Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes
Abstract
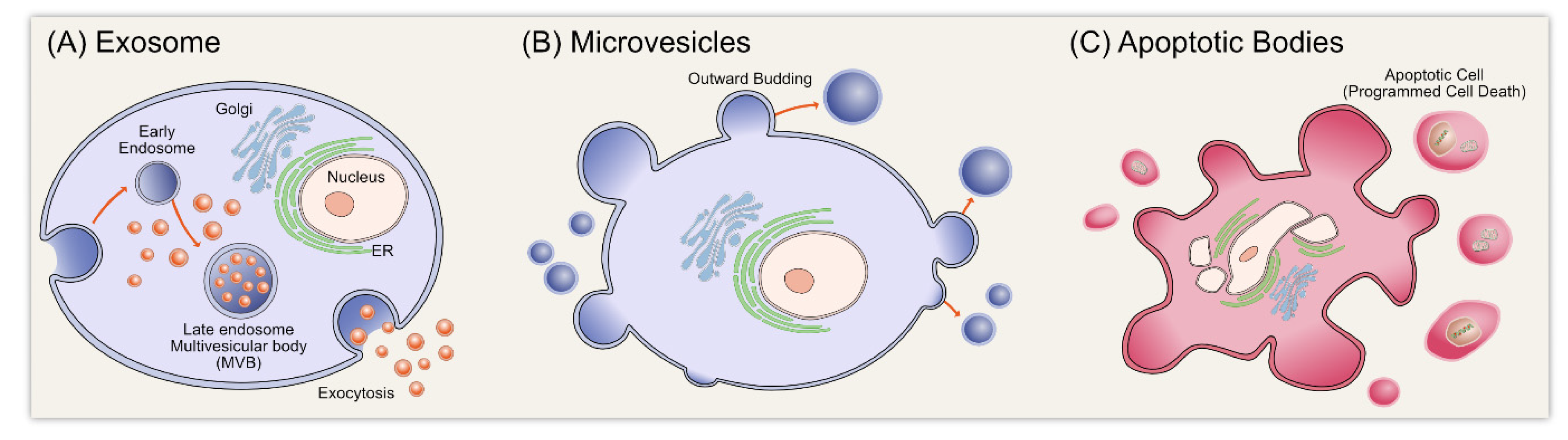
:1. Introduction
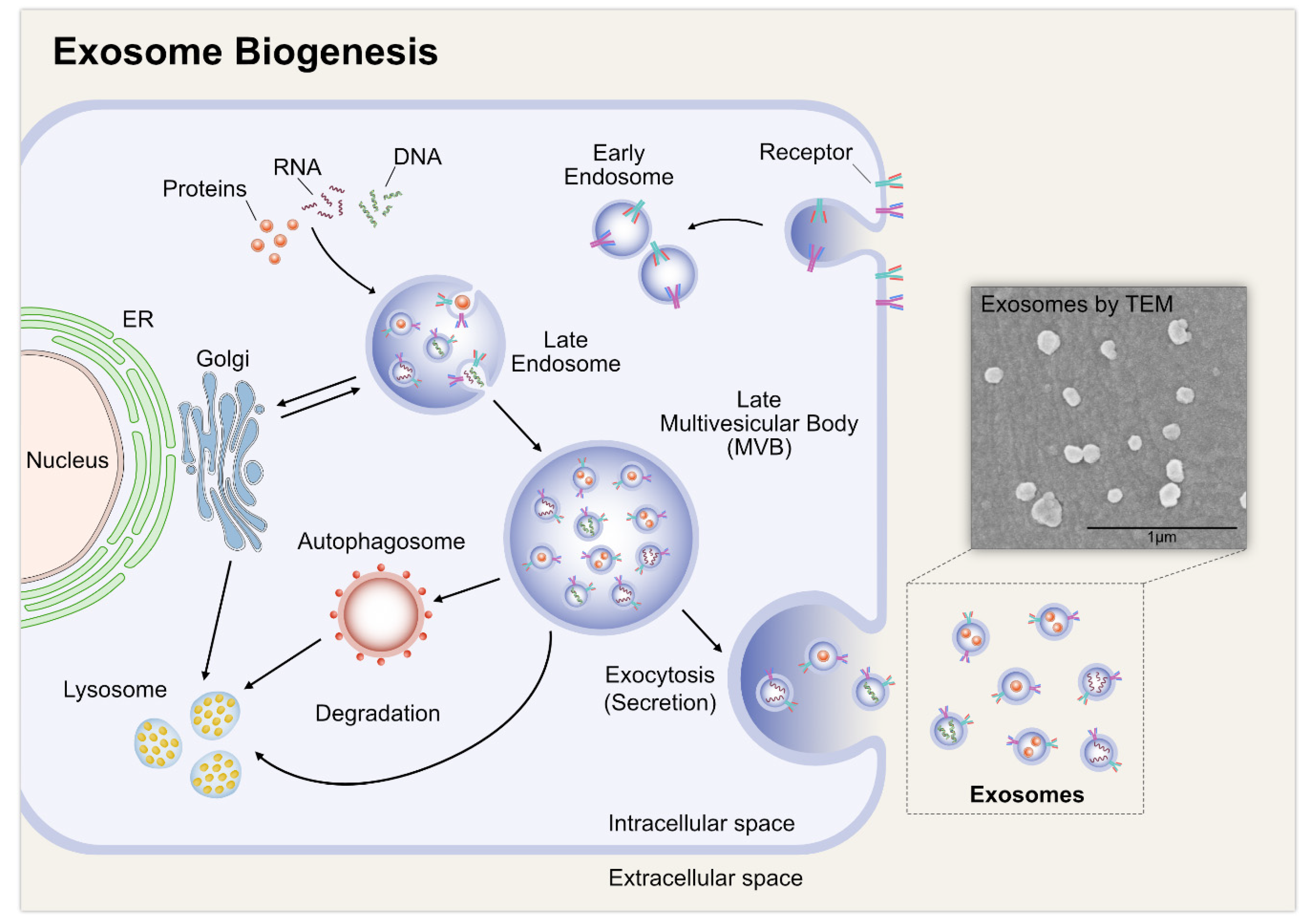
2. Biogenesis of Exosomes
3. Factors Influencing the Biogenesis of Exosomes
4. Isolation of Exosomes
4.1. Ultracentrifugation
4.2. Size-Based Filtration, Size-Exclusion Chromatography, and Polymer Precipitation
4.3. Immuno-Affinity Purification of Exosomes
4.4. Microfluidics-Based Isolation Techniques
5. Characterization of Exosomes
5.1. Nanoparticle Tracking Analysis
5.2. Dynamic Light Scattering
5.3. Resistive Pulse Sensing
5.4. Atomic Force Microscopy
5.5. Transmission Electron Microscopy
5.6. Flow Cytometry
6. Biological Functions of Exosomes
6.1. Role of Exosomes in Angiogenesis
6.2. Role of Exosomes in Apoptosis
6.3. Role of Exosomes in Antigen Presentation
6.4. Role of Exosomes in Inflammation
6.5. Exosomes as Biomarkers
6.6. Exosomes and Receptor Mediated Endocytosis
6.7. Role of Exosomes in Cell Proliferation and Differentiation
7. Therapeutic Applications of Exosomes
8. Technical and Biological Challenges of Using Exosomes
9. Conclusions and Future Perspectives
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef]
- Holme, P.A.; Solum, N.O.; Brosstad, F.; Roger, M.; Abdelnoor, M. Demonstration of platelet-derived microvesicles in blood from patients with activated coagulation and fibrinolysis using a filtration technique and western blotting. Thromb. Haemost. 1994, 72, 666–671. [Google Scholar] [PubMed]
- Hess, C.; Sadallah, S.; Hefti, A.; Landmann, R.; Schifferli, J.A. Ectosomes released by human neutrophils are specialized functional units. J. Immunol. 1999, 163, 4564–4573. [Google Scholar] [CrossRef]
- Cocucci, E.; Racchetti, G.; Meldolesi, J. Shedding microvesicles: Artefacts no more. Trends Cell Biol. 2009, 19, 43–51. [Google Scholar] [CrossRef] [PubMed]
- Gyorgy, B.; Modos, K.; Pallinger, E.; Paloczi, K.; Pasztoi, M.; Misjak, P.; Deli, M.A.; Sipos, A.; Szalai, A.; Voszka, I.; et al. Detection and isolation of cell-derived microparticles are compromised by protein complexes resulting from shared biophysical parameters. Blood 2011, 117, e39–e48. [Google Scholar] [CrossRef] [PubMed]
- Simons, M.; Raposo, G. Exosomes--vesicular carriers for intercellular communication. Curr. Opin. Cell Biol. 2009, 21, 575–581. [Google Scholar] [CrossRef]
- Thery, C.; Ostrowski, M.; Segura, E. Membrane vesicles as conveyors of immune responses. Nat. Rev. Immunol. 2009, 9, 581–593. [Google Scholar] [CrossRef] [PubMed]
- Simpson, R.J.; Jensen, S.S.; Lim, J.W. Proteomic profiling of exosomes: Current perspectives. Proteomics 2008, 8, 4083–4099. [Google Scholar] [CrossRef]
- Properzi, F.; Logozzi, M.; Fais, S. Exosomes: The future of biomarkers in medicine. Biomark. Med. 2013, 7, 769–778. [Google Scholar] [CrossRef]
- Thery, C.; Amigorena, S.; Raposo, G.; Clayton, A. Isolation and characterization of exosomes from cell culture supernatants and biological fluids. Curr. Protoc. Cell Biol. 2006, 30, 3–22. [Google Scholar] [CrossRef]
- Clayton, A.; Court, J.; Navabi, H.; Adams, M.; Mason, M.D.; Hobot, J.A.; Newman, G.R.; Jasani, B. Analysis of antigen presenting cell derived exosomes, based on immuno-magnetic isolation and flow cytometry. J. Immunol. Methods 2001, 247, 163–174. [Google Scholar] [CrossRef]
- Dragovic, R.A.; Gardiner, C.; Brooks, A.S.; Tannetta, D.S.; Ferguson, D.J.; Hole, P.; Carr, B.; Redman, C.W.; Harris, A.L.; Dobson, P.J.; et al. Sizing and phenotyping of cellular vesicles using Nanoparticle Tracking Analysis. Nanomedicine 2011, 7, 780–788. [Google Scholar] [CrossRef]
- van Niel, G.; Porto-Carreiro, I.; Simoes, S.; Raposo, G. Exosomes: A common pathway for a specialized function. J. Biochem. 2006, 140, 13–21. [Google Scholar] [CrossRef]
- Zoller, M. Tetraspanins: Push and pull in suppressing and promoting metastasis. Nat. Rev. Cancer 2009, 9, 40–55. [Google Scholar] [CrossRef]
- Thery, C.; Regnault, A.; Garin, J.; Wolfers, J.; Zitvogel, L.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Molecular characterization of dendritic cell-derived exosomes. Selective accumulation of the heat shock protein hsc73. J. Cell Biol. 1999, 147, 599–610. [Google Scholar] [CrossRef]
- Wubbolts, R.; Leckie, R.S.; Veenhuizen, P.T.; Schwarzmann, G.; Mobius, W.; Hoernschemeyer, J.; Slot, J.W.; Geuze, H.J.; Stoorvogel, W. Proteomic and biochemical analyses of human B cell-derived exosomes. Potential implications for their function and multivesicular body formation. J. Biol. Chem. 2003, 278, 10963–10972. [Google Scholar] [CrossRef]
- Wilson, H.L.; Francis, S.E.; Dower, S.K.; Crossman, D.C. Secretion of intracellular IL-1 receptor antagonist (type 1) is dependent on P2X7 receptor activation. J. Immunol. 2004, 173, 1202–1208. [Google Scholar] [CrossRef]
- Heijnen, H.F.; Schiel, A.E.; Fijnheer, R.; Geuze, H.J.; Sixma, J.J. Activated platelets release two types of membrane vesicles: Microvesicles by surface shedding and exosomes derived from exocytosis of multivesicular bodies and alpha-granules. Blood 1999, 94, 3791–3799. [Google Scholar]
- Obregon, C.; Rothen-Rutishauser, B.; Gitahi, S.K.; Gehr, P.; Nicod, L.P. Exovesicles from human activated dendritic cells fuse with resting dendritic cells, allowing them to present alloantigens. Am. J. Pathol. 2006, 169, 2127–2136. [Google Scholar] [CrossRef]
- Nolte-’t Hoen, E.N.; van der Vlist, E.J.; de Boer-Brouwer, M.; Arkesteijn, G.J.; Stoorvogel, W.; Wauben, M.H. Dynamics of dendritic cell-derived vesicles: High-resolution flow cytometric analysis of extracellular vesicle quantity and quality. J. Leukoc. Biol. 2013, 93, 395–402. [Google Scholar] [CrossRef]
- Savina, A.; Fader, C.M.; Damiani, M.T.; Colombo, M.I. Rab11 promotes docking and fusion of multivesicular bodies in a calcium-dependent manner. Traffic 2005, 6, 131–143. [Google Scholar] [CrossRef]
- Raposo, G.; Tenza, D.; Mecheri, S.; Peronet, R.; Bonnerot, C.; Desaymard, C. Accumulation of major histocompatibility complex class II molecules in mast cell secretory granules and their release upon degranulation. Mol. Biol. Cell 1997, 8, 2631–2645. [Google Scholar] [CrossRef]
- Rana, S.; Yue, S.; Stadel, D.; Zoller, M. Toward tailored exosomes: The exosomal tetraspanin web contributes to target cell selection. Int. J. Biochem. Cell Biol. 2012, 44, 1574–1584. [Google Scholar] [CrossRef]
- Hemler, M.E. Tetraspanin proteins mediate cellular penetration, invasion, and fusion events and define a novel type of membrane microdomain. Annu. Rev. Cell Dev. Biol. 2003, 19, 397–422. [Google Scholar] [CrossRef]
- Harding, C.V.; Heuser, J.E.; Stahl, P.D. Exosomes: Looking back three decades and into the future. J. Cell Biol. 2013, 200, 367–371. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Raposo, G.; Thery, C. Exosome secretion: Molecular mechanisms and roles in immune responses. Traffic 2011, 12, 1659–1668. [Google Scholar] [CrossRef]
- Rak, J. Microparticles in cancer. Semin. Thromb. Hemost. 2010, 36, 888–906. [Google Scholar] [CrossRef]
- Hood, J.L.; San, R.S.; Wickline, S.A. Exosomes released by melanoma cells prepare sentinel lymph nodes for tumor metastasis. Cancer Res. 2011, 71, 3792–3801. [Google Scholar] [CrossRef]
- Zhang, H.G.; Grizzle, W.E. Exosomes and cancer: A newly described pathway of immune suppression. Clin. Cancer Res. 2011, 17, 959–964. [Google Scholar] [CrossRef]
- Kujala, P.; Raymond, C.R.; Romeijn, M.; Godsave, S.F.; van Kasteren, S.I.; Wille, H.; Prusiner, S.B.; Mabbott, N.A.; Peters, P.J. Prion uptake in the gut: Identification of the first uptake and replication sites. PLoS Pathog. 2011, 7, e1002449. [Google Scholar] [CrossRef]
- Chargaff, E.; West, R. The biological significance of the thromboplastic protein of blood. J. Biol. Chem. 1946, 166, 189–197. [Google Scholar]
- De Broe, M.E.; Wieme, R.J.; Logghe, G.N.; Roels, F. Spontaneous shedding of plasma membrane fragments by human cells in vivo and in vitro. Clin. Chim. Acta 1977, 81, 237–245. [Google Scholar] [CrossRef]
- Wolf, P. The nature and significance of platelet products in human plasma. Br. J. Haematol. 1967, 13, 269–288. [Google Scholar] [CrossRef]
- Trams, E.G.; Lauter, C.J.; Salem, N., Jr.; Heine, U. Exfoliation of membrane ecto-enzymes in the form of micro-vesicles. Biochim. Biophys. Acta 1981, 645, 63–70. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Endocytosis and intracellular processing of transferrin and colloidal gold-transferrin in rat reticulocytes: Demonstration of a pathway for receptor shedding. Eur. J. Cell Biol. 1984, 35, 256–263. [Google Scholar]
- Pan, B.T.; Teng, K.; Wu, C.; Adam, M.; Johnstone, R.M. Electron microscopic evidence for externalization of the transferrin receptor in vesicular form in sheep reticulocytes. J. Cell Biol. 1985, 101, 942–948. [Google Scholar] [CrossRef]
- Raposo, G.; Nijman, H.W.; Stoorvogel, W.; Liejendekker, R.; Harding, C.V.; Melief, C.J.; Geuze, H.J. B lymphocytes secrete antigen-presenting vesicles. J. Exp. Med. 1996, 183, 1161–1172. [Google Scholar] [CrossRef]
- Zitvogel, L.; Regnault, A.; Lozier, A.; Wolfers, J.; Flament, C.; Tenza, D.; Ricciardi-Castagnoli, P.; Raposo, G.; Amigorena, S. Eradication of established murine tumors using a novel cell-free vaccine: Dendritic cell-derived exosomes. Nat. Med. 1998, 4, 594–600. [Google Scholar] [CrossRef]
- Harding, C.; Heuser, J.; Stahl, P. Receptor-mediated endocytosis of transferrin and recycling of the transferrin receptor in rat reticulocytes. J. Cell Biol. 1983, 97, 329–339. [Google Scholar] [CrossRef]
