Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches
Abstract
1. Introduction
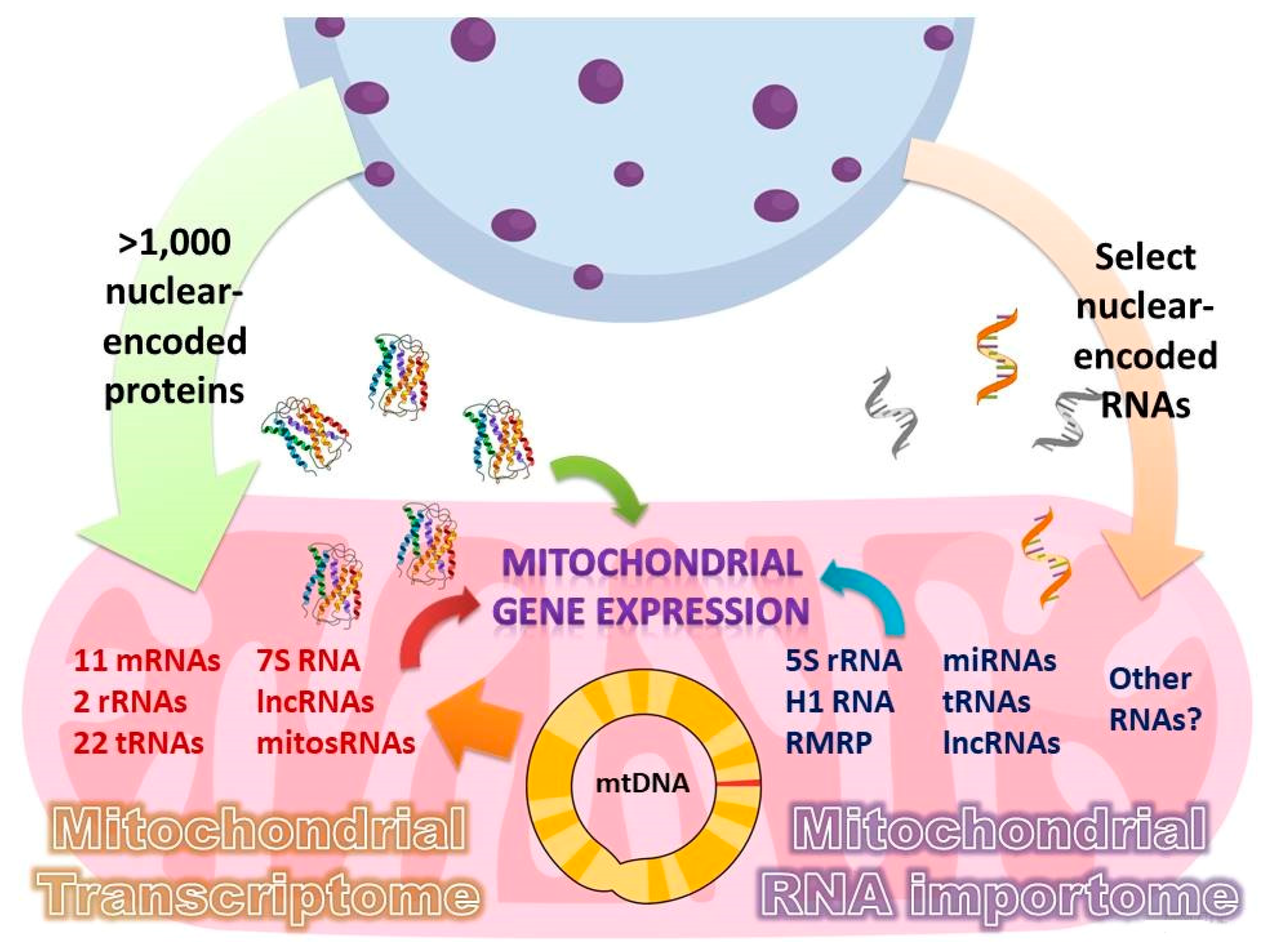
2. Mitochondrial RNA Importome
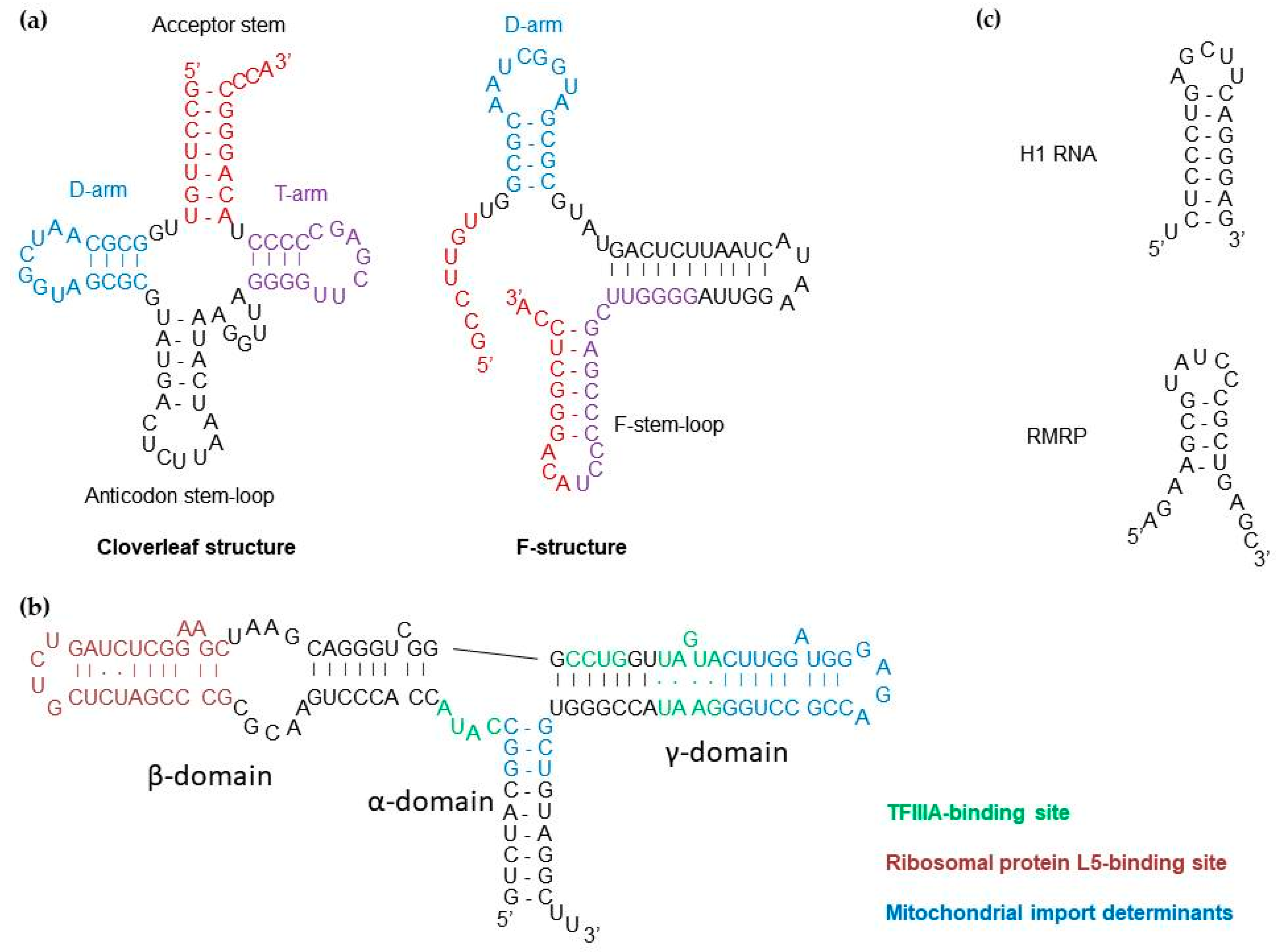
2.1. tRNAs
2.2. 5S rRNA
2.3. RNase P RNA Component (H1 RNA)
2.4. RNase MRP RNA Component (RMRP)
2.5. SAMMSON
2.6. hTERC
2.7. microRNAs
3. Mechanisms of RNA Import
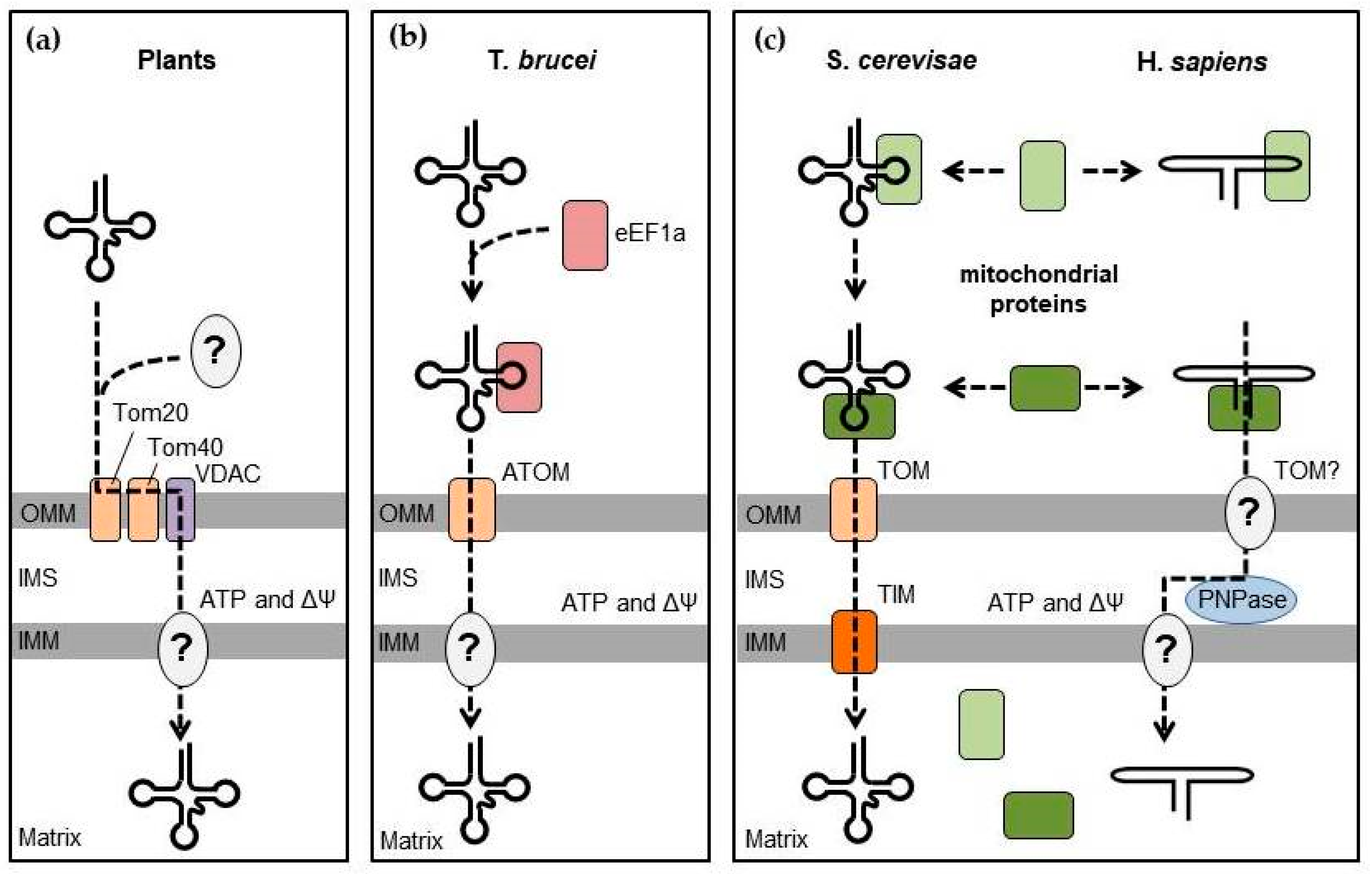
3.1. RNA Targeting to the Mitochondria in Various Eukaryotes
3.2. Mitochondrial RNA Targeting in Yeast and Human
3.3. Translocation through the Mitochondrial Membranes and the Role of Pnpase
4. Identification and Validation of Imported RNAs
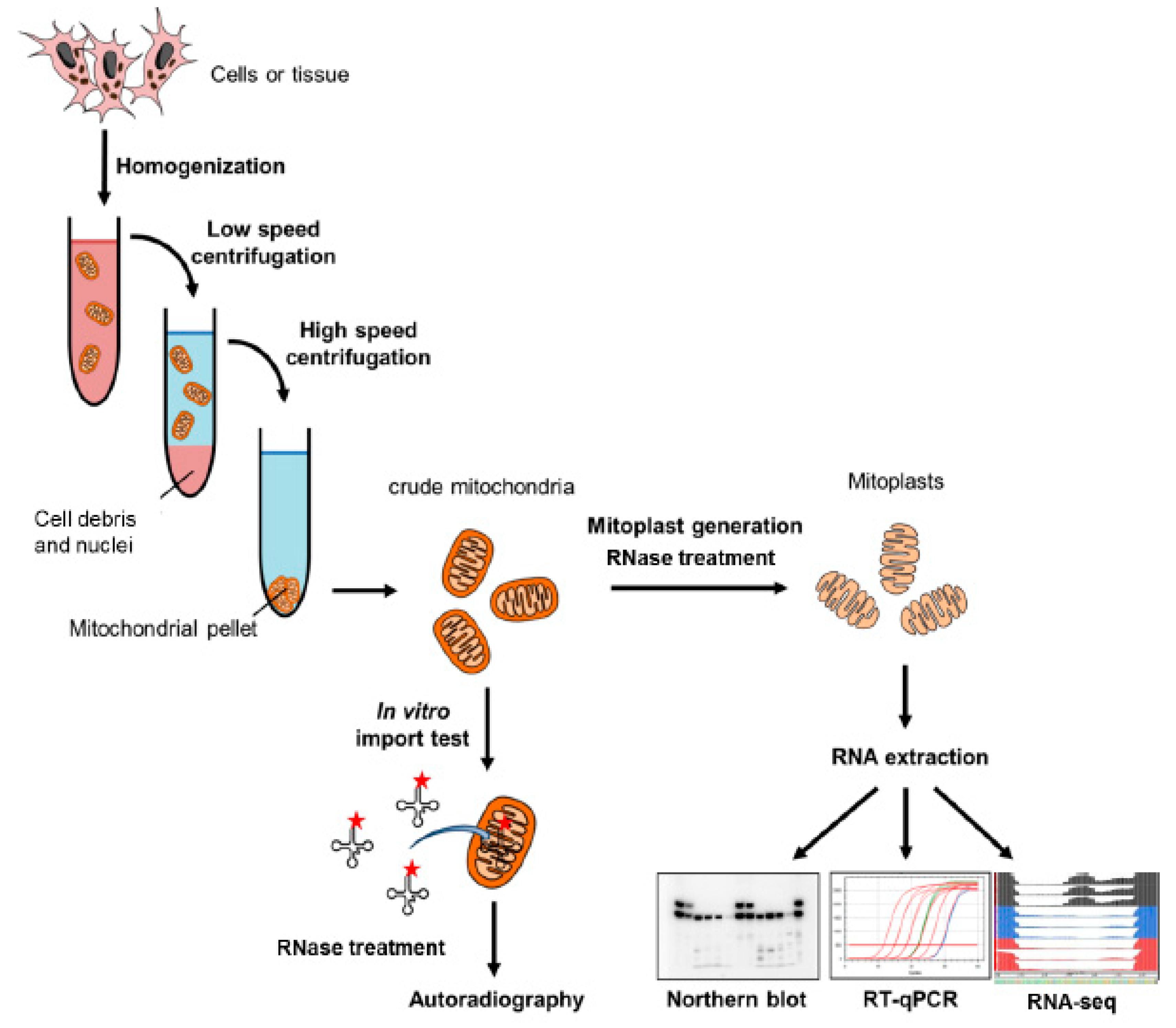
4.1. Classical Cellular Fractionation
4.1.1. Isolation and Purification of Mitochondria
4.1.2. Analysis of the Samples
4.2. In Vitro RNA Import into Isolated Mitochondria
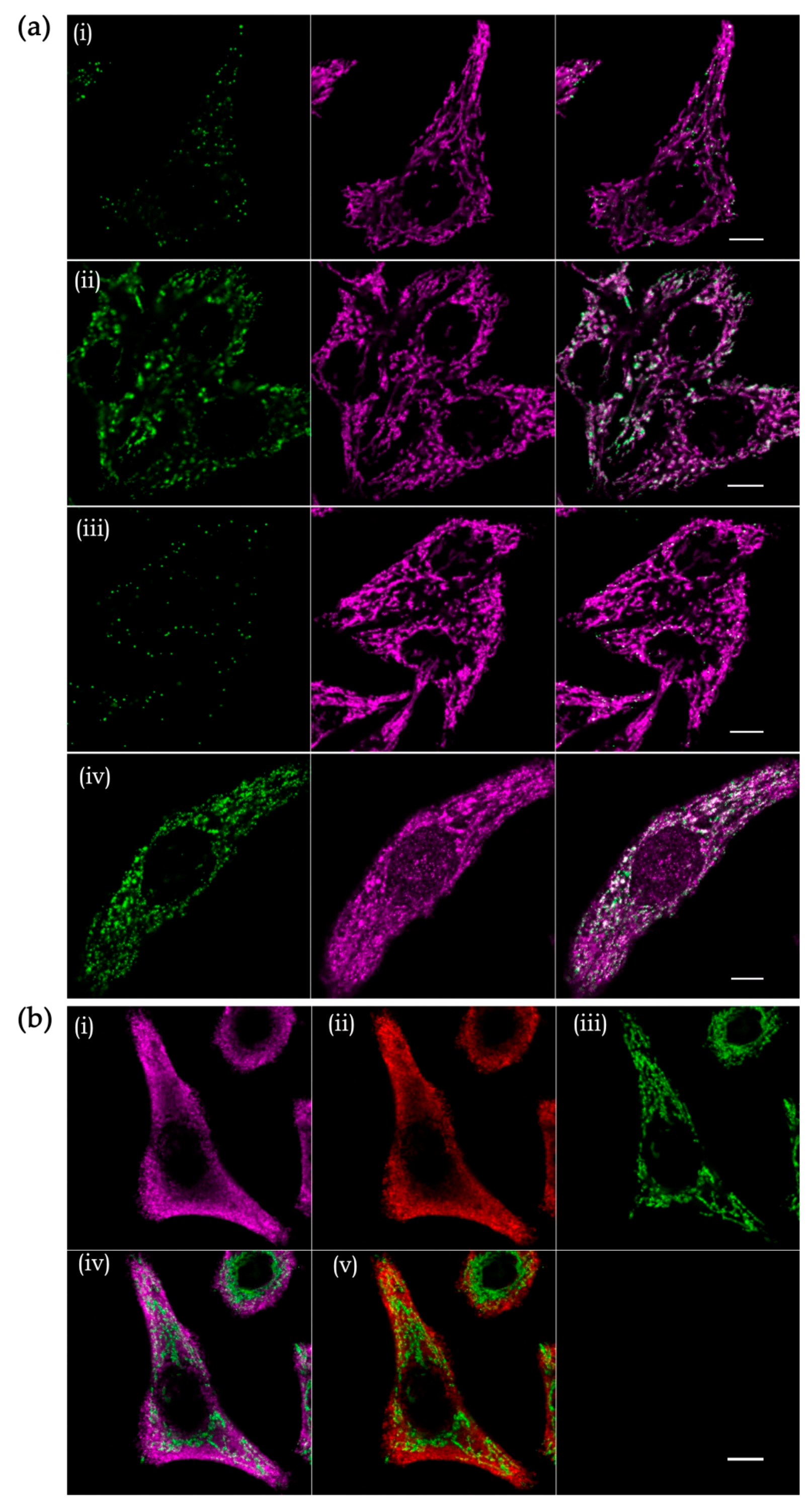
4.3. Imaging of Mitochondrial RNAs in Intact Cells
4.3.1. Recombinant RNA Probes
4.3.2. Imaging of Endogenous RNAs
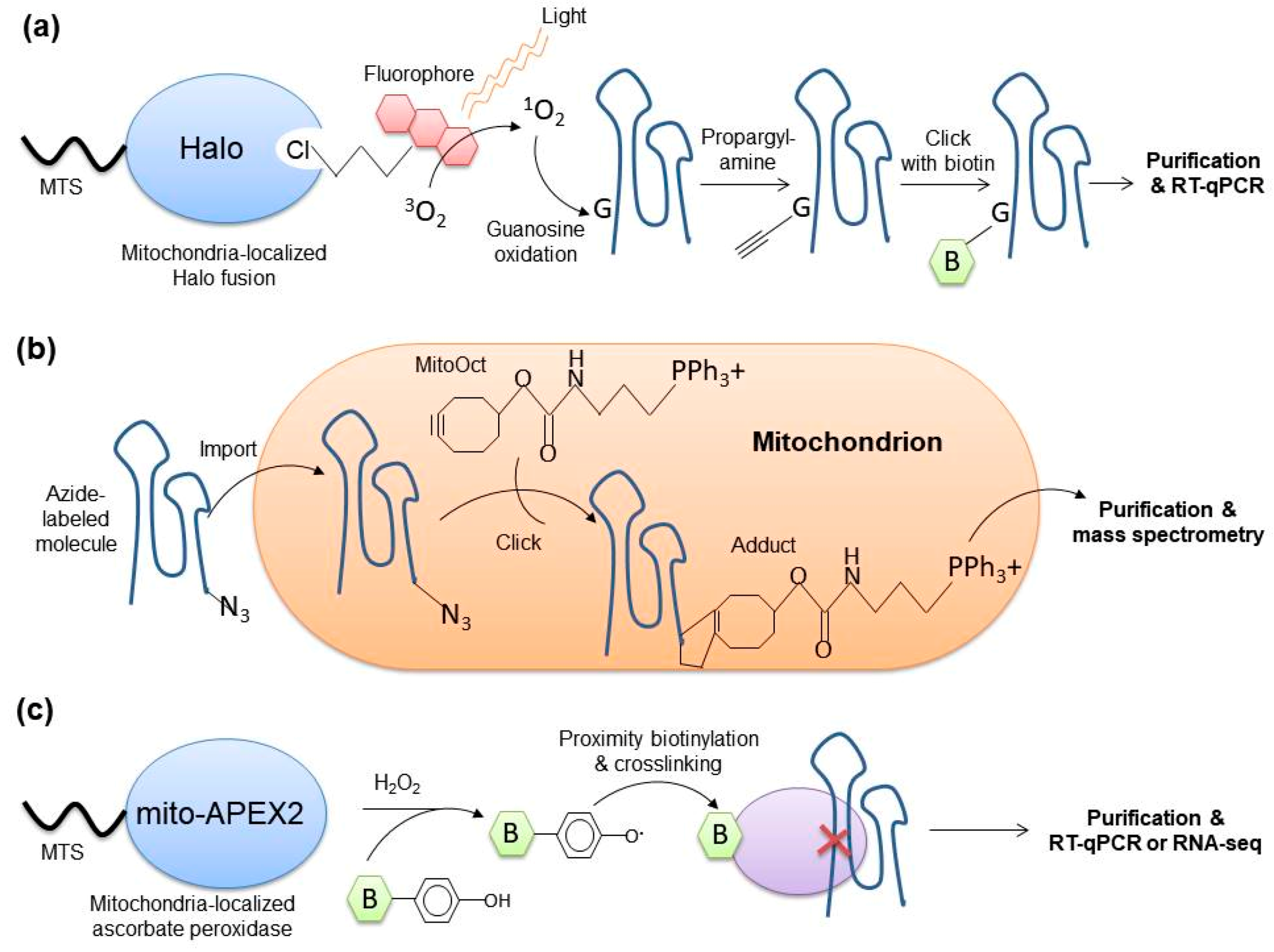
4.4. Mitochondria-Specific Tagging of RNAs
4.5. Spatially Restricted Crosslinking-Based Techniques
5. Unsolved Questions and Future Directions
Author Contributions
Funding
Conflicts of Interest
References
- Calvo, S.E.; Clauser, K.R.; Mootha, V.K. MitoCarta2.0: An updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res. 2016, 44, 1251–1257. [Google Scholar] [CrossRef]
- Sieber, F.; Duchene, A.M.; Marechal-Drouard, L. Mitochondrial RNA import: From diversity of natural mechanisms to potential applications. Int. Rev. Cell Mol. Biol. 2011, 287, 145–190. [Google Scholar] [CrossRef] [PubMed]
- Tarassov, I.; Chicherin, I.; Tonin, Y.; Smirnov, A.; Kamenski, P.; Entelis, N. Mitochondrial targeting of RNA and mitochondrial translation. In Translation in Mitochondria and Other Organelles; Duchene, A.M., Ed.; Springer-Verlag: Berlin, Heidelberg, 2013; pp. 85–109. [Google Scholar]
- Kim, K.M.; Noh, J.H.; Abdelmohsen, K.; Gorospe, M. Mitochondrial noncoding RNA transport. BMB Rep. 2017, 50, 164–174. [Google Scholar] [CrossRef] [PubMed]
- Hancock, K.; Hajduk, S.L. The mitochondrial tRNAs of Trypanosoma brucei are nuclear encoded. J. Biol. Chem. 1990, 265, 19208–19215. [Google Scholar] [PubMed]
- Simpson, L.; Shaw, J. RNA editing and the mitochondrial cryptogenes of kinetoplastid protozoa. Cell 1989, 57, 355–366. [Google Scholar] [CrossRef]
- Martin, R.P.; Schneller, J.M.; Stahl, A.J.; Dirheimer, G. Import of nuclear deoxyribonucleic acid coded lysine-accepting transfer ribonucleic acid (anticodon C-U-U) into yeast mitochondria. Biochemistry 1979, 18, 4600–4605. [Google Scholar] [CrossRef] [PubMed]
- Kamenski, P.; Kolesnikova, O.; Jubenot, V.; Entelis, N.; Krasheninnikov, I.A.; Martin, R.P.; Tarassov, I. Evidence for an adaptation mechanism of mitochondrial translation via tRNA import from the cytosol. Mol. Cell 2007, 26, 625–637. [Google Scholar] [CrossRef]
- Rubio, M.A.; Rinehart, J.J.; Krett, B.; Duvezin-Caubet, S.; Reichert, A.S.; Soll, D.; Alfonzo, J.D. Mammalian mitochondria have the innate ability to import tRNAs by a mechanism distinct from protein import. Proc. Natl. Acad. Sci. USA 2008, 105, 9186–9191. [Google Scholar] [CrossRef]
- Rubio, M.A.; Hopper, A.K. Transfer RNA travels from the cytoplasm to organelles. Wiley Interdiscip. Rev. RNA 2011, 2, 802–817. [Google Scholar] [CrossRef]
- Mercer, T.R.; Neph, S.; Dinger, M.E.; Crawford, J.; Smith, M.A.; Shearwood, A.M.; Haugen, E.; Bracken, C.P.; Rackham, O.; Stamatoyannopoulos, J.A.; et al. The human mitochondrial transcriptome. Cell 2011, 146, 645–658. [Google Scholar] [CrossRef]
- Yoshionari, S.; Koike, T.; Yokogawa, T.; Nishikawa, K.; Ueda, T.; Miura, K.; Watanabe, K. Existence of nuclear-encoded 5S-rRNA in bovine mitochondria. FEBS Lett. 1994, 338, 137–142. [Google Scholar] [CrossRef]
- Magalhaes, P.J.; Andreu, A.L.; Schon, E.A. Evidence for the presence of 5S rRNA in mammalian mitochondria. Mol. Biol. Cell 1998, 9, 2375–2382. [Google Scholar] [CrossRef]
- Entelis, N.S.; Kolesnikova, O.A.; Dogan, S.; Martin, R.P.; Tarassov, I.A. 5 S rRNA and tRNA import into human mitochondria. Comparison of in vitro requirements. J. Biol. Chem. 2001, 276, 45642–45653. [Google Scholar] [CrossRef] [PubMed]
- Autour, A.; Jeng, S.C.Y.; Cawte, A.D.C.; Abdolahzadeh, A.; Galli, A.; Panchapakesan, S.S.S.; Rueda, D.; Ryckelynck, M.; Unrau, P.J. Fluorogenic RNA Mango aptamers for imaging small non-coding RNAs in mammalian cells. Nat. Commun. 2018, 9, 656. [Google Scholar] [CrossRef] [PubMed]
- Zelenka, J.; Alan, L.; Jaburek, M.; Jezek, P. Import of desired nucleic acid sequences using addressing motif of mitochondrial ribosomal 5S-rRNA for fluorescent in vivo hybridization of mitochondrial DNA and RNA. J. Bioenerg. Biomembr. 2014, 46, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Bartkiewicz, M.; Gold, H.; Altman, S. Identification and characterization of an RNA molecule that copurifies with RNase P activity from HeLa cells. Genes Dev. 1989, 3, 488–499. [Google Scholar] [CrossRef] [PubMed]
- Puranam, R.S.; Attardi, G. The RNase P associated with HeLa cell mitochondria contains an essential RNA component identical in sequence to that of the nuclear RNase, P. Mol. Cell Biol. 2001, 21, 548–561. [Google Scholar] [CrossRef]
