Non-Redundant and Overlapping Oncogenic Readouts of Non-Canonical and Novel Colorectal Cancer KRAS and NRAS Mutants
Abstract
1. Introduction
2. Materials and Methods
2.1. Site-Directed Mutagenesis and Generation of KRAS and NRAS Mutant Constructs
2.2. Cell Culture and Transfection of NIH3T3 and HCT116 Cells
2.3. Cell Proliferation Assay
2.4. Caspase 3/7 Assay
2.5. Wound Healing Assay
2.6. Western Blotting
2.7. Actin Cytoskeleton Staining
2.8. Observation of Gross Morphology
2.9. ELK-TAD Luciferase Reporter Assay
2.10. Bioinformatics-Based Analysis of Potential Functional Impact of KRAS and NRAS Mutations
2.11. Statistical Analysis
3. Results
3.1. Cells Expressing Non-Canonical/Novel KRAS Mutants Exhibit Increased Proliferation Rates
3.2. The Rare Mutant NRAS A11V Confers Increased Resistance to Apoptosis
3.3. KRAS Mutants G12S, A59T, and Y137C Promote Cell Migration
3.4. KRAS Mutants G12S, A59T, and Y137C Upregulate the Mesenchymal Marker Vimentin
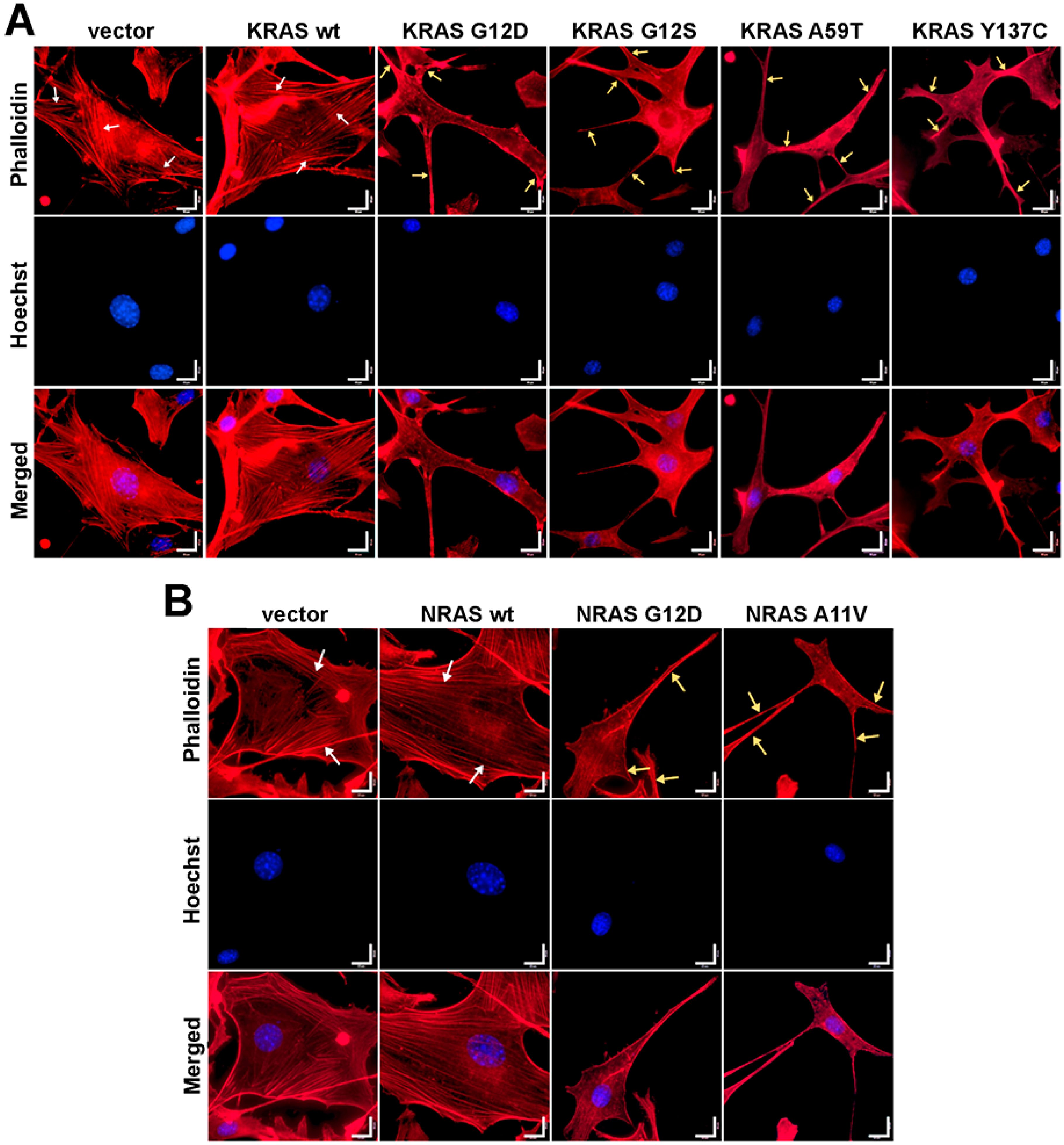
3.5. Cells Expressing KRAS and NRAS Mutants Display Altered F-Actin Cytoskeletal Organization
3.6. KRAS and NRAS Mutants Promote Cell Rounding, Refringency, and Cytoplasmic Shrinkage
3.7. Effects of KRAS and NRAS Mutations on ERK Phosphorylation and ELK-1 Transactivation
3.8. Bioinformatics-Based Modeling and Docking Simulations of the KRAS Mutants Predict Their Oncogenic Impact
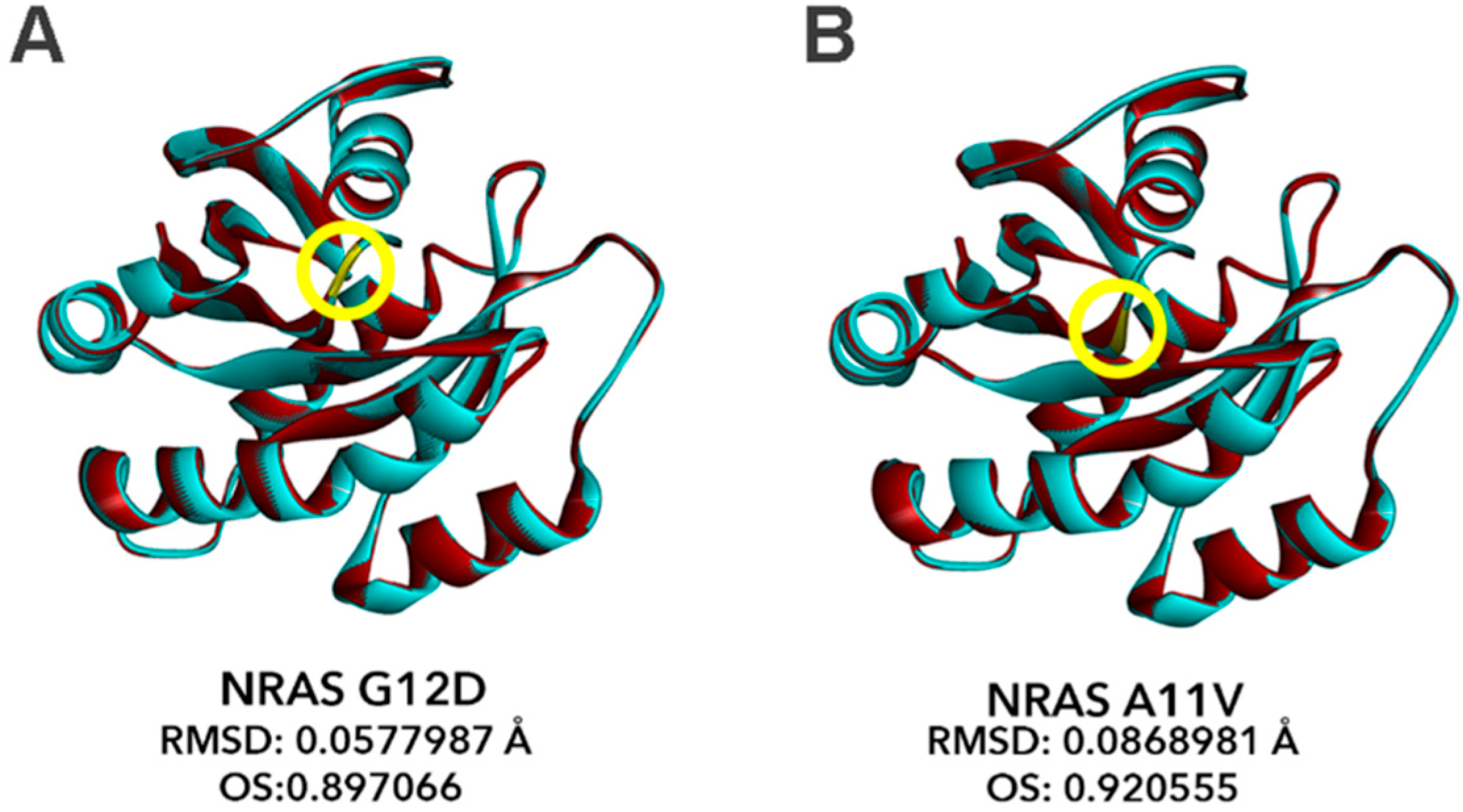
3.9. Bioinformatics-Based Modeling of the Novel NRAS A11V Mutant Predicts Larger Changes in Overall Protein Structure but Higher Similarity to NRAS-WT than G12D
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- De Stefano, A. Beyond KRAS: Predictive factors of the efficacy of anti-EGFR monoclonal antibodies in the treatment of metastatic colorectal cancer. World J. Gastroenterol. 2014, 20, 9732. [Google Scholar] [CrossRef]
- Castellano, E.; Santos, E. Functional Specificity of Ras Isoforms: So Similar but So Different. Genes Cancer 2011, 2, 216–231. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Ruiz, E.; Rueda, A.; Pereda, T.; Alcaide, J.; Bautista, D.; Rivas-Ruiz, F.; Villatoro, R.; Pérez, D.; Redondo, M. Involvement of K-RAS mutations and amino acid substitutions in the survival of metastatic colorectal cancer patients. Tumor Biol. 2012, 33, 1829–1835. [Google Scholar] [CrossRef]
