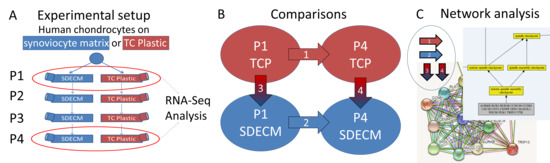
Transcriptome-Wide Analysis of Human Chondrocyte Expansion on Synoviocyte Matrix
Abstract
1. Introduction
2. Materials and Methods
3. Results
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Murphy, L.B.; Cisternas, M.G.; Pasta, D.J.; Helmick, C.G.; Yelin, E.H. Medical Expenditures and Earnings Losses Among US Adults With Arthritis in 2013. Arthritis Care Res. 2018, 70, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Giovannini, S.; Diaz-Romero, J.; Aigner, T.; Mainil-Varlet, P.; Nesic, D. Population doublings and percentage of S100-positive cells as predictors of in vitro chondrogenicity of expanded human articular chondrocytes. J. Cell. Physiol. 2010, 222, 411–420. [Google Scholar] [CrossRef] [PubMed]
- Kean, T.J.; Dennis, J.E. Synoviocyte Derived-Extracellular Matrix Enhances Human Articular Chondrocyte Proliferation and Maintains Re-Differentiation Capacity at Both Low and Atmospheric Oxygen Tensions. PLoS ONE 2015, 10, e0129961. [Google Scholar]
- Pei, M.; He, F. Extracellular matrix deposited by synovium-derived stem cells delays replicative senescent chondrocyte dedifferentiation and enhances redifferentiation. J. Cell. Physiol. 2012, 227, 2163–2174. [Google Scholar] [CrossRef] [PubMed]
- Everaert, C.; Luypaert, M.; Maag, J.L.V.; Cheng, Q.X.; Dinger, M.E.; Hellemans, J.; Mestdagh, P. Benchmarking of RNA-sequencing analysis workflows using whole-transcriptome RT-qPCR expression data. Sci Rep. 2017, 7, 1559. [Google Scholar] [CrossRef] [PubMed]
- Kean, T.J.; Koelewyn, S.; Ge, Z.; Li, Y.; Chen, R.; Dennis, J.E. Transcriptome-Wide Analysis of Human Chondrocyte Dedifferentiation on TC Plastic. In Proceedings of the Orthopedic Research Society Annual Meeting, Orlando, FL, USA, 5–8 March 2016. [Google Scholar]
- Langmead, B.; Trapnell, C.; Pop, M.; Salzberg, S.L. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009, 10. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Pachter, L.; Salzberg, S.L. TopHat: Discovering splice junctions with RNA-Seq. Bioinformatics 2009, 25. [Google Scholar] [CrossRef] [PubMed]
- Anders, S.; Pyl, P.T.; Huber, W. HTSeq—A Python framework to work with high-throughput sequencing data. Bioinformatics 2015, 31, 166–169. [Google Scholar] [CrossRef] [PubMed]
- Trapnell, C.; Williams, B.A.; Pertea, G.; Mortazavi, A.; Kwan, G.; van Baren, M.J.; Salzberg, S.L.; Wold, B.J.; Pachter, L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 2010, 28, 511–515. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing: Vienna, Austria, 2018. Available online: https://www.R-project.org/ (accessed on 9 December 2018).
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Morris, J.H.; Cook, H.; Kuhn, M.; Wyder, S.; Simonovic, M.; Santos, A.; Doncheva, N.T.; Roth, A.; Bork, P.; et al. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res. 2017, 45. [Google Scholar] [CrossRef] [PubMed]
- Boyle, E.I.; Weng, S.; Gollub, J.; Jin, H.; Botstein, D.; Cherry, J.M.; Sherlock, G. GO::TermFinder—Open source software for accessing Gene Ontology information and finding significantly enriched Gene Ontology terms associated with a list of genes. Bioinformatics 2004, 20, 3710–3715. [Google Scholar] [CrossRef] [PubMed]
- Jax. Gene Ontology Browser. 2018. Available online: http://www.informatics.jax.org/vocab/gene_ontology/ (accessed on 8 November 2018).