- Wang, G.; Chen, H.W.; Oktay, Y.; Zhang, J.; Allen, E.L.; Smith, G.M.; Fan, K.C.; Hong, J.S.; French, S.W.; McCaffery, J.M.; et al. PNPASE regulates RNA import into mitochondria. Cell 2010, 142, 456–467. [Google Scholar] [CrossRef]
- Chang, D.D.; Clayton, D.A. A mammalian mitochondrial RNA processing activity contains nucleus-encoded RNA. Science 1987, 235, 1178–1184. [Google Scholar] [CrossRef] [PubMed]
- Noh, J.H.; Kim, K.M.; Abdelmohsen, K.; Yoon, J.H.; Panda, A.C.; Munk, R.; Kim, J.; Curtis, J.; Moad, C.A.; Wohler, C.M.; et al. HuR and GRSF1 modulate the nuclear export and mitochondrial localization of the lncRNA RMRP. Genes Dev. 2016, 30, 1224–1239. [Google Scholar] [CrossRef]
- Li, K.; Smagula, C.S.; Parsons, W.J.; Richardson, J.A.; Gonzalez, M.; Hagler, H.K.; Williams, R.S. Subcellular partitioning of MRP RNA assessed by ultrastructural and biochemical analysis. J. Cell Biol. 1994, 124, 871–882. [Google Scholar] [CrossRef]
- Leucci, E.; Vendramin, R.; Spinazzi, M.; Laurette, P.; Fiers, M.; Wouters, J.; Radaelli, E.; Eyckerman, S.; Leonelli, C.; Vanderheyden, K.; et al. Melanoma addiction to the long non-coding RNA SAMMSON. Nature 2016, 531, 518–522. [Google Scholar] [CrossRef] [PubMed]
- Vendramin, R.; Verheyden, Y.; Ishikawa, H.; Goedert, L.; Nicolas, E.; Saraf, K.; Armaos, A.; Delli Ponti, R.; Izumikawa, K.; Mestdagh, P.; et al. SAMMSON fosters cancer cell fitness by concertedly enhancing mitochondrial and cytosolic translation. Nat. Struct. Mol. Biol. 2018, 25, 1035–1046. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Liu, P.; Zheng, Q.; Gao, G.; Yuan, J.; Wang, P.; Huang, J.; Xie, L.; Lu, X.; Tong, T.; et al. Mitochondrial Trafficking and Processing of Telomerase RNA TERC. Cell Rep. 2018, 24, 2589–2595. [Google Scholar] [CrossRef]
- Geiger, J.; Dalgaard, L.T. Interplay of mitochondrial metabolism and microRNAs. Cell Mol. Life Sci. 2017, 74, 631–646. [Google Scholar] [CrossRef] [PubMed]
- Barrey, E.; Saint-Auret, G.; Bonnamy, B.; Damas, D.; Boyer, O.; Gidrol, X. Pre-microRNA and mature microRNA in human mitochondria. PLoS ONE 2011, 6, e20220. [Google Scholar] [CrossRef]
- Sripada, L.; Tomar, D.; Prajapati, P.; Singh, R.; Singh, A.K.; Singh, R. Systematic analysis of small RNAs associated with human mitochondria by deep sequencing: Detailed analysis of mitochondrial associated miRNA. PLoS ONE 2012, 7, e44873. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Ferlito, M.; Kent, O.A.; Fox-Talbot, K.; Wang, R.; Liu, D.; Raghavachari, N.; Yang, Y.; Wheelan, S.J.; Murphy, E.; et al. Nuclear miRNA regulates the mitochondrial genome in the heart. Circ. Res. 2012, 110, 1596–1603. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Zuo, X.; Yang, B.; Li, Z.; Xue, Y.; Zhou, Y.; Huang, J.; Zhao, X.; Zhou, J.; Yan, Y.; et al. MicroRNA directly enhances mitochondrial translation during muscle differentiation. Cell 2014, 158, 607–619. [Google Scholar] [CrossRef]
- Bandiera, S.; Ruberg, S.; Girard, M.; Cagnard, N.; Hanein, S.; Chretien, D.; Munnich, A.; Lyonnet, S.; Henrion-Caude, A. Nuclear outsourcing of RNA interference components to human mitochondria. PLoS ONE 2011, 6, e20746. [Google Scholar] [CrossRef]
- Kren, B.T.; Wong, P.Y.; Sarver, A.; Zhang, X.; Zeng, Y.; Steer, C.J. MicroRNAs identified in highly purified liver-derived mitochondria may play a role in apoptosis. RNA Biol. 2009, 6, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Bian, Z.; Li, L.M.; Tang, R.; Hou, D.X.; Chen, X.; Zhang, C.Y.; Zen, K. Identification of mouse liver mitochondria-associated miRNAs and their potential biological functions. Cell Res. 2010, 20, 1076–1078. [Google Scholar] [CrossRef] [PubMed]
- Fan, S.; Tian, T.; Chen, W.; Lv, X.; Lei, X.; Zhang, H.; Sun, S.; Cai, L.; Pan, G.; He, L.; et al. Mitochondrial miRNA determines chemoresistance by reprogramming metabolism and regulating mitochondrial transcription. Cancer Res. 2019. [Google Scholar] [CrossRef] [PubMed]
- Szymanski, M.; Barciszewski, J. The genetic code--40 years on. Acta Biochim. Pol. 2007, 54, 51–54. [Google Scholar] [PubMed]
- Nagao, A.; Suzuki, T.; Katoh, T.; Sakaguchi, Y.; Suzuki, T. Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc. Natl. Acad. Sci. USA 2009, 106, 16209–16214. [Google Scholar] [CrossRef] [PubMed]
- Friederich, M.W.; Timal, S.; Powell, C.A.; Dallabona, C.; Kurolap, A.; Palacios-Zambrano, S.; Bratkovic, D.; Derks, T.G.J.; Bick, D.; Bouman, K.; et al. Pathogenic variants in glutamyl-tRNAGln amidotransferase subunits cause a lethal mitochondrial cardiomyopathy disorder. Nat. Commun. 2018, 1, 4065. [Google Scholar] [CrossRef]
- Kolesnikova, O.A.; Entelis, N.S.; Mireau, H.; Fox, T.D.; Martin, R.P.; Tarassov, I.A. Suppression of mutations in mitochondrial DNA by tRNAs imported from the cytoplasm. Science 2000, 289, 1931–1933. [Google Scholar] [CrossRef] [PubMed]
- Kolesnikova, O.A.; Entelis, N.S.; Jacquin-Becker, C.; Goltzene, F.; Chrzanowska-Lightowlers, Z.M.; Lightowlers, R.N.; Martin, R.P.; Tarassov, I. Nuclear DNA-encoded tRNAs targeted into mitochondria can rescue a mitochondrial DNA mutation associated with the MERRF syndrome in cultured human cells. Hum. Mol. Genet. 2004, 13, 2519–2534. [Google Scholar] [CrossRef] [PubMed]
