MiR-21, MiR-29a, GATA4, and MEF2c Expression Changes in Endothelin-1 and Angiotensin II Cardiac Hypertrophy Stimulated Isl-1+Sca-1+c-kit+ Porcine Cardiac Progenitor Cells In Vitro
Abstract
1. Introduction
2. Materials and Methods
2.1. Isolation of Porcine Cardiac Progenitor Cells from Heart Tissue
2.2. Characterization of pCPCs
2.2.1. Immunofluorescence Staining
2.2.2. Real-Time qPCR
2.2.3. Transfection with pMx-MGT
2.2.4. Induction of Hypertrophy in pCPC
2.3. Measurement of Hypertrophy-Induced Protein Expression and Cell Size
2.4. Quantification of mRNA and miRNA Gene Expression by Real-Time qPCR
2.5. Statistics
3. Results
3.1. Isolation and Characterizaton of Isl1+Sca1+ckit+ pCPC
3.2. Induction of Chemical Hypertrophy with Ang II and ET-1 Results in Hypertrophic Growth of Isl1+Sca1+ckit+ pCPC
3.3. Cardiac Reprogramming of pCPCs with pMx-MGT or Ang II Induced Hypertrophy Alone Leads to Increase of Intracellular MCP-1
3.4. Cx43 is Upregulated in pMx-MGT-Transfected and Ang II Stimulated Isl1+Sca1+ckit+ pCPCs
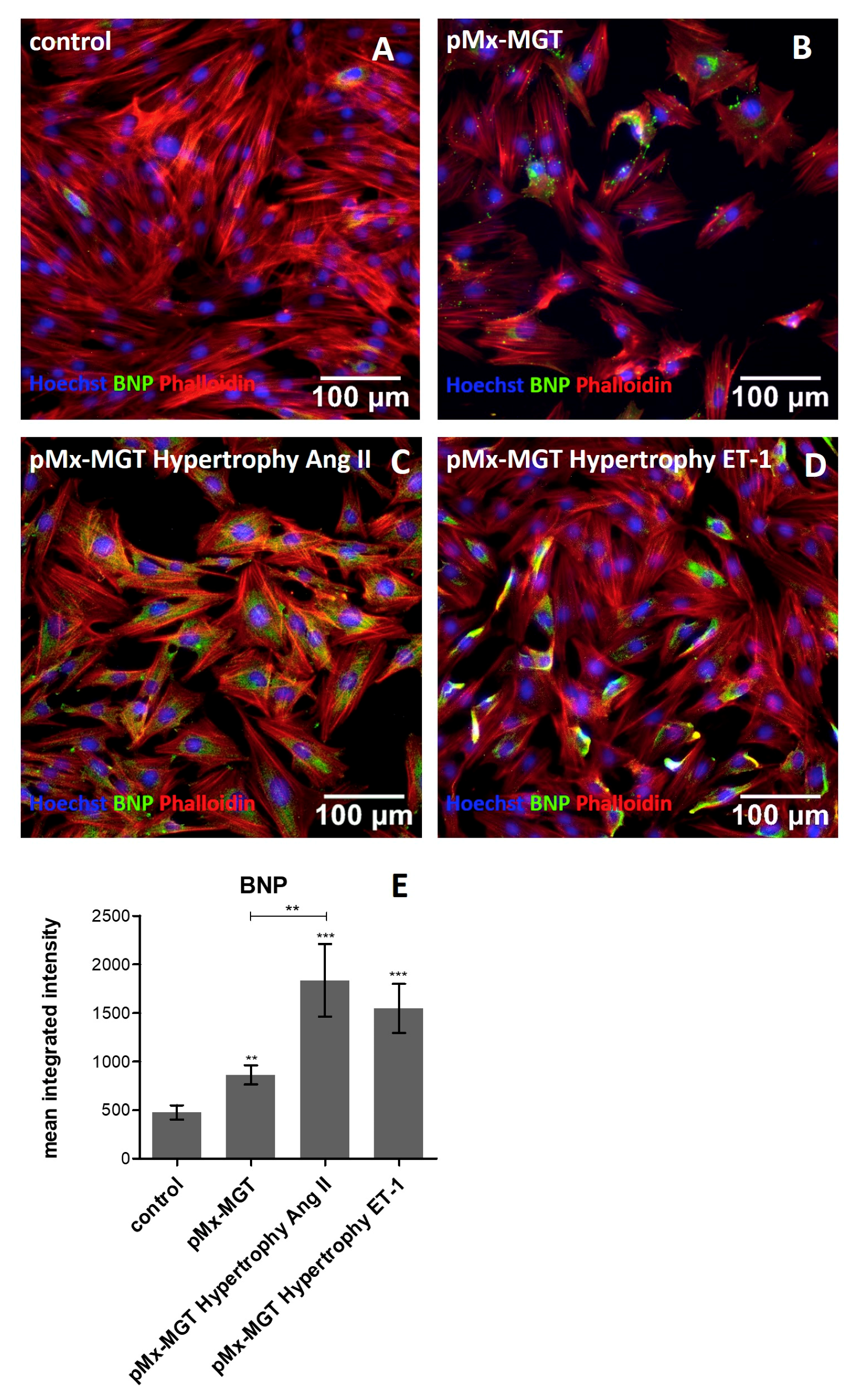
3.5. Increased Cellular BNP Expression Confirms Cardiac Hypertrophy
3.6. ET-1 and Ang II Induced Hypertrophy Leads to Overexpression of MEF2c and GATA4 in Isl1+Sca1+ckit+ pCPCs
3.7. miR-29a is Upregulated in Response to Ang II Stimulation
3.8. pMx-MGT-Transfected Ang II Stimulated Isl1+Sca1+ckit+ pCPCs Have a Significanly Higher miR-21 Gene Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Dorn, G.W.; Robbins, J.; Sugden, P.H. Phenotyping Hypertrophy. Circ. Res. 2003, 92, 1171–1175. [Google Scholar] [CrossRef] [PubMed]
- Kahan, T.; Bergfeldt, L. Left ventricular hypertrophy in hypertension: Its arrhythmogenic potential. Heart 2005, 91, 250–256. [Google Scholar] [CrossRef] [PubMed]
- Nakamura, M.; Sadoshima, J. Mechanisms of physiological and pathological cardiac hypertrophy. Nat. Rev. Cardiol. 2018, 15, 387–407. [Google Scholar] [CrossRef] [PubMed]
- Bisping, E.; Wakula, P.; Poteser, M.; Heinzel, F.R. Targeting cardiac hypertrophy: Toward a causal heart failure therapy. J. Cardiovasc. Pharmacol. 2014, 64, 293–305. [Google Scholar] [CrossRef] [PubMed]
- Barry, S.P.; Davidson, S.M.; Townsend, P.A. Molecular regulation of cardiac hypertrophy. Int. J. Biochem. Cell Biol. 2008, 40, 2023–2039. [Google Scholar] [CrossRef]
- Bernardo, B.C.; Weeks, K.L.; Pretorius, L.; McMullen, J.R. Molecular distinction between physiological and pathological cardiac hypertrophy: Experimental findings and therapeutic strategies. Pharmacol. Ther. 2010, 128, 191–227. [Google Scholar] [CrossRef]
- Matsa, E.; Burridge, P.W.; Wu, J.C. Human stem cells for modeling heart disease and for drug discovery. Sci. Transl. Med. 2014, 6, 239ps6. [Google Scholar] [CrossRef]
- Perino, M.G.; Yamanaka, S.; Li, J.; Wobus, A.M.; Boheler, K.R. Cardiomyogenic stem and progenitor cell plasticity and the dissection of cardiopoiesis. J. Mol. Cell. Cardiol. 2008, 45, 475–494. [Google Scholar] [CrossRef]
- Engleka, K.A.; Manderfield, L.J.; Brust, R.D.; Li, L.; Cohen, A.; Dymecki, S.M.; Epstein, J.A. Islet1 derivatives in the heart are of both neural crest and second heart field origin. Circ. Res. 2012, 110, 922–926. [Google Scholar] [CrossRef]
