A Software Tool for High-Throughput Real-Time Measurement of Intensity-Based Ratio-Metric FRET
Abstract
1. Introduction
2. Materials and Methods
2.1. Adult Mouse Cardiomyocyte Isolation
2.2. Neonatal Mouse Cardiomyocyte Isolation
2.3. Generating HiPSC-Derived Cardiomyocytes
2.4. Sensors Used
2.5. Transfection
2.6. Microscopy System
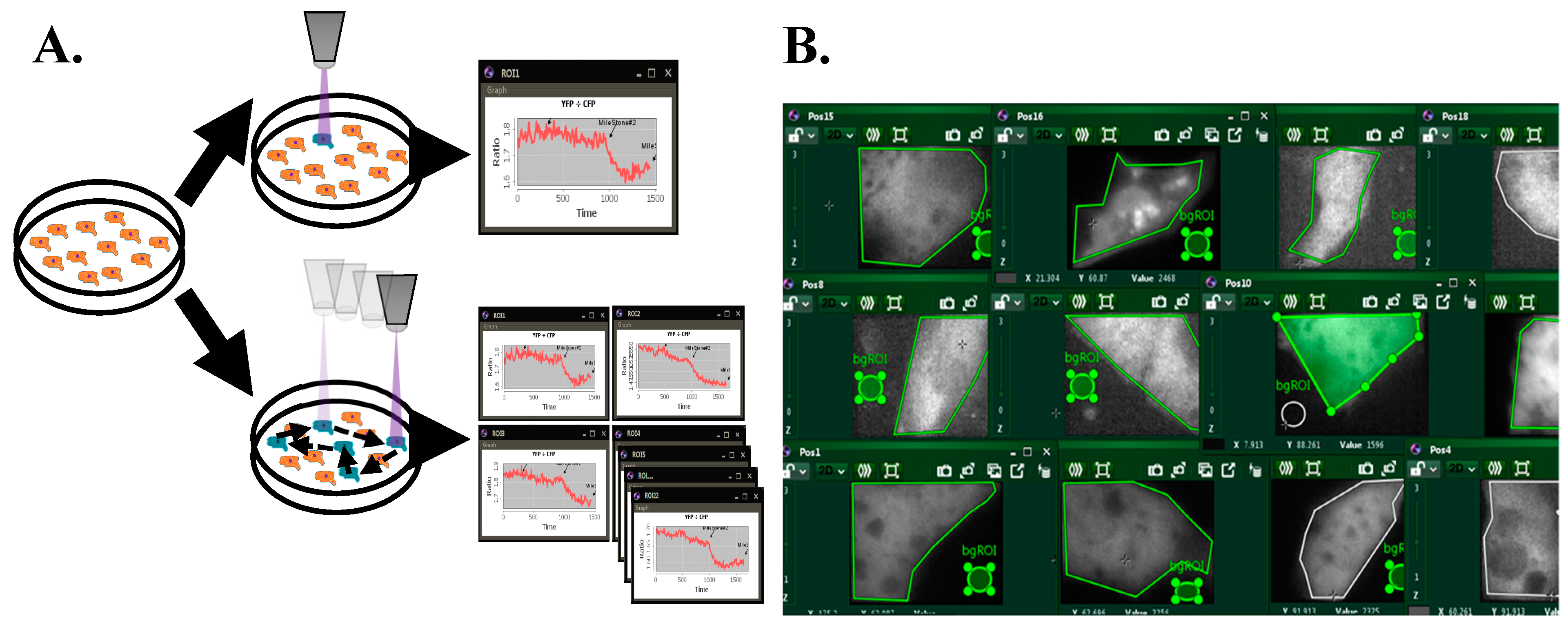
3. Results
3.1. Improvements Through New Real-Time FRET Software: “MultiFRET”
3.2. FRET Calculations
3.3. Software Validation
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Drummen, G.P.C. Fluorescent Probes and Fluorescence (Microscopy) Techniques—Illuminating Biological and Biomedical Research. Molecules 2012, 17, 14067–14090. [Google Scholar] [CrossRef]
- Förster, T. 10th Spiers Memorial Lecture. Transfer Mechanisms of Electronic Excitation. Discuss. Faraday Soc. 1959, 27, 7–17. [Google Scholar] [CrossRef]
- Selvin, P.R. The Renaissance of Fluorescence Resonance Energy Transfer. Nat. Struct. Mol. Biol. 2000, 7, 730–734. [Google Scholar] [CrossRef]
- Padilla-Parra, S.; Tramier, M. FRET Microscopy in the Living Cell: Different Approaches, Strengths and Weaknesses. BioEssays 2012, 34, 369–376. [Google Scholar] [CrossRef]
- Bajar, B.T.; Wang, E.S.; Zhang, S.; Lin, M.Z.; Chu, J. A Guide to Fluorescent Protein FRET Pairs. Sensors 2016, 16, 1488. [Google Scholar] [CrossRef]
- Kaminski, C.F.; Rees, E.J.; Schierle, G.S. A quantitative protocol for intensity-based live cell FRET imaging. In Fluorescence Spectroscopy and Microscopy; Humana Press: Totowa, NJ, USA, 2014; pp. 445–454. [Google Scholar]
- Shrestha, D.; Jenei, A.; Nagy, P.; Vereb, G.; Szöllősi, J. Understanding FRET as a Research Tool for Cellular Studies. Int. J. Mol. Sci. 2015, 16, 6718–6756. [Google Scholar] [CrossRef]
- Feige, J.N.; Sage, D.; Wahli, W.; Desvergne, B.; Gelman, L. PixFRET, an ImageJ Plug-in for FRET Calculation That Can Accommodate Variations in Spectral Bleed-Throughs. Microsc. Res. Tech. 2005, 68, 51–58. [Google Scholar] [CrossRef]
- Deplazes, E.; Jayatilaka, D.; Corry, B. ExiFRET: Flexible Tool for Understanding FRET in Complex Geometries. J. Biomed. Opt. 2012, 17, 011005. [Google Scholar] [CrossRef]
- Roszik, J.; Lisboa, D.; Szöllősi, J.; Vereb, G. Evaluation of Intensity-Based Ratiometric FRET in Image Cytometry—Approaches and a Software Solution. Cytom. Part A 2009, 75, 761–767. [Google Scholar] [CrossRef]
- Stepensky, D. FRETcalc Plugin for Calculation of FRET in Non-Continuous Intracellular Compartments. Biochem. Biophys. Res. Commun. 2007, 359, 752–758. [Google Scholar] [CrossRef]
