Targeting Telomeres and Telomerase: Studies in Aging and Disease Utilizing CRISPR/Cas9 Technology
Abstract
1. Introduction
1.1. Telomeres and Telomerase
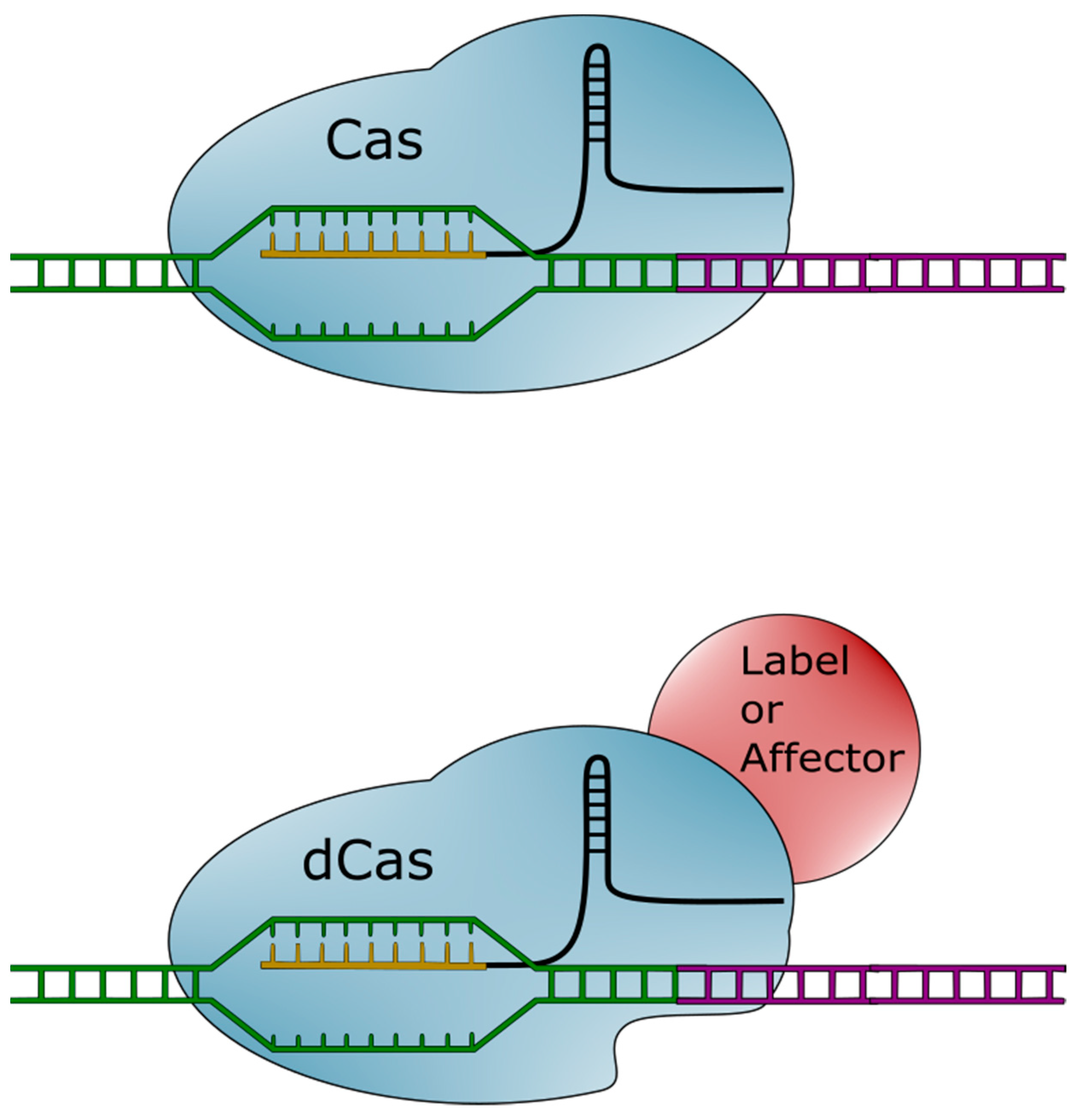
1.2. The CRISPR-Cas System
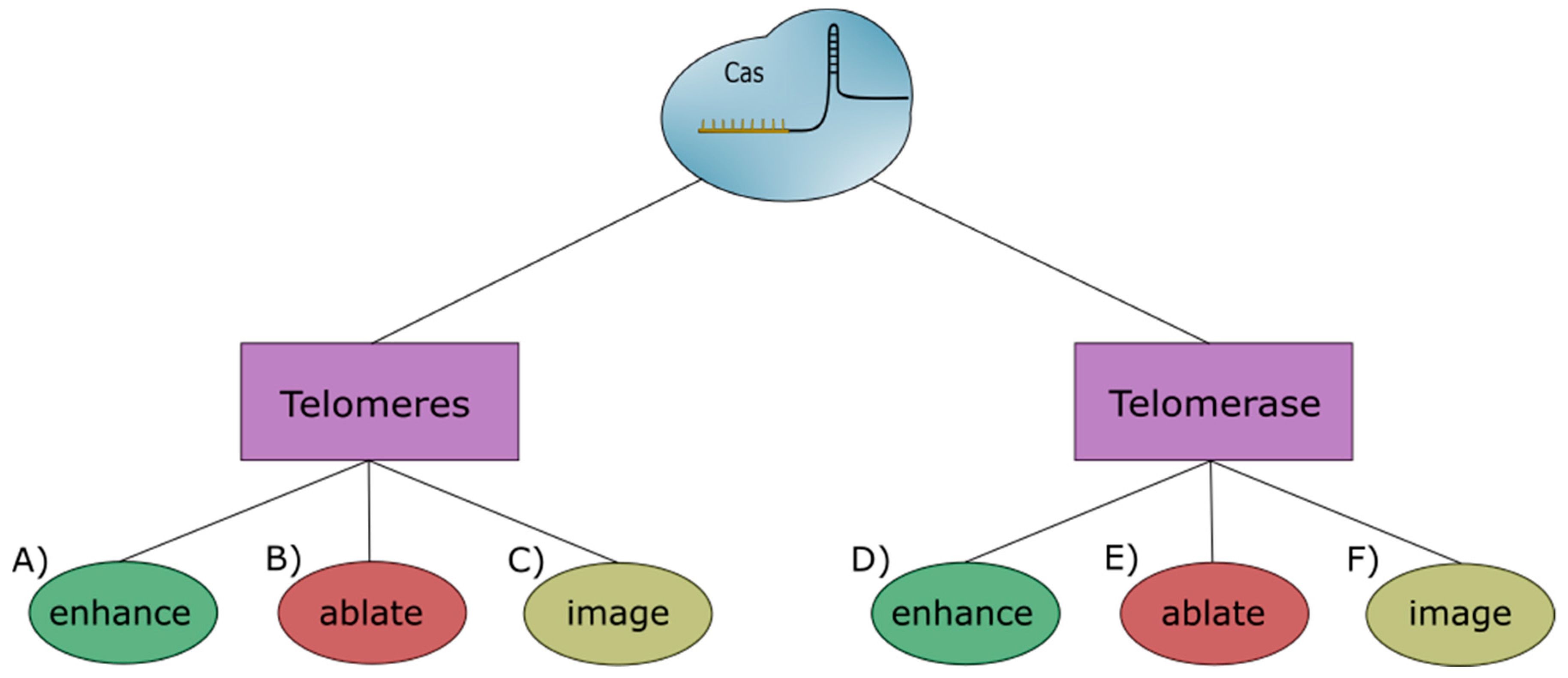
2. Telomeres—Imaging
3. Telomeres—Editing
4. Telomerase—Imaging
5. Telomerase—Editing
6. Genes that Affect Telomeres and Telomerase
7. Epigenetics—Editing
8. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Muller, H.J. The remaking of chromosomes. Collect. Net 1938, 13, 181–198. [Google Scholar]
- Blackburn, E.H.; Gall, J.G. A Tandemly Repeated Sequence at the Termini of the Extrachromosomal Ribosomal RNA Genes in Tetrahymena. J. Mol. Biol. 1978, 120, 33–53. [Google Scholar] [CrossRef]
- Moyzis, R.K.; Buckingham, J.M.; Cram, L.S.; Dani, M.; Deaven, L.L.; Jones, M.D.; Meyne, J.; Ratliff, R.L.; Wu, J.R. A highly conserved repetitive DNA sequence, (TTAGGG)n, present at the telomeres of human chromosomes. Proc. Natl. Acad. Sci. USA 1988, 85, 6622–6626. [Google Scholar] [CrossRef]
- Szostak, J.W.; Blackburn, E.H. Cloning Yeast Telomeres on Linear Plasmid Vectors. Cell 1982, 29, 245–255. [Google Scholar] [CrossRef]
- Harley, C.B.; Vaziri, H.; Counter, C.M.; Allsopp, R.C. The Telomere Hypothesis of Cellular Aging. Exp. Gerontol. 1992, 27, 375–382. [Google Scholar] [CrossRef]
- Greider, C.W.; Blackburn, E.H. Identification of a Specific Telomere Terminal Transferase Activity in Tetrahymena Extracts. Cell 1985, 43, 405–413. [Google Scholar] [CrossRef]
- Nugent, C.I.; Lundblad, V. The telomerase reverse transcriptase: Components and regulation. Cold Spring Harb. Lab. Press. 1998, 12, 1073–1085. [Google Scholar] [CrossRef]
- Nguyen, T.H.D.; Tam, J.; Wu, R.A.; Greber, B.J.; Toso, D.; Nogales, E.; Collins, K. Cryo-EM structure of substrate-bound human telomerase holoenzyme. Nature 2018, 557, 190–195. [Google Scholar] [CrossRef]
