An Update on Mitochondrial Ribosome Biology: The Plant Mitoribosome in the Spotlight
Abstract
:1. Introduction
2. The Plant Mitoribosome Is Structurally and Compositionally Distinct from both Prokaryotic and Non-Plant Mitochondrial Ribosomes
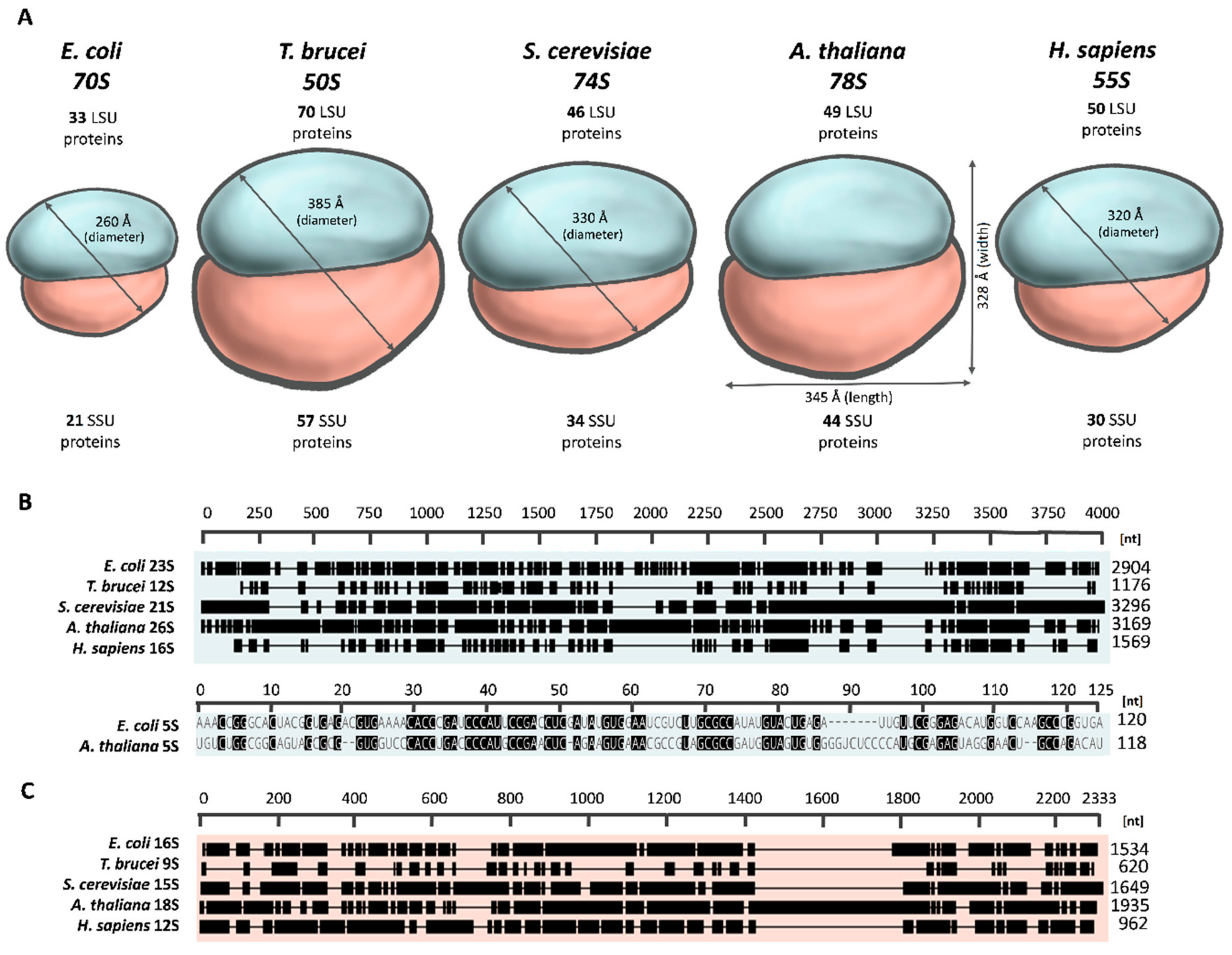
2.1. The Overall Architecture of the Mitoribosome
2.2. Variation in Mitoribosomal rRNAs
2.3. Variation in Mitoribosomal Protein Composition
2.4. Association of Mitoribosomes with the Inner Mitochondrial Membrane
3. Heterogeneity of the Mitoribosome Composition
4. Mitoribosome Features Revealed by Ribosomal Profiling
5. Mitoribosomes in Expressosome-Like Assemblies
6. Conclusions
Funding
Conflicts of Interest
References
- Gray, M.W. Mitochondrial Evolution. Science 1999, 283, 1476–1481. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Desmond, E.; Brochier-Armanet, C.; Forterre, P.; Gribaldo, S. On the last common ancestor and early evolution of eukaryotes: Reconstructing the history of mitochondrial ribosomes. Res. Microbiol. 2011, 162, 53–70. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yusupova, G.; Yusupov, M. Crystal structure of eukaryotic ribosome and its complexes with inhibitors. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372, 20160184. [Google Scholar] [CrossRef] [PubMed]
- Ban, N.; Nissen, P.; Hansen, J.; Moore, P.B.; Steitz, T.A. The complete atomic structure of the large ribosomal subunit at 2.4 A resolution. Science 2000, 289, 905–920. [Google Scholar] [CrossRef] [PubMed]
- Amunts, A.; Brown, A.; Toots, J.; Scheres, S.H.W.; Ramakrishnan, V. Ribosome. The structure of the human mitochondrial ribosome. Science 2015, 348, 95–98. [Google Scholar] [CrossRef] [Green Version]
- Desai, N.; Brown, A.; Amunts, A.; Ramakrishnan, V. The structure of the yeast mitochondrial ribosome. Science 2017, 355, 528–531. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramrath, D.J.F.; Niemann, M.; Leibundgut, M.; Bieri, P.; Prange, C.; Horn, E.K.; Leitner, A.; Boehringer, D.; Schneider, A.; Ban, N. Evolutionary shift toward protein-based architecture in trypanosomal mitochondrial ribosomes. Science 2018, 362, eaau7735. [Google Scholar] [CrossRef]
- Waltz, F.; Nguyen, T.T.; Arrivé, M.; Bochler, A.; Chicher, J.; Hammann, P.; Kuhn, L.; Quadrado, M.; Mireau, H.; Hashem, Y.; et al. Small is big in Arabidopsis mitochondrial ribosome. Nat. Plants 2019, 5, 106–117. [Google Scholar] [CrossRef]
- Smits, P.; Smeitink, J.A.M.; van den Heuvel, L.P.; Huynen, M.A.; Ettema, T.J.G. Reconstructing the evolution of the mitochondrial ribosomal proteome. Nucleic Acids Res. 2007, 35, 4686–4703. [Google Scholar] [CrossRef]