- Marketou, M.E.; Parthenakis, F.; Vardas, P.E. Pathological Left Ventricular Hypertrophy and Stem Cells: Current Evidence and New Perspectives. Stem Cells Int. 2016, 2016, 5720758. [Google Scholar] [CrossRef]
- Santini, M.P.; Forte, E.; Harvey, R.P.; Kovacic, J.C. Developmental origin and lineage plasticity of endogenous cardiac stem cells. Dev. Camb. Engl. 2016, 143, 1242–1258. [Google Scholar] [CrossRef] [PubMed]
- Cai, C.-L.; Liang, X.; Shi, Y.; Chu, P.-H.; Pfaff, S.L.; Chen, J.; Evans, S. Isl1 Identifies a Cardiac Progenitor Population that Proliferates Prior to Differentiation and Contributes a Majority of Cells to the Heart. Dev. Cell 2003, 5, 877–889. [Google Scholar] [CrossRef]
- Kwon, C.; Qian, L.; Cheng, P.; Nigam, V.; Arnold, J.; Srivastava, D. A Regulatory Pathway Involving Notch1/β-Catenin/Isl1 Determines Cardiac Progenitor Cell Fate. Nat. Cell Biol. 2009, 11, 951–957. [Google Scholar] [CrossRef] [PubMed]
- Dodou, E.; Verzi, M.P.; Anderson, J.P.; Xu, S.-M.; Black, B.L. Mef2c is a direct transcriptional target of ISL1 and GATA factors in the anterior heart field during mouse embryonic development. Dev. Camb. Engl. 2004, 131, 3931–3942. [Google Scholar] [CrossRef] [PubMed]
- Herzig, T.C.; Jobe, S.M.; Aoki, H.; Molkentin, J.D.; Cowley, A.W.; Izumo, S.; Markham, B.E. Angiotensin II type1a receptor gene expression in the heart: AP-1 and GATA-4 participate in the response to pressure overload. Proc. Natl. Acad. Sci. USA 1997, 94, 7543–7548. [Google Scholar] [CrossRef]
- Hasegawa, K.; Lee, S.J.; Jobe, S.M.; Markham, B.E.; Kitsis, R.N. cis-Acting sequences that mediate induction of beta-myosin heavy chain gene expression during left ventricular hypertrophy due to aortic constriction. Circulation 1997, 96, 3943–3953. [Google Scholar] [CrossRef]
- Molkentin, J.D.; Kalvakolanu, D.V.; Markham, B.E. Transcription factor GATA-4 regulates cardiac muscle-specific expression of the alpha-myosin heavy-chain gene. Mol. Cell. Biol. 1994, 14, 4947–4957. [Google Scholar] [CrossRef]
- McGrew, M.J.; Bogdanova, N.; Hasegawa, K.; Hughes, S.H.; Kitsis, R.N.; Rosenthal, N. Distinct gene expression patterns in skeletal and cardiac muscle are dependent on common regulatory sequences in the MLC1/3 locus. Mol. Cell. Biol. 1996, 16, 4524–4534. [Google Scholar] [CrossRef][Green Version]
- Ip, H.S.; Wilson, D.B.; Heikinheimo, M.; Tang, Z.; Ting, C.N.; Simon, M.C.; Leiden, J.M.; Parmacek, M.S. The GATA-4 transcription factor transactivates the cardiac muscle-specific troponin C promoter-enhancer in nonmuscle cells. Mol. Cell. Biol. 1994, 14, 7517–7526. [Google Scholar] [CrossRef]
- Murphy, A.M.; Thompson, W.R.; Peng, L.F.; Jones, L. Regulation of the rat cardiac troponin I gene by the transcription factor GATA-4. Biochem. J. 1997, 322 Pt 2, 393–401. [Google Scholar] [CrossRef]
- Montgomery, M.O.; Litvin, J. The cardiac-muscle specific enhancer-promoter of slow/cardiac troponin C binds HMG-2. Gene 1997, 187, 159–164. [Google Scholar] [CrossRef]
- Durocher, D.; Charron, F.; Warren, R.; Schwartz, R.J.; Nemer, M. The cardiac transcription factors Nkx2-5 and GATA-4 are mutual cofactors. EMBO J. 1997, 16, 5687–5696. [Google Scholar] [CrossRef] [PubMed]
