2 |
| GO:1901363 | heterocyclic compound binding | 0.0000018894696087891545 | ADAMTS17,PRDM16,SCRT2,MEIS3,HIC1,DMRTB1,PRRX1,TBX18,GLIS1,LHX4,GLI2,HIVEP3,NFATC4,EBF4,ALX4,ELF4,HES3,ZBTB7C,PAX7,HEYL,ZNF536,DPF3,GLIS2 |
| GO:0097159 | organic cyclic compound binding | 0.0000025120194207196536 | ADAMTS17,PRDM16,SCRT2,MEIS3,HIC1,DMRTB1,PRRX1,TBX18,GLIS1,LHX4,GLI2,HIVEP3,NFATC4,EBF4,ALX4,ELF4,HES3,ZBTB7C,PAX7,HEYL,ZNF536,DPF3,GLIS2 |
| GO:0000978 | RNA polymerase II proximal promoter sequence-specific DNA binding | 0.00011929626986549908 | SCRT2,PRRX1,GLI2,NFATC4,ELF4,HES3,HEYL,ZNF536 |
| GO:0000987 | proximal promoter sequence-specific DNA binding | 0.00014630761558464278 | SCRT2,PRRX1,GLI2,NFATC4,ELF4,HES3,HEYL,ZNF536 |
| GO:0001227 | DNA-binding transcription repressor activity, RNA polymerase II-specific | 0.006154396615738856 | SCRT2,HIC1,GLIS1,NFATC4 |
| GO:0046872 | metal ion binding | 0.03598926908109106 | ADAMTS17,PRDM16,SCRT2,HIC1,DMRTB1,FBN1,GLIS1,LHX4,GLI2,HIVEP3,EBF4,ZBTB7C,ZNF536,DPF3,GLIS2 |
| GO:0003712 | transcription coregulator activity | 0.037118154816446446 | PRDM16,PRRX1,TBX18,NFATC4,HES3,HEYL |
| GO:0001216 | DNA-binding transcription activator activity | 0.03790861793015616 | GLIS1,LHX4,EBF4,ALX4 |
| GO:0001228 | DNA-binding transcription activator activity, RNA polymerase II-specific | 0.03790861793015616 | GLIS1,LHX4,EBF4,ALX4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tchurikov, N.A.; Fedoseeva, D.M.; Klushevskaya, E.S.; Slovohotov, I.Y.; Chechetkin, V.R.; Kravatsky, Y.V.; Kretova, O.V. rDNA Clusters Make Contact with Genes that Are Involved in Differentiation and Cancer and Change Contacts after Heat Shock Treatment. Cells 2019, 8, 1393. https://doi.org/10.3390/cells8111393
Tchurikov NA, Fedoseeva DM, Klushevskaya ES, Slovohotov IY, Chechetkin VR, Kravatsky YV, Kretova OV. rDNA Clusters Make Contact with Genes that Are Involved in Differentiation and Cancer and Change Contacts after Heat Shock Treatment. Cells. 2019; 8(11):1393. https://doi.org/10.3390/cells8111393
Chicago/Turabian StyleTchurikov, Nickolai A., Daria M. Fedoseeva, Elena S. Klushevskaya, Ivan Y. Slovohotov, Vladimir R. Chechetkin, Yuri V. Kravatsky, and Olga V. Kretova. 2019. "rDNA Clusters Make Contact with Genes that Are Involved in Differentiation and Cancer and Change Contacts after Heat Shock Treatment" Cells 8, no. 11: 1393. https://doi.org/10.3390/cells8111393
APA StyleTchurikov, N. A., Fedoseeva, D. M., Klushevskaya, E. S., Slovohotov, I. Y., Chechetkin, V. R., Kravatsky, Y. V., & Kretova, O. V. (2019). rDNA Clusters Make Contact with Genes that Are Involved in Differentiation and Cancer and Change Contacts after Heat Shock Treatment. Cells, 8(11), 1393. https://doi.org/10.3390/cells8111393