- Karicheva, O.Z.; Kolesnikova, O.A.; Schirtz, T.; Vysokikh, M.Y.; Mager-Heckel, A.M.; Lombes, A.; Boucheham, A.; Krasheninnikov, I.A.; Martin, R.P.; Entelis, N.; et al. Correction of the consequences of mitochondrial 3243A > G mutation in the MT-TL1 gene causing the MELAS syndrome by tRNA import into mitochondria. Nucleic Acids Res. 2011, 39, 8173–8186. [Google Scholar] [CrossRef] [PubMed]
- Greber, B.J.; Ban, N. Structure and function of the mitochondrial ribosome. Annu. Rev. Biochem. 2016, 85, 103–132. [Google Scholar] [CrossRef] [PubMed]
- Bogdanov, A.A.; Dontsova, O.A.; Dokudovskaya, S.S.; Lavrik, I.N. Structure and function of 5S rRNA in the ribosome. Biochem. Cell Biol. 1995, 73, 869–876. [Google Scholar] [CrossRef] [PubMed]
- Kouvela, E.C.; Gerbanas, G.V.; Xaplanteri, M.A.; Petropoulos, A.D.; Dinos, G.P.; Kalpaxis, D.L. Changes in the conformation of 5S rRNA cause alterations in principal functions of the ribosomal nanomachine. Nucleic Acids Res. 2007, 35, 5108–5119. [Google Scholar] [CrossRef]
- Smirnov, A.V.; Entelis, N.S.; Krasheninnikov, I.A.; Martin, R.; Tarassov, I.A. Specific features of 5S rRNA structure—Its interactions with macromolecules and possible functions. Biochemistry 2008, 73, 1418–1437. [Google Scholar] [CrossRef]
- Greber, B.J.; Bieri, P.; Leibundgut, M.; Leitner, A.; Aebersold, R.; Boehringer, D.; Ban, N. The complete structure of the 55S mammalian mitochondrial ribosome. Science 2015, 348, 303–308. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.; Amunts, A.; Bai, X.C.; Sugimoto, Y.; Edwards, P.C.; Murshudov, G.; Scheres, S.H.; Ramakrishnan, V. Structure of the large ribosomal subunit from human mitochondria. Science 2014, 346, 718–722. [Google Scholar] [CrossRef] [PubMed]
- Rorbach, J.; Gao, F.; Powell, C.A.; D’Souza, A.; Lightowlers, R.N.; Minczuk, M.; Chrzanowska-Lightowlers, Z.M. Human mitochondrial ribosomes can switch their structural RNA composition. Proc. Natl. Acad. Sci. USA 2016, 113, 12198–12201. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Comte, C.; Mager-Heckel, A.M.; Addis, V.; Krasheninnikov, I.A.; Martin, R.P.; Entelis, N.; Tarassov, I. Mitochondrial enzyme rhodanese is essential for 5 S ribosomal RNA import into human mitochondria. J. Biol. Chem. 2010, 285, 30792–30803. [Google Scholar] [CrossRef]
- Smirnov, A.; Entelis, N.; Martin, R.P.; Tarassov, I. Biological significance of 5S rRNA import into human mitochondria: Role of ribosomal protein MRP-L18. Genes Dev. 2011, 25, 1289–1305. [Google Scholar] [CrossRef] [PubMed]
- Ojala, D.; Montoya, J.; Attardi, G. tRNA punctuation model of RNA processing in human mitochondria. Nature 1981, 290, 470–474. [Google Scholar] [CrossRef]
- Rackham, O.; Busch, J.D.; Matic, S.; Siira, S.J.; Kuznetsova, I.; Atanassov, I.; Ermer, J.A.; Shearwood, A.M.; Richman, T.R.; Stewart, J.B.; et al. Hierarchical RNA processing is required for mitochondrial ribosome assembly. Cell Rep. 2016, 16, 1874–1890. [Google Scholar] [CrossRef]
- Klemm, B.P.; Wu, N.; Chen, Y.; Liu, X.; Kaitany, K.J.; Howard, M.J.; Fierke, C.A. The Diversity of ribonuclease P: Protein and RNA catalysts with analogous biological functions. Biomolecules 2016, 6, 27. [Google Scholar] [CrossRef]
- Lechner, M.; Rossmanith, W.; Hartmann, R.K.; Thölken, C.; Gutmann, B.; Giegé, P.; Gobert, A. Distribution of ribonucleoprotein and protein-only RNase P in Eukarya. Mol. Biol. Evol. 2015, 32, 3186–3193. [Google Scholar] [CrossRef]
- Holzmann, J.; Frank, P.; Loffler, E.; Bennett, K.L.; Gerner, C.; Rossmanith, W. RNase P without RNA: Identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 2008, 135, 462–474. [Google Scholar] [CrossRef] [PubMed]
- Nickel, A.I.; Waber, N.B.; Gossringer, M.; Lechner, M.; Linne, U.; Toth, U.; Rossmanith, W.; Hartmann, R.K. Minimal and RNA-free RNase P in Aquifex aeolicus. Proc. Natl. Acad. Sci. USA 2017, 114, 11121–11126. [Google Scholar] [CrossRef]
- Esakova, O.; Krasilnikov, A.S. Of proteins and RNA: The RNase P/MRP family. RNA 2010, 16, 1725–1747. [Google Scholar] [CrossRef]
- Schmitt, M.E.; Clayton, D.A. Nuclear RNase MRP is required for correct processing of pre-5.8S rRNA in Saccharomyces cerevisiae. Mol. Cell Biol. 1993, 13, 7935–7941. [Google Scholar] [CrossRef] [PubMed]
- Gill, T.; Cai, T.; Aulds, J.; Wierzbicki, S.; Schmitt, M.E. RNase MRP cleaves the CLB2 mRNA to promote cell cycle progression: Novel method of mRNA degradation. Mol. Cell Biol. 2004, 24, 945–953. [Google Scholar] [CrossRef] [PubMed]
- Maida, Y.; Yasukawa, M.; Furuuchi, M.; Lassmann, T.; Possemato, R.; Okamoto, N.; Kasim, V.; Hayashizaki, Y.; Hahn, W.C.; Masutomi, K. An RNA-dependent RNA polymerase formed by TERT and the RMRP RNA. Nature 2009, 461, 230–235. [Google Scholar] [CrossRef]
- Chang, D.D.; Clayton, D.A. Mouse RNAase MRP RNA is encoded by a nuclear gene and contains a decamer sequence complementary to a conserved region of mitochondrial RNA substrate. Cell 1989, 56, 131–139. [Google Scholar] [CrossRef]
- Agaronyan, K.; Morozov, Y.I.; Anikin, M.; Temiakov, D. Mitochondrial biology. Replication-transcription switch in human mitochondria. Science 2015, 347, 548–551. [Google Scholar] [CrossRef]