- Thuerauf, D.J.; Hanford, D.S.; Glembotski, C.C. Regulation of rat brain natriuretic peptide transcription. A potential role for GATA-related transcription factors in myocardial cell gene expression. J. Biol. Chem. 1994, 269, 17772–17775. [Google Scholar] [PubMed]
- Pikkarainen, S.; Tokola, H.; Majalahti-Palviainen, T.; Kerkelä, R.; Hautala, N.; Bhalla, S.S.; Charron, F.; Nemer, M.; Vuolteenaho, O.; Ruskoaho, H. GATA-4 Is a Nuclear Mediator of Mechanical Stretch-activated Hypertrophic Program. J. Biol. Chem. 2003, 278, 23807–23816. [Google Scholar] [CrossRef] [PubMed]
- Liang, Q.; De Windt, L.J.; Witt, S.A.; Kimball, T.R.; Markham, B.E.; Molkentin, J.D. The transcription factors GATA4 and GATA6 regulate cardiomyocyte hypertrophy in vitro and in vivo. J. Biol. Chem. 2001, 276, 30245–30253. [Google Scholar] [CrossRef]
- Stansfield, W.E.; Ranek, M.; Pendse, A.; Schisler, J.C.; Wang, S.; Pulinilkunnil, T.; Willis, M.S. Chapter 4—The Pathophysiology of Cardiac Hypertrophy and Heart Failure. In Cellular and Molecular Pathobiology of Cardiovascular Disease; Willis, M.S., Homeister, J.W., Stone, J.R., Eds.; Academic Press: San Diego, CA, USA, 2014; pp. 51–78. ISBN 978-0-12-405206-2. [Google Scholar]
- Muñoz, J.P.; Collao, A.; Chiong, M.; Maldonado, C.; Adasme, T.; Carrasco, L.; Ocaranza, P.; Bravo, R.; Gonzalez, L.; Díaz-Araya, G.; et al. The transcription factor MEF2C mediates cardiomyocyte hypertrophy induced by IGF-1 signaling. Biochem. Biophys. Res. Commun. 2009, 388, 155–160. [Google Scholar] [CrossRef]
- Wei, J.; Joshi, S.; Speransky, S.; Crowley, C.; Jayathilaka, N.; Lei, X.; Wu, Y.; Gai, D.; Jain, S.; Hoosien, M.; et al. Reversal of pathological cardiac hypertrophy via the MEF2-coregulator interface. JCI Insight 2 2017, 2, 91068. [Google Scholar] [CrossRef]
- Hata, A. Functions of microRNAs in cardiovascular biology and disease. Annu. Rev. Physiol. 2013, 75, 69–93. [Google Scholar] [CrossRef]
- Callis, T.E.; Pandya, K.; Seok, H.Y.; Tang, R.-H.; Tatsuguchi, M.; Huang, Z.-P.; Chen, J.-F.; Deng, Z.; Gunn, B.; Shumate, J.; et al. MicroRNA-208a is a regulator of cardiac hypertrophy and conduction in mice. J. Clin. Invest. 2009, 119, 2772–2786. [Google Scholar] [CrossRef]
- Lin, Z.; Murtaza, I.; Wang, K.; Jiao, J.; Gao, J.; Li, P.-F. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc. Natl. Acad. Sci. USA 2009, 106, 12103–12108. [Google Scholar] [CrossRef]
- Thum, T.; Gross, C.; Fiedler, J.; Fischer, T.; Kissler, S.; Bussen, M.; Galuppo, P.; Just, S.; Rottbauer, W.; Frantz, S.; et al. MicroRNA-21 contributes to myocardial disease by stimulating MAP kinase signalling in fibroblasts. Nature 2008, 456, 980–984. [Google Scholar] [CrossRef] [PubMed]
- Van Rooij, E.; Sutherland, L.B.; Thatcher, J.E.; DiMaio, J.M.; Naseem, R.H.; Marshall, W.S.; Hill, J.A.; Olson, E.N. Dysregulation of microRNAs after myocardial infarction reveals a role of miR-29 in cardiac fibrosis. Proc. Natl. Acad. Sci. USA 2008, 105, 13027–13032. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.-P.; Wu, J.; Wang, X.; Sartor, M.A.; Jones, K.; Qian, J.; Nicolaou, P.; Pritchard, T.J.; Fan, G.-C. MicroRNA-320 is involved in the regulation of cardiac ischemia/reperfusion injury by targeting heat-shock protein 20. Circulation 2009, 119, 2357–2366. [Google Scholar] [CrossRef] [PubMed]