- Christoph, D.C.; Eberhardt, W.E.E. ESMO Handbook of Cancer in the Senior Patient; Schrijvers, D., Aapro, M., Zakotnik, B., Audisio, R., Eds.; CRC Press: Boca Raton, FL, USA, 2010; ISBN 9780429110252. [Google Scholar]
- De Roock, W.; Claes, B.; Bernasconi, D.; De Schutter, J.; Biesmans, B.; Fountzilas, G.; Kalogeras, K.T.; Kotoula, V.; Papamichael, D.; Laurent-Puig, P.; et al. Effects of KRAS, BRAF, NRAS, and PIK3CA mutations on the efficacy of cetuximab plus chemotherapy in chemotherapy-refractory metastatic colorectal cancer: A retrospective consortium analysis. Lancet Oncol. 2010, 11, 753–762. [Google Scholar] [CrossRef]
- Bronte, G.; Silvestris, N.; Castiglia, M.; Galvano, A.; Passiglia, F.; Sortino, G.; Cicero, G.; Rolfo, C.; Peeters, M.; Bazan, V.; et al. New findings on primary and acquired resistance to anti-EGFR therapy in metastatic colorectal cancer: Do all roads lead to RAS? Oncotarget 2015, 6, 24780–24796. [Google Scholar] [CrossRef]
- Cox, A.D.; Fesik, S.W.; Kimmelman, A.C.; Luo, J.; Der, C.J. Drugging the undruggable RAS: Mission Possible? Nat. Rev. Drug Discov. 2014, 13, 828–851. [Google Scholar] [CrossRef]
- Hobbs, G.A.; Der, C.J.; Rossman, K.L. RAS isoforms and mutations in cancer at a glance. J. Cell Sci. 2016, 129, 1287–1292. [Google Scholar] [CrossRef]
- Buhrman, G.; Wink, G.; Mattos, C. Transformation Efficiency of RasQ61 Mutants Linked to Structural Features of the Switch Regions in the Presence of Raf. Structure 2007, 15, 1618–1629. [Google Scholar] [CrossRef]
- Burd, C.E.; Liu, W.; Huynh, M.V.; Waqas, M.A.; Gillahan, J.E.; Clark, K.S.; Fu, K.; Martin, B.L.; Jeck, W.R.; Souroullas, G.P.; et al. Mutation-Specific RAS Oncogenicity Explains NRAS Codon 61 Selection in Melanoma. Cancer Discov. 2014, 4, 1418–1429. [Google Scholar] [CrossRef]
- Smith, M.J.; Neel, B.G.; Ikura, M. NMR-based functional profiling of RASopathies and oncogenic RAS mutations. Proc. Natl. Acad. Sci. USA 2013, 110, 4574–4579. [Google Scholar] [CrossRef]
- Vagaja, N.N.; Parry, J.; McCallum, D.; Thomas, M.A.; Bentel, J.M. Are all RAS mutations the same? Coexisting KRAS and NRAS mutations in a caecal adenocarcinoma and contiguous tubulovillous adenoma. J. Clin. Pathol. 2015, 68, 657–660. [Google Scholar] [CrossRef] [PubMed]