- Colombo, M.; Raposo, G.; Thery, C. Biogenesis, secretion, and intercellular interactions of exosomes and other extracellular vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Batista, B.S.; Eng, W.S.; Pilobello, K.T.; Hendricks-Munoz, K.D.; Mahal, L.K. Identification of a conserved glycan signature for microvesicles. J. Proteome Res. 2011, 10, 4624–4633. [Google Scholar] [CrossRef]
- Cocucci, E.; Meldolesi, J. Ectosomes and exosomes: Shedding the confusion between extracellular vesicles. Trends Cell Biol. 2015, 25, 364–372. [Google Scholar] [CrossRef]
- Hessvik, N.P.; Llorente, A. Current knowledge on exosome biogenesis and release. Cell. Mol. Life Sci. 2018, 75, 193–208. [Google Scholar] [CrossRef]
- Henne, W.M.; Stenmark, H.; Emr, S.D. Molecular mechanisms of the membrane sculpting ESCRT pathway. Cold Spring Harb. Perspect. Biol. 2013, 5, a016766. [Google Scholar] [CrossRef]
- Zhu, H.; Guariglia, S.; Yu, R.Y.; Li, W.; Brancho, D.; Peinado, H.; Lyden, D.; Salzer, J.; Bennett, C.; Chow, C.W. Mutation of SIMPLE in Charcot-Marie-Tooth 1C alters production of exosomes. Mol. Biol. Cell 2013, 24, 1619–1637. [Google Scholar] [CrossRef]
- Stuffers, S.; Sem Wegner, C.; Stenmark, H.; Brech, A. Multivesicular endosome biogenesis in the absence of ESCRTs. Traffic 2009, 10, 925–937. [Google Scholar] [CrossRef]
- McMahon, H.T.; Boucrot, E. Membrane curvature at a glance. J. Cell Sci. 2015, 128, 1065–1070. [Google Scholar] [CrossRef]
- Trajkovic, K.; Hsu, C.; Chiantia, S.; Rajendran, L.; Wenzel, D.; Wieland, F.; Schwille, P.; Brugger, B.; Simons, M. Ceramide triggers budding of exosome vesicles into multivesicular endosomes. Science 2008, 319, 1244–1247. [Google Scholar] [CrossRef]
- Conde-Vancells, J.; Rodriguez-Suarez, E.; Embade, N.; Gil, D.; Matthiesen, R.; Valle, M.; Elortza, F.; Lu, S.C.; Mato, J.M.; Falcon-Perez, J.M. Characterization and comprehensive proteome profiling of exosomes secreted by hepatocytes. J. Proteome Res. 2008, 7, 5157–5166. [Google Scholar] [CrossRef]
- Subra, C.; Grand, D.; Laulagnier, K.; Stella, A.; Lambeau, G.; Paillasse, M.; De Medina, P.; Monsarrat, B.; Perret, B.; Silvente-Poirot, S.; et al. Exosomes account for vesicle-mediated transcellular transport of activatable phospholipases and prostaglandins. J. Lipid Res. 2010, 51, 2105–2120. [Google Scholar] [CrossRef]
- Alvarez-Erviti, L.; Seow, Y.; Yin, H.; Betts, C.; Lakhal, S.; Wood, M.J. Delivery of siRNA to the mouse brain by systemic injection of targeted exosomes. Nat. Biotechnol. 2011, 29, 341–345. [Google Scholar] [CrossRef]
- Gurunathan, S.; Chapman-Shimshoni, D.; Trajkovic, S.; Gerst, J.E. Yeast exocytic v-SNAREs confer endocytosis. Mol. Biol. Cell 2000, 11, 3629–3643. [Google Scholar] [CrossRef]
- Gurunathan, S.; Marash, M.; Weinberger, A.; Gerst, J.E. t-SNARE phosphorylation regulates endocytosis in yeast. Mol. Biol. Cell 2002, 13, 1594–1607. [Google Scholar] [CrossRef]
- Mobius, W.; van Donselaar, E.; Ohno-Iwashita, Y.; Shimada, Y.; Heijnen, H.F.; Slot, J.W.; Geuze, H.J. Recycling compartments and the internal vesicles of multivesicular bodies harbor most of the cholesterol found in the endocytic pathway. Traffic 2003, 4, 222–231. [Google Scholar] [CrossRef]
- Valadi, H.; Ekstrom, K.; Bossios, A.; Sjostrand, M.; Lee, J.J.; Lotvall, J.O. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007, 9, 654–659. [Google Scholar] [CrossRef]
- Keerthikumar, S.; Chisanga, D.; Ariyaratne, D.; Al Saffar, H.; Anand, S.; Zhao, K.; Samuel, M.; Pathan, M.; Jois, M.; Chilamkurti, N.; et al. ExoCarta: A Web-Based Compendium of Exosomal Cargo. J. Mol. Biol. 2016, 428, 688–692. [Google Scholar] [CrossRef]
- Dickens, A.M.; Tovar, Y.R.L.B.; Yoo, S.W.; Trout, A.L.; Bae, M.; Kanmogne, M.; Megra, B.; Williams, D.W.; Witwer, K.W.; Gacias, M.; et al. Astrocyte-shed extracellular vesicles regulate the peripheral leukocyte response to inflammatory brain lesions. Sci. Signal. 2017, 10, eaai7696. [Google Scholar] [CrossRef]
- Howitt, J.; Hill, A.F. Exosomes in the Pathology of Neurodegenerative Diseases. J. Biol. Chem. 2016, 291, 26589–26597. [Google Scholar] [CrossRef]
- Tian, Y.; Li, S.; Song, J.; Ji, T.; Zhu, M.; Anderson, G.J.; Wei, J.; Nie, G. A doxorubicin delivery platform using engineered natural membrane vesicle exosomes for targeted tumor therapy. Biomaterials 2014, 35, 2383–2390. [Google Scholar] [CrossRef]
- Chen, T.S.; Arslan, F.; Yin, Y.; Tan, S.S.; Lai, R.C.; Choo, A.B.; Padmanabhan, J.; Lee, C.N.; de Kleijn, D.P.; Lim, S.K. Enabling a robust scalable manufacturing process for therapeutic exosomes through oncogenic immortalization of human ESC-derived MSCs. J. Transl. Med. 2011, 9, 47. [Google Scholar] [CrossRef]
- Yeo, R.W.; Lai, R.C.; Zhang, B.; Tan, S.S.; Yin, Y.; Teh, B.J.; Lim, S.K. Mesenchymal stem cell: An efficient mass producer of exosomes for drug delivery. Adv. Drug Deliv. Rev. 2013, 65, 336–341. [Google Scholar] [CrossRef]
- Gal, D.; MacDonald, P.C.; Porter, J.C.; Smith, J.W.; Simpson, E.R. Effect of cell density and confluency on cholesterol metabolism in cancer cells in monolayer culture. Cancer Res. 1981, 41, 473–477. [Google Scholar]
- Steinman, R.A.; Wentzel, A.; Lu, Y.; Stehle, C.; Grandis, J.R. Activation of Stat3 by cell confluence reveals negative regulation of Stat3 by cdk2. Oncogene 2003, 22, 3608–3615. [Google Scholar] [CrossRef]
- Hayes, O.; Ramos, B.; Rodriguez, L.L.; Aguilar, A.; Badia, T.; Castro, F.O. Cell confluency is as efficient as serum starvation for inducing arrest in the G0/G1 phase of the cell cycle in granulosa and fibroblast cells of cattle. Anim. Reprod. Sci. 2005, 87, 181–192. [Google Scholar] [CrossRef]
- Savina, A.; Furlan, M.; Vidal, M.; Colombo, M.I. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J. Biol. Chem. 2003, 278, 20083–20090. [Google Scholar] [CrossRef]
- Svensson, K.J.; Kucharzewska, P.; Christianson, H.C.; Skold, S.; Lofstedt, T.; Johansson, M.C.; Morgelin, M.; Bengzon, J.; Ruf, W.; Belting, M. Hypoxia triggers a proangiogenic pathway involving cancer cell microvesicles and PAR-2-mediated heparin-binding EGF signaling in endothelial cells. Proc. Natl. Acad. Sci. USA 2011, 108, 13147–13152. [Google Scholar] [CrossRef]
- King, H.W.; Michael, M.Z.; Gleadle, J.M. Hypoxic enhancement of exosome release by breast cancer cells. BMC Cancer 2012, 12, 421. [Google Scholar] [CrossRef]
- Kucharzewska, P.; Christianson, H.C.; Welch, J.E.; Svensson, K.J.; Fredlund, E.; Ringner, M.; Morgelin, M.; Bourseau-Guilmain, E.; Bengzon, J.; Belting, M. Exosomes reflect the hypoxic status of glioma cells and mediate hypoxia-dependent activation of vascular cells during tumor development. Proc. Natl. Acad. Sci. USA 2013, 110, 7312–7317. [Google Scholar] [CrossRef]
- Koumangoye, R.B.; Sakwe, A.M.; Goodwin, J.S.; Patel, T.; Ochieng, J. Detachment of breast tumor cells induces rapid secretion of exosomes which subsequently mediate cellular adhesion and spreading. PLoS ONE 2011, 6, e24234. [Google Scholar] [CrossRef]
- Lund, P.; Pilgaard, L.; Duroux, M.; Fink, T.; Zachar, V. Effect of growth media and serum replacements on the proliferation and differentiation of adipose-derived stem cells. Cytotherapy 2009, 11, 189–197. [Google Scholar] [CrossRef]
- Bryan, N.; Andrews, K.D.; Loughran, M.J.; Rhodes, N.P.; Hunt, J.A. Elucidating the contribution of the elemental composition of fetal calf serum to antigenic expression of primary human umbilical-vein endothelial cells in vitro. Biosci. Rep. 2011, 31, 199–210. [Google Scholar] [CrossRef]
- de Jong, O.G.; Verhaar, M.C.; Chen, Y.; Vader, P.; Gremmels, H.; Posthuma, G.; Schiffelers, R.M.; Gucek, M.; van Balkom, B.W. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles 2012, 1, 18396. [Google Scholar] [CrossRef]
- Li, J.; Lee, Y.; Johansson, H.J.; Mager, I.; Vader, P.; Nordin, J.Z.; Wiklander, O.P.; Lehtio, J.; Wood, M.J.; Andaloussi, S.E. Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J. Extracell. Vesicles 2015, 4, 26883. [Google Scholar] [CrossRef]
- Peterson, M.F.; Otoc, N.; Sethi, J.K.; Gupta, A.; Antes, T.J. Integrated systems for exosome investigation. Methods 2015, 87, 31–45. [Google Scholar] [CrossRef]
- Witwer, K.W.; Buzas, E.I.; Bemis, L.T.; Bora, A.; Lasser, C.; Lotvall, J.; Nolte-’t Hoen, E.N.; Piper, M.G.; Sivaraman, S.; Skog, J.; et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles 2013, 2, 20360. [Google Scholar] [CrossRef]
- Van Deun, J.; Mestdagh, P.; Sormunen, R.; Cocquyt, V.; Vermaelen, K.; Vandesompele, J.; Bracke, M.; De Wever, O.; Hendrix, A. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles 2014, 3, 24858. [Google Scholar] [CrossRef]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Zeringer, E.; Barta, T.; Li, M.; Vlassov, A.V. Strategies for isolation of exosomes. Cold Spring Harb. Protoc. 2015, 2015, 319–323. [Google Scholar] [CrossRef]
- Momen-Heravi, F.; Balaj, L.; Alian, S.; Trachtenberg, A.J.; Hochberg, F.H.; Skog, J.; Kuo, W.P. Impact of biofluid viscosity on size and sedimentation efficiency of the isolated microvesicles. Front. Physiol. 2012, 3, 162. [Google Scholar] [CrossRef]
- Sabapatha, A.; Gercel-Taylor, C.; Taylor, D.D. Specific isolation of placenta-derived exosomes from the circulation of pregnant women and their immunoregulatory consequences. Am. J. Reprod. Immunol. 2006, 56, 345–355. [Google Scholar] [CrossRef]
- Alvarez, M.L.; Khosroheidari, M.; Kanchi Ravi, R.; DiStefano, J.K. Comparison of protein, microRNA, and mRNA yields using different methods of urinary exosome isolation for the discovery of kidney disease biomarkers. Kidney Int. 2012, 82, 1024–1032. [Google Scholar] [CrossRef]