- Jourdain, A.A.; Koppen, M.; Wydro, M.; Rodley, C.D.; Lightowlers, R.N.; Chrzanowska-Lightowlers, Z.M.; Martinou, J.C. GRSF1 regulates RNA processing in mitochondrial RNA granules. Cell Metab. 2013, 17, 399–410. [Google Scholar] [CrossRef]
- Antonicka, H.; Sasarman, F.; Nishimura, T.; Paupe, V.; Shoubridge, E.A. The mitochondrial RNA-binding protein GRSF1 localizes to RNA granules and is required for posttranscriptional mitochondrial gene expression. Cell Metab. 2013, 17, 386–398. [Google Scholar] [CrossRef]
- Fogal, V.; Richardson, A.D.; Karmali, P.P.; Scheffler, I.E.; Smith, J.W.; Ruoslahti, E. Mitochondrial p32 protein is a critical regulator of tumor metabolism via maintenance of oxidative phosphorylation. Mol. Cell Biol. 2010, 30, 1303–1318. [Google Scholar] [CrossRef]
- McGee, A.M.; Douglas, D.L.; Liang, Y.; Hyder, S.M.; Baines, C.P. The mitochondrial protein C1qbp promotes cell proliferation, migration and resistance to cell death. Cell Cycle 2011, 10, 4119–4127. [Google Scholar] [CrossRef] [PubMed]
- Yagi, M.; Uchiumi, T.; Takazaki, S.; Okuno, B.; Nomura, M.; Yoshida, S.; Kanki, T.; Kang, D. p32/gC1qR is indispensable for fetal development and mitochondrial translation: Importance of its RNA-binding ability. Nucleic Acids Res. 2012, 40, 9717–9737. [Google Scholar] [CrossRef]
- Hu, M.; Crawford, S.A.; Henstridge, D.C.; Ng, I.H.; Boey, E.J.; Xu, Y.; Febbraio, M.A.; Jans, D.A.; Bogoyevitch, M.A. p32 protein levels are integral to mitochondrial and endoplasmic reticulum morphology, cell metabolism and survival. Biochem. J. 2013, 453, 381–391. [Google Scholar] [CrossRef] [PubMed]
- Muta, T.; Kang, D.; Kitajima, S.; Fujiwara, T.; Hamasaki, N. p32 protein, a splicing factor 2-associated protein, is localized in mitochondrial matrix and is functionally important in maintaining oxidative phosphorylation. J. Biol. Chem. 1997, 272, 24363–24370. [Google Scholar] [CrossRef] [PubMed]
- Gall, J.G. Telomerase RNA: Tying up the loose ends. Nature 1990, 344, 108–109. [Google Scholar] [CrossRef]
- Zheng, Q.; Liu, P.; Gao, G.; Yuan, J.; Wang, P.; Huang, J.; Xie, L.; Lu, X.; Di, F.; Tong, T.; et al. Mitochondrion-processed TERC regulates senescence without affecting telomerase activities. Protein Cell 2019. [Google Scholar] [CrossRef]
- Huntzinger, E.; Izaurralde, E. Gene silencing by microRNAs: Contributions of translational repression and mRNA decay. Nat. Rev. Genet 2011, 12, 99–110. [Google Scholar] [CrossRef]
- Fabian, M.R.; Sonenberg, N.; Filipowicz, W. Regulation of mRNA translation and stability by microRNAs. Annu. Rev. Biochem. 2010, 79, 351–379. [Google Scholar] [CrossRef] [PubMed]
- Guerra-Assuncao, J.A.; Enright, A.J. Large-scale analysis of microRNA evolution. BMC Genom. 2012, 13, 218. [Google Scholar] [CrossRef]
- Bianchessi, V.; Badi, I.; Bertolotti, M.; Nigro, P.; D’Alessandra, Y.; Capogrossi, M.C.; Zanobini, M.; Pompilio, G.; Raucci, A.; Lauri, A. The mitochondrial lncRNA ASncmtRNA-2 is induced in aging and replicative senescence in Endothelial Cells. J. Mol. Cell. Cardiol. 2015, 81, 62–70. [Google Scholar] [CrossRef] [PubMed]
- Lobos-Gonzalez, L.; Silva, V.; Araya, M.; Restovic, F.; Echenique, J.; Oliveira-Cruz, L.; Fitzpatrick, C.; Briones, M.; Villegas, J.; Villota, C.; et al. Targeting antisense mitochondrial ncRNAs inhibits murine melanoma tumor growth and metastasis through reduction in survival and invasion factors. Oncotarget 2016, 7, 58331–58350. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Kohr, M.; Dunkerly-Eyring, B.; Lee, D.I.; Bedja, D.; Kent, O.A.; Leung, A.K.; Henao-Mejia, J.; Flavell, R.A.; Steenbergen, C. Divergent effects of miR-181 family members on myocardial function through protective cytosolic and detrimental mitochondrial microRNA targets. J. Am. Heart Assoc. 2017, 6. [Google Scholar] [CrossRef]
- Jagannathan, R.; Thapa, D.; Nichols, C.E.; Shepherd, D.L.; Stricker, J.C.; Croston, T.L.; Baseler, W.A.; Lewis, S.E.; Martinez, I.; Hollander, J.M. Translational regulation of the mitochondrial genome following redistribution of mitochondrial microRNA in the diabetic heart. Circ. Cardiovasc. Genet 2015, 8, 785–802. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Mollet, S.; Souquere, S.; Le Roy, F.; Ernoult-Lange, M.; Pierron, G.; Dautry, F.; Weil, D. Mitochondria associate with P-bodies and modulate microRNA-mediated RNA interference. J. Biol. Chem. 2011, 286, 24219–24230. [Google Scholar] [CrossRef] [PubMed]
- Salinas-Giege, T.; Giege, R.; Giege, P. tRNA biology in mitochondria. Int. J. Mol. Sci. 2015, 16, 4518–4559. [Google Scholar] [CrossRef]
- Schneider, A. Mitochondrial tRNA import and its consequences for mitochondrial translation. Annu. Rev. Biochem. 2011, 80, 1033–1053. [Google Scholar] [CrossRef]
- Sieber, F.; Placido, A.; El Farouk-Ameqrane, S.; Duchene, A.M.; Marechal-Drouard, L. A protein shuttle system to target RNA into mitochondria. Nucleic Acids Res. 2011, 39, e96. [Google Scholar] [CrossRef]
- Delage, L.; Dietrich, A.; Cosset, A.; Marechal-Drouard, L. In vitro import of a nuclearly encoded tRNA into mitochondria of Solanum tuberosum. Mol. Cell Biol. 2003, 23, 4000–4012. [Google Scholar] [CrossRef] [PubMed]