- Cohen, S.B.; Graham, M.E.; Lovrecz, G.O.; Bache, N.; Robinson, P.J.; Reddel, R.R. Protein Composition of Catalytically Active Human Telomerase from Immortal Cells. Science 2007, 315, 1850–1853. [Google Scholar] [CrossRef]
- Li, C.; Wu, M.; Liang, Y.; Wu, X. Correlation between expression of human telomerase subunits and telomerase activity in esophageal squamous cell carcinoma. World J. Gastroenterol. 2003, 9, 2395–2399. [Google Scholar] [CrossRef]
- Herrera, E.; Samper, E.; Martin-Caballero, J.; Flores, J.M.; Lee, H.W.; Blasco, M.A. Disease states associated with telomerase deficiency appear earlier in mice with short telomeres. Embo J. 1999, 18, 2950–2960. [Google Scholar] [CrossRef]
- Martinez, P.; Blasco, M.A. Telomere-driven diseases and telomere-targeting therapies. J. Cell Biol. 2017, 216, 875–887. [Google Scholar] [CrossRef]
- Carneiro, M.C.; Henriques, C.M.; Nabais, J.; Ferreira, T.; Carvalho, T.; Ferreira, M.G. Short Telomeres in Key Tissues Initiate Local and Systemic Aging in Zebrafish. Plos Genet. 2016, 12, e1005798. [Google Scholar] [CrossRef]
- Bernadotte, A.; Mikhelson, V.M.; Spivak, I.M. Markers of cellular senescence. Telomere shortening as a marker of cellular senescence. Aging 2016, 8, 3–11. [Google Scholar] [CrossRef]
- Flores, I.; Cayuela, M.L.; Blasco, M.A. Effects of telomerase and telomere length on epidermal stem cell behavior. Science 2005, 309, 1253–1256. [Google Scholar] [CrossRef]
- Sharpless, N.E.; DePinho, R.A. How stem cells age and why this makes us grow old. Nat. Rev. Mol. Cell Biol. 2007, 8, 703–713. [Google Scholar] [CrossRef]
- Ullah, M.; Sun, Z. Klotho deficiency accelerates stem cells aging by impairing telomerase activity. J. Gerontol. Ser. A 2018. [Google Scholar] [CrossRef]
- Ibañez-Cabellos, J.S.; Perez-Machado, G.; Seco-Cervera, M.; Berenguer-Pascual, E.; Garcia-Gimenez, J.L.; Palllardo, F.V. Acute telomerase components depletion triggers oxidative stress as an early event previous to telomeric shortening. Redox Biol. 2018, 14, 398–408. [Google Scholar] [CrossRef]