- Marlovits, S.; Hombauer, M.; Truppe, M.; Vecsei, V.; Schlegel, W. Changes in the ratio of type-I and type-II collagen expression during monolayer culture of human chondrocytes. J. Bone Jt. Surg. J. 2004, 86, 286–295. [Google Scholar] [CrossRef] [PubMed]
- Melrose, J.; Smith, S.; Cake, M.; Read, R.; Whitelock, J. Comparative spatial and temporal localisation of perlecan, aggrecan and type I.; II and IV collagen in the ovine meniscus: An ageing study. Histochem. Cell Biol. 2005, 124, 225–235. [Google Scholar] [CrossRef] [PubMed]
- Rosen, N.; Chalifa-Caspi, V.; Shmueli, O.; Adato, A.; Lapidot, M.; Stampnitzky, J.; Safran, M.; Lancet, D. GeneLoc: Exon-based integration of human genome maps. Bioinformatics 2003, 19, i222–i224. [Google Scholar] [CrossRef] [PubMed]
- Peffers, M.; Liu, X.; Clegg, P. Transcriptomic signatures in cartilage ageing. Arthritis Res. Ther. 2013, 15, R98. [Google Scholar] [CrossRef] [PubMed]
- Cuzzocrea, S.; Mazzon, E.; Dugo, L.; Patel, N.S.; Serraino, I.; Di Paola, R.; Genovese, T.; Britti, D.; De Maio, M.; Caputi, A.P.; et al. Reduction in the evolution of murine type II collagen-induced arthritis by treatment with rosiglitazone, a ligand of the peroxisome proliferator-activated receptor gamma. Arthritis Rheum. 2003, 48, 3544–3556. [Google Scholar] [CrossRef]
- Xu, J.; Lv, S.; Hou, Y.; Xu, K.; Sun, D.; Zheng, Y.; Zhang, Z.; Li, X.; Li, Y.; Chi, G. miR-27b promotes type II collagen expression by targetting peroxisome proliferator-activated receptor-gamma2 during rat articular chondrocyte differentiation. Biosci. Rep. 2018, 38. [Google Scholar] [CrossRef]
- Liu, C.F.; Lefebvre, V. The transcription factors SOX9 and SOX5/SOX6 cooperate genome-wide through super-enhancers to drive chondrogenesis. Nucleic Acids Res. 2015, 43, 8183–8203. [Google Scholar] [CrossRef]