- Salinas, T.; Duchene, A.M.; Delage, L.; Nilsson, S.; Glaser, E.; Zaepfel, M.; Marechal-Drouard, L. The voltage-dependent anion channel, a major component of the tRNA import machinery in plant mitochondria. Proc. Natl. Acad. Sci. USA 2006, 103, 18362–18367. [Google Scholar] [CrossRef] [PubMed]
- Salinas, T.; El Farouk-Ameqrane, S.; Ubrig, E.; Sauter, C.; Duchene, A.M.; Marechal-Drouard, L. Molecular basis for the differential interaction of plant mitochondrial VDAC proteins with tRNAs. Nucleic Acids Res. 2014, 42, 9937–9948. [Google Scholar] [CrossRef] [PubMed]
- Crausaz Esseiva, A.; Marechal-Drouard, L.; Cosset, A.; Schneider, A. The T-stem determines the cytosolic or mitochondrial localization of trypanosomal tRNAsMet. Mol. Biol. Cell 2004, 15, 2750–2757. [Google Scholar] [CrossRef] [PubMed]
- Bouzaidi-Tiali, N.; Aeby, E.; Charriere, F.; Pusnik, M.; Schneider, A. Elongation factor 1a mediates the specificity of mitochondrial tRNA import in T. brucei. EMBO J. 2007, 26, 4302–4312. [Google Scholar] [CrossRef]
- Niemann, M.; Harsman, A.; Mani, J.; Peikert, C.D.; Oeljeklaus, S.; Warscheid, B.; Wagner, R.; Schneider, A. tRNAs and proteins use the same import channel for translocation across the mitochondrial outer membrane of trypanosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E7679–E7687. [Google Scholar] [CrossRef]
- Entelis, N.; Brandina, I.; Kamenski, P.; Krasheninnikov, I.A.; Martin, R.P.; Tarassov, I. A glycolytic enzyme, enolase, is recruited as a cofactor of tRNA targeting toward mitochondria in Saccharomyces cerevisiae. Genes Dev. 2006, 20, 1609–1620. [Google Scholar] [CrossRef] [PubMed]
- Kazakova, H.A.; Entelis, N.S.; Martin, R.P.; Tarassov, I.A. The aminoacceptor stem of the yeast tRNA(Lys) contains determinants of mitochondrial import selectivity. FEBS Lett. 1999, 442, 193–197. [Google Scholar] [CrossRef]
- Kolesnikova, O.; Kazakova, H.; Comte, C.; Steinberg, S.; Kamenski, P.; Martin, R.P.; Tarassov, I.; Entelis, N. Selection of RNA aptamers imported into yeast and human mitochondria. RNA 2010, 16, 926–941. [Google Scholar] [CrossRef]
- Kamenski, P.; Smirnova, E.; Kolesnikova, O.; Krasheninnikov, I.A.; Martin, R.P.; Entelis, N.; Tarassov, I. tRNA mitochondrial import in yeast: Mapping of the import determinants in the carrier protein, the precursor of mitochondrial lysyl-tRNA synthetase. Mitochondrion 2010, 10, 284–293. [Google Scholar] [CrossRef] [PubMed]
- Baleva, M.; Gowher, A.; Kamenski, P.; Tarassov, I.; Entelis, N.; Masquida, B. A Moonlighting human protein is involved in mitochondrial import of tRNA. Int. J. Mol. Sci. 2015, 16, 9354–9367. [Google Scholar] [CrossRef] [PubMed]
- Gowher, A.; Smirnov, A.; Tarassov, I.; Entelis, N. Induced tRNA import into human mitochondria: Implication of a host aminoacyl-tRNA-synthetase. PLoS ONE 2013, 8, e66228. [Google Scholar] [CrossRef]
- Tonin, Y.; Heckel, A.M.; Vysokikh, M.; Dovydenko, I.; Meschaninova, M.; Rotig, A.; Munnich, A.; Venyaminova, A.; Tarassov, I.; Entelis, N. Modeling of antigenomic therapy of mitochondrial diseases by mitochondrially addressed RNA targeting a pathogenic point mutation in mitochondrial DNA. J. Biol. Chem. 2014, 289, 13323–13334. [Google Scholar] [CrossRef] [PubMed]
- Smirnov, A.; Tarassov, I.; Mager-Heckel, A.M.; Letzelter, M.; Martin, R.P.; Krasheninnikov, I.A.; Entelis, N. Two distinct structural elements of 5S rRNA are needed for its import into human mitochondria. RNA 2008, 14, 749–759. [Google Scholar] [CrossRef]
- Steitz, J.A.; Berg, C.; Hendrick, J.P.; La Branche-Chabot, H.; Metspalu, A.; Rinke, J.; Yario, T. A 5S rRNA/L5 complex is a precursor to ribosome assembly in mammalian cells. J. Cell Biol. 1988, 106, 545–556. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, X.; Coots, R.A.; Conn, C.S.; Liu, B.; Qian, S.B. Translational control of the cytosolic stress response by mitochondrial ribosomal protein L18. Nat. Struct. Mol. Biol. 2015, 22, 404–410. [Google Scholar] [CrossRef]
- Zabezhinsky, D.; Slobodin, B.; Rapaport, D.; Gerst, J.E. An essential role for COPI in mRNA localization to mitochondria and mitochondrial function. Cell Rep. 2016, 15, 540–549. [Google Scholar] [CrossRef]
- Schatton, D.; Rugarli, E.I. A concert of RNA-binding proteins coordinates mitochondrial function. Crit. Rev. Biochem. Mol. Biol. 2018, 53, 652–666. [Google Scholar] [CrossRef]
- Tarassov, I.; Entelis, N.; Martin, R.P. An intact protein translocating machinery is required for mitochondrial import of a yeast cytoplasmic tRNA. J. Mol. Biol. 1995, 245, 315–323. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.W.; Rainey, R.N.; Balatoni, C.E.; Dawson, D.W.; Troke, J.J.; Wasiak, S.; Hong, J.S.; McBride, H.M.; Koehler, C.M.; Teitell, M.A.; et al. Mammalian polynucleotide phosphorylase is an intermembrane space RNase that maintains mitochondrial homeostasis. Mol. Cell Biol. 2006, 26, 8475–8487. [Google Scholar] [CrossRef] [PubMed]
- Borowski, L.S.; Dziembowski, A.; Hejnowicz, M.S.; Stepien, P.P.; Szczesny, R.J. Human mitochondrial RNA decay mediated by PNPase-hSuv3 complex takes place in distinct foci. Nucleic Acids Res. 2013, 41, 1223–1240. [Google Scholar] [CrossRef]
- Nagaike, T.; Suzuki, T.; Katoh, T.; Ueda, T. Human mitochondrial mRNAs are stabilized with polyadenylation regulated by mitochondria-specific poly(A) polymerase and polynucleotide phosphorylase. J. Biol. Chem. 2005, 280, 19721–19727. [Google Scholar] [CrossRef]