- Qian, L.; Van Laake, L.W.; Huang, Y.; Liu, S.; Wendland, M.F.; Srivastava, D. miR-24 inhibits apoptosis and represses Bim in mouse cardiomyocytes. J. Exp. Med. 2011, 208, 549–560. [Google Scholar] [CrossRef] [PubMed]
- Rane, S.; He, M.; Sayed, D.; Vashistha, H.; Malhotra, A.; Sadoshima, J.; Vatner, D.E.; Vatner, S.F.; Abdellatif, M. Downregulation of miR-199a derepresses hypoxia-inducible factor-1alpha and Sirtuin 1 and recapitulates hypoxia preconditioning in cardiac myocytes. Circ. Res. 2009, 104, 879–886. [Google Scholar] [CrossRef] [PubMed]
- Roncarati, R.; Viviani Anselmi, C.; Losi, M.A.; Papa, L.; Cavarretta, E.; Da Costa Martins, P.; Contaldi, C.; Saccani Jotti, G.; Franzone, A.; Galastri, L.; et al. Circulating miR-29a, Among Other Up-Regulated MicroRNAs, Is the Only Biomarker for Both Hypertrophy and Fibrosis in Patients With Hypertrophic Cardiomyopathy. J. Am. Coll. Cardiol. 2014, 63, 920–927. [Google Scholar] [CrossRef]
- Shi, J.-Y.; Chen, C.; Xu, X.; Lu, Q. miR-29a promotes pathological cardiac hypertrophy by targeting the PTEN/AKT/mTOR signalling pathway and suppressing autophagy. Acta Physiol. Oxf. Engl. 2019, 2019, e13323. [Google Scholar] [CrossRef]
- Adiarto, S.; Heiden, S.; Vignon-Zellweger, N.; Nakayama, K.; Yagi, K.; Yanagisawa, M.; Emoto, N. ET-1 from endothelial cells is required for complete angiotensin II-induced cardiac fibrosis and hypertrophy. Life Sci. 2012, 91, 651–657. [Google Scholar] [CrossRef]
- Moreau, P.; d’Uscio, L.V.; Shaw, S.; Takase, H.; Barton, M.; Lüscher, T.F. Angiotensin II Increases Tissue Endothelin and Induces Vascular Hypertrophy. Circulation 1997, 96, 1593–1597. [Google Scholar] [CrossRef]
- Irukayama-Tomobe, Y.; Miyauchi, T.; Sakai, S.; Kasuya, Y.; Ogata, T.; Takanashi, M.; Iemitsu, M.; Sudo, T.; Goto, K.; Yamaguchi, I. Endothelin-1–Induced Cardiac Hypertrophy Is Inhibited by Activation of Peroxisome Proliferator–Activated Receptor-α Partly Via Blockade of c-Jun NH2-Terminal Kinase Pathway. Circulation 2004, 109, 904–910. [Google Scholar] [CrossRef]
- Bupha-Intr, T.; Haizlip, K.M.; Janssen, P.M.L. Role of Endothelin in the Induction of Cardiac Hypertrophy In Vitro. PLoS ONE 2012, 7, e43179. [Google Scholar] [CrossRef]
- Gray, M.O.; Long, C.S.; Kalinyak, J.E.; Li, H.T.; Karliner, J.S. Angiotensin II stimulates cardiac myocyte hypertrophy via paracrine release of TGF-beta 1 and endothelin-1 from fibroblasts. Cardiovasc. Res. 1998, 40, 352–363. [Google Scholar] [CrossRef]
- Suzuki, Y.J. Cell signalling pathways for the regulation of GATA4 transcription factor: Implications for cell growth and apoptosis. Cell. Signal. 2011, 23, 1094–1099. [Google Scholar] [CrossRef] [PubMed]
- Hautala, N.; Tokola, H.; Luodonpää, M.; Puhakka, J.; Romppanen, H.; Vuolteenaho, O.; Ruskoaho, H. Pressure overload increases GATA4 binding activity via endothelin-1. Circulation 2001, 103, 730–735. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, T.; Hasegawa, K.; Kaburagi, S.; Kakita, T.; Wada, H.; Yanazume, T.; Sasayama, S. Phosphorylation of GATA-4 is involved in alpha 1-adrenergic agonist-responsive transcription of the endothelin-1 gene in cardiac myocytes. J. Biol. Chem. 2000, 275, 13721–13726. [Google Scholar] [CrossRef]