- Van Cutsem, E.; Cervantes, A.; Adam, R.; Sobrero, A.; Van Krieken, J.H.; Aderka, D.; Aranda Aguilar, E.; Bardelli, A.; Benson, A.; Bodoky, G.; et al. ESMO consensus guidelines for the management of patients with metastatic colorectal cancer. Ann. Oncol. 2016, 27, 1386–1422. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, N.; YAmada, Y.; Taniguchi, H.; Akiyoshi, K.; Honma, Y.; Kato, S.I.; Hamaguchi, T.; Shimada, Y. Mutations in NRAS codon 61, KRAS codon 146, and BRAF V600E as prognostic factors in patients who received anti-EGFR antibody for metastatic colorectal cancer. J. Clin. Oncol. 2012, 30, e14126. [Google Scholar]
- Malapit, J.R.P.; Canlas, K.K.V.; Rigor, J.P.T.; Padilla, C.D.; Cutiongco-de la Paz, E.M.; Garcia, R.L. Next generation targeted sequencing reveals novel, non-canonical mutations in downstream effectors of the EGFR pathway in Filipino colorectal cancer patients. Unpublished work.
- Margonis, G.A.; Kim, Y.; Spolverato, G.; Ejaz, A.; Gupta, R.; Cosgrove, D.; Anders, R.; Karagkounis, G.; Choti, M.A.; Pawlik, T.M. Association Between Specific Mutations in KRAS Codon 12 and Colorectal Liver Metastasis. JAMA Surg. 2015, 150, 722. [Google Scholar] [CrossRef]
- Lou, E.; D’Souza, D.; Nelson, A.C. Therapeutic Response of Metastatic Colorectal Cancer Harboring a KRAS Missense Mutation After Combination Chemotherapy with the EGFR Inhibitor Panitumumab. J. Natl. Compr. Cancer Netw. 2017, 15, 427–432. [Google Scholar] [CrossRef][Green Version]
- Shihab, H.A.; Gough, J.; Cooper, D.N.; Stenson, P.D.; Barker, G.L.A.; Edwards, K.J.; Day, I.N.M.; Gaunt, T.R. Predicting the Functional, Molecular, and Phenotypic Consequences of Amino Acid Substitutions using Hidden Markov Models. Hum. Mutat. 2013, 34, 57–65. [Google Scholar] [CrossRef]
- Angeles, A.; Yu, R.; Cutiongco-De la Paz, E.; Garcia, R. Phenotypic characterization of the novel, non-hotspot oncogenic KRAS mutants E31D and E63K. Oncol. Lett. 2019, 18, 420–432. [Google Scholar] [CrossRef] [PubMed]
- Petty, E.M. Cellular cancer markers. Am. J. Med. Genet. 1997, 68, 492–493. [Google Scholar] [CrossRef]