- De Crombrugghe, B.; Lefebvre, V.; Behringer, R.R.; Bi, W.; Murakami, S.; Huang, W. Transcriptional mechanisms of chondrocyte differentiation. Matrix Biol. 2000, 19, 389–394. [Google Scholar] [CrossRef]
- Cozzolino, A.M.; Noce, V.; Battistelli, C.; Marchetti, A.; Grassi, G.; Cicchini, C.; Tripodi, M.; Amicone, L. Modulating the Substrate Stiffness to Manipulate Differentiation of Resident Liver Stem Cells and to Improve the Differentiation State of Hepatocytes. Stem Cells Int. 2016, 2016, 5481493. [Google Scholar] [CrossRef] [PubMed]
- Recha-Sancho, L.; Moutos, F.T.; Abella, J.; Guilak, F.; Semino, C.E. Dedifferentiated Human Articular Chondrocytes Redifferentiate to a Cartilage-Like Tissue Phenotype in a Poly(epsilon-Caprolactone)/Self-Assembling Peptide Composite Scaffold. Materials 2016, 9, 472. [Google Scholar] [CrossRef] [PubMed]
- Zhang, T.; Gong, T.; Xie, J.; Lin, S.; Liu, Y.; Zhou, T.; Lin, Y. Softening Substrates Promote Chondrocytes Phenotype via RhoA/ROCK Pathway. ACS Appl. Mater. Interfaces 2016, 8, 22884–22891. [Google Scholar] [CrossRef] [PubMed]
- Callahan, L.A.; Ganios, A.M.; Childers, E.P.; Weiner, S.D.; Becker, M.L. Primary human chondrocyte extracellular matrix formation and phenotype maintenance using RGD-derivatized PEGDM hydrogels possessing a continuous Young’s modulus gradient. Acta Biomater. 2013, 9, 6095–6104. [Google Scholar] [CrossRef] [PubMed]
- Efrat, S. Mechanisms of adult human beta-cell in vitro dedifferentiation and redifferentiation. Diabetes Obes. Metab. 2016, 18 (Suppl. 1), 97–101. [Google Scholar] [CrossRef] [PubMed]
- Barta, T.; Dolezalova, D.; Holubcova, Z.; Hampl, A. Cell cycle regulation in human embryonic stem cells: Links to adaptation to cell culture. Exp. Biol. Med. 2013, 238, 271–275. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. Geroconversion: Irreversible step to cellular senescence. Cell Cycle 2014, 13, 3628–3635. [Google Scholar] [CrossRef]