- Kockskämper, J.; von Lewinski, D.; Khafaga, M.; Elgner, A.; Grimm, M.; Eschenhagen, T.; Gottlieb, P.A.; Sachs, F.; Pieske, B. The slow force response to stretch in atrial and ventricular myocardium from human heart: Functional relevance and subcellular mechanisms. Prog. Biophys. Mol. Biol. 2008, 97, 250–267. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, Z.; Xiao, W. MicroRNA-26a protects against cardiac hypertrophy via inhibiting GATA4 in rat model and cultured cardiomyocytes. Mol. Med. Rep. 2016, 14, 2860–2866. [Google Scholar] [CrossRef]
- Lu, D.; Wang, J.; Li, J.; Guan, F.; Zhang, X.; Dong, W.; Liu, N.; Gao, S.; Zhang, L. Meox1 accelerates myocardial hypertrophic decompensation through Gata4. Cardiovasc. Res. 2018, 114, 300–311. [Google Scholar] [CrossRef]
- Ovchinnikova, E.; Hoes, M.; Ustyantsev, K.; Bomer, N.; de Jong, T.V.; van der Mei, H.; Berezikov, E.; van der Meer, P. Modeling Human Cardiac Hypertrophy in Stem Cell-Derived Cardiomyocytes. Stem Cell Rep. 2018, 10, 794–807. [Google Scholar] [CrossRef]
- Aggarwal, P.; Turner, A.; Matter, A.; Kattman, S.J.; Stoddard, A.; Lorier, R.; Swanson, B.J.; Arnett, D.K.; Broeckel, U. RNA expression profiling of human iPSC-derived cardiomyocytes in a cardiac hypertrophy model. PLoS ONE 2014, 9, e108051. [Google Scholar] [CrossRef]
- Milani-Nejad, N.; Janssen, P.M.L. Small and large animal models in cardiac contraction research: Advantages and disadvantages. Pharmacol. Ther. 2014, 141, 235–249. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Liu, Z.; Yin, C.; Asfour, H.; Chen, O.; Li, Y.; Bursac, N.; Liu, J.; Qian, L. Stoichiometry of Gata4, Mef2c, and Tbx5 influences the efficiency and quality of induced cardiac myocyte reprogramming. Circ. Res. 2015, 116, 237–244. [Google Scholar] [CrossRef] [PubMed]
- Sergeeva, I.A.; Christoffels, V.M. Regulation of expression of atrial and brain natriuretic peptide, biomarkers for heart development and disease. Biochim. Biophys. Acta BBA-Mol. Basis Dis. 2013, 1832, 2403–2413. [Google Scholar] [CrossRef] [PubMed]
- Kerkelä, R.; Ulvila, J.; Magga, J. Natriuretic Peptides in the Regulation of Cardiovascular Physiology and Metabolic Events. J. Am. Heart Assoc. Cardiovasc. Cerebrovasc. Dis. 2015, 4, e002423. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Bergmann, O.; Bhardwaj, R.D.; Bernard, S.; Zdunek, S.; Barnabe-Heider, F.; Walsh, S.; Zupicich, J.; Alkass, K.; Buchholz, B.A.; Druid, H.; et al. Evidence for cardiomyocyte renewal in humans. Science 2009, 324, 98–102. [Google Scholar] [CrossRef] [PubMed]
- Beltrami, A.P.; Barlucchi, L.; Torella, D.; Baker, M.; Limana, F.; Chimenti, S.; Kasahara, H.; Rota, M.; Musso, E.; Urbanek, K.; et al. Adult cardiac stem cells are multipotent and support myocardial regeneration. Cell 2003, 114, 763–776. [Google Scholar] [CrossRef]
- Urbanek, K.; Torella, D.; Sheikh, F.; De Angelis, A.; Nurzynska, D.; Silvestri, F.; Beltrami, C.A.; Bussani, R.; Beltrami, A.P.; Quaini, F.; et al. Myocardial regeneration by activation of multipotent cardiac stem cells in ischemic heart failure. Proc. Natl. Acad. Sci. USA 2005, 102, 8692–8697. [Google Scholar] [CrossRef]