- Wang, D.D.; Guo, X.E.; Modrek, A.S.; Chen, C.F.; Chen, P.L.; Lee, W.H. Helicase SUV3, polynucleotide phosphorylase, and mitochondrial polyadenylation polymerase form a transient complex to modulate mitochondrial mRNA polyadenylated tail lengths in response to energetic changes. J. Biol. Chem. 2014, 289, 16727–16735. [Google Scholar] [CrossRef]
- Jourdain, A.A.; Koppen, M.; Rodley, C.D.; Maundrell, K.; Gueguen, N.; Reynier, P.; Guaras, A.M.; Enriquez, J.A.; Anderson, P.; Simarro, M.; et al. A mitochondria-specific isoform of FASTK is present in mitochondrial RNA granules and regulates gene expression and function. Cell Rep. 2015, 10, 1110–1121. [Google Scholar] [CrossRef]
- Pietras, Z.; Wojcik, M.A.; Borowski, L.S.; Szewczyk, M.; Kulinski, T.M.; Cysewski, D.; Stepien, P.P.; Dziembowski, A.; Szczesny, R.J. Dedicated surveillance mechanism controls G-quadruplex forming non-coding RNAs in human mitochondria. Nat. Commun. 2018, 9, 2558. [Google Scholar] [CrossRef] [PubMed]
- Silva, S.; Camino, L.P.; Aguilera, A. Human mitochondrial degradosome prevents harmful mitochondrial R loops and mitochondrial genome instability. Proc. Natl. Acad. Sci. USA 2018, 115, 11024–11029. [Google Scholar] [CrossRef] [PubMed]
- Dhir, A.; Dhir, S.; Borowski, L.S.; Jimenez, L.; Teitell, M.; Rotig, A.; Crow, Y.J.; Rice, G.I.; Duffy, D.; Tamby, C.; et al. Mitochondrial double-stranded RNA triggers antiviral signalling in humans. Nature 2018, 560, 238–242. [Google Scholar] [CrossRef]
- Liu, X.; Fu, R.; Pan, Y.; Meza-Sosa, K.F.; Zhang, Z.; Lieberman, J. PNPT1 release from mitochondria during apoptosis triggers decay of poly(A) RNAs. Cell 2018, 174, 187–201.e112. [Google Scholar] [CrossRef]
- Von Ameln, S.; Wang, G.; Boulouiz, R.; Rutherford, M.A.; Smith, G.M.; Li, Y.; Pogoda, H.M.; Nurnberg, G.; Stiller, B.; Volk, A.E.; et al. A mutation in PNPT1, encoding mitochondrial-RNA-import protein PNPase, causes hereditary hearing loss. Am. J. Hum. Genet 2012, 91, 919–927. [Google Scholar] [CrossRef]
- Vedrenne, V.; Gowher, A.; De Lonlay, P.; Nitschke, P.; Serre, V.; Boddaert, N.; Altuzarra, C.; Mager-Heckel, A.M.; Chretien, F.; Entelis, N.; et al. Mutation in PNPT1, which encodes a polyribonucleotide nucleotidyltransferase, impairs RNA import into mitochondria and causes respiratory-chain deficiency. Am. J. Hum. Genet 2012, 91, 912–918. [Google Scholar] [CrossRef] [PubMed]
- Matilainen, S.; Carroll, C.J.; Richter, U.; Euro, L.; Pohjanpelto, M.; Paetau, A.; Isohanni, P.; Suomalainen, A. Defective mitochondrial RNA processing due to PNPT1 variants causes Leigh syndrome. Hum. Mol. Genet 2017, 26, 3352–3361. [Google Scholar] [CrossRef]
- Golzarroshan, B.; Lin, C.L.; Li, C.L.; Yang, W.Z.; Chu, L.Y.; Agrawal, S.; Yuan, H.S. Crystal structure of dimeric human PNPase reveals why disease-linked mutants suffer from low RNA import and degradation activities. Nucleic Acids Res. 2018, 46, 8630–8640. [Google Scholar] [CrossRef]
- Cameron, T.A.; Matz, L.M.; De Lay, N.R. Polynucleotide phosphorylase: Not merely an RNase but a pivotal post-transcriptional regulator. PLoS Genet 2018, 14, e1007654. [Google Scholar] [CrossRef] [PubMed]
- Lin, C.L.; Wang, Y.T.; Yang, W.Z.; Hsiao, Y.Y.; Yuan, H.S. Crystal structure of human polynucleotide phosphorylase: Insights into its domain function in RNA binding and degradation. Nucleic Acids Res. 2012, 40, 4146–4157. [Google Scholar] [CrossRef] [PubMed]
- Mager-Heckel, A.M.; Entelis, N.; Brandina, I.; Kamenski, P.; Krasheninnikov, I.A.; Martin, R.P.; Tarassov, I. The analysis of tRNA import into mammalian mitochondria. Methods Mol. Biol. 2007, 372, 235–253. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Shimada, E.; Nili, M.; Koehler, C.M.; Teitell, M.A. Mitochondria-targeted RNA import. Methods Mol. Biol. 2015, 1264, 107–116. [Google Scholar] [CrossRef]
- Claude, A. Fractionation of mammalian liver cells by differential centrifugation; experimental procedures and results. J. Exp. Med. 1946, 84, 61–89. [Google Scholar] [CrossRef]
- Clayton, D.A.; Shadel, G.S. Purification of mitochondria by sucrose step density gradient centrifugation. Cold Spring Harb. Protoc. 2014, 10. [Google Scholar] [CrossRef]
- Franko, A.; Baris, O.R.; Bergschneider, E.; von Toerne, C.; Hauck, S.M.; Aichler, M.; Walch, A.K.; Wurst, W.; Wiesner, R.J.; Johnston, I.C.; et al. Efficient isolation of pure and functional mitochondria from mouse tissues using automated tissue disruption and enrichment with anti-TOM22 magnetic beads. PLoS ONE 2013, 8, e82392. [Google Scholar] [CrossRef]
- Marchi, S.; Patergnani, S.; Pinton, P. The endoplasmic reticulum-mitochondria connection: One touch, multiple functions. Biochim. Biophys. Acta 2014, 1837, 461–469. [Google Scholar] [CrossRef] [PubMed]
- Aravin, A.A.; Chan, D.C. piRNAs meet mitochondria. Dev. Cell 2011, 20, 287–288. [Google Scholar] [CrossRef] [PubMed]
- Cande, C.; Vahsen, N.; Metivier, D.; Tourriere, H.; Chebli, K.; Garrido, C.; Tazi, J.; Kroemer, G. Regulation of cytoplasmic stress granules by apoptosis-inducing factor. J. Cell Sci. 2004, 117, 4461–4468. [Google Scholar] [CrossRef]