- Yu, J.L.; May, L.; Lhotak, V.; Shahrzad, S.; Shirasawa, S.; Weitz, J.I.; Coomber, B.L.; Mackman, N.; Rak, J.W. Oncogenic events regulate tissue factor expression in colorectal cancer cells: Implications for tumor progression and angiogenesis. Blood 2005, 105, 1734–1741. [Google Scholar] [CrossRef]
- Gebäck, T.; Schulz, M.M.P.; Koumoutsakos, P.; Detmar, M. TScratch: A novel and simple software tool for automated analysis of monolayer wound healing assays. Biotechniques 2009, 46, 265–274. [Google Scholar] [CrossRef]
- Gilda, J.E.; Gomes, A.V. Stain-Free total protein staining is a superior loading control to b-actin for Western blots. Anal. Biochem. 2013, 440, 186–188. [Google Scholar] [CrossRef] [PubMed]
- Aldridge, G.M.; Podrebarac, D.M.; Greenough, W.T.; Weiler, I.J. The use of total protein stains as loading controls: An alternative to high-abundance single-protein controls in semi-quantitative immunoblotting. J. Neurosci. Methods 2008, 172, 250–254. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Adzhubei, I.A.; Schmidt, S.; Peshkin, L.; Ramensky, V.E.; Gerasimova, A.; Bork, P.; Kondrashov, A.S.; Sunyaev, S.R. A method and server for predicting damaging missense mutations. Nat. Methods 2010, 7, 248–249. [Google Scholar] [CrossRef]
- Kumar, P.; Henikoff, S.; Ng, P.C. Predicting the effects of coding non-synonymous variants on protein function using the SIFT algorithm. Nat. Protoc. 2009, 4, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Tavtigian, S.V. Comprehensive statistical study of 452 BRCA1 missense substitutions with classification of eight recurrent substitutions as neutral. J. Med. Genet. 2005, 43, 295–305. [Google Scholar] [CrossRef]
- Zhou, B.; Der, C.J.; Cox, A.D. The role of wild type RAS isoforms in cancer. Semin. Cell Dev. Biol. 2016, 58, 60–69. [Google Scholar] [CrossRef]
- Cox, A.D.; Der, C.J. Ras history. Small GTPases 2010, 1, 2–27. [Google Scholar] [CrossRef]
- Pylayeva-Gupta, Y.; Grabocka, E.; Bar-Sagi, D. RAS oncogenes: Weaving a tumorigenic web. Nat. Rev. Cancer 2011, 11, 761–774. [Google Scholar] [CrossRef]
- Karnoub, A.E.; Weinberg, R.A. Ras oncogenes: Split personalities. Nat. Rev. Mol. Cell Biol. 2008, 9, 517–531. [Google Scholar] [CrossRef]
- Fernández-Medarde, A.; Santos, E. Ras in cancer and developmental diseases. Genes Cancer 2011, 2, 344–358. [Google Scholar] [CrossRef] [PubMed]