- Meta Imaging Series® MetaFluor. 2004. Available online: https://www.well.ox.ac.uk/files-library/metafluor-user-guide.pdf (accessed on 25 October 2019).
- Sprenger, J.U.; Perera, R.K.; Götz, K.R.; Nikolaev, V.O. Fret Microscopy for Real-Time Monitoring of Signaling Events in Live Cells Using Unimolecular Biosensors. J. Vis. Exp. 2012, 66, e4081. [Google Scholar] [CrossRef]
- Leroy, J.; Vandecasteele, G.; Fischmeister, R. Cyclic AMP Signaling in Cardiac Myocytes. Curr. Opin. Physiol. 2018, 1, 161–171. [Google Scholar] [CrossRef]
- Peter, W. The Regionality of Cardiac Beta-2-Adrenoceptor Signalling; Imperial College London: London, UK, 2014; Available online: https://spiral.imperial.ac.uk:8443/handle/10044/1/15514 (accessed on 28 November 2019).
- Burridge, P.W.; Anderson, D.; Priddle, H.; Barbadillo Munoz, M.D.; Chamberlain, S.; Allegrucci, C.; Young, L.E.; Denning, C. Improved Human Embryonic Stem Cell Embryoid Body Homogeneity and Cardiomyocyte Differentiation from a Novel V-96 Plate Aggregation System Highlights Interline Variability. Stem Cells 2007, 25, 929–938. [Google Scholar] [CrossRef]
- Nikolaev, V.O.; Bünemann, M.; Hein, L.; Hannawacker, A.; Lohse, M.J. Novel Single Chain cAMP Sensors for Receptor-Induced Signal Propagation. J. Biol. Chem. 2004, 279, 37215–37218. [Google Scholar] [CrossRef]
- Perera, R.K.; Sprenger, J.U.; Steinbrecher, J.H.; Hübscher, D.; Lehnart, S.E.; Abesser, M.; Schuh, K.; El-Armouche, A.; Nikolaev, V.O. Microdomain Switch of cGMP-Regulated Phosphodiesterases Leads to ANP-Induced Augmentation of β-Adrenoceptor-Stimulated Contractility in Early Cardiac Hypertrophy. Circ. Res. 2015, 116, 1304–1311. [Google Scholar] [CrossRef]
- Sprenger, J.U.; Perera, R.K.; Steinbrecher, J.H.; Lehnart, S.E.; Maier, L.S.; Hasenfuss, G.; Nikolaev, V.O. In Vivo Model with Targeted cAMP Biosensor Reveals Changes in Receptor-Microdomain Communication in Cardiac Disease. Nat. Commun. 2015, 6, 6965. [Google Scholar] [CrossRef]
- Klarenbeek, J.B.; Goedhart, J.; Hink, M.A.; Gadella, T.W.; Jalink, K. A mTurquoise-Based cAMP Sensor for Both FLIM and Ratiometric Read-Out Has Improved Dynamic Range. PLoS ONE 2011, 6, e19170. [Google Scholar] [CrossRef]
- Surdo, N.C.; Berrera, M.; Koschinski, A.; Brescia, M.; Machado, M.R.; Carr, C.; Wright, P.; Gorelik, J.; Morotti, S.; Grandi, E.; et al. FRET Biosensor Uncovers cAMP Nano-Domains at β-Adrenergic Targets That Dictate Precise Tuning of Cardiac Contractility. Nat. Commun. 2017, 8, 15031. [Google Scholar] [CrossRef]