- Rackham, O.; Filipovska, A. Supernumerary proteins of mitochondrial ribosomes. Biochim. Biophys. Acta—Gen. Subj. 2014, 1840, 1227–1232. [Google Scholar] [CrossRef]
- Rugen, N.; Straube, H.; Franken, L.E.; Braun, H.-P.; Eubel, H. Complexome Profiling Reveals Association of PPR Proteins with Ribosomes in the Mitochondria of Plants. Mol. Cell. Proteomics 2019, 18, 1345–1362. [Google Scholar] [CrossRef] [PubMed]
- Bieri, P.; Greber, B.J.; Ban, N. High-resolution structures of mitochondrial ribosomes and their functional implications. Curr. Opin. Struct. Biol. 2018, 49, 44–53. [Google Scholar] [CrossRef] [PubMed]
- Selmer, M.; Dunham, C.M.; Murphy, F.V.; Weixlbaumer, A.; Petry, S.; Kelley, A.C.; Weir, J.R.; Ramakrishnan, V. Structure of the 70S ribosome complexed with mRNA and tRNA. Science 2006, 313, 1935–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ridlon, L.; Škodová, I.; Pan, S.; Lukeš, J.; Maslov, D.A. The importance of the 45 S ribosomal small subunit-related complex for mitochondrial translation in Trypanosoma brucei. J. Biol. Chem. 2013, 288, 32963–32978. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Greber, B.J.; Bieri, P.; Leibundgut, M.; Leitner, A.; Aebersold, R.; Boehringer, D.; Ban, N. The complete structure of the 55S mammalian mitochondrial ribosome. Nature 2014, 348, 303–308. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Waltz, F.; Soufari, H.; Bochler, A.; Giegé, P.; Hashem, Y. Cryo-EM structure of the RNA-rich plant mitochondrial ribosome. bioRxiv 2019, 777342. [Google Scholar]
- Amunts, A.; Brown, A.; Bai, X.-C.; Llácer, J.L.; Hussain, T.; Emsley, P.; Long, F.; Murshudov, G.; Scheres, S.H.W.; Ramakrishnan, V. Structure of the yeast mitochondrial large ribosomal subunit. Science 2014, 343, 1485–1489. [Google Scholar] [CrossRef] [Green Version]
- Greber, B.J.; Boehringer, D.; Leibundgut, M.; Bieri, P.; Leitner, A.; Schmitz, N.; Aebersold, R.; Ban, N. The complete structure of the large subunit of the mammalian mitochondrial ribosome. Nature 2014, 515, 283–286. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.; Amunts, A.; Bai, X.-C.; Sugimoto, Y.; Edwards, P.C.; Murshudov, G.; Scheres, S.H.W.; Ramakrishnan, V. Structure of the large ribosomal subunit from human mitochondria. Science 2014, 346, 718–722. [Google Scholar] [CrossRef] [Green Version]
- Agrawal, R.K.; Sharma, M.R.; Yassin, A.; Lahiri, I.; Spremulli, L. Structure and function of organellar ribosomes as revealed by cryo-EM. In Ribosomes; Springer Vienna: Vienna, Austria, 2011; pp. 83–96. [Google Scholar]