- Tojkander, S.; Gateva, G.; Lappalainen, P. Actin stress fibers—Assembly, dynamics and biological roles. J. Cell Sci. 2012, 125, 1855–1864. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, M.; Christofori, G. EMT, the cytoskeleton, and cancer cell invasion. Cancer Metastasis Rev. 2009, 28, 15–33. [Google Scholar] [CrossRef] [PubMed]
- Lombardi, L.; Ballinari, D.; Bongarzone, I.; Migliari, M.; Mondellini, P.; Traversai, C.; Modina, S. Ultrastructural cytoskeleton alterations and modification of actin expression in the NIH/3T3 cell line after transformation with Ha-ras-Activated oncogene. Cell Motil. Cytoskel. 1990, 15, 220–229. [Google Scholar] [CrossRef] [PubMed]
- Nobes, C.D.; Hall, A. Rho GTPases Control Polarity, Protrusion, and Adhesion during Cell Movement. J. Cell Biol. 1999, 144, 1235–1244. [Google Scholar] [CrossRef] [PubMed]
- Pawlak, G.; Helfman, D.M. Cytoskeletal changes in cell transformation and tumorigenesis. Curr. Opin. Genet. Dev. 2001, 11, 41–47. [Google Scholar] [CrossRef]
- Basu, S.K.; Lee, S.; Salotti, J.; Basu, S.; Sakchaisri, K.; Xiao, Z.; Walia, V.; Westlake, C.J.; Morrison, D.K.; Johnson, P.F. Oncogenic RAS-Induced Perinuclear Signaling Complexes Requiring KSR1 Regulate Signal Transmission to Downstream Targets. Cancer Res. 2018, 78, 891–908. [Google Scholar] [CrossRef]
- Liu, F.; Yang, X.; Geng, M.; Huang, M. Targeting ERK, an Achilles’ Heel of the MAPK pathway, in cancer therapy. Acta Pharm. Sin. B 2018, 8, 552–562. [Google Scholar] [CrossRef]
- Whitmarsh, A.J. Regulation of gene transcription by mitogen-activated protein kinase signaling pathways. Biochim. Biophys. Acta - Mol. Cell Res. 2007, 1773, 1285–1298. [Google Scholar] [CrossRef]
- Lu, S.; Jang, H.; Nussinov, R.; Zhang, J. The Structural Basis of Oncogenic Mutations G12, G13 and Q61 in Small GTPase K-Ras4B. Sci. Rep. 2016, 6, 21949. [Google Scholar] [CrossRef]
- Lu, S.; Jang, H.; Muratcioglu, S.; Gursoy, A.; Keskin, O.; Nussinov, R.; Zhang, J. Ras Conformational Ensembles, Allostery, and Signaling. Chem. Rev. 2016, 116, 6607–6665. [Google Scholar] [CrossRef] [PubMed]