- Lai, R.C.; Arslan, F.; Lee, M.M.; Sze, N.S.; Choo, A.; Chen, T.S.; Salto-Tellez, M.; Timmers, L.; Lee, C.N.; El Oakley, R.M.; et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010, 4, 214–222. [Google Scholar] [CrossRef] [PubMed]
- Ibrahim, A.G.; Cheng, K.; Marban, E. Exosomes as critical agents of cardiac regeneration triggered by cell therapy. Stem Cell Rep. 2014, 2, 606–619. [Google Scholar] [CrossRef] [PubMed]
- Tauro, B.J.; Greening, D.W.; Mathias, R.A.; Ji, H.; Mathivanan, S.; Scott, A.M.; Simpson, R.J. Comparison of ultracentrifugation, density gradient separation, and immunoaffinity capture methods for isolating human colon cancer cell line LIM1863-derived exosomes. Methods 2012, 56, 293–304. [Google Scholar] [CrossRef]
- Grasso, L.; Wyss, R.; Weidenauer, L.; Thampi, A.; Demurtas, D.; Prudent, M.; Lion, N.; Vogel, H. Molecular screening of cancer-derived exosomes by surface plasmon resonance spectroscopy. Anal. Bioanal. Chem. 2015, 407, 5425–5432. [Google Scholar] [CrossRef]
- Zarovni, N.; Corrado, A.; Guazzi, P.; Zocco, D.; Lari, E.; Radano, G.; Muhhina, J.; Fondelli, C.; Gavrilova, J.; Chiesi, A. Integrated isolation and quantitative analysis of exosome shuttled proteins and nucleic acids using immunocapture approaches. Methods 2015, 87, 46–58. [Google Scholar] [CrossRef]
- Hong, C.S.; Muller, L.; Boyiadzis, M.; Whiteside, T.L. Isolation and characterization of CD34+ blast-derived exosomes in acute myeloid leukemia. PLoS ONE 2014, 9, e103310. [Google Scholar] [CrossRef]
- Lee, K.; Shao, H.; Weissleder, R.; Lee, H. Acoustic purification of extracellular microvesicles. ACS Nano 2015, 9, 2321–2327. [Google Scholar] [CrossRef]
- Davies, R.T.; Kim, J.; Jang, S.C.; Choi, E.J.; Gho, Y.S.; Park, J. Microfluidic filtration system to isolate extracellular vesicles from blood. Lab Chip 2012, 12, 5202–5210. [Google Scholar] [CrossRef] [PubMed]
- Contreras-Naranjo, J.C.; Wu, H.J.; Ugaz, V.M. Microfluidics for exosome isolation and analysis: Enabling liquid biopsy for personalized medicine. Lab Chip 2017, 17, 3558–3577. [Google Scholar] [CrossRef]
- Chen, C.; Skog, J.; Hsu, C.H.; Lessard, R.T.; Balaj, L.; Wurdinger, T.; Carter, B.S.; Breakefield, X.O.; Toner, M.; Irimia, D. Microfluidic isolation and transcriptome analysis of serum microvesicles. Lab Chip 2010, 10, 505–511. [Google Scholar] [CrossRef]
- Wang, Z.; Wu, H.J.; Fine, D.; Schmulen, J.; Hu, Y.; Godin, B.; Zhang, J.X.; Liu, X. Ciliated micropillars for the microfluidic-based isolation of nanoscale lipid vesicles. Lab Chip 2013, 13, 2879–2882. [Google Scholar] [CrossRef] [PubMed]
- Kanwar, S.S.; Dunlay, C.J.; Simeone, D.M.; Nagrath, S. Microfluidic device (ExoChip) for on-chip isolation, quantification and characterization of circulating exosomes. Lab Chip 2014, 14, 1891–1900. [Google Scholar] [CrossRef]
- Ashcroft, B.A.; de Sonneville, J.; Yuana, Y.; Osanto, S.; Bertina, R.; Kuil, M.E.; Oosterkamp, T.H. Determination of the size distribution of blood microparticles directly in plasma using atomic force microscopy and microfluidics. Biomed. Microdevices 2012, 14, 641–649. [Google Scholar] [CrossRef]
- Shao, H.; Chung, J.; Issadore, D. Diagnostic technologies for circulating tumour cells and exosomes. Biosci. Rep. 2015, 36, e00292. [Google Scholar] [CrossRef] [PubMed]
- de Necochea-Campion, R.; Gonda, A.; Kabagwira, J.; Mirshahidi, S.; Cao, H.; Reeves, M.E.; Wall, N.R. A practical approach to extracellular vesicle characterization among similar biological samples. Biomed. Phys. Eng. Express 2018, 4, 065013. [Google Scholar] [CrossRef]
- Szatanek, R.; Baj-Krzyworzeka, M.; Zimoch, J.; Lekka, M.; Siedlar, M.; Baran, J. The Methods of Choice for Extracellular Vesicles (EVs) Characterization. Int. J. Mol. Sci. 2017, 18. [Google Scholar] [CrossRef] [PubMed]
- Bryant, G.; Abeynayake, C.; Thomas, J.C. Improved particle size distribution measurements using multiangle dynamic light scattering. 2. Refinements and applications. Langmuir 1996, 12, 6224–6228. [Google Scholar] [CrossRef]
- Hoo, C.M.; Starostin, N.; West, P.; Mecartney, M.L. A comparison of atomic force microscopy (AFM) and dynamic light scattering (DLS) methods to characterize nanoparticle size distributions. J. Nanopart. Res. 2008, 10, 89–96. [Google Scholar] [CrossRef]
- Lawrie, A.S.; Albanyan, A.; Cardigan, R.A.; Mackie, I.J.; Harrison, P. Microparticle sizing by dynamic light scattering in fresh-frozen plasma. Vox Sang. 2009, 96, 206–212. [Google Scholar] [CrossRef]
- Gercel-Taylor, C.; Atay, S.; Tullis, R.H.; Kesimer, M.; Taylor, D.D. Nanoparticle analysis of circulating cell-derived vesicles in ovarian cancer patients. Anal. Biochem. 2012, 428, 44–53. [Google Scholar] [CrossRef]
- Anderson, W.; Lane, R.; Korbie, D.; Trau, M. Observations of Tunable Resistive Pulse Sensing for Exosome Analysis: Improving System Sensitivity and Stability. Langmuir 2015, 31, 6577–6587. [Google Scholar] [CrossRef]
- Vogel, R.; Pal, A.K.; Jambhrunkar, S.; Patel, P.; Thakur, S.S.; Reategui, E.; Parekh, H.S.; Saa, P.; Stassinopoulos, A.; Broom, M.F. High-Resolution Single Particle Zeta Potential Characterisation of Biological Nanoparticles using Tunable Resistive Pulse Sensing. Sci. Rep. 2017, 7, 17479. [Google Scholar] [CrossRef]
- Patko, D.; Gyorgy, B.; Nemeth, A.; Szabó-Taylor, K.; Kittel, A.; Buzás, E.I.; Horvath, R. Label-free optical monitoring of surface adhesion of extracellular vesicles by grating coupled interferometry. Sens. Actuators B Chem. 2013, 188, 697–701. [Google Scholar] [CrossRef]
- Shimbo, K.; Miyaki, S.; Ishitobi, H.; Kato, Y.; Kubo, T.; Shimose, S.; Ochi, M. Exosome-formed synthetic microRNA-143 is transferred to osteosarcoma cells and inhibits their migration. Biochem. Biophys. Res. Commun. 2014, 445, 381–387. [Google Scholar] [CrossRef]
- Katsuda, T.; Kosaka, N.; Takeshita, F.; Ochiya, T. The therapeutic potential of mesenchymal stem cell-derived extracellular vesicles. Proteomics 2013, 13, 1637–1653. [Google Scholar] [CrossRef]
- Binnig, G.; Quate, C.F.; Gerber, C. Atomic force microscope. Phys. Rev. Lett. 1986, 56, 930–933. [Google Scholar] [CrossRef]
- Yuana, Y.; Oosterkamp, T.H.; Bahatyrova, S.; Ashcroft, B.; Garcia Rodriguez, P.; Bertina, R.M.; Osanto, S. Atomic force microscopy: A novel approach to the detection of nanosized blood microparticles. J. Thromb. Haemost. 2010, 8, 315–323. [Google Scholar] [CrossRef]
- Sharma, S.; Zuniga, F.; Rice, G.E.; Perrin, L.C.; Hooper, J.D.; Salomon, C. Tumor-derived exosomes in ovarian cancer—Liquid biopsies for early detection and real-time monitoring of cancer progression. Oncotarget 2017, 8, 104687–104703. [Google Scholar] [CrossRef]
- Hardij, J.; Cecchet, F.; Berquand, A.; Gheldof, D.; Chatelain, C.; Mullier, F.; Chatelain, B.; Dogne, J.M. Characterisation of tissue factor-bearing extracellular vesicles with AFM: Comparison of air-tapping-mode AFM and liquid Peak Force AFM. J. Extracell. Vesicles 2013, 2. [Google Scholar] [CrossRef]
- Sharma, S.; Rasool, H.I.; Palanisamy, V.; Mathisen, C.; Schmidt, M.; Wong, D.T.; Gimzewski, J.K. Structural-mechanical characterization of nanoparticle exosomes in human saliva, using correlative AFM, FESEM, and force spectroscopy. ACS Nano 2010, 4, 1921–1926. [Google Scholar] [CrossRef]
- Zaborowski, M.P.; Balaj, L.; Breakefield, X.O.; Lai, C.P. Extracellular Vesicles: Composition, Biological Relevance, and Methods of Study. Bioscience 2015, 65, 783–797. [Google Scholar] [CrossRef]
- Pospichalova, V.; Svoboda, J.; Dave, Z.; Kotrbova, A.; Kaiser, K.; Klemova, D.; Ilkovics, L.; Hampl, A.; Crha, I.; Jandakova, E.; et al. Simplified protocol for flow cytometry analysis of fluorescently labeled exosomes and microvesicles using dedicated flow cytometer. J. Extracell. Vesicles 2015, 4, 25530. [Google Scholar] [CrossRef]
- Suarez, H.; Gamez-Valero, A.; Reyes, R.; Lopez-Martin, S.; Rodriguez, M.J.; Carrascosa, J.L.; Cabanas, C.; Borras, F.E.; Yanez-Mo, M. A bead-assisted flow cytometry method for the semi-quantitative analysis of Extracellular Vesicles. Sci. Rep. 2017, 7, 11271. [Google Scholar] [CrossRef]
- van der Vlist, E.J.; Nolte-’t Hoen, E.N.; Stoorvogel, W.; Arkesteijn, G.J.; Wauben, M.H. Fluorescent labeling of nano-sized vesicles released by cells and subsequent quantitative and qualitative analysis by high-resolution flow cytometry. Nat. Protoc. 2012, 7, 1311–1326. [Google Scholar] [CrossRef]
- Erdbrugger, U.; Rudy, C.K.; Etter, M.E.; Dryden, K.A.; Yeager, M.; Klibanov, A.L.; Lannigan, J. Imaging flow cytometry elucidates limitations of microparticle analysis by conventional flow cytometry. Cytom. A 2014, 85, 756–770. [Google Scholar] [CrossRef]
- Chandler, W.L.; Yeung, W.; Tait, J.F. A new microparticle size calibration standard for use in measuring smaller microparticles using a new flow cytometer. J. Thromb. Haemost. 2011, 9, 1216–1224. [Google Scholar] [CrossRef]
- Orozco, A.F.; Lewis, D.E. Flow cytometric analysis of circulating microparticles in plasma. Cytom. A 2010, 77, 502–514. [Google Scholar] [CrossRef]
- Smith, Z.J.; Lee, C.; Rojalin, T.; Carney, R.P.; Hazari, S.; Knudson, A.; Lam, K.; Saari, H.; Ibanez, E.L.; Viitala, T.; et al. Single exosome study reveals subpopulations distributed among cell lines with variability related to membrane content. J. Extracell. Vesicles 2015, 4, 28533. [Google Scholar] [CrossRef]