- Matsumoto, S.; Uchiumi, T.; Saito, T.; Yagi, M.; Takazaki, S.; Kanki, T.; Kang, D. Localization of mRNAs encoding human mitochondrial oxidative phosphorylation proteins. Mitochondrion 2012, 12, 391–398. [Google Scholar] [CrossRef]
- Gold, V.A.; Chroscicki, P.; Bragoszewski, P.; Chacinska, A. Visualization of cytosolic ribosomes on the surface of mitochondria by electron cryo-tomography. EMBO Rep. 2017, 18, 1786–1800. [Google Scholar] [CrossRef]
- Petit, P.X.; Goubern, M.; Diolez, P.; Susin, S.A.; Zamzami, N.; Kroemer, G. Disruption of the outer mitochondrial membrane as a result of large amplitude swelling: The impact of irreversible permeability transition. FEBS Lett. 1998, 426, 111–116. [Google Scholar] [CrossRef]
- Zischka, H.; Larochette, N.; Hoffmann, F.; Hamoller, D.; Jagemann, N.; Lichtmannegger, J.; Jennen, L.; Muller-Hocker, J.; Roggel, F.; Gottlicher, M.; et al. Electrophoretic analysis of the mitochondrial outer membrane rupture induced by permeability transition. Anal. Chem. 2008, 80, 5051–5058. [Google Scholar] [CrossRef] [PubMed]
- Cannon, M.V.; Irwin, M.H.; Pinkert, C.A. Mitochondrially-imported RNA in drug discovery. Drug Dev. Res. 2015, 76, 61–71. [Google Scholar] [CrossRef] [PubMed]
- Fuller, K.M.; Arriaga, E.A. Capillary electrophoresis monitors changes in the electrophoretic behavior of mitochondrial preparations. J. Chromatogr. B Analyt. Technol. Biomed. Life Sci. 2004, 806, 151–159. [Google Scholar] [CrossRef] [PubMed]
- Aryani, A.; Denecke, B. In vitro application of ribonucleases: Comparison of the effects on mRNA and miRNA stability. BMC Res. Notes 2015, 8, 164. [Google Scholar] [CrossRef]
- Hrdlickova, R.; Toloue, M.; Tian, B. RNA-Seq methods for transcriptome analysis. Wiley Interdiscip. Rev. RNA 2017, 8. [Google Scholar] [CrossRef]
- Crosetto, N.; Bienko, M.; van Oudenaarden, A. Spatially resolved transcriptomics and beyond. Nat. Rev. Genet 2015, 16, 57–66. [Google Scholar] [CrossRef] [PubMed]
- Urbanek, M.O.; Nawrocka, A.U.; Krzyzosiak, W.J. Small RNA detection by in situ hybridization methods. Int. J. Mol. Sci. 2015, 16, 13259–13286. [Google Scholar] [CrossRef]
- Yoshimura, H. Live Cell imaging of endogenous RNAs using Pumilio homology domain mutants: Principles and applications. Biochemistry 2018, 57, 200–208. [Google Scholar] [CrossRef] [PubMed]
- Bertrand, E.; Chartrand, P.; Schaefer, M.; Shenoy, S.M.; Singer, R.H.; Long, R.M. Localization of ASH1 mRNA particles in living yeast. Mol. Cell 1998, 2, 437–445. [Google Scholar] [CrossRef]
- Heinrich, S.; Sidler, C.L.; Azzalin, C.M.; Weis, K. Stem-loop RNA labeling can affect nuclear and cytoplasmic mRNA processing. RNA 2017, 23, 134–141. [Google Scholar] [CrossRef] [PubMed]
- Babendure, J.R.; Adams, S.R.; Tsien, R.Y. Aptamers switch on fluorescence of triphenylmethane dyes. J. Am. Chem. Soc. 2003, 125, 14716–14717. [Google Scholar] [CrossRef]
- Huang, H.; Suslov, N.B.; Li, N.S.; Shelke, S.A.; Evans, M.E.; Koldobskaya, Y.; Rice, P.A.; Piccirilli, J.A. A G-quadruplex-containing RNA activates fluorescence in a GFP-like fluorophore. Nat. Chem. Biol. 2014, 10, 686–691. [Google Scholar] [CrossRef]
- Paige, J.S.; Wu, K.Y.; Jaffrey, S.R. RNA mimics of green fluorescent protein. Science 2011, 333, 642–646. [Google Scholar] [CrossRef]
- Filonov, G.S.; Moon, J.D.; Svensen, N.; Jaffrey, S.R. Broccoli: Rapid selection of an RNA mimic of green fluorescent protein by fluorescence-based selection and directed evolution. J. Am. Chem. Soc. 2014, 136, 16299–16308. [Google Scholar] [CrossRef]
- Ozawa, T.; Natori, Y.; Sato, M.; Umezawa, Y. Imaging dynamics of endogenous mitochondrial RNA in single living cells. Nat. Methods 2007, 4, 413–419. [Google Scholar] [CrossRef]
- Lecuyer, E.; Yoshida, H.; Parthasarathy, N.; Alm, C.; Babak, T.; Cerovina, T.; Hughes, T.R.; Tomancak, P.; Krause, H.M. Global analysis of mRNA localization reveals a prominent role in organizing cellular architecture and function. Cell 2007, 131, 174–187. [Google Scholar] [CrossRef] [PubMed]
- Chatre, L.; Ricchetti, M. Large heterogeneity of mitochondrial DNA transcription and initiation of replication exposed by single-cell imaging. J. Cell Sci. 2013, 126, 914–926. [Google Scholar] [CrossRef] [PubMed]
- Femino, A.M.; Fay, F.S.; Fogarty, K.; Singer, R.H. Visualization of single RNA transcripts in situ. Science 1998, 280, 585–590. [Google Scholar] [CrossRef] [PubMed]
- Player, A.N.; Shen, L.P.; Kenny, D.; Antao, V.P.; Kolberg, J.A. Single-copy gene detection using branched DNA (bDNA) in situ hybridization. J. Histochem. Cytochem. 2001, 49, 603–612. [Google Scholar] [CrossRef] [PubMed]
- Battich, N.; Stoeger, T.; Pelkmans, L. Image-based transcriptomics in thousands of single human cells at single-molecule resolution. Nat. Methods 2013, 10, 1127–1133. [Google Scholar] [CrossRef] [PubMed]
- Huang, B.; Bates, M.; Zhuang, X. Super-resolution fluorescence microscopy. Annu. Rev. Biochem. 2009, 78, 993–1016. [Google Scholar] [CrossRef] [PubMed]
- Jakobs, S.; Wurm, C.A. Super-resolution microscopy of mitochondria. Curr. Opin. Chem. Biol. 2014, 20, 9–15. [Google Scholar] [CrossRef] [PubMed]
- Brown, T.A.; Tkachuk, A.N.; Shtengel, G.; Kopek, B.G.; Bogenhagen, D.F.; Hess, H.F.; Clayton, D.A. Superresolution fluorescence imaging of mitochondrial nucleoids reveals their spatial range, limits, and membrane interaction. Mol. Cell Biol. 2011, 31, 4994–5010. [Google Scholar] [CrossRef] [PubMed]