- Bonen, L. Translational Machinery in Plant Organelles. In Molecular Biology and Biotechnology of Plant Organelles; Springer Netherlands: Dordrecht, The Netherlands, 2004; pp. 323–345. [Google Scholar]
- Szymanski, M.; Barciszewska, M.Z.; Erdmann, V.A.; Barciszewski, J. 5S Ribosomal RNA Database. Nucleic Acids Res. 2002, 30, 176–178. [Google Scholar] [CrossRef]
- Rorbach, J.; Gao, F.; Powell, C.A.; D’Souza, A.; Lightowlers, R.N.; Minczuk, M.; Chrzanowska-Lightowlers, Z.M. Human mitochondrial ribosomes can switch their structural RNA composition. Proc. Natl. Acad. Sci. USA 2016, 113, 12198–12201. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scharff, L.B.; Ehrnthaler, M.; Janowski, M.; Childs, L.H.; Hasse, C.; Gremmels, J.; Ruf, S.; Zoschke, R.; Bock, R. Shine-Dalgarno Sequences Play an Essential Role in the Translation of Plastid mRNAs in Tobacco. Plant Cell 2017, 29, 3085–3101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bieri, P.; Leibundgut, M.; Saurer, M.; Boehringer, D.; Ban, N. The complete structure of the chloroplast 70S ribosome in complex with translation factor pY. EMBO J. 2017, 36, 475–486. [Google Scholar] [CrossRef] [PubMed]
- Greber, B.J.; Ban, N. Structure and Function of the Mitochondrial Ribosome. Annu. Rev. Biochem. 2016, 85, 103–132. [Google Scholar] [CrossRef] [PubMed]
- Boore, J.L. Animal mitochondrial genomes. Nucleic Acids Res. 1999, 27, 1767–1780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Silva, D.; Tu, Y.T.; Amunts, A.; Fontanesi, F.; Barrientos, A. Mitochondrial ribosome assembly in health and disease. Cell Cycle 2015, 14, 2226–2250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Adams, K. Evolution of mitochondrial gene content: Gene loss and transfer to the nucleus. Mol. Phylogenet. Evol. 2003, 29, 380–395. [Google Scholar] [CrossRef]
- Bonen, L.; Calixte, S. Comparative Analysis of Bacterial-Origin Genes for Plant Mitochondrial Ribosomal Proteins. Mol. Biol. Evol. 2006, 23, 701–712. [Google Scholar] [CrossRef] [Green Version]
- Barkan, A.; Small, I. Pentatricopeptide Repeat Proteins in Plants. Annu. Rev. Plant Biol. 2014, 65, 415–442. [Google Scholar] [CrossRef]
- Davies, S.M.K.; Lopez Sanchez, M.I.G.; Narsai, R.; Shearwood, A.-M.J.; Razif, M.F.M.; Small, I.D.; Whelan, J.; Rackham, O.; Filipovska, A. MRPS27 is a pentatricopeptide repeat domain protein required for the translation of mitochondrially encoded proteins. FEBS Lett. 2012, 586, 3555–3561. [Google Scholar] [CrossRef] [Green Version]