- Davis, D.R.; Zhang, Y.; Smith, R.R.; Cheng, K.; Terrovitis, J.; Malliaras, K.; Li, T.-S.; White, A.; Makkar, R.; Marbán, E. Validation of the Cardiosphere Method to Culture Cardiac Progenitor Cells from Myocardial Tissue. PLoS ONE 2009, 4, e7195. [Google Scholar] [CrossRef]
- Bu, L.; Jiang, X.; Martin-Puig, S.; Caron, L.; Zhu, S.; Shao, Y.; Roberts, D.J.; Huang, P.L.; Domian, I.J.; Chien, K.R. Human ISL1 heart progenitors generate diverse multipotent cardiovascular cell lineages. Nature 2009, 460, 113–117. [Google Scholar] [CrossRef]
- Valente, M.; Nascimento, D.S.; Cumano, A.; Pinto-do-Ó, P. Sca-1+ Cardiac Progenitor Cells and Heart-Making: A Critical Synopsis. Stem Cells Dev. 2014, 23, 2263–2273. [Google Scholar] [CrossRef] [PubMed]
- Mauretti, A.; Spaans, S.; Bax, N.A.M.; Sahlgren, C.; Bouten, C.V.C. Cardiac Progenitor Cells and the Interplay with Their Microenvironment. Stem Cells Int. 2017, 2017, 7471582. [Google Scholar] [CrossRef] [PubMed]
- Bearzi, C.; Rota, M.; Hosoda, T.; Tillmanns, J.; Nascimbene, A.; De Angelis, A.; Yasuzawa-Amano, S.; Trofimova, I.; Siggins, R.W.; LeCapitaine, N.; et al. Human cardiac stem cells. Proc. Natl. Acad. Sci. USA 2007, 104, 14068–14073. [Google Scholar] [CrossRef] [PubMed]
- Al-Maqtari, T.; Hong, K.U.; Vajravelu, B.N.; Moktar, A.; Cao, P.; Moore, J.B.; Bolli, R. Transcription factor-induced activation of cardiac gene expression in human c-kit+ cardiac progenitor cells. PLoS ONE 2017, 12, e0174242. [Google Scholar] [CrossRef]
- Ong, S.-B.; Lee, W.H.; Shao, N.-Y.; Ismail, N.I.; Katwadi, K.; Lim, M.-M.; Kwek, X.-Y.; Michel, N.A.; Li, J.; Newson, J.; et al. Calpain Inhibition Restores Autophagy and Prevents Mitochondrial Fragmentation in a Human iPSC Model of Diabetic Endotheliopathy. Stem Cell Rep. 2019, 12, 597–610. [Google Scholar] [CrossRef]
- Hu, J.; Verzi, M.P.; Robinson, A.S.; Tang, P.L.-F.; Hua, L.L.; Xu, S.-M.; Kwok, P.-Y.; Black, B.L. Endothelin signaling activates Mef2c expression in the neural crest through a MEF2C-dependent positive-feedback transcriptional pathway. Dev. Camb. Engl. 2015, 142, 2775–2780. [Google Scholar] [CrossRef]
- Yelamanchili, S.V.; Chaudhuri, A.D.; Chen, L.-N.; Xiong, H.; Fox, H.S. MicroRNA-21 dysregulates the expression of MEF2C in neurons in monkey and human SIV/HIV neurological disease. Cell Death Dis. 2010, 1, e77. [Google Scholar] [CrossRef]
- Wang, J.; Duan, L.; Gao, Y.; Zhou, S.; Liu, Y.; wei, S.; An, S.; Liu, J.; Tian, L.; Wang, S. Angiotensin II receptor blocker valsartan ameliorates cardiac fibrosis partly by inhibiting miR-21 expression in diabetic nephropathy mice. Mol. Cell. Endocrinol. 2018, 472, 149–158. [Google Scholar] [CrossRef]
- Adam, O.; Löhfelm, B.; Thum, T.; Gupta, S.K.; Puhl, S.-L.; Schäfers, H.-J.; Böhm, M.; Laufs, U. Role of miR-21 in the pathogenesis of atrial fibrosis. Basic Res. Cardiol. 2012, 107, 278. [Google Scholar] [CrossRef]
- Ning, Z.-W.; Luo, X.-Y.; Wang, G.-Z.; Li, Y.; Pan, M.-X.; Yang, R.-Q.; Ling, X.-G.; Huang, S.; Ma, X.-X.; Jin, S.-Y.; et al. MicroRNA-21 mediates angiotensin II-induced liver fibrosis by activating NLRP3 Inflammasome/IL-1β axis via targeting Smad7 and Spry1. Antioxid. Redox Signal. 2017, 27, 1–20. [Google Scholar] [CrossRef]