- Blagosklonny, M.V. Cell cycle arrest is not senescence. Aging 2011, 3, 94–101. [Google Scholar] [CrossRef]
- Leontieva, O.V.; Blagosklonny, M.V. Gerosuppression in confluent cells. Aging 2014, 6, 1010–1018. [Google Scholar] [CrossRef]
- De Luna-Preitschopf, A.; Zwickl, H.; Nehrer, S.; Hengstschlager, M.; Mikula, M. Rapamycin Maintains the Chondrocytic Phenotype and Interferes with Inflammatory Cytokine Induced Processes. Int. J. Mol. Sci. 2017, 18, 1494. [Google Scholar] [CrossRef]
- Yan, B.; Zhang, Z.; Jin, D.; Cai, C.; Jia, C.; Liu, W.; Wang, T.; Li, S.; Zhang, H.; Huang, B.; et al. mTORC1 regulates PTHrP to coordinate chondrocyte growth, proliferation and differentiation. Nat. Commun. 2016, 7, 11151. [Google Scholar] [CrossRef] [PubMed]
- Preitschopf, A.; Schorghofer, D.; Kinslechner, K.; Schutz, B.; Zwickl, H.; Rosner, M.; Joo, J.G.; Nehrer, S.; Hengstschlager, M.; Mikula, M. Rapamycin-Induced Hypoxia Inducible Factor 2A Is Essential for Chondrogenic Differentiation of Amniotic Fluid Stem Cells. Stem Cells Transl. Med. 2016, 5, 580–590. [Google Scholar] [CrossRef] [PubMed]
- Morales, T.I. Chondrocyte moves: Clever strategies? Osteoarthr. Cartil. 2007, 15, 861–871. [Google Scholar] [CrossRef] [PubMed]
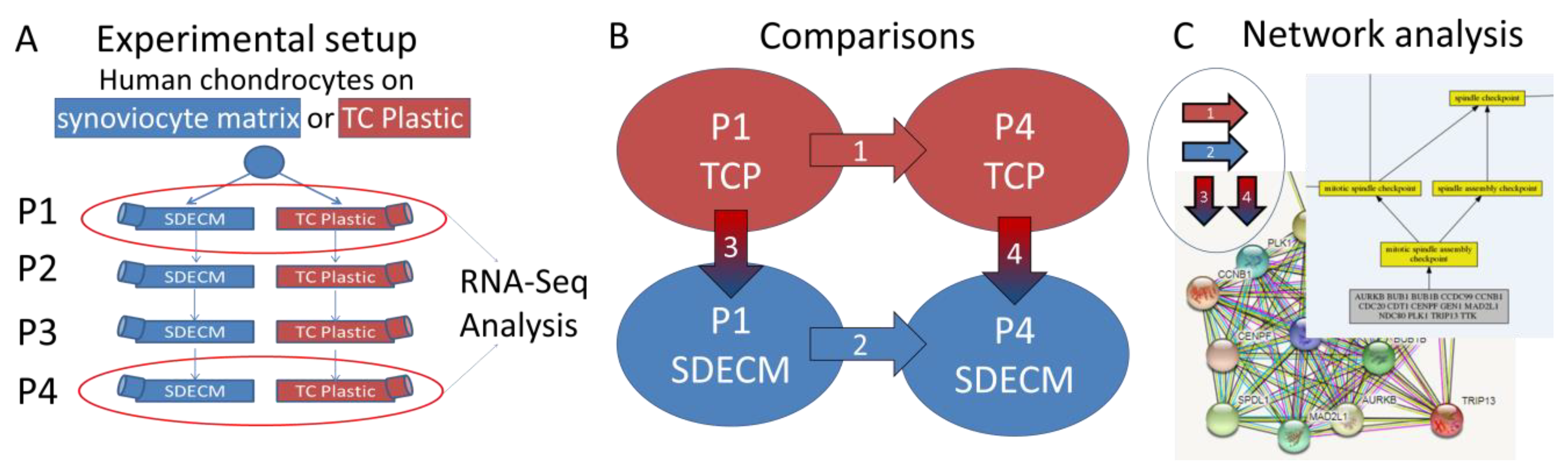



 ) indicates that gene expression increased and a blue downward-pointing arrow (
) indicates that gene expression increased and a blue downward-pointing arrow ( ) that it decreased.
) that it decreased.
 ) indicates that gene expression increased and a blue downward-pointing arrow (
) indicates that gene expression increased and a blue downward-pointing arrow ( ) that it decreased.
) that it decreased.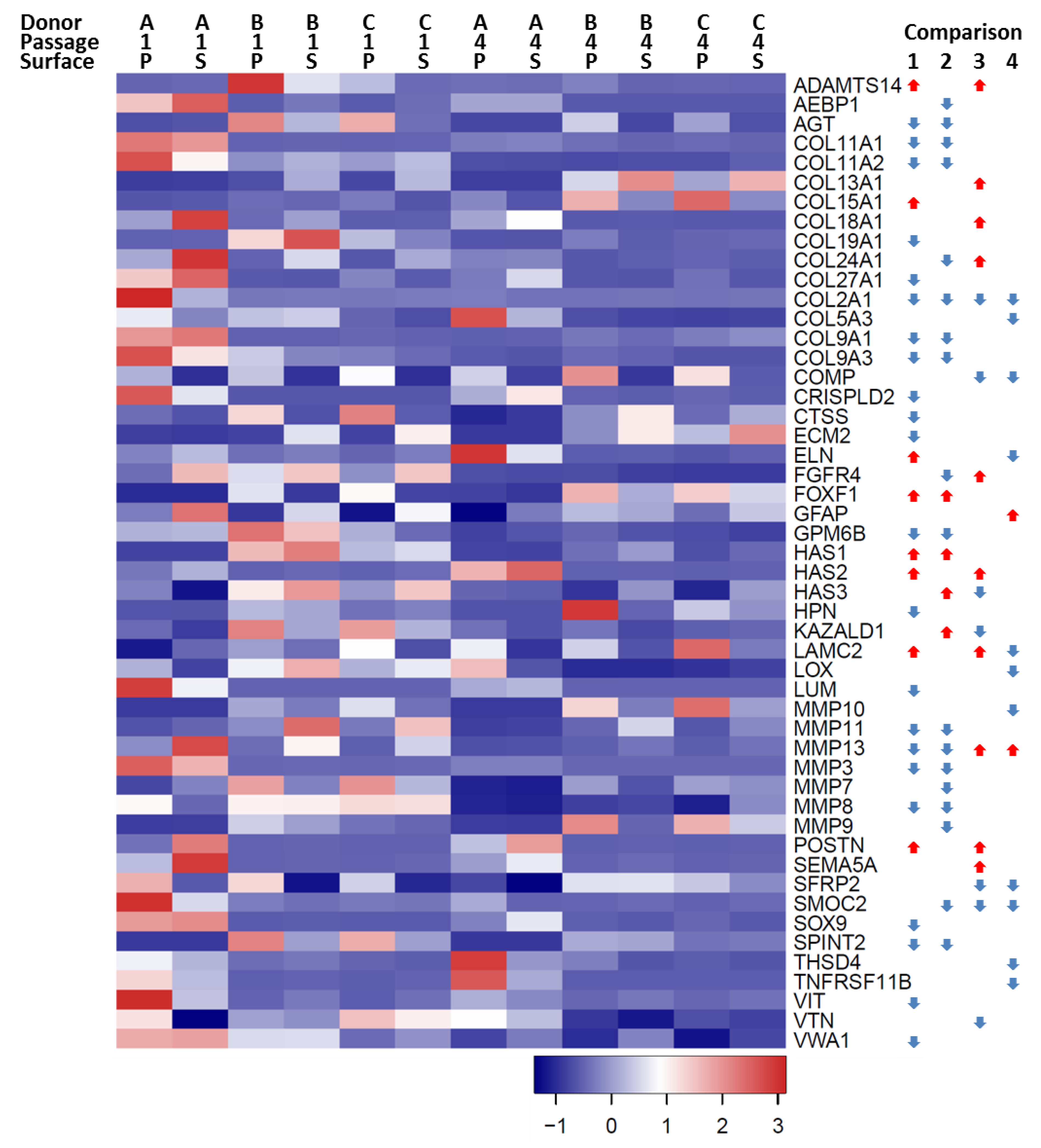
 ) indicates that gene expression increased and a blue downward-pointing arrow (