- Davies, S.M.K.; Rackham, O.; Shearwood, A.-M.J.; Hamilton, K.L.; Narsai, R.; Whelan, J.; Filipovska, A. Pentatricopeptide repeat domain protein 3 associates with the mitochondrial small ribosomal subunit and regulates translation. FEBS Lett. 2009, 583, 1853–1858. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aphasizheva, I.; Maslov, D.A.; Qian, Y.; Huang, L.; Wang, Q.; Costello, C.E.; Aphasizhev, R. Ribosome-associated pentatricopeptide repeat proteins function as translational activators in mitochondria of trypanosomes. Mol. Microbiol. 2016, 99, 1043–1058. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Delage, L.; Giegé, P.; Sakamoto, M.; Maréchal-Drouard, L. Four paralogues of RPL12 are differentially associated to ribosome in plant mitochondria. Biochimie 2007, 89, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Fiori, A.; Mason, T.L.; Fox, T.D. Evidence that Synthesis of the Saccharomyces cerevisiae Mitochondrially Encoded Ribosomal Protein Var1p May Be Membrane Localized. Eukaryot. Cell 2003, 2, 651–653. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pfeffer, S.; Woellhaf, M.W.; Herrmann, J.M.; Förster, F. Organization of the mitochondrial translation machinery studied in situ by cryoelectron tomography. Nat. Commun. 2015, 6, 6019. [Google Scholar] [CrossRef] [Green Version]
- Englmeier, R.; Pfeffer, S.; Förster, F. Structure of the Human Mitochondrial Ribosome Studied In Situ by Cryoelectron Tomography. Structure 2017, 25, 1574–1581. [Google Scholar] [CrossRef] [Green Version]
- Guo, H. Specialized ribosomes and the control of translation. Biochem. Soc. Trans. 2018, 46, 855–869. [Google Scholar] [CrossRef]
- Genuth, N.R.; Barna, M. The Discovery of Ribosome Heterogeneity and Its Implications for Gene Regulation and Organismal Life. Mol. Cell 2018, 71, 364–374. [Google Scholar] [CrossRef] [Green Version]
- Xue, S.; Barna, M. Specialized ribosomes: A new frontier in gene regulation and organismal biology. Nat. Rev. Mol. Cell Biol. 2012, 13, 355–369. [Google Scholar] [CrossRef] [Green Version]
- Lilleorg, S.; Reier, K.; Pulk, A.; Liiv, A.; Tammsalu, T.; Peil, L.; Cate, J.H.D.; Remme, J. Bacterial ribosome heterogeneity: Changes in ribosomal protein composition during transition into stationary growth phase. Biochimie 2019, 156, 169–180. [Google Scholar] [CrossRef]