- He, M.; Crow, J.; Roth, M.; Zeng, Y.; Godwin, A.K. Integrated immunoisolation and protein analysis of circulating exosomes using microfluidic technology. Lab Chip 2014, 14, 3773–3780. [Google Scholar] [CrossRef]
- Ramirez, S.H.; Andrews, A.M.; Paul, D.; Pachter, J.S. Extracellular vesicles: Mediators and biomarkers of pathology along CNS barriers. Fluids Barriers CNS 2018, 15, 19. [Google Scholar] [CrossRef]
- Thery, C.; Zitvogel, L.; Amigorena, S. Exosomes: Composition, biogenesis and function. Nat. Rev. Immunol. 2002, 2, 569–579. [Google Scholar] [CrossRef]
- Adams, R.H.; Alitalo, K. Molecular regulation of angiogenesis and lymphangiogenesis. Nat. Rev. Mol. Cell Biol. 2007, 8, 464–478. [Google Scholar] [CrossRef]
- Bazigou, E.; Makinen, T. Flow control in our vessels: Vascular valves make sure there is no way back. Cell. Mol. Life Sci. 2013, 70, 1055–1066. [Google Scholar] [CrossRef]
- Kaur, S.; Chang, T.; Singh, S.P.; Lim, L.; Mannan, P.; Garfield, S.H.; Pendrak, M.L.; Soto-Pantoja, D.R.; Rosenberg, A.Z.; Jin, S.; et al. CD47 signaling regulates the immunosuppressive activity of VEGF in T cells. J. Immunol. 2014, 193, 3914–3924. [Google Scholar] [CrossRef] [PubMed]
- Hood, J.L.; Pan, H.; Lanza, G.M.; Wickline, S.A. Paracrine induction of endothelium by tumor exosomes. Lab. Investig. 2009, 89, 1317–1328. [Google Scholar] [CrossRef] [PubMed]
- Zhou, W.; Fong, M.Y.; Min, Y.; Somlo, G.; Liu, L.; Palomares, M.R.; Yu, Y.; Chow, A.; O’Connor, S.T.; Chin, A.R.; et al. Cancer-secreted miR-105 destroys vascular endothelial barriers to promote metastasis. Cancer Cell 2014, 25, 501–515. [Google Scholar] [CrossRef]
- Ma, T.; Chen, Y.; Chen, Y.; Meng, Q.; Sun, J.; Shao, L.; Yu, Y.; Huang, H.; Hu, Y.; Yang, Z. MicroRNA-132, Delivered by Mesenchymal Stem Cell-Derived Exosomes, Promote Angiogenesis in Myocardial Infarction. Stem Cells Int. 2018, 2018, 3290372. [Google Scholar] [CrossRef]
- Atay, S.; Gercel-Taylor, C.; Suttles, J.; Mor, G.; Taylor, D.D. Trophoblast-derived exosomes mediate monocyte recruitment and differentiation. Am. J. Reprod. Immunol. 2011, 65, 65–77. [Google Scholar] [CrossRef]
- Sidhu, S.S.; Mengistab, A.T.; Tauscher, A.N.; LaVail, J.; Basbaum, C. The microvesicle as a vehicle for EMMPRIN in tumor-stromal interactions. Oncogene 2004, 23, 956–963. [Google Scholar] [CrossRef]
- Grange, C.; Tapparo, M.; Collino, F.; Vitillo, L.; Damasco, C.; Deregibus, M.C.; Tetta, C.; Bussolati, B.; Camussi, G. Microvesicles released from human renal cancer stem cells stimulate angiogenesis and formation of lung premetastatic niche. Cancer Res. 2011, 71, 5346–5356. [Google Scholar] [CrossRef]
- Vrijsen, K.R.; Sluijter, J.P.; Schuchardt, M.W.; van Balkom, B.W.; Noort, W.A.; Chamuleau, S.A.; Doevendans, P.A. Cardiomyocyte progenitor cell-derived exosomes stimulate migration of endothelial cells. J. Cell Mol. Med. 2010, 14, 1064–1070. [Google Scholar] [CrossRef]
- Zhu, W.; Huang, L.; Li, Y.; Zhang, X.; Gu, J.; Yan, Y.; Xu, X.; Wang, M.; Qian, H.; Xu, W. Exosomes derived from human bone marrow mesenchymal stem cells promote tumor growth in vivo. Cancer Lett. 2012, 315, 28–37. [Google Scholar] [CrossRef]
- Cantaluppi, V.; Biancone, L.; Figliolini, F.; Beltramo, S.; Medica, D.; Deregibus, M.C.; Galimi, F.; Romagnoli, R.; Salizzoni, M.; Tetta, C.; et al. Microvesicles derived from endothelial progenitor cells enhance neoangiogenesis of human pancreatic islets. Cell Transplant. 2012, 21, 1305–1320. [Google Scholar] [CrossRef]
- Chen, T.S.; Lai, R.C.; Lee, M.M.; Choo, A.B.; Lee, C.N.; Lim, S.K. Mesenchymal stem cell secretes microparticles enriched in pre-microRNAs. Nucleic Acids Res. 2010, 38, 215–224. [Google Scholar] [CrossRef]
- Yoo, J.K.; Kim, J.; Choi, S.J.; Noh, H.M.; Kwon, Y.D.; Yoo, H.; Yi, H.S.; Chung, H.M.; Kim, J.K. Discovery and characterization of novel microRNAs during endothelial differentiation of human embryonic stem cells. Stem Cells Dev. 2012, 21, 2049–2057. [Google Scholar] [CrossRef]
- Zhang, B.; Wang, M.; Gong, A.; Zhang, X.; Wu, X.; Zhu, Y.; Shi, H.; Wu, L.; Zhu, W.; Qian, H. HucMSC-exosome mediated-Wnt4 signaling is required for cutaneous wound healing. Stem Cells 2015, 33, 2158–2168. [Google Scholar] [CrossRef]
- Nakamura, Y.; Miyaki, S.; Ishitobi, H.; Matsuyama, S.; Nakasa, T.; Kamei, N.; Akimoto, T.; Higashi, Y.; Ochi, M. Mesenchymal-stem-cell-derived exosomes accelerate skeletal muscle regeneration. FEBS Lett. 2015, 589, 1257–1265. [Google Scholar] [CrossRef]
- Atienzar-Aroca, S.; Flores-Bellver, M.; Serrano-Heras, G.; Martinez-Gil, N.; Barcia, J.M.; Aparicio, S.; Perez-Cremades, D.; Garcia-Verdugo, J.M.; Diaz-Llopis, M.; Romero, F.J.; et al. Oxidative stress in retinal pigment epithelium cells increases exosome secretion and promotes angiogenesis in endothelial cells. J. Cell Mol. Med. 2016, 20, 1457–1466. [Google Scholar] [CrossRef]
- Ribatti, D. Cancer stem cells and tumor angiogenesis. Cancer Lett. 2012, 321, 13–17. [Google Scholar] [CrossRef]
- Sugimori, M.; Hayakawa, Y.; Boman, B.M.; Fields, J.Z.; Awaji, M.; Kozano, H.; Tamura, R.; Yamamoto, S.; Ogata, T.; Yamada, M.; et al. Discovery of Power-Law Growth in the Self-Renewal of Heterogeneous Glioma Stem Cell Populations. PLoS ONE 2015, 10, e0135760. [Google Scholar] [CrossRef]
- Sun, X.; Ma, X.; Wang, J.; Zhao, Y.; Wang, Y.; Bihl, J.C.; Chen, Y.; Jiang, C. Glioma stem cells-derived exosomes promote the angiogenic ability of endothelial cells through miR-21/VEGF signal. Oncotarget 2017, 8, 36137–36148. [Google Scholar] [CrossRef]
- Conigliaro, A.; Costa, V.; Lo Dico, A.; Saieva, L.; Buccheri, S.; Dieli, F.; Manno, M.; Raccosta, S.; Mancone, C.; Tripodi, M.; et al. CD90+ liver cancer cells modulate endothelial cell phenotype through the release of exosomes containing H19 lncRNA. Mol. Cancer 2015, 14, 155. [Google Scholar] [CrossRef]
- Merino-Gonzalez, C.; Zuniga, F.A.; Escudero, C.; Ormazabal, V.; Reyes, C.; Nova-Lamperti, E.; Salomon, C.; Aguayo, C. Mesenchymal Stem Cell-Derived Extracellular Vesicles Promote Angiogenesis: Potencial Clinical Application. Front. Physiol. 2016, 7, 24. [Google Scholar] [CrossRef]
- Kerr, J.F.; Wyllie, A.H.; Currie, A.R. Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 1972, 26, 239–257. [Google Scholar] [CrossRef]
- Thery, C.; Boussac, M.; Veron, P.; Ricciardi-Castagnoli, P.; Raposo, G.; Garin, J.; Amigorena, S. Proteomic analysis of dendritic cell-derived exosomes: A secreted subcellular compartment distinct from apoptotic vesicles. J. Immunol. 2001, 166, 7309–7318. [Google Scholar] [CrossRef]
- Gregory, C.D.; Pound, J.D. Microenvironmental influences of apoptosis in vivo and in vitro. Apoptosis 2010, 15, 1029–1049. [Google Scholar] [CrossRef]
- Caruso, S.; Poon, I.K.H. Apoptotic Cell-Derived Extracellular Vesicles: More Than Just Debris. Front. Immunol. 2018, 9, 1486. [Google Scholar] [CrossRef]
- Atkin-Smith, G.K.; Poon, I.K.H. Disassembly of the Dying: Mechanisms and Functions. Trends Cell Biol. 2017, 27, 151–162. [Google Scholar] [CrossRef]
- Atkin-Smith, G.K.; Tixeira, R.; Paone, S.; Mathivanan, S.; Collins, C.; Liem, M.; Goodall, K.J.; Ravichandran, K.S.; Hulett, M.D.; Poon, I.K. A novel mechanism of generating extracellular vesicles during apoptosis via a beads-on-a-string membrane structure. Nat. Commun. 2015, 6, 7439. [Google Scholar] [CrossRef]
- Vallabhaneni, K.C.; Hassler, M.Y.; Abraham, A.; Whitt, J.; Mo, Y.Y.; Atfi, A.; Pochampally, R. Mesenchymal Stem/Stromal Cells under Stress Increase Osteosarcoma Migration and Apoptosis Resistance via Extracellular Vesicle Mediated Communication. PLoS ONE 2016, 11, e0166027. [Google Scholar] [CrossRef]
- Zaharie, F.; Muresan, M.S.; Petrushev, B.; Berce, C.; Gafencu, G.A.; Selicean, S.; Jurj, A.; Cojocneanu-Petric, R.; Lisencu, C.I.; Pop, L.A.; et al. Exosome-Carried microRNA-375 Inhibits Cell Progression and Dissemination via Bcl-2 Blocking in Colon Cancer. J. Gastrointest. Liver Dis. 2015, 24, 435–443. [Google Scholar] [CrossRef]
- Clayton, A.; Mitchell, J.P.; Court, J.; Linnane, S.; Mason, M.D.; Tabi, Z. Human tumor-derived exosomes down-modulate NKG2D expression. J. Immunol. 2008, 180, 7249–7258. [Google Scholar] [CrossRef]
- Andreola, G.; Rivoltini, L.; Castelli, C.; Huber, V.; Perego, P.; Deho, P.; Squarcina, P.; Accornero, P.; Lozupone, F.; Lugini, L.; et al. Induction of lymphocyte apoptosis by tumor cell secretion of FasL-bearing microvesicles. J. Exp. Med. 2002, 195, 1303–1316. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Paine, M.S.; Brooks, A.M.; McCubrey, J.A.; Renegar, R.H.; Wang, R.; Terrian, D.M. Senescence-associated exosome release from human prostate cancer cells. Cancer Res. 2008, 68, 7864–7871. [Google Scholar] [CrossRef]
- Aharon, A.; Tamari, T.; Brenner, B. Monocyte-derived microparticles and exosomes induce procoagulant and apoptotic effects on endothelial cells. Thromb. Haemost. 2008, 100, 878–885. [Google Scholar]
- Bruno, S.; Collino, F.; Deregibus, M.C.; Grange, C.; Tetta, C.; Camussi, G. Microvesicles derived from human bone marrow mesenchymal stem cells inhibit tumor growth. Stem Cells Dev. 2013, 22, 758–771. [Google Scholar] [CrossRef]
- Shenoda, B.B.; Ajit, S.K. Modulation of Immune Responses by Exosomes Derived from Antigen-Presenting Cells. Clin. Med. Insights Pathol. 2016, 9, 1–8. [Google Scholar] [CrossRef]
- Bianchi, M.E. DAMPs, PAMPs and alarmins: All we need to know about danger. J. Leukoc. Biol. 2007, 81, 1–5. [Google Scholar] [CrossRef]
- Mogensen, T.H. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin. Microbiol. Rev. 2009, 22, 240–273. [Google Scholar] [CrossRef]