- Kim, N.W.; Piatyszek, M.A.; Prowse, K.R.; Harley, C.B.; West, M.D.; Ho, P.L.; Coviello, G.M.; Wright, W.E.; Weinrich, S.L.; Shay, J.W. Specific association of human telomerase activity with immortal cells and cancer. Science 1994, 266, 2001–2015. [Google Scholar] [CrossRef]
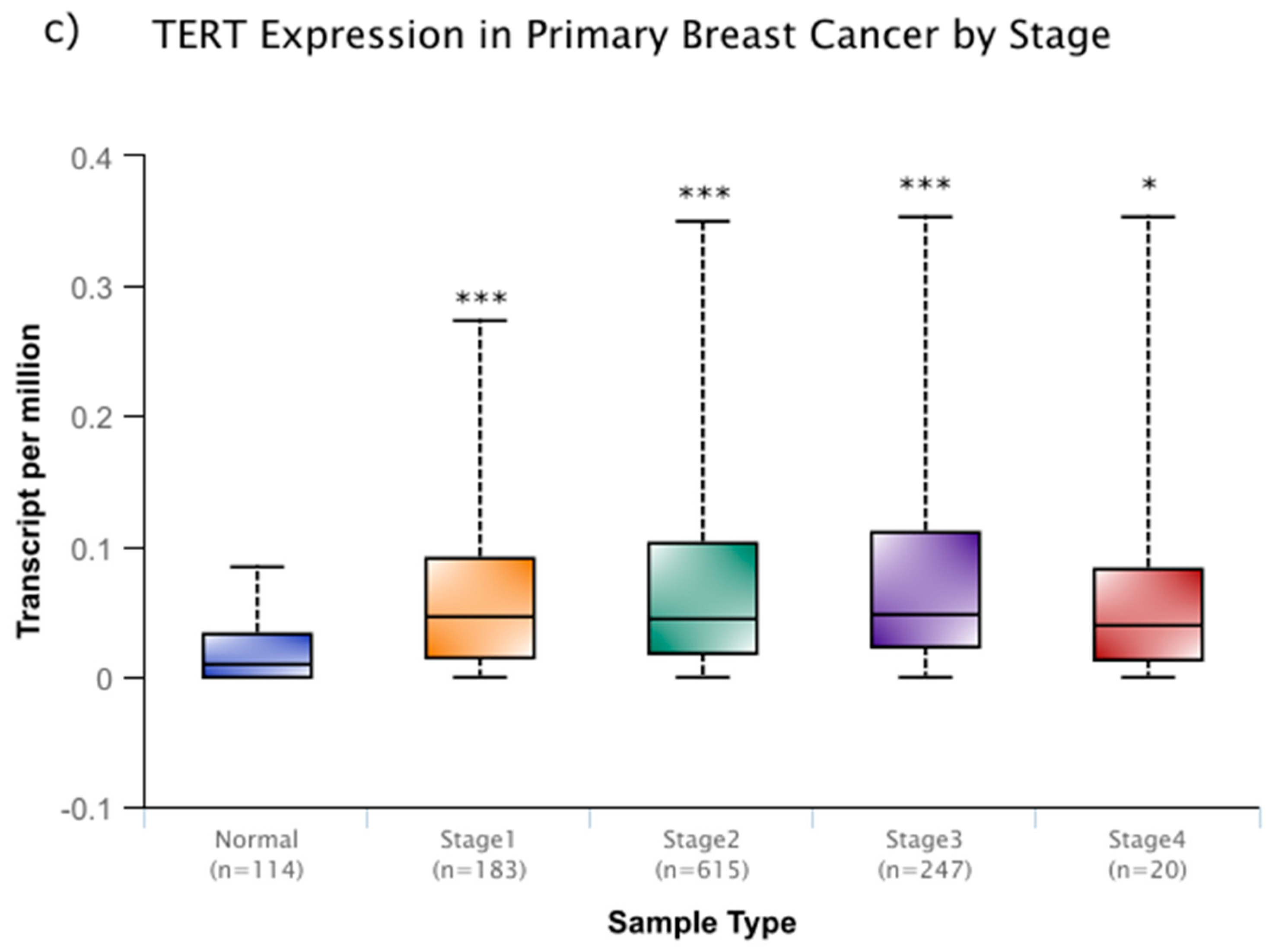
- Chandrashekar, D.S.; Bashel, B.; Balasubramanya, S.A.H.; Creighton, C.J.; Rodriguez, I.P.; Chakravarthi, B.V.S.K.; Varambally, S. UALCAN: A portal for facilitating tumor subgroup gene expression and survival analyses. Neoplasia 2017, 19, 649–658. [Google Scholar] [CrossRef]
- Kar, A.; Chowdhury, S. Inhibition of telomerase activity by NME2: Impact on metastasis suppression? Naunyn Schmiedebergs Arch Pharmacol. 2015, 388, 235–241. [Google Scholar] [CrossRef]
- Mojica, F.J.M.; Diez-Villaseñor, C.; Garcia-Martinez, J.; Soria, E. Intervening Sequences of Regularly Spaced Prokaryotic Repeats Derive from Foreign Genetic Elements. J. Mol. Evol. 2005, 60, 174–182. [Google Scholar] [CrossRef]
- Mali, P.; Yang, L.; Esvelt, K.M.; Aach, J.; Guell, M.; DiCarlo, J.E.; Norville, J.E.; Church, G.M. RNA-guided human genome engineering via Cas9. Science 2013, 339, 823–826. [Google Scholar] [CrossRef]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and Applications of CRISPR-Cas9 for Genome Engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef]
- Komor, A.C.; Badran, A.H.; Liu, D.R. CRISPR-Based Technologies for the Manipulation of Eukaryotic Genomes. Cell 2017, 168, 20–36. [Google Scholar] [CrossRef]
- Gupta, R.M.; Musunuru, K. Expanding the genetic editing tool kit: ZFNs, TALENs, and CRISPR-Cas9. J. Clin. Investig. 2014, 124, 4154–4161. [Google Scholar] [CrossRef]