) indicates that gene expression increased and a blue downward-pointing arrow ( ) that it decreased.
) that it decreased.
 ) indicates that gene expression increased and a blue downward-pointing arrow (
) indicates that gene expression increased and a blue downward-pointing arrow ( ) that it decreased.
) that it decreased.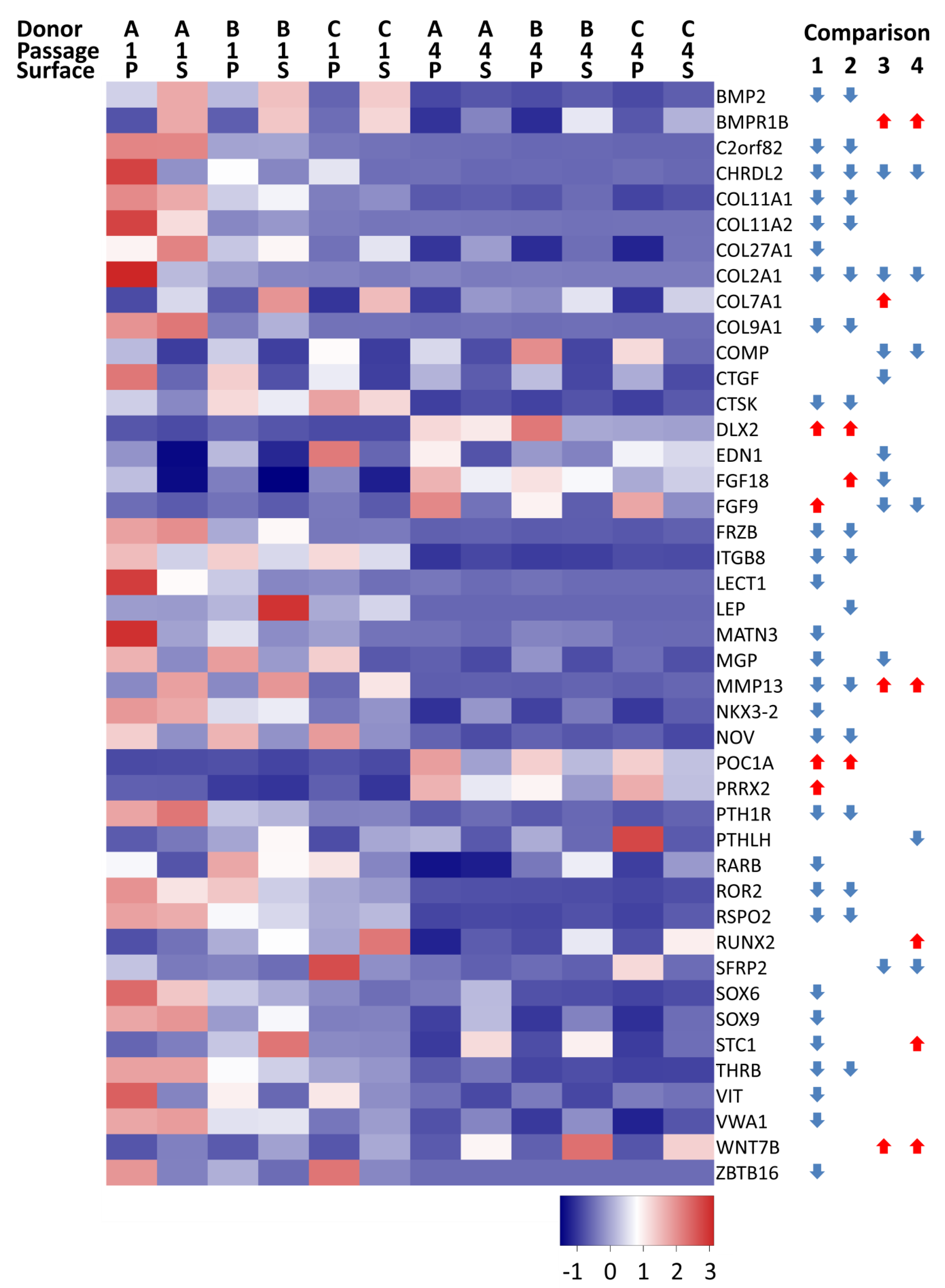
| Comparison | Upregulated (%) | Downregulated (%) | Total | Common 3 |
|---|---|---|---|---|
| (1) P1 vs. P4 on TCP 1 | 404 (39) | 620 (61) | 1024 | 512 |
| (2) P1 vs. P4 on SDECM 1 | 316 (41) | 447 (59) | 763 | |
| (3) TCP vs. SDECM P1 2 | 151 (46) | 180 (54) | 331 | 107 |
| (4) TCP vs. SDECM P4 2 | 162 (48) | 177 (52) | 339 |
| Comparison 1 | Comparison 2 | ||||
|---|---|---|---|---|---|
| Gene Symbol | Gene Name | FC 1 | Gene Symbol | Gene Name | FC 1 |
| MAFB | MAF BZIP transcription factor B | 25.5 D | APLN | Apelin | 21.8 D |
| PLK1 | Polo-like kinase 1 | 23.1 U | MMP1 | Matrix metallopeptidase 1 | 26.6 D |
| CENPF | Centromere protein F | 12.0 U | KIF20A | Kinesin family member 20A | 27.6 U |
| SLC40A1 | Solute carrier family 40 member 1 | 166. D | TOP2A | DNA topoisomerase II alpha | 16.3 U |
| TOP2A | DNA topoisomerase II alpha | 13.0 U | CDC20 | Cell division cycle 20 | 21.1 U |
| MKI67 | Marker of proliferation Ki-67 | 14.5 U | TPX2 | TPX2, microtubule nucleation factor | 16.0 U |
| CCNB1 | Cyclin B1 | 15.0 U | FOXM1 | Forkhead box M1 | 12.3 U |
| FGFR2 | Fibroblast growth factor receptor 2 | 19.0 D | BIRC5 | Baculoviral IAP repeat containing 5 | 19.8 U |
| ISM1 | Isthmin 1 | 25.5 D | MKI67 | Marker of proliferation Ki-67 | 21.4 U |
| CDC20 | Cell division cycle 20 | 19.6 U | PRC1 | Protein regulator of cytokinesis 1 | 12.6 U |
| PRC1 | Protein regulator of cytokinesis 1 | 12.1 U | DLGAP5 | DL-associated protein 5 | 23.6 U |