- Foglia, M.J.; Poss, K.D. Building and re-building the heart by cardiomyocyte proliferation. Development 2016, 143, 729–740. [Google Scholar] [CrossRef] [PubMed]
- Yutzey, K.E. Cardiomyocyte Proliferation. Circ. Res. 2017, 120, 627–629. [Google Scholar] [CrossRef] [PubMed]
- Soonpaa, M.H.; Kim, K.K.; Pajak, L.; Franklin, M.; Field, L.J. Cardiomyocyte DNA synthesis and binucleation during murine development. Am. J. Physiol. 1996, 271, H2183–H2189. [Google Scholar] [CrossRef] [PubMed]
- Aiello Robert, J.; Bourassa Patricia-Ann, K.; Lindsey, S.; Weng, W.; Natoli, E.; Rollins, B.J.; Milos, P.M. Monocyte Chemoattractant Protein-1 Accelerates Atherosclerosis in Apolipoprotein E-Deficient Mice. Arterioscler. Thromb. Vasc. Biol. 1999, 19, 1518–1525. [Google Scholar] [CrossRef]
- Behr Thomas, M.; Wang, X.; Aiyar, N.; Coatney, R.W.; Li, X.; Koster, P.; Angermann, C.E.; Ohlstein, E.; Feuerstein, G.Z.; Winaver, J. Monocyte Chemoattractant Protein-1 is upregulated in rats with volume-overload congestive heart failure. Circulation 2000, 102, 1315–1322. [Google Scholar] [CrossRef]
- Niu, J.; Kolattukudy, P.E. Role of MCP-1 in cardiovascular disease: Molecular mechanisms and clinical implications. Clin. Sci. 2009, 117, 95–109. [Google Scholar] [CrossRef]
- Iwai, M.; Mogi, M.; Horiuchi, M. Role of NAD(P)H oxidase and its regulation in chronic hypertension. Hypertens. Res. Off. J. Jpn. Soc. Hypertens. 2006, 29, 743–744. [Google Scholar] [CrossRef][Green Version]
- Harrison, D.G.; Gongora, M.C. Oxidative stress and hypertension. Med. Clin. N. Am. 2009, 93, 621–635. [Google Scholar] [CrossRef]
- Harrison, D.G.; Guzik, T.J.; Lob, H.; Madhur, M.; Marvar, P.J.; Thabet, S.; Vinh, A.; Weyand, C. Inflammation, Immunity and Hypertension. Hypertension 2011, 57, 132–140. [Google Scholar] [CrossRef]
- Matsuda, S.; Umemoto, S.; Yoshimura, K.; Itoh, S.; Murata, T.; Fukai, T.; Matsuzaki, M. Angiotensin II Activates MCP-1 and Induces Cardiac Hypertrophy and Dysfunction via Toll-like Receptor 4. J. Atheroscler. Thromb. 2015, 22, 833–844. [Google Scholar] [CrossRef]
- Lo Cecilia, W. Role of Gap Junctions in Cardiac Conduction and Development. Circ. Res. 2000, 87, 346–348. [Google Scholar]
- Manias, J.L.; Plante, I.; Gong, X.-Q.; Shao, Q.; Churko, J.; Bai, D.; Laird, D.W. Fate of connexin43 in cardiac tissue harbouring a disease-linked connexin43 mutant. Cardiovasc. Res. 2008, 80, 385–395. [Google Scholar] [CrossRef] [PubMed]
- Kostin, S.; Dammer, S.; Hein, S.; Klovekorn, W.P.; Bauer, E.P.; Schaper, J. Connexin 43 expression and distribution in compensated and decompensated cardiac hypertrophy in patients with aortic stenosis. Cardiovasc. Res. 2004, 62, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Teunissen, B.E.J.; Jongsma, H.J.; Bierhuizen, M.F.A. Regulation of myocardial connexins during hypertrophic remodelling. Eur. Heart J. 2004, 25, 1979–1989. [Google Scholar] [CrossRef] [PubMed]
- Bupha-Intr, T.; Haizlip, K.M.; Janssen, P.M.L. Temporal changes in expression of connexin 43 after load-induced hypertrophy in vitro. Am. J. Physiol.-Heart Circ. Physiol. 2009, 296, H806–H814. [Google Scholar] [CrossRef]