- McCall, C.E.; El Gazzar, M.; Liu, T.; Vachharajani, V.; Yoza, B. Epigenetics, bioenergetics, and microRNA coordinate gene-specific reprogramming during acute systemic inflammation. J. Leukoc. Biol. 2011, 90, 439–446. [Google Scholar] [CrossRef]
- McDonald, M.K.; Tian, Y.; Qureshi, R.A.; Gormley, M.; Ertel, A.; Gao, R.; Aradillas Lopez, E.; Alexander, G.M.; Sacan, A.; Fortina, P.; et al. Functional significance of macrophage-derived exosomes in inflammation and pain. Pain 2014, 155, 1527–1539. [Google Scholar] [CrossRef]
- Choudhuri, K.; Llodra, J.; Roth, E.W.; Tsai, J.; Gordo, S.; Wucherpfennig, K.W.; Kam, L.C.; Stokes, D.L.; Dustin, M.L. Polarized release of T-cell-receptor-enriched microvesicles at the immunological synapse. Nature 2014, 507, 118–123. [Google Scholar] [CrossRef]
- Yanez-Mo, M.; Siljander, P.R.; Andreu, Z.; Zavec, A.B.; Borras, F.E.; Buzas, E.I.; Buzas, K.; Casal, E.; Cappello, F.; Carvalho, J.; et al. Biological properties of extracellular vesicles and their physiological functions. J. Extracell. Vesicles 2015, 4, 27066. [Google Scholar] [CrossRef]
- Segura, E.; Amigorena, S.; Thery, C. Mature dendritic cells secrete exosomes with strong ability to induce antigen-specific effector immune responses. Blood. Cells Mol. Dis. 2005, 35, 89–93. [Google Scholar] [CrossRef] [PubMed]
- Admyre, C.; Johansson, S.M.; Paulie, S.; Gabrielsson, S. Direct exosome stimulation of peripheral human T cells detected by ELISPOT. Eur. J. Immunol. 2006, 36, 1772–1781. [Google Scholar] [CrossRef]
- Lindenbergh, M.F.S.; Stoorvogel, W. Antigen Presentation by Extracellular Vesicles from Professional Antigen-Presenting Cells. Annu. Rev. Immunol. 2018, 36, 435–459. [Google Scholar] [CrossRef]
- Colino, J.; Snapper, C.M. Exosomes from bone marrow dendritic cells pulsed with diphtheria toxoid preferentially induce type 1 antigen-specific IgG responses in naive recipients in the absence of free antigen. J. Immunol. 2006, 177, 3757–3762. [Google Scholar] [CrossRef]
- Del Cacho, E.; Gallego, M.; Lee, S.H.; Lillehoj, H.S.; Quilez, J.; Lillehoj, E.P.; Sanchez-Acedo, C. Induction of protective immunity against Eimeria tenella infection using antigen-loaded dendritic cells (DC) and DC-derived exosomes. Vaccine 2011, 29, 3818–3825. [Google Scholar] [CrossRef]
- Giri, P.K.; Schorey, J.S. Exosomes derived from M. Bovis BCG infected macrophages activate antigen-specific CD4+ and CD8+ T cells in vitro and in vivo. PLoS ONE 2008, 3, e2461. [Google Scholar] [CrossRef]
- Ramachandra, L.; Qu, Y.; Wang, Y.; Lewis, C.J.; Cobb, B.A.; Takatsu, K.; Boom, W.H.; Dubyak, G.R.; Harding, C.V. Mycobacterium tuberculosis synergizes with ATP to induce release of microvesicles and exosomes containing major histocompatibility complex class II molecules capable of antigen presentation. Infect. Immun. 2010, 78, 5116–5125. [Google Scholar] [CrossRef]
- Asano, K.; Nabeyama, A.; Miyake, Y.; Qiu, C.H.; Kurita, A.; Tomura, M.; Kanagawa, O.; Fujii, S.; Tanaka, M. CD169-positive macrophages dominate antitumor immunity by crosspresenting dead cell-associated antigens. Immunity 2011, 34, 85–95. [Google Scholar] [CrossRef] [PubMed]
- Cheng, Y.; Schorey, J.S. Exosomes carrying mycobacterial antigens can protect mice against Mycobacterium tuberculosis infection. Eur. J. Immunol. 2013, 43, 3279–3290. [Google Scholar] [CrossRef] [PubMed]
- Schorey, J.S.; Harding, C.V. Extracellular vesicles and infectious diseases: New complexity to an old story. J. Clin. Investig. 2016, 126, 1181–1189. [Google Scholar] [CrossRef]
- Van Niel, G.; Mallegol, J.; Bevilacqua, C.; Candalh, C.; Brugiere, S.; Tomaskovic-Crook, E.; Heath, J.K.; Cerf-Bensussan, N.; Heyman, M. Intestinal epithelial exosomes carry MHC class II/peptides able to inform the immune system in mice. Gut 2003, 52, 1690–1697. [Google Scholar] [CrossRef]
- Mallegol, J.; Van Niel, G.; Lebreton, C.; Lepelletier, Y.; Candalh, C.; Dugave, C.; Heath, J.K.; Raposo, G.; Cerf-Bensussan, N.; Heyman, M. T84-intestinal epithelial exosomes bear MHC class II/peptide complexes potentiating antigen presentation by dendritic cells. Gastroenterology 2007, 132, 1866–1876. [Google Scholar] [CrossRef] [PubMed]
- Buning, J.; von Smolinski, D.; Tafazzoli, K.; Zimmer, K.P.; Strobel, S.; Apostolaki, M.; Kollias, G.; Heath, J.K.; Ludwig, D.; Gebert, A. Multivesicular bodies in intestinal epithelial cells: Responsible for MHC class II-restricted antigen processing and origin of exosomes. Immunology 2008, 125, 510–521. [Google Scholar] [CrossRef]
- Monleon, I.; Martinez-Lorenzo, M.J.; Monteagudo, L.; Lasierra, P.; Taules, M.; Iturralde, M.; Pineiro, A.; Larrad, L.; Alava, M.A.; Naval, J.; et al. Differential secretion of Fas ligand- or APO2 ligand/TNF-related apoptosis-inducing ligand-carrying microvesicles during activation-induced death of human T cells. J. Immunol. 2001, 167, 6736–6744. [Google Scholar] [CrossRef]
- Alonso, R.; Rodriguez, M.C.; Pindado, J.; Merino, E.; Merida, I.; Izquierdo, M. Diacylglycerol kinase alpha regulates the secretion of lethal exosomes bearing Fas ligand during activation-induced cell death of T lymphocytes. J. Biol. Chem. 2005, 280, 28439–28450. [Google Scholar] [CrossRef]
- Busch, A.; Quast, T.; Keller, S.; Kolanus, W.; Knolle, P.; Altevogt, P.; Limmer, A. Transfer of T cell surface molecules to dendritic cells upon CD4+ T cell priming involves two distinct mechanisms. J. Immunol. 2008, 181, 3965–3973. [Google Scholar] [CrossRef]
- Sugimoto, M.A.; Sousa, L.P.; Pinho, V.; Perretti, M.; Teixeira, M.M. Resolution of Inflammation: What Controls Its Onset? Front. Immunol. 2016, 7, 160. [Google Scholar] [CrossRef] [PubMed]
- Scrivo, R.; Vasile, M.; Bartosiewicz, I.; Valesini, G. Inflammation as “common soil” of the multifactorial diseases. Autoimmun. Rev. 2011, 10, 369–374. [Google Scholar] [CrossRef] [PubMed]
- Console, L.; Scalise, M.; Indiveri, C. Exosomes in inflammation and role as biomarkers. Clin. Chim. Acta 2019, 488, 165–171. [Google Scholar] [CrossRef]
- Colotta, F.; Allavena, P.; Sica, A.; Garlanda, C.; Mantovani, A. Cancer-related inflammation, the seventh hallmark of cancer: Links to genetic instability. Carcinogenesis 2009, 30, 1073–1081. [Google Scholar] [CrossRef] [PubMed]
- Grivennikov, S.I.; Greten, F.R.; Karin, M. Immunity, inflammation, and cancer. Cell 2010, 140, 883–899. [Google Scholar] [CrossRef]
- Wu, L.; Zhang, X.; Zhang, B.; Shi, H.; Yuan, X.; Sun, Y.; Pan, Z.; Qian, H.; Xu, W. Exosomes derived from gastric cancer cells activate NF-kappaB pathway in macrophages to promote cancer progression. Tumour Biol. 2016, 37, 12169–12180. [Google Scholar] [CrossRef] [PubMed]
- Chow, A.; Zhou, W.; Liu, L.; Fong, M.Y.; Champer, J.; Van Haute, D.; Chin, A.R.; Ren, X.; Gugiu, B.G.; Meng, Z. Macrophage immunomodulation by breast cancer-derived exosomes requires Toll-like receptor 2-mediated activation of NF-κB. Sci. Rep. 2014, 4, 5750. [Google Scholar] [CrossRef] [PubMed]
- Fabbri, M.; Paone, A.; Calore, F.; Galli, R.; Gaudio, E.; Santhanam, R.; Lovat, F.; Fadda, P.; Mao, C.; Nuovo, G.J.; et al. MicroRNAs bind to Toll-like receptors to induce prometastatic inflammatory response. Proc. Natl. Acad. Sci. USA 2012, 109, E2110–E2116. [Google Scholar] [CrossRef] [PubMed]
- Abusamra, A.J.; Zhong, Z.; Zheng, X.; Li, M.; Ichim, T.E.; Chin, J.L.; Min, W.P. Tumor exosomes expressing Fas ligand mediate CD8+ T-cell apoptosis. Blood. Cells Mol. Dis. 2005, 35, 169–173. [Google Scholar] [CrossRef] [PubMed]
- Ye, S.B.; Li, Z.L.; Luo, D.H.; Huang, B.J.; Chen, Y.S.; Zhang, X.S.; Cui, J.; Zeng, Y.X.; Li, J. Tumor-derived exosomes promote tumor progression and T-cell dysfunction through the regulation of enriched exosomal microRNAs in human nasopharyngeal carcinoma. Oncotarget 2014, 5, 5439–5452. [Google Scholar] [CrossRef]
- Dupaul-Chicoine, J.; Dagenais, M.; Saleh, M. Crosstalk between the intestinal microbiota and the innate immune system in intestinal homeostasis and inflammatory bowel disease. Inflamm. Bowel. Dis. 2013, 19, 2227–2237. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, S.; Feldbrugge, L.; Csizmadia, E.; Mitsuhashi, M.; Robson, S.C.; Moss, A.C. Luminal Extracellular Vesicles (EVs) in Inflammatory Bowel Disease (IBD) Exhibit Proinflammatory Effects on Epithelial Cells and Macrophages. Inflamm. Bowel. Dis. 2016, 22, 1587–1595. [Google Scholar] [CrossRef]
- Lv, L.L.; Feng, Y.; Wen, Y.; Wu, W.J.; Ni, H.F.; Li, Z.L.; Zhou, L.T.; Wang, B.; Zhang, J.D.; Crowley, S.D.; et al. Exosomal CCL2 from Tubular Epithelial Cells Is Critical for Albumin-Induced Tubulointerstitial Inflammation. J. Am. Soc. Nephrol. 2018, 29, 919–935. [Google Scholar] [CrossRef] [PubMed]
- Deng, Z.B.; Poliakov, A.; Hardy, R.W.; Clements, R.; Liu, C.; Liu, Y.; Wang, J.; Xiang, X.; Zhang, S.; Zhuang, X.; et al. Adipose tissue exosome-like vesicles mediate activation of macrophage-induced insulin resistance. Diabetes 2009, 58, 2498–2505. [Google Scholar] [CrossRef] [PubMed]
- Ferrante, S.C.; Nadler, E.P.; Pillai, D.K.; Hubal, M.J.; Wang, Z.; Wang, J.M.; Gordish-Dressman, H.; Koeck, E.; Sevilla, S.; Wiles, A.A.; et al. Adipocyte-derived exosomal miRNAs: A novel mechanism for obesity-related disease. Pediatr. Res. 2015, 77, 447–454. [Google Scholar] [CrossRef] [PubMed]
- Kato, T.; Miyaki, S.; Ishitobi, H.; Nakamura, Y.; Nakasa, T.; Lotz, M.K.; Ochi, M. Exosomes from IL-1beta stimulated synovial fibroblasts induce osteoarthritic changes in articular chondrocytes. Arthritis Res. Ther. 2014, 16, R163. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Barres, B.A. Microglia and macrophages in brain homeostasis and disease. Nat. Rev. Immunol. 2018, 18, 225–242. [Google Scholar] [CrossRef] [PubMed]