- Robles, P.; Quesada, V. Emerging Roles of Mitochondrial Ribosomal Proteins in Plant Development. Int. J. Mol. Sci. 2017, 18, 2595. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; Luo, M.; Day, R.C.; Talbot, M.J.; Ivanova, A.; Ashton, A.R.; Chaudhury, A.M.; Macknight, R.C.; Hrmova, M.; Koltunow, A.M. Developmentally regulated HEART STOPPER, a mitochondrially targeted L18 ribosomal protein gene, is required for cell division, differentiation, and seed development in Arabidopsis. J. Exp. Bot. 2015, 66, 5867–5880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qi, W.; Lu, L.; Huang, S.; Song, R. Maize Dek44 Encodes Mitochondrial Ribosomal Protein L9 and Is Required for Seed Development. Plant Physiol. 2019, 180, 2106–2119. [Google Scholar] [CrossRef] [PubMed]
- Kwasniak, M.; Majewski, P.; Skibior, R.; Adamowicz, A.; Czarna, M.; Sliwinska, E.; Janska, H. Silencing of the Nuclear RPS10 Gene Encoding Mitochondrial Ribosomal Protein Alters Translation in Arabidopsis Mitochondria. Plant Cell 2013, 25, 1855–1867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nouws, J.; Goswami, A.V.; Bestwick, M.; McCann, B.J.; Surovtseva, Y.V.; Shadel, G.S. Mitochondrial ribosomal protein L12 Is required for POLRMT stability and exists as two forms generated by alternative proteolysis during import. J. Biol. Chem. 2016, 291, 989–997. [Google Scholar] [CrossRef] [Green Version]
- Surovtseva, Y.V.; Shutt, T.E.; Cotney, J.; Cimen, H.; Chen, S.Y.; Koc, E.C.; Shadel, G.S. Mitochondrial ribosomal protein L12 selectively associates with human mitochondrial RNA polymerase to activate transcription. Proc. Natl. Acad. Sci. USA 2011, 108, 17921–17926. [Google Scholar] [CrossRef] [Green Version]
- Janska, H.; Kwasniak, M. Mitoribosomal regulation of OXPHOS biogenesis in plants. Front. Plant Sci. 2014, 5, 79. [Google Scholar] [CrossRef] [Green Version]
- Kwasniak-Owczarek, M.; Kazmierczak, U.; Tomal, A.; Mackiewicz, P.; Janska, H. Deficiency of mitoribosomal S10 protein affects translation and splicing in Arabidopsis mitochondria. Nucleic Acids Res. 2019. [Google Scholar] [CrossRef] [Green Version]
- Mays, J.-N.; Camacho-Villasana, Y.; Garcia-Villegas, R.; Perez-Martinez, X.; Barrientos, A.; Fontanesi, F. The mitoribosome-specific protein mS38 is preferentially required for synthesis of cytochrome c oxidase subunits. Nucleic Acids Res. 2019, 47, 5746–5760. [Google Scholar] [CrossRef] [Green Version]
- Mauro, V.P.; Edelman, G.M. The Ribosome Filter Redux. Cell Cycle 2007, 6, 2246–2251. [Google Scholar] [CrossRef] [Green Version]