| A2M | Alpha-2-macroglobulin | 21.9 D | PLK1 | Polo-like kinase 1 | 17.7 U |
| HMMR | Hyaluronan-mediated motility receptor | 20.9 U | SLC40A1 | Solute carrier family 40 member 1 | 25.9 D |
| DLGAP5 | DLG-associated protein 5 | 17.1 U | ASPM | Abnormal spindle microtubule assembly | 10.9 U |
| AURKA | Aurora kinase A | 20.0 U | CCNB1 | Cyclin B1 | 11.7 U |
| CCNB2 | Cyclin B2 | 17.2 U | PRR11 | Proline rich 11 | 9.43 U |
| TPX2 | TPX2, microtubule nucleation factor | 14.2 U | CENPF | Centromere protein F | 12.6 U |
| STEAP4 | STEAP4 metalloreductase | 14.1 D | KIF23 | Kinesin family member 23 | 13.6 U |
| ELN | Elastin | 7.66 U | CEP55 | Centrosomal protein 55 | 14.6 U |
| PRR11 | Proline rich 11 | 11.9 U | ANLN | Anillin actin binding protein | 12.2 U |
| Comparison 3 | Comparison 4 | ||||
|---|---|---|---|---|---|
| Gene Symbol | Gene Name | FC 1 | Gene Symbol | Gene Name | FC 1 |
| POSTN | Periostin | 16.9 U | COLEC12 | Collectin subfamily member 12 | 32.3 U |
| COMP | Cartilage oligomeric matrix protein | 28.5 D | MME | Membrane metalloendopeptidase | 9.11 U |
| CLEC3B | C-type lectin domain family 3 member B | 14.7 D | CRLF1 | Cytokine receptor-like factor 1 | 7.62 D |
| DCN | Decorin | 5.87 D | MFAP5 | Tetratricopeptide repeat domain 9 | 14.5 D |
| PODN | Podocan | 8.89 D | TTC9 | Microfibril-associated protein 5 | 12.1 D |
| CTGF | Connective tissue growth factor | 5.52 D | IGFBP1 | Insulin-like growth factor binding protein 1 | 11.9 D |
| ADAMTS5 | ADAM metallopeptidase thrombospondin type 1 motif 5 | 9.99 D | CLEC3B | C-type lectin domain family 3 member B | 6.48 D |
| PPAP2B | Phospholipid phosphatase 3 | 5.57 D | DSP | Desmoplakin | 10.1 D |
| TAGLN | Transgelin | 7.23 D | LOX | Lysyl oxidase | 4.02 D |