- Gupta, A.; Pulliam, L. Exosomes as mediators of neuroinflammation. J. Neuroinflamm. 2014, 11, 68. [Google Scholar] [CrossRef] [PubMed]
- Logozzi, M.; De Milito, A.; Lugini, L.; Borghi, M.; Calabro, L.; Spada, M.; Perdicchio, M.; Marino, M.L.; Federici, C.; Iessi, E.; et al. High levels of exosomes expressing CD63 and caveolin-1 in plasma of melanoma patients. PLoS ONE 2009, 4, e5219. [Google Scholar] [CrossRef]
- Welker, M.W.; Reichert, D.; Susser, S.; Sarrazin, C.; Martinez, Y.; Herrmann, E.; Zeuzem, S.; Piiper, A.; Kronenberger, B. Soluble serum CD81 is elevated in patients with chronic hepatitis C and correlates with alanine aminotransferase serum activity. PLoS ONE 2012, 7, e30796. [Google Scholar] [CrossRef]
- Skog, J.; Wurdinger, T.; van Rijn, S.; Meijer, D.H.; Gainche, L.; Sena-Esteves, M.; Curry, W.T., Jr.; Carter, B.S.; Krichevsky, A.M.; Breakefield, X.O. Glioblastoma microvesicles transport RNA and proteins that promote tumour growth and provide diagnostic biomarkers. Nat. Cell Biol. 2008, 10, 1470–1476. [Google Scholar] [CrossRef]
- Rajendran, L.; Honsho, M.; Zahn, T.R.; Keller, P.; Geiger, K.D.; Verkade, P.; Simons, K. Alzheimer’s disease beta-amyloid peptides are released in association with exosomes. Proc. Natl. Acad. Sci. USA 2006, 103, 11172–11177. [Google Scholar] [CrossRef]
- Saman, S.; Kim, W.; Raya, M.; Visnick, Y.; Miro, S.; Saman, S.; Jackson, B.; McKee, A.C.; Alvarez, V.E.; Lee, N.C.; et al. Exosome-associated tau is secreted in tauopathy models and is selectively phosphorylated in cerebrospinal fluid in early Alzheimer disease. J. Biol. Chem. 2012, 287, 3842–3849. [Google Scholar] [CrossRef]
- Goetzl, E.J.; Boxer, A.; Schwartz, J.B.; Abner, E.L.; Petersen, R.C.; Miller, B.L.; Kapogiannis, D. Altered lysosomal proteins in neural-derived plasma exosomes in preclinical Alzheimer disease. Neurology 2015, 85, 40–47. [Google Scholar] [CrossRef]
- Zhou, H.; Cheruvanky, A.; Hu, X.; Matsumoto, T.; Hiramatsu, N.; Cho, M.E.; Berger, A.; Leelahavanichkul, A.; Doi, K.; Chawla, L.S.; et al. Urinary exosomal transcription factors, a new class of biomarkers for renal disease. Kidney Int. 2008, 74, 613–621. [Google Scholar] [CrossRef]
- Smalley, D.M.; Sheman, N.E.; Nelson, K.; Theodorescu, D. Isolation and identification of potential urinary microparticle biomarkers of bladder cancer. J. Proteome Res. 2008, 7, 2088–2096. [Google Scholar] [CrossRef]
- Melo, S.A.; Luecke, L.B.; Kahlert, C.; Fernandez, A.F.; Gammon, S.T.; Kaye, J.; LeBleu, V.S.; Mittendorf, E.A.; Weitz, J.; Rahbari, N.; et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature 2015, 523, 177–182. [Google Scholar] [CrossRef]
- Chen, X.; Liang, H.; Zhang, J.; Zen, K.; Zhang, C.Y. Secreted microRNAs: A new form of intercellular communication. Trends Cell Biol. 2012, 22, 125–132. [Google Scholar] [CrossRef]
- Calin, G.A.; Dumitru, C.D.; Shimizu, M.; Bichi, R.; Zupo, S.; Noch, E.; Aldler, H.; Rattan, S.; Keating, M.; Rai, K.; et al. Frequent deletions and down-regulation of micro- RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc. Natl. Acad. Sci. USA 2002, 99, 15524–15529. [Google Scholar] [CrossRef]
- Taylor, D.D.; Gercel-Taylor, C. MicroRNA signatures of tumor-derived exosomes as diagnostic biomarkers of ovarian cancer. Gynecol. Oncol. 2008, 110, 13–21. [Google Scholar] [CrossRef]
- Rabinowits, G.; Gercel-Taylor, C.; Day, J.M.; Taylor, D.D.; Kloecker, G.H. Exosomal microRNA: A diagnostic marker for lung cancer. Clin. Lung. Cancer 2009, 10, 42–46. [Google Scholar] [CrossRef] [PubMed]
- Silva, J.; Garcia, V.; Zaballos, A.; Provencio, M.; Lombardia, L.; Almonacid, L.; Garcia, J.M.; Dominguez, G.; Pena, C.; Diaz, R.; et al. Vesicle-related microRNAs in plasma of nonsmall cell lung cancer patients and correlation with survival. Eur. Respir. J. 2011, 37, 617–623. [Google Scholar] [CrossRef]
- Mitchell, P.S.; Parkin, R.K.; Kroh, E.M.; Fritz, B.R.; Wyman, S.K.; Pogosova-Agadjanyan, E.L.; Peterson, A.; Noteboom, J.; O’Briant, K.C.; Allen, A.; et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl. Acad. Sci. USA 2008, 105, 10513–10518. [Google Scholar] [CrossRef]
- Lau, C.; Kim, Y.; Chia, D.; Spielmann, N.; Eibl, G.; Elashoff, D.; Wei, F.; Lin, Y.L.; Moro, A.; Grogan, T.; et al. Role of pancreatic cancer-derived exosomes in salivary biomarker development. J. Biol. Chem. 2013, 288, 26888–26897. [Google Scholar] [CrossRef]
- Keller, S.; Rupp, C.; Stoeck, A.; Runz, S.; Fogel, M.; Lugert, S.; Hager, H.D.; Abdel-Bakky, M.S.; Gutwein, P.; Altevogt, P. CD24 is a marker of exosomes secreted into urine and amniotic fluid. Kidney Int. 2007, 72, 1095–1102. [Google Scholar] [CrossRef]
- Shi, W.; Yang, J.; Li, S.; Shan, X.; Liu, X.; Hua, H.; Zhao, C.; Feng, Z.; Cai, Z.; Zhang, L.; et al. Potential involvement of miR-375 in the premalignant progression of oral squamous cell carcinoma mediated via transcription factor KLF5. Oncotarget 2015, 6, 40172–40185. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, S.; Sakata, Y.; Suna, S.; Nakatani, D.; Usami, M.; Hara, M.; Kitamura, T.; Hamasaki, T.; Nanto, S.; Kawahara, Y.; et al. Circulating p53-responsive microRNAs are predictive indicators of heart failure after acute myocardial infarction. Circ. Res. 2013, 113, 322–326. [Google Scholar] [CrossRef]
- Li, J.; Yu, J.; Zhang, H.; Wang, B.; Guo, H.; Bai, J.; Wang, J.; Dong, Y.; Zhao, Y.; Wang, Y. Exosomes-Derived MiR-302b Suppresses Lung Cancer Cell Proliferation and Migration via TGFbetaRII Inhibition. Cell. Physiol. Biochem. 2016, 38, 1715–1726. [Google Scholar] [CrossRef] [PubMed]
- Cazzoli, R.; Buttitta, F.; Di Nicola, M.; Malatesta, S.; Marchetti, A.; Rom, W.N.; Pass, H.I. microRNAs derived from circulating exosomes as noninvasive biomarkers for screening and diagnosing lung cancer. J. Thorac. Oncol. 2013, 8, 1156–1162. [Google Scholar] [CrossRef] [PubMed]
- Thind, A.; Wilson, C. Exosomal miRNAs as cancer biomarkers and therapeutic targets. J. Extracell. Vesicles 2016, 5, 31292. [Google Scholar] [CrossRef]
- Conner, S.D.; Schmid, S.L. Regulated portals of entry into the cell. Nature 2003, 422, 37–44. [Google Scholar] [CrossRef] [PubMed]
- Grapp, M.; Wrede, A.; Schweizer, M.; Hüwel, S.; Galla, H.-J.; Snaidero, N.; Simons, M.; Bückers, J.; Low, P.S.; Urlaub, H.; et al. Choroid plexus transcytosis and exosome shuttling deliver folate into brain parenchyma. Nat. Commun. 2013, 4, 2123. [Google Scholar] [CrossRef] [PubMed]
- Feng, D.; Zhao, W.-L.; Ye, Y.-Y.; Bai, X.-C.; Liu, R.-Q.; Chang, L.-F.; Zhou, Q.; Sui, S.-F. Cellular Internalization of Exosomes Occurs Through Phagocytosis. Traffic 2010, 11, 675–687. [Google Scholar] [CrossRef] [PubMed]
- Svensson, K.J.; Christianson, H.C.; Wittrup, A.; Bourseau-Guilmain, E.; Lindqvist, E.; Svensson, L.M.; Morgelin, M.; Belting, M. Exosome uptake depends on ERK1/2-heat shock protein 27 signaling and lipid Raft-mediated endocytosis negatively regulated by caveolin-1. J. Biol. Chem. 2013, 288, 17713–17724. [Google Scholar] [CrossRef]
- Tian, T.; Zhu, Y.L.; Zhou, Y.Y.; Liang, G.F.; Wang, Y.Y.; Hu, F.H.; Xiao, Z.D. Exosome uptake through clathrin-mediated endocytosis and macropinocytosis and mediating miR-21 delivery. J. Biol. Chem. 2014, 289, 22258–22267. [Google Scholar] [CrossRef] [PubMed]
- Nakase, I.; Kobayashi, N.B.; Takatani-Nakase, T.; Yoshida, T. Active macropinocytosis induction by stimulation of epidermal growth factor receptor and oncogenic Ras expression potentiates cellular uptake efficacy of exosomes. Sci. Rep. 2015, 5, 10300. [Google Scholar] [CrossRef]
- Huang, A.; Dong, J.; Li, S.; Wang, C.; Ding, H.; Li, H.; Su, X.; Ge, X.; Sun, L.; Bai, C.; et al. Exosomal transfer of vasorin expressed in hepatocellular carcinoma cells promotes migration of human umbilical vein endothelial cells. Int. J. Biol. Sci. 2015, 11, 961–969. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Chopp, M.; Liu, X.S.; Katakowski, M.; Wang, X.; Tian, X.; Wu, D.; Zhang, Z.G. Exosomes Derived from Mesenchymal Stromal Cells Promote Axonal Growth of Cortical Neurons. Mol. Neurobiol. 2017, 54, 2659–2673. [Google Scholar] [CrossRef] [PubMed]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH is a key factor for exosome traffic in tumor cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef]
- Chivet, M.; Javalet, C.; Laulagnier, K.; Blot, B.; Hemming, F.J.; Sadoul, R. Exosomes secreted by cortical neurons upon glutamatergic synapse activation specifically interact with neurons. J. Extracell. Vesicles 2014, 3, 24722. [Google Scholar] [CrossRef]
- Montecalvo, A.; Larregina, A.T.; Shufesky, W.J.; Stolz, D.B.; Sullivan, M.L.; Karlsson, J.M.; Baty, C.J.; Gibson, G.A.; Erdos, G.; Wang, Z.; et al. Mechanism of transfer of functional microRNAs between mouse dendritic cells via exosomes. Blood 2012, 119, 756–766. [Google Scholar] [CrossRef]
- Banizs, A.B.; Huang, T.; Nakamoto, R.K.; Shi, W.; He, J. Endocytosis Pathways of Endothelial Cell Derived Exosomes. Mol. Pharm. 2018. [Google Scholar] [CrossRef]
- Maia, J.; Caja, S.; Strano Moraes, M.C.; Couto, N.; Costa-Silva, B. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol. 2018, 6, 18. [Google Scholar] [CrossRef]
- Janowska-Wieczorek, A.; Wysoczynski, M.; Kijowski, J.; Marquez-Curtis, L.; Machalinski, B.; Ratajczak, J.; Ratajczak, M.Z. Microvesicles derived from activated platelets induce metastasis and angiogenesis in lung cancer. Int. J. Cancer 2005, 113, 752–760. [Google Scholar] [CrossRef]