- Ikeda, M.; Matsuyama, S.; Akagi, S.; Ohkoshi, K.; Nakamura, S.; Minabe, S.; Kimura, K.; Hosoe, M. Correction of a Disease Mutation using CRISPR/Cas9-assisted Genome Editing in Japanese Black Cattle. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef]
- Chen, B.; Gilbert, L.A.; Cimini, B.A.; Schnitzbauer, J.; Zhang, W.; Li, G.; Park, J.; Blackburn, E.H.; Weissman, J.S.; Qi, L.S.; et al. Dynamic Imaging of Genomic Loci in Living Human Cells by an Optimized CRISPR/Cas System. Cell 2013, 155, 1479–1491. [Google Scholar] [CrossRef]
- Deng, W.; Shi, X.; Tjian, R.; Lionnet, T.; Singer, R.H. CASFISH: CRISPR/Cas9-mediated in situ labeling of genomic loci in fixed cells. PNAS 2015, 112, 11870–11875. [Google Scholar] [CrossRef]
- Zhang, S.; Song, Z. Aio-Casilio: A robust CRISPR–Cas9–Pumilio system for chromosome labeling. J. Mol. Hist. 2017, 48, 293–299. [Google Scholar] [CrossRef]
- Ge, J.; Wood, D.K.; Weingeist, D.M.; Prasongtanakij, S.; Navasumrit, P.; Ruchirawat, M.; Engelward, B.P. Standard fluorescent imaging of live cells is highly genotoxic. J. Quant. Cell Sci. 2013, 83A, 552–560. [Google Scholar] [CrossRef]
- Shao, S.; Zhang, W.; Hu, H.; Xue, B.; Qin, J.; Sun, C.; Sun, Y.; Wei, W.; Sun, Y. Long-term dual-color tracking of genomic loci by modified sgRNAs of the CRISPR/Cas9 system. Nucleic Acids Res. 2016, 4, e86. [Google Scholar] [CrossRef]
- Dreissig, S.; Schiml, S.; Schindele, P.; Weiss, O.; Rutten, T.; Schubert, V.; Mette, M.F.; Puchta, H.; Houben, A. Live-cell CRISPR imaging in plants reveals dynamic telomere movements. Plant J. 2017, 91, 565–573. [Google Scholar] [CrossRef]
- Duan, J.; Lu, G.; Hong, Y.; Hu, Q.; Mai, X.; Guo, J.; Si, X.; Wang, F.; Zhang, Y. Live imaging and tracking of genome regions in CRISPR/dCas9 knock-in mice. Genome Biol. 2018, 19, 1–7. [Google Scholar] [CrossRef]
- Mao, P.; Liu, J.; Zhang, Z.; Zhang, H.; Liu, H.; Gao, S.; Rong, Y.; Zhao, Y. Homologous recombination-dependent repair of telomeric DSBs in proliferating human cells. Nature Commun. 2016, 7. [Google Scholar] [CrossRef]