| SFRP4 | Secreted frizzled-related protein 4 | 9.26 D | IGFBP5 | Insulin-like growth factor binding protein 5 | 4.61 D |
| CAMK2N1 | Calcium/calmodulin-dependent protein kinase II inhibitor 1 | 11.3 U | C6orf132 | Chromosome 6 open reading frame 132 | 9.53 D |
| FHL1 | Four and a half LIM domains 1 | 5.66 D | MOXD1 | Monooxygenase DBH-like 1 | 7.35 U |
| OMD | Osteomodulin | 4.92 D | CAMK2N1 | Calcium/calmodulin-dependent protein kinase II inhibitor 1 | 20.1 U |
| COL18A1 | Collagen type XVIII alpha 1 chain | 6.22 U | PHLDA1 | Pleckstrin homology-like domain family A Member 1 | 4.67 U |
| SEMA5A | Semaphorin 5A | 4.22 U | EMB | Embigin | 6.48 U |
| SCD | Stearoyl-CoA desaturase | 5.78 U | PITPNM3 | PITPNM family member 3 | 4.76 D |
| MYL9 | Myosin light chain 9 | 6.15 D | INHBB | Inhibin subunit beta B | 10.0 D |
| CPM | Carboxypeptidase M | 6.23 U | ACTA2 | Actin, alpha 2, smooth muscle, aorta | 4.66 D |
| ITIH5 | Inter-alpha-trypsin inhibitor heavy chain family member 5 | 6.58 D | LOC100505633 | Long intergenic non-protein coding RNA 1133 | 5.19 D |
| AP1S3 | Adaptor-related protein complex 1 subunit sigma 3 | 11.3 D | FOXQ1 | Forkhead box Q1 | 4.96 U |
| Gene Symbol | Gene Name | TCP FC | TCP padj | SDECM FC | SDECM padj |
|---|---|---|---|---|---|
| BRCA1 | BRCA1, DNA repair associated | 4.60 | 3.9 × 10−38 | 6.54 | 2.1 × 10−31 |
| CEBPA | CCAAT enhancer binding protein alpha | 2.11 | 2.7 × 10−1 | 4.82 | 8.0 × 10−4 |
| E2F8 | E2F transcription factor 8 | 8.16 | 1.1 × 10−22 | 21.21 | 3.2 × 10−22 |
| EZH2 | Enhancer of zeste 2 polycomb repressive complex 2 subunit | 5.76 | 1.5 × 10−46 | 3.87 | 1.1 × 10−18 |
| FOSL1 | FOS-like 1, AP-1 transcription factor subunit | 4.82 | 5.0 × 10−43 | 1.10 | 6.5 × 10−1 |
| MXD3 | MAX dimerization protein 3 | 4.79 | 1.8 ×10−24 | 4.43 | 2.7 × 10−11 |
| Gene Symbol | Gene Name | Fold Change | padj |
|---|---|---|---|