- Falanga, A.; Marchetti, M.; Vignoli, A.; Balducci, D. Clotting mechanisms and cancer: Implications in thrombus formation and tumor progression. Clin. Adv. Hematol. Oncol. 2003, 1, 673–678. [Google Scholar]
- Raimondo, S.; Saieva, L.; Corrado, C.; Fontana, S.; Flugy, A.; Rizzo, A.; De Leo, G.; Alessandro, R. Chronic myeloid leukemia-derived exosomes promote tumor growth through an autocrine mechanism. Cell Commun. Signal. 2015, 13, 8. [Google Scholar] [CrossRef]
- Qu, J.L.; Qu, X.J.; Zhao, M.F.; Teng, Y.E.; Zhang, Y.; Hou, K.Z.; Jiang, Y.H.; Yang, X.H.; Liu, Y.P. Gastric cancer exosomes promote tumour cell proliferation through PI3K/Akt and MAPK/ERK activation. Dig. Liver Dis. 2009, 41, 875–880. [Google Scholar] [CrossRef]
- Pan, L.; Liang, W.; Fu, M.; Huang, Z.H.; Li, X.; Zhang, W.; Zhang, P.; Qian, H.; Jiang, P.C.; Xu, W.R.; et al. Exosomes-mediated transfer of long noncoding RNA ZFAS1 promotes gastric cancer progression. J. Cancer Res. Clin. Oncol. 2017, 143, 991–1004. [Google Scholar] [CrossRef]
- Yang, L.; Wu, X.H.; Wang, D.; Luo, C.L.; Chen, L.X. Bladder cancer cell-derived exosomes inhibit tumor cell apoptosis and induce cell proliferation in vitro. Mol. Med. Rep. 2013, 8, 1272–1278. [Google Scholar] [CrossRef]
- Matsumoto, A.; Takahashi, Y.; Nishikawa, M.; Sano, K.; Morishita, M.; Charoenviriyakul, C.; Saji, H.; Takakura, Y. Accelerated growth of B16BL6 tumor in mice through efficient uptake of their own exosomes by B16BL6 cells. Cancer Sci. 2017, 108, 1803–1810. [Google Scholar] [CrossRef]
- Ramteke, A.; Ting, H.; Agarwal, C.; Mateen, S.; Somasagara, R.; Hussain, A.; Graner, M.; Frederick, B.; Agarwal, R.; Deep, G. Exosomes secreted under hypoxia enhance invasiveness and stemness of prostate cancer cells by targeting adherens junction molecules. Mol. Carcinog. 2015, 54, 554–565. [Google Scholar] [CrossRef]
- Gutkin, A.; Uziel, O.; Beery, E.; Nordenberg, J.; Pinchasi, M.; Goldvaser, H.; Henick, S.; Goldberg, M.; Lahav, M. Tumor cells derived exosomes contain hTERT mRNA and transform nonmalignant fibroblasts into telomerase positive cells. Oncotarget 2016, 7, 59173–59188. [Google Scholar] [CrossRef]
- Takasugi, M.; Okada, R.; Takahashi, A.; Virya Chen, D.; Watanabe, S.; Hara, E. Small extracellular vesicles secreted from senescent cells promote cancer cell proliferation through EphA2. Nat. Commun. 2017, 8, 15729. [Google Scholar] [CrossRef]
- Zhao, H.; Yang, L.; Baddour, J.; Achreja, A.; Bernard, V.; Moss, T.; Marini, J.C.; Tudawe, T.; Seviour, E.G.; San Lucas, F.A.; et al. Tumor microenvironment derived exosomes pleiotropically modulate cancer cell metabolism. eLife 2016, 5, e10250. [Google Scholar] [CrossRef]
- Huang, Z.; Feng, Y. Exosomes Derived from Hypoxic Colorectal Cancer Cells Promote Angiogenesis Through Wnt4-Induced beta-Catenin Signaling in Endothelial Cells. Oncol. Res. 2017, 25, 651–661. [Google Scholar] [CrossRef]
- Rashed, H.M.; Bayraktar, E.; KHelal, G.; Abd-Ellah, M.; Amero, P.; Chavez-Reyes, A.; Rodriguez-Aguayo, C. Exosomes: From Garbage Bins to Promising Therapeutic Targets. Int. J. Mol. Sci. 2017, 18, 538. [Google Scholar] [CrossRef]
- Narayanan, R.; Huang, C.C.; Ravindran, S. Hijacking the Cellular Mail: Exosome Mediated Differentiation of Mesenchymal Stem Cells. Stem Cells Int. 2016, 2016, 3808674. [Google Scholar] [CrossRef]
- Ismail, N.; Wang, Y.; Dakhlallah, D.; Moldovan, L.; Agarwal, K.; Batte, K.; Shah, P.; Wisler, J.; Eubank, T.D.; Tridandapani, S.; et al. Macrophage microvesicles induce macrophage differentiation and miR-223 transfer. Blood 2013, 121, 984–995. [Google Scholar] [CrossRef]
- Ekström, K.; Omar, O.; Granéli, C.; Wang, X.; Vazirisani, F.; Thomsen, P. Monocyte Exosomes Stimulate the Osteogenic Gene Expression of Mesenchymal Stem Cells. PLoS ONE 2013, 8, e75227. [Google Scholar] [CrossRef]
- Gutzeit, C.; Nagy, N.; Gentile, M.; Lyberg, K.; Gumz, J.; Vallhov, H.; Puga, I.; Klein, E.; Gabrielsson, S.; Cerutti, A.; et al. Exosomes derived from Burkitt’s lymphoma cell lines induce proliferation, differentiation, and class-switch recombination in B cells. J. Immunol. 2014, 192, 5852–5862. [Google Scholar] [CrossRef]
- Solberg, L.B.; Stang, E.; Brorson, S.H.; Andersson, G.; Reinholt, F.P. Tartrate-resistant acid phosphatase (TRAP) co-localizes with receptor activator of NF-KB ligand (RANKL) and osteoprotegerin (OPG) in lysosomal-associated membrane protein 1 (LAMP1)-positive vesicles in rat osteoblasts and osteocytes. Histochem. Cell Biol. 2015, 143, 195–207. [Google Scholar] [CrossRef]
- Huynh, N.; VonMoss, L.; Smith, D.; Rahman, I.; Felemban, M.F.; Zuo, J.; Rody, W.J., Jr.; McHugh, K.P.; Holliday, L.S. Characterization of Regulatory Extracellular Vesicles from Osteoclasts. J. Dent. Res. 2016, 95, 673–679. [Google Scholar] [CrossRef]
- Cui, Y.; Luan, J.; Li, H.; Zhou, X.; Han, J. Exosomes derived from mineralizing osteoblasts promote ST2 cell osteogenic differentiation by alteration of microRNA expression. FEBS Lett. 2016, 590, 185–192. [Google Scholar] [CrossRef]
- Wang, X.; Omar, O.; Vazirisani, F.; Thomsen, P.; Ekstrom, K. Mesenchymal stem cell-derived exosomes have altered microRNA profiles and induce osteogenic differentiation depending on the stage of differentiation. PLoS ONE 2018, 13, e0193059. [Google Scholar] [CrossRef]
- Liao, F.-L.; Tan, L.; Liu, H.; Wang, J.-J.; Ma, X.-t.; Zhao, B.; Chen, Y.; Bihl, J.; Yang, Y.; Chen, R.-L. Hematopoietic stem cell-derived exosomes promote hematopoietic differentiation of mouse embryonic stem cells in vitro via inhibiting the miR126/Notch1 pathway. Acta Pharmacol. Sin. 2017, 39, 552. [Google Scholar] [CrossRef]
- Goulet, C.R.; Champagne, A.; Bernard, G.; Vandal, D.; Chabaud, S.; Pouliot, F.; Bolduc, S. Cancer-associated fibroblasts induce epithelial–mesenchymal transition of bladder cancer cells through paracrine IL-6 signalling. BMC Cancer 2019, 19, 137. [Google Scholar] [CrossRef]
- Xu, T.; Xu, M.; Bai, J.; Lin, J.; Yu, B.; Liu, Y.; Guo, X.; Shen, J.; Sun, H.; Hao, Y.; et al. Tenocyte-derived exosomes induce the tenogenic differentiation of mesenchymal stem cells through TGF-beta. Cytotechnology 2019, 71, 57–65. [Google Scholar] [CrossRef]
- Willis, G.R.; Kourembanas, S.; Mitsialis, S.A. Toward Exosome-Based Therapeutics: Isolation, Heterogeneity, and Fit-for-Purpose Potency. Front. Cardiovasc. Med. 2017, 4, 63. [Google Scholar] [CrossRef]
- Teng, X.; Chen, L.; Chen, W.; Yang, J.; Yang, Z.; Shen, Z. Mesenchymal Stem Cell-Derived Exosomes Improve the Microenvironment of Infarcted Myocardium Contributing to Angiogenesis and Anti-Inflammation. Cell. Physiol. Biochem. 2015, 37, 2415–2424. [Google Scholar] [CrossRef]
- Lou, G.; Chen, Z.; Zheng, M.; Liu, Y. Mesenchymal stem cell-derived exosomes as a new therapeutic strategy for liver diseases. Exp. Mol. Med. 2017, 49, e346. [Google Scholar] [CrossRef]
- Lee, C.; Mitsialis, S.A.; Aslam, M.; Vitali, S.H.; Vergadi, E.; Konstantinou, G.; Sdrimas, K.; Fernandez-Gonzalez, A.; Kourembanas, S. Exosomes mediate the cytoprotective action of mesenchymal stromal cells on hypoxia-induced pulmonary hypertension. Circulation 2012, 126, 2601–2611. [Google Scholar] [CrossRef]
- Xin, H.; Li, Y.; Cui, Y.; Yang, J.J.; Zhang, Z.G.; Chopp, M. Systemic administration of exosomes released from mesenchymal stromal cells promote functional recovery and neurovascular plasticity after stroke in rats. J. Cereb. Blood Flow Metab. 2013, 33, 1711–1715. [Google Scholar] [CrossRef]
- Yu, B.; Kim, H.W.; Gong, M.; Wang, J.; Millard, R.W.; Wang, Y.; Ashraf, M.; Xu, M. Exosomes secreted from GATA-4 overexpressing mesenchymal stem cells serve as a reservoir of anti-apoptotic microRNAs for cardioprotection. Int. J. Cardiol. 2015, 182, 349–360. [Google Scholar] [CrossRef]
- Wang, Y.; Zhang, L.; Li, Y.; Chen, L.; Wang, X.; Guo, W.; Zhang, X.; Qin, G.; He, S.H.; Zimmerman, A.; et al. Exosomes/microvesicles from induced pluripotent stem cells deliver cardioprotective miRNAs and prevent cardiomyocyte apoptosis in the ischemic myocardium. Int. J. Cardiol. 2015, 192, 61–69. [Google Scholar] [CrossRef]
- Khan, M.; Nickoloff, E.; Abramova, T.; Johnson, J.; Verma, S.K.; Krishnamurthy, P.; Mackie, A.R.; Vaughan, E.; Garikipati, V.N.; Benedict, C.; et al. Embryonic stem cell-derived exosomes promote endogenous repair mechanisms and enhance cardiac function following myocardial infarction. Circ. Res. 2015, 117, 52–64. [Google Scholar] [CrossRef]
- Vrijsen, K.R.; Maring, J.A.; Chamuleau, S.A.; Verhage, V.; Mol, E.A.; Deddens, J.C.; Metz, C.H.; Lodder, K.; van Eeuwijk, E.C.; van Dommelen, S.M.; et al. Exosomes from Cardiomyocyte Progenitor Cells and Mesenchymal Stem Cells Stimulate Angiogenesis Via EMMPRIN. Adv. Healthc. Mater. 2016, 5, 2555–2565. [Google Scholar] [CrossRef]
- Baglio, S.R.; Devescovi, V.; Granchi, D.; Baldini, N. MicroRNA expression profiling of human bone marrow mesenchymal stem cells during osteogenic differentiation reveals Osterix regulation by miR-31. Gene 2013, 527, 321–331. [Google Scholar] [CrossRef]
- Arntz, O.J.; Pieters, B.C.; Oliveira, M.C.; Broeren, M.G.; Bennink, M.B.; de Vries, M.; van Lent, P.L.; Koenders, M.I.; van den Berg, W.B.; van der Kraan, P.M.; et al. Oral administration of bovine milk derived extracellular vesicles attenuates arthritis in two mouse models. Mol. Nutr. Food Res. 2015, 59, 1701–1712. [Google Scholar] [CrossRef]