- Kuzmenko, A.; Tankov, S.; English, B.P.; Tarassov, I.; Tenson, T.; Kamenski, P.; Elf, J.; Hauryliuk, V. Single molecule tracking fluorescence microscopy in mitochondria reveals highly dynamic but confined movement of Tom40. Sci. Rep. 2011, 1, 195. [Google Scholar] [CrossRef] [PubMed]
- Fei, J.; Singh, D.; Zhang, Q.; Park, S.; Balasubramanian, D.; Golding, I.; Vanderpool, C.K.; Ha, T. RNA biochemistry. Determination of in vivo target search kinetics of regulatory noncoding RNA. Science 2015, 347, 1371–1374. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Aggarwal, M.B.; Nguyen, K.; Ke, K.; Spitale, R.C. Assaying RNA localization in situ with spatially restricted nucleobase oxidation. ACS Chem. Biol. 2017, 12, 2709–2714. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Aggarwal, M.B.; Ke, K.; Nguyen, K.; Spitale, R.C. Improved analysis of RNA localization by spatially restricted oxidation of RNA-protein complexes. Biochemistry 2018, 57, 1577–1581. [Google Scholar] [CrossRef]
- Hoogewijs, K.; James, A.M.; Smith, R.A.; Gait, M.J.; Murphy, M.P.; Lightowlers, R.N. Assessing the delivery of molecules to the mitochondrial matrix using click chemistry. Chembiochem 2016, 17, 1312–1316. [Google Scholar] [CrossRef] [PubMed]
- Kaewsapsak, P.; Shechner, D.M.; Mallard, W.; Rinn, J.L.; Ting, A.Y. Live-cell mapping of organelle-associated RNAs via proximity biotinylation combined with protein-RNA crosslinking. Elife 2017, 6. [Google Scholar] [CrossRef] [PubMed]
- Benhalevy, D.; Anastasakis, D.G.; Hafner, M. Proximity-CLIP provides a snapshot of protein-occupied RNA elements in subcellular compartments. Nat. Methods 2018, 15, 1074–1082. [Google Scholar] [CrossRef] [PubMed]
- Rhee, H.W.; Zou, P.; Udeshi, N.D.; Martell, J.D.; Mootha, V.K.; Carr, S.A.; Ting, A.Y. Proteomic mapping of mitochondria in living cells via spatially restricted enzymatic tagging. Science 2013, 339, 1328–1331. [Google Scholar] [CrossRef] [PubMed]
- Jourdain, A.A.; Boehm, E.; Maundrell, K.; Martinou, J.C. Mitochondrial RNA granules: Compartmentalizing mitochondrial gene expression. J. Cell Biol. 2016, 212, 611–614. [Google Scholar] [CrossRef]
- Pearce, S.F.; Rebelo-Guiomar, P.; D’Souza, A.R.; Powell, C.A.; Van Haute, L.; Minczuk, M. Regulation of mammalian mitochondrial gene expression: Recent advances. Trends Biochem. Sci. 2017, 42, 625–639. [Google Scholar] [CrossRef]
- Smirnov, A.; Schneider, C.; Hör, J.; Vogel, J. Discovery of new RNA classes and global RNA-binding proteins. Curr. Opin. Microbiol. 2017, 39, 152–160. [Google Scholar] [CrossRef]
- Gambarotto, D.; Zwettler, F.U.; Le Guennec, M.; Schmidt-Cernohorska, M.; Fortun, D.; Borgers, S.; Heine, J.; Schloetel, J.G.; Reuss, M.; Unser, M.; et al. Imaging cellular ultrastructures using expansion microscopy (U-ExM). Nat. Methods 2019, 16, 71–74. [Google Scholar] [CrossRef] [PubMed]
- Gammage, P.A.; Moraes, C.T.; Minczuk, M. Mitochondrial genome engineering: The revolution may not be CRISPR-ized. Trends Genet 2018, 34, 101–110. [Google Scholar] [CrossRef] [PubMed]
- Schimmel, P. The emerging complexity of the tRNA world: Mammalian tRNAs beyond protein synthesis. Nat. Rev. Mol. Cell Biol. 2018, 19, 45–58. [Google Scholar] [CrossRef] [PubMed]

| RNA | Cytosolic Function | Evidence for Mitochondrial Localisation | Proposed Function in Mitochondria | References |
|---|---|---|---|---|
| Select tRNAs (including tRNALeuUAA, tRNAGlnUUG, tRNAGlnCUG) | Translation |
| Mitochondrial translation under normal or stress conditions | [9,11] |
| 5S rRNA | Component of the cytosolic ribosome |
| Related to mitochondrial translation? | [11,12,13,14,15,16] |
| H1 RNA | Component of the nuclear RNase P required for pre-tRNA processing |
| Pre-tRNA processing? | [11,17,18,19] |
| RMRP | 5.8S rRNA processing |
| Mitochondrial RNA metabolism? | [11,19,20,21,22] |
| SAMMSON | Facilitates p32 targeting to the mitochondria in melanoma cells |
| Unknown | [23,24] |
| hTERC | RNA component of telomerase |
| Mitochondria-cytosol communication | [25] |
| Various miRNAs (including miR-1, miR-181c, miR-378) and pre-miRNAs | Repression of mRNA translation |
| Repression or activation of mRNA translation, repression of transcription | [11,26,27,28,29,30,31,32,33,34] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jeandard, D.; Smirnova, A.; Tarassov, I.; Barrey, E.; Smirnov, A.; Entelis, N. Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches. Cells 2019, 8, 286. https://doi.org/10.3390/cells8030286
Jeandard D, Smirnova A, Tarassov I, Barrey E, Smirnov A, Entelis N. Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches. Cells. 2019; 8(3):286. https://doi.org/10.3390/cells8030286
Chicago/Turabian StyleJeandard, Damien, Anna Smirnova, Ivan Tarassov, Eric Barrey, Alexandre Smirnov, and Nina Entelis. 2019. "Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches" Cells 8, no. 3: 286. https://doi.org/10.3390/cells8030286
APA StyleJeandard, D., Smirnova, A., Tarassov, I., Barrey, E., Smirnov, A., & Entelis, N. (2019). Import of Non-Coding RNAs into Human Mitochondria: A Critical Review and Emerging Approaches. Cells, 8(3), 286. https://doi.org/10.3390/cells8030286