| FOSL1 | FOS Like 1, AP-1 transcription factor subunit | 4.81 U | 5.0 × 10−43 |
| STC1 | Stanniocalcin 1 | 4.73 D | 2.2 × 10−40 |
| CDH2 | Cadherin 2 | 11.7 U | 1.0 × 10−27 |
| NXPH3 | Neurexophilin 3 | 4.46 D | 7.1 × 10−19 |
| POM121L9P | POM121 transmembrane nucleoporin-like 9, pseudogene | 10.9 D | 9.6 × 10−19 |
| MSC | Musculin | 4.62 U | 4.3 × 10−12 |
| POSTN | Periostin | 7.64 U | 1.2 × 10−11 |
| CCRL1 | Atypical chemokine receptor 4 | 8.45 D | 4.7 × 10−10 |
| ADAMTS14 | ADAM metallopeptidase with thrombospondin type 1 motif 14 | 4.60 U | 8.6 × 10−10 |
| ACSS1 | Acyl-CoA synthetase short chain family member 1 | 4.24 D | 1.5 × 10−8 |
| SH3TC2 | SH3 domain and tetratricopeptide repeats 2 | 5.20 U | 1.1 × 10−6 |
| NTF3 | Neurotrophin 3 | 4.00 U | 2.1 × 10−6 |
| MYBPH | Myosin binding protein H | 6.36 D | 1.2 × 10−5 |
| ELOVL2 | ELOVL fatty acid elongase 2 | 9.81 U | 1.2 × 10−4 |
| GRIK5 | Glutamate ionotropic receptor kainate type subunit 5 | 4.46 D | 9.2 × 10−3 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kean, T.J.; Ge, Z.; Li, Y.; Chen, R.; Dennis, J.E. Transcriptome-Wide Analysis of Human Chondrocyte Expansion on Synoviocyte Matrix. Cells 2019, 8, 85. https://doi.org/10.3390/cells8020085
Kean TJ, Ge Z, Li Y, Chen R, Dennis JE. Transcriptome-Wide Analysis of Human Chondrocyte Expansion on Synoviocyte Matrix. Cells. 2019; 8(2):85. https://doi.org/10.3390/cells8020085
Chicago/Turabian StyleKean, Thomas J., Zhongqi Ge, Yumei Li, Rui Chen, and James E. Dennis. 2019. "Transcriptome-Wide Analysis of Human Chondrocyte Expansion on Synoviocyte Matrix" Cells 8, no. 2: 85. https://doi.org/10.3390/cells8020085
APA StyleKean, T. J., Ge, Z., Li, Y., Chen, R., & Dennis, J. E. (2019). Transcriptome-Wide Analysis of Human Chondrocyte Expansion on Synoviocyte Matrix. Cells, 8(2), 85. https://doi.org/10.3390/cells8020085