- Couvillion, M.T.; Soto, I.C.; Shipkovenska, G.; Churchman, L.S. Synchronized mitochondrial and cytosolic translation programs. Nature 2016, 533, 499–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rooijers, K.; Loayza-Puch, F.; Nijtmans, L.G.; Agami, R. Ribosome profiling reveals features of normal and disease-associated mitochondrial translation. Nat. Commun. 2013, 4, 2886. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chotewutmontri, P.; Stiffler, N.; Watkins, K.P.; Barkan, A. Ribosome Profiling in Maize. Methods Mol Biol. 2018, 1676, 165–183. [Google Scholar] [PubMed]
- Planchard, N.; Bertin, P.; Quadrado, M.; Dargel-Graffin, C.; Hatin, I.; Namy, O.; Mireau, H. The translational landscape of Arabidopsis mitochondria. Nucleic Acids Res. 2018, 46, 6218–6228. [Google Scholar] [CrossRef] [Green Version]
- Fuchs, P.; Rugen, N.; Carrie, C.; Elsässer, M.; Finkemeier, I.; Giese, J.; Hildebrandt, T.M.; Kühn, K.; Maurino, V.G.; Ruberti, C.; et al. Single organelle function and organization as estimated from Arabidopsis mitochondrial proteomics. Plant J. 2019, 14534. [Google Scholar] [CrossRef] [Green Version]
- Small, I.D.; Schallenberg-Rüdinger, M.; Takenaka, M.; Mireau, H.; Ostersetzer-Biran, O. Plant organellar RNA editing: What 30 years of research has revealed. Plant J. 2019, 14578. [Google Scholar] [CrossRef]
- Jourdain, A.A.; Koppen, M.; Wydro, M.; Rodley, C.D.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M.; Martinou, J.-C. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013, 17, 399–410. [Google Scholar] [CrossRef] [Green Version]
- Buchan, J.R. mRNP granules. Assembly, function, and connections with disease. RNA Biol. 2014, 11, 1019–1030. [Google Scholar] [CrossRef] [Green Version]
- Antonicka, H.; Shoubridge, E.A. Mitochondrial RNA Granules Are Centers for Posttranscriptional RNA Processing and Ribosome Biogenesis. Cell Rep. 2015, 10, 920–932. [Google Scholar] [CrossRef] [Green Version]
- Barchiesi, A.; Vascotto, C. Transcription, Processing, and Decay of Mitochondrial RNA in Health and Disease. Int. J. Mol. Sci. 2019, 20, 2221. [Google Scholar] [CrossRef] [Green Version]
- Hensen, F.; Potter, A.; van Esveld, S.L.; Tarrés-Solé, A.; Chakraborty, A.; Solà, M.; Spelbrink, J.N. Mitochondrial RNA granules are critically dependent on mtDNA replication factors Twinkle and mtSSB. Nucleic Acids Res. 2019, 47, 3680–3698. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ott, M.; Amunts, A.; Brown, A. Organization and Regulation of Mitochondrial Protein Synthesis. Annu. Rev. Biochem. 2016, 85, 77–101. [Google Scholar] [CrossRef] [PubMed]