- Kim, H.; Ham, S.; Jo, M.; Lee, G.H.; Lee, Y.; Shin, J.; Lee, Y. CRISPR-Cas9 Mediated Telomere Removal Leads to Mitochondrial Stress and Protein Aggregation. Int. J. Mol. Sci. 2017, 18, 2093. [Google Scholar] [CrossRef]
- Beishline, K.; Vladimirova, O.; Tutton, S.; Wang, Z.; Deng, Z.; Lieberman, P.M. CTCF driven TERRA transcription facilitates completion of telomere DNA replication. Nat. Commun. 2017, 8. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Zaug, A.J.; Cech, T.R. Live Cell Imaging Reveals the Dynamics of Telomerase Recruitment to Telomeres. Cell 2016, 166, 1188–1197. [Google Scholar] [CrossRef]
- Schmidt, J.C.; Zaug, A.J.; Kufer, R.; Cech, T.R. Dynamics of human telomerase recruitment depend on template-telomere base pairing. MBoC 2018, 29, 869–880. [Google Scholar] [CrossRef]
- Chiba, K.; Johnson, J.Z.; Vogan, J.M.; Wagner, T.; Boyle, J.M.; Hockemeyer, D. Cancer-associated TERT promoter mutations abrogate telomerase silencing. eLife 2015, 4. [Google Scholar] [CrossRef]
- Chiba, K.; Vogan, J.M.; Wu, R.A.; Gill, M.S.; Zhang, X.; Collins, K.; Hockemeyer, D. Endogenous Telomerase Reverse Transcriptase N-Terminal Tagging Affects Human Telomerase Function at Telomeres In Vivo. Mol. Cell. Biol. 2016, 37, e00541–16. [Google Scholar] [CrossRef]
- Xi, L.; Schmidt, J.C.; Zaug, A.J.; Ascarrunz, D.R.; Cech, T.R. A novel two-step genome editing strategy with CRISPR-Cas9 provides new insights into telomerase action and TERT gene expression. Genome Biol. 2015, 16. [Google Scholar] [CrossRef]
- Akincilar, S.C.; Khattar, E.; Boon, P.L.; Unal, B.; Fullwood, M.J.; Tergaonkar, V. Long-Range Chromatin Interactions Drive Mutant TERT Promoter Activation. Cancer Discov. 2016, 6, 1276–1291. [Google Scholar] [CrossRef]