- Cao, L.; Chen, Y.; Lu, L.; Liu, Y.; Wang, Y.; Fan, J.; Yin, Y. Angiotensin II upregulates fibroblast-myofibroblast transition through Cx43-dependent CaMKII and TGF-β1 signaling in neonatal rat cardiac fibroblasts. Acta Biochim. Biophys. Sin. 2018, 50, 843–852. [Google Scholar] [CrossRef]
- Dodge, S.M.; Beardslee, M.A.; Darrow, B.J.; Green, K.G.; Beyer, E.C.; Saffitz, J.E. Effects of angiotensin II on expression of the gap junction channel protein connexin43 in neonatal rat ventricular myocytes. J. Am. Coll. Cardiol. 1998, 32, 800–807. [Google Scholar] [CrossRef]
- Spinella, F.; Rosanò, L.; Castro, V.D.; Nicotra, M.R.; Natali, P.G.; Bagnato, A. Endothelin-1 Decreases Gap Junctional Intercellular Communication by Inducing Phosphorylation of Connexin 43 in Human Ovarian Carcinoma Cells. J. Biol. Chem. 2003, 278, 41294–41301. [Google Scholar] [CrossRef]






| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| ISL-1 | ACATGACGGTGGCTTACAGG | ATGTCACTCTGCAAGGCGAA |
| SCA-1/Ly6a | AGCTCAGGGACTGGAGTGTT | ATCAGGGTAGGGGCAGGTAA |
| KIT (c-kit) | GCCTGTGACTATCTGGGCTC | GCCTTATTCATGCCCGGAGA |
| BNP | CGCAGTAGCATCTTCCAAGTC | ACCTCCTGAGCACATTGCAG |
| ACTB (HK) | TCAACACCCCAGCCATGTAC | CTCCGGAGTCCATCACGATG |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| MEF2c | TAACATGCCGCCATCCGCCC | ATCCTCTCGGTCGCTGCCGT |
| GATA4 | AGAAAACGGAAGCCCAAGAAC | CCACACTGCTGGAGTTGCTG |
| miR-29a | CGGACCTAGCACCATCTGAA | miScript Universal Primer (Qiagen) |
| miR-21 | CGTAGCTAGCTTATCAGACTG | miScript Universal Primer (Qiagen) |
| ACTB (HK) | TCAACACCCCAGCCATGTAC | CTCCGGAGTCCATCACGATG |
| let7a (HK) | GCAGTGAGGTAGTAGGTTGT | miScript Universal Primer (Qiagen) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zlabinger, K.; Spannbauer, A.; Traxler, D.; Gugerell, A.; Lukovic, D.; Winkler, J.; Mester-Tonczar, J.; Podesser, B.; Gyöngyösi, M. MiR-21, MiR-29a, GATA4, and MEF2c Expression Changes in Endothelin-1 and Angiotensin II Cardiac Hypertrophy Stimulated Isl-1+Sca-1+c-kit+ Porcine Cardiac Progenitor Cells In Vitro. Cells 2019, 8, 1416. https://doi.org/10.3390/cells8111416
Zlabinger K, Spannbauer A, Traxler D, Gugerell A, Lukovic D, Winkler J, Mester-Tonczar J, Podesser B, Gyöngyösi M. MiR-21, MiR-29a, GATA4, and MEF2c Expression Changes in Endothelin-1 and Angiotensin II Cardiac Hypertrophy Stimulated Isl-1+Sca-1+c-kit+ Porcine Cardiac Progenitor Cells In Vitro. Cells. 2019; 8(11):1416. https://doi.org/10.3390/cells8111416
Chicago/Turabian StyleZlabinger, Katrin, Andreas Spannbauer, Denise Traxler, Alfred Gugerell, Dominika Lukovic, Johannes Winkler, Julia Mester-Tonczar, Bruno Podesser, and Mariann Gyöngyösi. 2019. "MiR-21, MiR-29a, GATA4, and MEF2c Expression Changes in Endothelin-1 and Angiotensin II Cardiac Hypertrophy Stimulated Isl-1+Sca-1+c-kit+ Porcine Cardiac Progenitor Cells In Vitro" Cells 8, no. 11: 1416. https://doi.org/10.3390/cells8111416
APA StyleZlabinger, K., Spannbauer, A., Traxler, D., Gugerell, A., Lukovic, D., Winkler, J., Mester-Tonczar, J., Podesser, B., & Gyöngyösi, M. (2019). MiR-21, MiR-29a, GATA4, and MEF2c Expression Changes in Endothelin-1 and Angiotensin II Cardiac Hypertrophy Stimulated Isl-1+Sca-1+c-kit+ Porcine Cardiac Progenitor Cells In Vitro. Cells, 8(11), 1416. https://doi.org/10.3390/cells8111416