- Andre, F.; Chaput, N.; Schartz, N.E.; Flament, C.; Aubert, N.; Bernard, J.; Lemonnier, F.; Raposo, G.; Escudier, B.; Hsu, D.H.; et al. Exosomes as potent cell-free peptide-based vaccine. I. Dendritic cell-derived exosomes transfer functional MHC class I/peptide complexes to dendritic cells. J. Immunol. 2004, 172, 2126–2136. [Google Scholar] [CrossRef]
- Morishita, M.; Takahashi, Y.; Matsumoto, A.; Nishikawa, M.; Takakura, Y. Exosome-based tumor antigens-adjuvant co-delivery utilizing genetically engineered tumor cell-derived exosomes with immunostimulatory CpG DNA. Biomaterials 2016, 111, 55–65. [Google Scholar] [CrossRef]
- Morishita, M.; Takahashi, Y.; Nishikawa, M.; Ariizumi, R.; Takakura, Y. Enhanced Class I Tumor Antigen Presentation via Cytosolic Delivery of Exosomal Cargos by Tumor-Cell-Derived Exosomes Displaying a pH-Sensitive Fusogenic Peptide. Mol. Pharm. 2017, 14, 4079–4086. [Google Scholar] [CrossRef] [PubMed]
- Doeppner, T.R.; Herz, J.; Gorgens, A.; Schlechter, J.; Ludwig, A.K.; Radtke, S.; de Miroschedji, K.; Horn, P.A.; Giebel, B.; Hermann, D.M. Extracellular Vesicles Improve Post-Stroke Neuroregeneration and Prevent Postischemic Immunosuppression. Stem Cells Transl. Med. 2015, 4, 1131–1143. [Google Scholar] [CrossRef] [PubMed]
- Bruno, S.; Grange, C.; Deregibus, M.C.; Calogero, R.A.; Saviozzi, S.; Collino, F.; Morando, L.; Busca, A.; Falda, M.; Bussolati, B.; et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol. 2009, 20, 1053–1067. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Yan, Y.; Wang, B.; Qian, H.; Zhang, X.; Shen, L.; Wang, M.; Zhou, Y.; Zhu, W.; Li, W.; et al. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells Dev. 2013, 22, 845–854. [Google Scholar] [CrossRef]
- Lamparski, H.G.; Metha-Damani, A.; Yao, J.Y.; Patel, S.; Hsu, D.H.; Ruegg, C.; Le Pecq, J.B. Production and characterization of clinical grade exosomes derived from dendritic cells. J. Immunol. Methods 2002, 270, 211–226. [Google Scholar] [CrossRef]
- Gutierrez-Vazquez, C.; Villarroya-Beltri, C.; Mittelbrunn, M.; Sanchez-Madrid, F. Transfer of extracellular vesicles during immune cell-cell interactions. Immunol. Rev. 2013, 251, 125–142. [Google Scholar] [CrossRef]
- Hadla, M.; Palazzolo, S.; Corona, G.; Caligiuri, I.; Canzonieri, V.; Toffoli, G.; Rizzolio, F. Exosomes increase the therapeutic index of doxorubicin in breast and ovarian cancer mouse models. Nanomedicine 2016, 11, 2431–2441. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef]
- Lou, G.; Song, X.; Yang, F.; Wu, S.; Wang, J.; Chen, Z.; Liu, Y. Exosomes derived from miR-122-modified adipose tissue-derived MSCs increase chemosensitivity of hepatocellular carcinoma. J. Hematol. Oncol. 2015, 8, 122. [Google Scholar] [CrossRef] [PubMed]
- Katakowski, M.; Buller, B.; Zheng, X.; Lu, Y.; Rogers, T.; Osobamiro, O.; Shu, W.; Jiang, F.; Chopp, M. Exosomes from marrow stromal cells expressing miR-146b inhibit glioma growth. Cancer Lett. 2013, 335, 201–204. [Google Scholar] [CrossRef] [PubMed]
- Shtam, T.A.; Kovalev, R.A.; Varfolomeeva, E.Y.; Makarov, E.M.; Kil, Y.V.; Filatov, M.V. Exosomes are natural carriers of exogenous siRNA to human cells in vitro. Cell Commun. Signal. 2013, 11, 88. [Google Scholar] [CrossRef]
- Jiang, Z.Z.; Liu, Y.M.; Niu, X.; Yin, J.Y.; Hu, B.; Guo, S.C.; Fan, Y.; Wang, Y.; Wang, N.S. Exosomes secreted by human urine-derived stem cells could prevent kidney complications from type I diabetes in rats. Stem Cell Res. Ther. 2016, 7, 24. [Google Scholar] [CrossRef]
- Venkat, P.; Chopp, M.; Zacharek, A.; Cui, C.; Zhang, L.; Li, Q.; Lu, M.; Zhang, T.; Liu, A.; Chen, J. White matter damage and glymphatic dysfunction in a model of vascular dementia in rats with no prior vascular pathologies. Neurobiol. Aging 2017, 50, 96–106. [Google Scholar] [CrossRef]
- Nakano, M.; Nagaishi, K.; Konari, N.; Saito, Y.; Chikenji, T.; Mizue, Y.; Fujimiya, M. Bone marrow-derived mesenchymal stem cells improve diabetes-induced cognitive impairment by exosome transfer into damaged neurons and astrocytes. Sci. Rep. 2016, 6, 24805. [Google Scholar] [CrossRef]
- Arslan, F.; Lai, R.C.; Smeets, M.B.; Akeroyd, L.; Choo, A.; Aguor, E.N.; Timmers, L.; van Rijen, H.V.; Doevendans, P.A.; Pasterkamp, G.; et al. Mesenchymal stem cell-derived exosomes increase ATP levels, decrease oxidative stress and activate PI3K/Akt pathway to enhance myocardial viability and prevent adverse remodeling after myocardial ischemia/reperfusion injury. Stem Cell Res. 2013, 10, 301–312. [Google Scholar] [CrossRef]
- Chen, L.; Wang, Y.; Pan, Y.; Zhang, L.; Shen, C.; Qin, G.; Ashraf, M.; Weintraub, N.; Ma, G.; Tang, Y. Cardiac progenitor-derived exosomes protect ischemic myocardium from acute ischemia/reperfusion injury. Biochem. Biophys. Res. Commun. 2013, 431, 566–571. [Google Scholar] [CrossRef]
- Munagala, R.; Aqil, F.; Jeyabalan, J.; Gupta, R.C. Bovine milk-derived exosomes for drug delivery. Cancer Lett. 2016, 371, 48–61. [Google Scholar] [CrossRef]
- Sun, D.; Zhuang, X.; Xiang, X.; Liu, Y.; Zhang, S.; Liu, C.; Barnes, S.; Grizzle, W.; Miller, D.; Zhang, H.G. A novel nanoparticle drug delivery system: The anti-inflammatory activity of curcumin is enhanced when encapsulated in exosomes. Mol. Ther. 2010, 18, 1606–1614. [Google Scholar] [CrossRef]
- Yamashita, T.; Takahashi, Y.; Takakura, Y. Possibility of Exosome-Based Therapeutics and Challenges in Production of Exosomes Eligible for Therapeutic Application. Biol. Pharm. Bull. 2018, 41, 835–842. [Google Scholar] [CrossRef]
- Yamashita, T.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation. Eur. J. Pharm. Biopharm. 2016, 98, 1–8. [Google Scholar] [CrossRef]
- Charoenviriyakul, C.; Takahashi, Y.; Morishita, M.; Matsumoto, A.; Nishikawa, M.; Takakura, Y. Cell type-specific and common characteristics of exosomes derived from mouse cell lines: Yield, physicochemical properties, and pharmacokinetics. Eur. J. Pharm. Sci. 2017, 96, 316–322. [Google Scholar] [CrossRef]
- Ramirez, M.I.; Amorim, M.G.; Gadelha, C.; Milic, I.; Welsh, J.A.; Freitas, V.M.; Nawaz, M.; Akbar, N.; Couch, Y.; Makin, L.; et al. Technical challenges of working with extracellular vesicles. Nanoscale 2018, 10, 881–906. [Google Scholar] [CrossRef] [PubMed]
- Watson, D.C.; Bayik, D.; Srivatsan, A.; Bergamaschi, C.; Valentin, A.; Niu, G.; Bear, J.; Monninger, M.; Sun, M.; Morales-Kastresana, A.; et al. Efficient production and enhanced tumor delivery of engineered extracellular vesicles. Biomaterials 2016, 105, 195–205. [Google Scholar] [CrossRef]
- Harmati, M.; Tarnai, Z.; Decsi, G.; Kormondi, S.; Szegletes, Z.; Janovak, L.; Dekany, I.; Saydam, O.; Gyukity-Sebestyen, E.; Dobra, G.; et al. Stressors alter intercellular communication and exosome profile of nasopharyngeal carcinoma cells. J. Oral Pathol. Med. 2017, 46, 259–266. [Google Scholar] [CrossRef]
- Bobrie, A.; Colombo, M.; Krumeich, S.; Raposo, G.; Thery, C. Diverse subpopulations of vesicles secreted by different intracellular mechanisms are present in exosome preparations obtained by differential ultracentrifugation. J. Extracell. Vesicles 2012, 1. [Google Scholar] [CrossRef] [PubMed]
- Lesniak, A.; Fenaroli, F.; Monopoli, M.P.; Aberg, C.; Dawson, K.A.; Salvati, A. Effects of the presence or absence of a protein corona on silica nanoparticle uptake and impact on cells. ACS Nano 2012, 6, 5845–5857. [Google Scholar] [CrossRef]
- Willms, E.; Johansson, H.J.; Mager, I.; Lee, Y.; Blomberg, K.E.; Sadik, M.; Alaarg, A.; Smith, C.I.; gLehtio, J.; El Andaloussi, S.; et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep. 2016, 6, 22519. [Google Scholar] [CrossRef] [PubMed]
- Kowal, J.; Arras, G.; Colombo, M.; Jouve, M.; Morath, J.P.; Primdal-Bengtson, B.; Dingli, F.; Loew, D.; Tkach, M.; Thery, C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA 2016, 113, E968–E977. [Google Scholar] [CrossRef] [PubMed]
- Bosch, S.; de Beaurepaire, L.; Allard, M.; Mosser, M.; Heichette, C.; Chretien, D.; Jegou, D.; Bach, J.M. Trehalose prevents aggregation of exosomes and cryodamage. Sci. Rep. 2016, 6, 36162. [Google Scholar] [CrossRef]
- Lamichhane, T.N.; Leung, C.A.; Douti, L.Y.; Jay, S.M. Ethanol Induces Enhanced Vascularization Bioactivity of Endothelial Cell-Derived Extracellular Vesicles via Regulation of MicroRNAs and Long Non-Coding RNAs. Sci. Rep. 2017, 7, 13794. [Google Scholar] [CrossRef]
- Cui, G.H.; Wu, J.; Mou, F.F.; Xie, W.H.; Wang, F.B.; Wang, Q.L.; Fang, J.; Xu, Y.W.; Dong, Y.R.; Liu, J.R.; et al. Exosomes derived from hypoxia-preconditioned mesenchymal stromal cells ameliorate cognitive decline by rescuing synaptic dysfunction and regulating inflammatory responses in APP/PS1 mice. FASEB J. 2018, 32, 654–668. [Google Scholar] [CrossRef]





© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Qasim, M.; Kim, J.-H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells 2019, 8, 307. https://doi.org/10.3390/cells8040307
Gurunathan S, Kang M-H, Jeyaraj M, Qasim M, Kim J-H. Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells. 2019; 8(4):307. https://doi.org/10.3390/cells8040307
Chicago/Turabian StyleGurunathan, Sangiliyandi, Min-Hee Kang, Muniyandi Jeyaraj, Muhammad Qasim, and Jin-Hoi Kim. 2019. "Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes" Cells 8, no. 4: 307. https://doi.org/10.3390/cells8040307
APA StyleGurunathan, S., Kang, M.-H., Jeyaraj, M., Qasim, M., & Kim, J.-H. (2019). Review of the Isolation, Characterization, Biological Function, and Multifarious Therapeutic Approaches of Exosomes. Cells, 8(4), 307. https://doi.org/10.3390/cells8040307