- Möller-Hergt, B.V.; Carlström, A.; Stephan, K.; Imhof, A.; Ott, M. The ribosome receptors Mrx15 and Mba1 jointly organize cotranslational insertion and protein biogenesis in mitochondria. Mol. Biol. Cell 2018, 29, 2386–2396. [Google Scholar] [CrossRef] [PubMed]
- Kehrein, K.; Schilling, R.; Möller-Hergt, B.V.; Wurm, C.A.; Jakobs, S.; Lamkemeyer, T.; Langer, T.; Ott, M. Organization of Mitochondrial Gene Expression in Two Distinct Ribosome-Containing Assemblies. Cell Rep. 2015, 10, 843–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kehrein, K.; Möller-Hergt, B.V.; Ott, M. The MIOREX complex—lean management of mitochondrial gene expression. Oncotarget 2015, 6, 16806–16807. [Google Scholar] [CrossRef]
- Oldenkott, B.; Yang, Y.; Lesch, E.; Knoop, V.; Schallenberg-Rüdinger, M. Plant-type pentatricopeptide repeat proteins with a DYW domain drive C-to-U RNA editing in Escherichia coli. Commun. Biol. 2019, 2, 85. [Google Scholar] [CrossRef]
| Higher Plants (A. thaliana) [8] | Arabidopsis Gene ID | Bacteria (E. coli) [13] | Kinetoplastids (T. brucei) [7] | Yeast (S. cerevisiae) [6] | Mammals (H. sapiens) [5] |
|---|---|---|---|---|---|
| Small Subunit | |||||
| uS2m | AT3G03600 | ✓ | ✓ | ✓ | |
| uS3m | ATMG00090 | ✓ | ✓ | ✓ | ✓ |
| uS4m | ATMG00290 | ✓ | ✓ | ||
| uS5m | AT1G64880 | ✓ | ✓ | ✓ | ✓ |
| bS6m | AT3G18760 | ✓ | ✓ | ✓ | ✓ |
| uS7m | ATMG01270 | ✓ | ✓ | ✓ | |
| uS8m 1 | AT4G29430 AT2G19720 | ✓ | ✓ | ✓ | |
| uS9m | AT3G49080 | ✓ | ✓ | ✓ | ✓ |
| uS10m | AT3G22300 | ✓ | ✓ | ✓ | ✓ |
| uS11m | AT1G31817 | ✓ | ✓ | ✓ | ✓ |
| uS12m | ATMG00980 | ✓ | ✓ | ✓ | ✓ |
| uS13m | AT1G77750 | ✓ | ✓ | ||
| uS14m | AT2G34520 | ✓ | ✓ | ✓ | ✓ |
| uS15m 1 | AT1G15810 AT1G80620 | ✓ | ✓ | ✓ | ✓ |
| bS16m | AT5G56940 | ✓ | ✓ | ✓ | ✓ |
| uS17m | AT1G49400 | ✓ | ✓ | ✓ | ✓ |
| bS18m | AT1G07210 | ✓ | ✓ | ✓ | ✓ |
| uS19m | AT5G47320 | ✓ | ✓ | ✓ | |
| bS21m | AT3G26360 | ✓ | ✓ | ✓ | ✓ |
| bTHXm 2 | AT2G21290 | ✓ | |||
| mS22 | AT1G64600 | ✓ | ✓ | ||
| mS23 | AT1G26750 | ✓ | ✓ | ✓ | |
| mS29 | AT1G16870 | ✓ | ✓ | ✓ | |
| mS33 | AT5G44710 | ✓ | ✓ | ✓ | |
| mS34 | AT5G52370 | ✓ | ✓ | ✓ | |
| mS35 1 | AT3G18240 AT4G21460 | ✓ | ✓ | ✓ | |
| mS47 | AT4G31810 | ✓ | ✓ | ||
| mS75 | AT5G62270 | ||||
| mS76 (rPPR1 3) | AT1G61870 | ||||
| mS77 (rPPR2 3) | AT1G19520 | ||||
| mS78 (rPPR3a 3) | AT1G55890 | ||||
| mS79 (rPPR3b 3) | AT3G13160 | ||||
| mS80 (rPPR6 3) | AT3G02650 | ||||
| mS81 (rPPR8 3) | AT5G15980 | ||||
| mS82 | AT4G22000 | ||||
| mS83 | AT4G15640 AT3G21465 | ||||
| mS84 | AT1G53645 | ||||
| mS85 | AT1G18630 | ||||
| mS86 | AT1G47278 | ||||
| mS87 | AT5G26800 | ||||
| Large Subunit | |||||