- Forbes, S.A.; Beare, D.; Boutselakis, H.; Bamford, S.; Bindal, N.; Tate, J.; Cole, C.G.; Ward, S.; Dawson, E.; Ponting, L.; et al. COSMIC: Somatic cancer genetics at high-resolution. Nucleic Acids Res. 2017, 45, D777–D783. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.H.; Lee, J.W.; Soung, Y.H.; Kim, H.S.; Park, W.S.; Kim, S.Y.; Lee, J.H.; Park, J.Y.; Cho, Y.G.; Kim, C.J.; et al. BRAF and KRAS mutations in stomach cancer. Oncogene 2003, 22, 6942–6945. [Google Scholar] [CrossRef] [PubMed]
- Cros, J.; Sbidian, E.; Hans, S.; Roussel, H.; Scotte, F.; Tartour, E.; Brasnu, D.; Laurent-Puig, P.; Bruneval, P.; Blons, H.; et al. Expression and mutational status of treatment-relevant targets and key oncogenes in 123 malignant salivary gland tumours. Ann. Oncol. 2013, 24, 2624–2629. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Xiang, L.; Pei, X.; He, T.; Shen, X.; Wu, X.; Yang, H. Mutational analysis of KRAS and its clinical implications in cervical cancer patients. J. Gynecol. Oncol. 2018, 29. [Google Scholar] [CrossRef] [PubMed]
- Sforza, V.; Martinelli, E.; Ciardiello, F.; Gambardella, V.; Napolitano, S.; Martini, G.; della Corte, C.; Cardone, C.; Ferrara, M.L.; Reginelli, A.; et al. Mechanisms of resistance to anti-epidermal growth factor receptor inhibitors in metastatic colorectal cancer. World J. Gastroenterol. 2016, 22, 6345. [Google Scholar] [CrossRef]
- Cercek, A.; Capanu, M.; Hechtman, J.F.; Kemeny, N.; Saltz, L.; Braghiroli, M.I.; Yaeger, R.; Chou, J.F. Clinical Features and Outcomes of Patients with Colorectal Cancers Harboring NRAS Mutations. Clin. Cancer Res. 2017, 23, 4753–4760. [Google Scholar] [CrossRef]
- Nakhaeizadeh, H.; Amin, E.; Nakhaei-Rad, S.; Dvorsky, R.; Ahmadian, M.R. The RAS-effector interface: Isoform-specific differences in the effector binding regions. PLoS ONE 2016, 11, e0167145. [Google Scholar] [CrossRef]





| Designation | Sequence (5′-3′) |
|---|---|
| KRAS-WT-F | GCCGTCCTGAAGCTTCGCCGGATGACTGAATATAAACTTG |
| KRAS-WT-R | GTCGTGCCGCGGCCGCGTGCCGTTACATAATTACACACTTTG |
| KRAS-G12D-F | GCCGTCCTGAAGCTTCGCCGGATGACTGAATATAAACTTGTGGTAGTTGGAGCTGA*TGGCGTAGG |
| KRAS-G12S-F | GACTGTGGATCCATGACTGAATATAAACTTGTGGTAGTTGGAGCTA*GTG |
| KRAS-A59T-F | GGATATTCTCGACACAA*CAGGTCAAGAG GAGTAC |
| KRAS-A59T-R | GTACTCCTCTTGACCTGTTGTGTCGAGAATATCC |
| KRAS-Y137C-F | GGACTTAGCAAGAAGTTG*TGGAATTCCTTTTATTG |
| KRAS-Y137C-R | CAATAAAAGGAATTCCACAACTTCTTGCTAAGTCC |
| Designation | Sequence (5′-3′) |
|---|---|
| NRAS-WT-F | ATGACTGAGTACAAACTGGTGGTGGTTGGAG |