- Hu, Y.; Shi, G.; Zhang, L.; Li, F.; Jiang, Y.; Jiang, S.; Ma, W.; Zhao, Y.; Songyang, Z.; Huang, J. Switch telomerase to ALT mechanism by inducing telomeric DNA damages and dysfunction of ATRX and DAXX. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Min, J.; Wright, W.E.; Shay, J.W. Alternative lengthening of telomeres can be maintained by preferential elongation of lagging strands. Nucleic Acids Res. 2017, 45, 2615–2628. [Google Scholar] [CrossRef]
- Wang, S.; Liu, Z.; Ye, Y.; Li, B.; Liu, T.; Zhang, W.; Liu, G.; Zhang, Y.A.; Qu, J.; Xu, D.; et al. Ectopic hTERT expression facilitates reprograming of fibroblasts derived from patients with Werner syndrome as a WS cellular model. Cell Death Dis. 2018, 9. [Google Scholar] [CrossRef]
- Stanley, S.E.; Gable, D.L.; Wagner, C.L.; Carlile, T.M.; Hanumanthu, V.S.; Podlevsky, J.D.; Khalil, S.E.; DeZern, A.E.; Rojas-Duran, M.F.; Applegate, C.D.; et al. Loss-of-function mutations in the RNA biogenesis factor NAF1 predispose to pulmonary fibrosis–emphysema. Sci. Transl. Med. 2016, 8. [Google Scholar] [CrossRef]
- Zhang, Y.; Wu, Y.; Mao, P.; Li, F.; Han, X.; Zhang, Y.; Jiang, S.; Chen, Y.; Huang, J.; Liu, D.; et al. Cold-inducible RNA-binding protein CIRP/hnRNP A18 regulates telomerase activity in a temperature-dependent manner. Nucleic Acids Res. 2015, 44, 761–775. [Google Scholar] [CrossRef]
- Wanlada, S.; Mandasari, M.; Aida, J.; Morita, K.; Kayamori, K.; Ikeda, T.; Sakamoto, K. Loss of Notch1 predisposes oro-esophageal epithelium to tumorigenesis. Exp. Cell Res. 2018. [Google Scholar] [CrossRef]
- Lobry, C.; Oh, P.; Aifantis, I. Oncogenic and tumor suppressor functions of Notch in cancer: it’s NOTCH what you think. J. Exp. Med. 2011, 208, 1931–1935. [Google Scholar] [CrossRef]
- Gu, P.; Jia, S.; Takasugi, T.; Smith, E.; Nandakumar, J.; Hendrickson, E.; Chang, S. CTC1-STN1 coordinates G- and C-strand synthesis to regulate telomere length. Aging Cell 2018, 17. [Google Scholar] [CrossRef]