| uL1m | AT2G42710 | ✓ | ✓ | ✓ | |
| uL2m | AT2G44065 | ✓ | ✓ | ✓ | |
| uL3m | AT3G17465 | ✓ | ✓ | ✓ | ✓ |
| uL4m | AT2G20060 | ✓ | ✓ | ✓ | ✓ |
| uL5m | ATMG00210 | ✓ | ✓ | ||
| uL6m | AT2G18400 | ✓ | ✓ | ||
| bL9m | AT5G53070 | ✓ | ✓ | ✓ | ✓ |
| uL10m | AT3G12370 | ✓ | ✓ | ✓ | ✓ |
| uL11m | AT4G35490 | ✓ | ✓ | ✓ | ✓ |
| bL12m 1 | AT3G06040 AT1G70190 AT4G37660 | ✓ | ✓ | ✓ | ✓ |
| uL13m | AT3G01790 | ✓ | ✓ | ✓ | ✓ |
| uL14m | AT5G46160 | ✓ | ✓ | ✓ | |
| uL15m | AT5G64670 | ✓ | ✓ | ✓ | ✓ |
| uL16m | ATMG00080 | ✓ | ✓ | ✓ | ✓ |
| bL17m 1 | AT5G09770 AT5G64650 | ✓ | ✓ | ✓ | ✓ |
| uL18m | AT5G27820 | ✓ | ✓ | ||
| bL19m | AT1G24240 | ✓ | ✓ | ✓ | ✓ |
| bL20m | AT1G16740 | ✓ | ✓ | ✓ | |
| bL21m | AT4G30930 | ✓ | ✓ | ✓ | ✓ |
| uL22m 1 | AT1G52370 AT4G28360 | ✓ | ✓ | ✓ | ✓ |
| uL23m | AT4G39880 | ✓ | ✓ | ✓ | ✓ |
| uL24m | AT5G23535 | ✓ | ✓ | ✓ | ✓ |
| bL25m 1 | AT4G23620 AT5G66860 | ✓ | |||
| bL27m | AT2G16930 | ✓ | ✓ | ✓ | ✓ |
| bL28m | AT4G31460 | ✓ | ✓ | ✓ | ✓ |
| uL29m | AT1G07830 | ✓ | ✓ | ✓ | ✓ |
| uL30m | AT5G55140 | ✓ | ✓ | ✓ | ✓ |
| bL31m 1 | AT5G55125 AT1G27435 | ✓ | ✓ | ✓ | |
| bL33m | AT5G18790 | ✓ | ✓ | ✓ | ✓ |
| bL36m | AT5G20180 | ✓ | ✓ | ✓ | ✓ |
| mL40 | AT4G05400 | ✓ | ✓ | ✓ | |
| mL41 1 | AT5G40080 AT5G39800 | ✓ | ✓ | ✓ | |
| mL43 | AT3G59650 | ✓ | ✓ | ✓ | |
| mL46 | AT1G14620 | ✓ | ✓ | ✓ | |
| mL53 | AT5G39600 | ✓ | ✓ | ✓ | |
| mL54 | AT3G01740 | ✓ | ✓ | ||
| mL101 (rPPR4 3) | AT1G60770 | ||||
| mL102 (rPPR5 3) | AT2G37230 | ||||
| mL103 (rPPR7 3) | AT4G36680 | ||||
| mL104 (rPPR9 3) | AT5G60960 | ||||
| mL105 | AT3G51010 | ||||
| mL106 | AT1G73940 | ||||
| Assignment to either subunit unclear | |||||
| mrpX | AT5G49210 | ||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tomal, A.; Kwasniak-Owczarek, M.; Janska, H. An Update on Mitochondrial Ribosome Biology: The Plant Mitoribosome in the Spotlight. Cells 2019, 8, 1562. https://doi.org/10.3390/cells8121562
Tomal A, Kwasniak-Owczarek M, Janska H. An Update on Mitochondrial Ribosome Biology: The Plant Mitoribosome in the Spotlight. Cells. 2019; 8(12):1562. https://doi.org/10.3390/cells8121562
Chicago/Turabian StyleTomal, Artur, Malgorzata Kwasniak-Owczarek, and Hanna Janska. 2019. "An Update on Mitochondrial Ribosome Biology: The Plant Mitoribosome in the Spotlight" Cells 8, no. 12: 1562. https://doi.org/10.3390/cells8121562
APA StyleTomal, A., Kwasniak-Owczarek, M., & Janska, H. (2019). An Update on Mitochondrial Ribosome Biology: The Plant Mitoribosome in the Spotlight. Cells, 8(12), 1562. https://doi.org/10.3390/cells8121562