- Sprenger, J.U.; Bork, N.I.; Herting, J.; Fischer, T.H.; Nikolaev, V.O. Interactions of Calcium Fluctuations during Cardiomyocyte Contraction with Real-Time cAMP Dynamics Detected by FRET. PLoS ONE 2016, 11, e0167974. [Google Scholar] [CrossRef]
- Börner, S.; Schwede, F.; Schlipp, A.; Berisha, F.; Calebiro, D.; Lohse, M.J.; Nikolaev, V.O. FRET Measurements of Intracellular cAMP Concentrations and cAMP Analog Permeability in Intact Cells. Nat. Protoc. 2011, 6, 427–438. [Google Scholar] [CrossRef]
- Spiering, D.; Bravo-Cordero, J.J.; Moshfegh, Y.; Miskolci, V.; Hodgson, L. Quantitative Ratiometric Imaging of FRET-Biosensors in Living Cells. Methods Cell Biol. 2013, 114, 593–609. [Google Scholar] [CrossRef] [PubMed]

| Sensor | Binding Domain | EC50 | Reference |
|---|---|---|---|
| Epac1-camps | Epac1 | 2.4 µM | V.O. Nikolaev et al., J. Biol. Chem. 2004 [17] |
| pmEpac1 | Epac1 | 9.1 µM | R.K. Perera et al., Circ. Res. 2015 [18] |
| Epac1-PLN | Epac1 | 5.3 µM | J.U. Sprenger et al., Nat. Commun. 2015 [19] |
| Epac-SH74 | Epac1 | 11–14 µM | J. Klarenbeek et al., PLoS ONE 2011 [20] |
| AKAP79-CUTie | Prkar2β | 7.4 µM | N. Surdo et al., Nature Commun 2017 [21] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ramuz, M.; Hasan, A.; Gruscheski, L.; Diakonov, I.; Pavlaki, N.; Nikolaev, V.O.; Harding, S.; Dunsby, C.; Gorelik, J. A Software Tool for High-Throughput Real-Time Measurement of Intensity-Based Ratio-Metric FRET. Cells 2019, 8, 1541. https://doi.org/10.3390/cells8121541
Ramuz M, Hasan A, Gruscheski L, Diakonov I, Pavlaki N, Nikolaev VO, Harding S, Dunsby C, Gorelik J. A Software Tool for High-Throughput Real-Time Measurement of Intensity-Based Ratio-Metric FRET. Cells. 2019; 8(12):1541. https://doi.org/10.3390/cells8121541
Chicago/Turabian StyleRamuz, Masoud, Alveera Hasan, Lena Gruscheski, Ivan Diakonov, Nikoleta Pavlaki, Viacheslav O. Nikolaev, Sian Harding, Chris Dunsby, and Julia Gorelik. 2019. "A Software Tool for High-Throughput Real-Time Measurement of Intensity-Based Ratio-Metric FRET" Cells 8, no. 12: 1541. https://doi.org/10.3390/cells8121541
APA StyleRamuz, M., Hasan, A., Gruscheski, L., Diakonov, I., Pavlaki, N., Nikolaev, V. O., Harding, S., Dunsby, C., & Gorelik, J. (2019). A Software Tool for High-Throughput Real-Time Measurement of Intensity-Based Ratio-Metric FRET. Cells, 8(12), 1541. https://doi.org/10.3390/cells8121541