| NRAS-WT-R | TTACATCACCACACATGGCAATCCCATAC |
| NRAS-G12D-F | ATGACTGAGTACAAACTGGTGGTGGTTGGAGCAGA*TGGT |
| NRAS-A11V-F | ATGACTGAGTACAAACTGGTGGTGGTTGGAGT*AGGTGG |
| Protein | Mutation | Classification | Polyphen-2 (score) | SIFT (score) | Align GVGD (GD Score) |
|---|---|---|---|---|---|
| KRAS | G12D | canonical | Possibly damaging (0.517) | Damaging (0.00) | Class C65 (93.77) |
| KRAS | G12S | COSMIC | Possibly damaging (0.682) | Damaging (0.01) | Class C55 (55.27) |
| KRAS | A59T | COSMIC | Possibly damaging (0.936) | Damaging (0.03) | Class C55 (58.02) |
| KRAS | Y137C | novel | Benign (0.353) | Damaging (0.00) | Class C65 (193.72) |
| NRAS | G12D | canonical | Possibly damaging (0.488) | Damaging (0.00) | Class C65 (93.77) |
| NRAS | A11V | novel | Possibly damaging (0.922) | Damaging (0.02) | Class C55 (64.43) |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alcantara, K.M.M.; Malapit, J.R.P.; Yu, R.T.D.; Garrido, J.A.M.G.; Rigor, J.P.T.; Angeles, A.K.J.; Cutiongco-de la Paz, E.M.; Garcia, R.L. Non-Redundant and Overlapping Oncogenic Readouts of Non-Canonical and Novel Colorectal Cancer KRAS and NRAS Mutants. Cells 2019, 8, 1557. https://doi.org/10.3390/cells8121557
Alcantara KMM, Malapit JRP, Yu RTD, Garrido JAMG, Rigor JPT, Angeles AKJ, Cutiongco-de la Paz EM, Garcia RL. Non-Redundant and Overlapping Oncogenic Readouts of Non-Canonical and Novel Colorectal Cancer KRAS and NRAS Mutants. Cells. 2019; 8(12):1557. https://doi.org/10.3390/cells8121557
Chicago/Turabian StyleAlcantara, Krizelle Mae M., Joshua Reginald P. Malapit, Ryan Timothy D. Yu, Jose Antonio Ma. G. Garrido, John Paul T. Rigor, Arlou Kristina J. Angeles, Eva Maria Cutiongco-de la Paz, and Reynaldo L. Garcia. 2019. "Non-Redundant and Overlapping Oncogenic Readouts of Non-Canonical and Novel Colorectal Cancer KRAS and NRAS Mutants" Cells 8, no. 12: 1557. https://doi.org/10.3390/cells8121557
APA StyleAlcantara, K. M. M., Malapit, J. R. P., Yu, R. T. D., Garrido, J. A. M. G., Rigor, J. P. T., Angeles, A. K. J., Cutiongco-de la Paz, E. M., & Garcia, R. L. (2019). Non-Redundant and Overlapping Oncogenic Readouts of Non-Canonical and Novel Colorectal Cancer KRAS and NRAS Mutants. Cells, 8(12), 1557. https://doi.org/10.3390/cells8121557