- Zhou, Z.; Wang, L.; Ge, F.; Gong, P.; Wang, H.; Wang, F.; Chen, L.; Liu, L. Pold3 is required for genomic stability and telomere integrity in embryonic stem cells and meiosis. Nucleic Acids Res. 2018, 46, 3468–3486. [Google Scholar] [CrossRef]
- Maciejowski, J.; Li, Y.; Bosco, N.; Campbell, P.J.; Lange, T. Chromothripsis and kataegis induced by telomere crisis. Cell 2015, 163, 1641–1654. [Google Scholar] [CrossRef]
- Jeltsch, A.; Ehrenhofer-Murray, A.; Jurkowski, T.P.; Lyko, F.; Reuter, G.; Ankri, S.; Nellen, W.; Schaefer, M.; Helm, M. Mechanism and biological role of Dnmt2 in Nucleic Acid Methylation. RNA Biol. 2017, 14, 1108–1123. [Google Scholar] [CrossRef]
- Lewinska, A.; Adamczyk-Grochala, J.; Kwasniewicz, E.; Wnuk, M. Downregulation of methyltransferase Dnmt2 results in condition-dependent telomere shortening and senescence or apoptosis in mouse fibroblasts. J. Cell. Physiol. 2017, 232, 3714–3726. [Google Scholar] [CrossRef]
- Lu, F.; Liu, Y.; Jiang, L.; Yamaguchi, S.; Zhang, Y. Role of Tet proteins in enhancer activity and telomere elongation. Genes Dev. 2014, 28, 2103–2119. [Google Scholar] [CrossRef]

© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brane, A.C.; Tollefsbol, T.O. Targeting Telomeres and Telomerase: Studies in Aging and Disease Utilizing CRISPR/Cas9 Technology. Cells 2019, 8, 186. https://doi.org/10.3390/cells8020186
Brane AC, Tollefsbol TO. Targeting Telomeres and Telomerase: Studies in Aging and Disease Utilizing CRISPR/Cas9 Technology. Cells. 2019; 8(2):186. https://doi.org/10.3390/cells8020186
Chicago/Turabian StyleBrane, Andrew C., and Trygve O. Tollefsbol. 2019. "Targeting Telomeres and Telomerase: Studies in Aging and Disease Utilizing CRISPR/Cas9 Technology" Cells 8, no. 2: 186. https://doi.org/10.3390/cells8020186
APA StyleBrane, A. C., & Tollefsbol, T. O. (2019). Targeting Telomeres and Telomerase: Studies in Aging and Disease Utilizing CRISPR/Cas9 Technology. Cells, 8(2), 186. https://doi.org/10.3390/cells8020186




