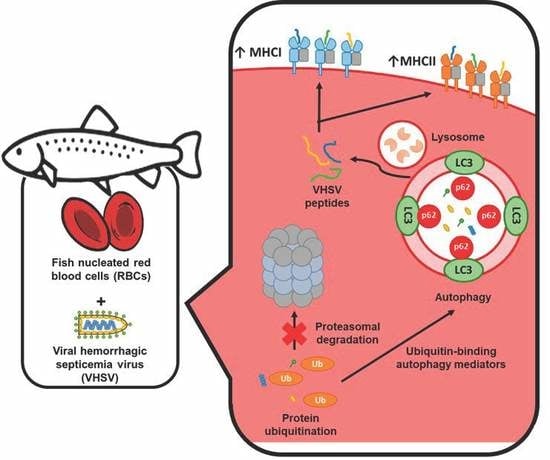
Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-presenting Cell Markers
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Cell Cultures and Virus
2.3. Antibodies
2.4. Viral Exposure Assays
2.5. Rainbow Trout Challenge with VHSV
2.6. Proteasome Activity Assay
2.7. RNA Isolation and cDNA Synthesis
2.8. Transcriptome Analysis
2.9. Proteome Analysis
2.10. Pathway Enrichment Analysis
2.11. Semi-quantitative PCR
2.12. Agarose Gel Electrophoresis
2.13. Gene Expression by RT-qPCR
2.14. Extracellular Immunofluorescence Staining
2.15. Intracellular Immunofluorescence Staining
2.16. Transmission Electron Microscopy (TEM)
2.17. In situ Proximity Ligation Assay (PLA)
2.18. Western Blot
2.19. Software and Statistics
3. Results
3.1. Transcriptomic Analysis Indicated Up-Regulation of Antigen-Processing-Related Molecules in Ex Vivo VHSV-Exposed Rainbow Trout RBCs
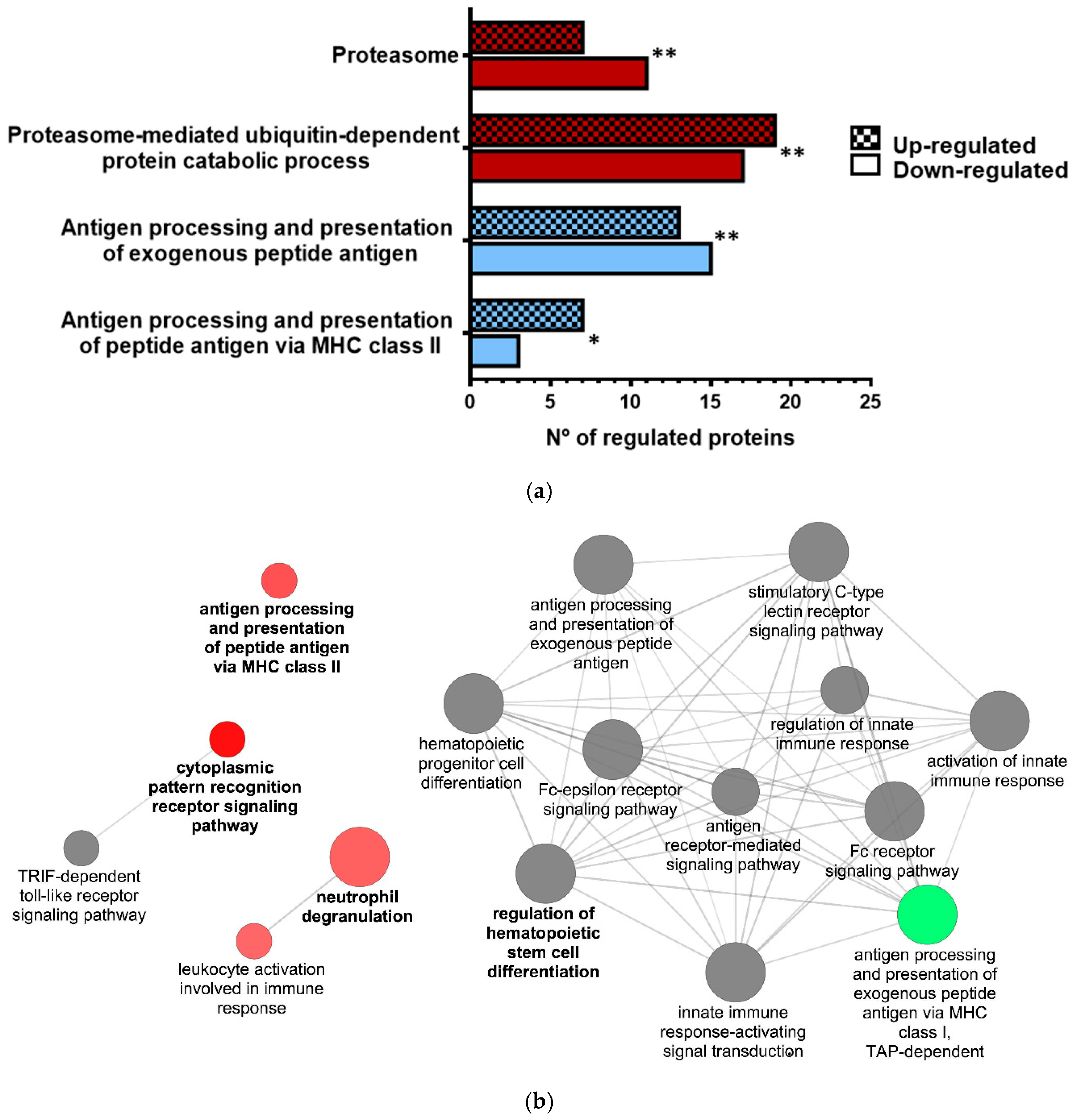
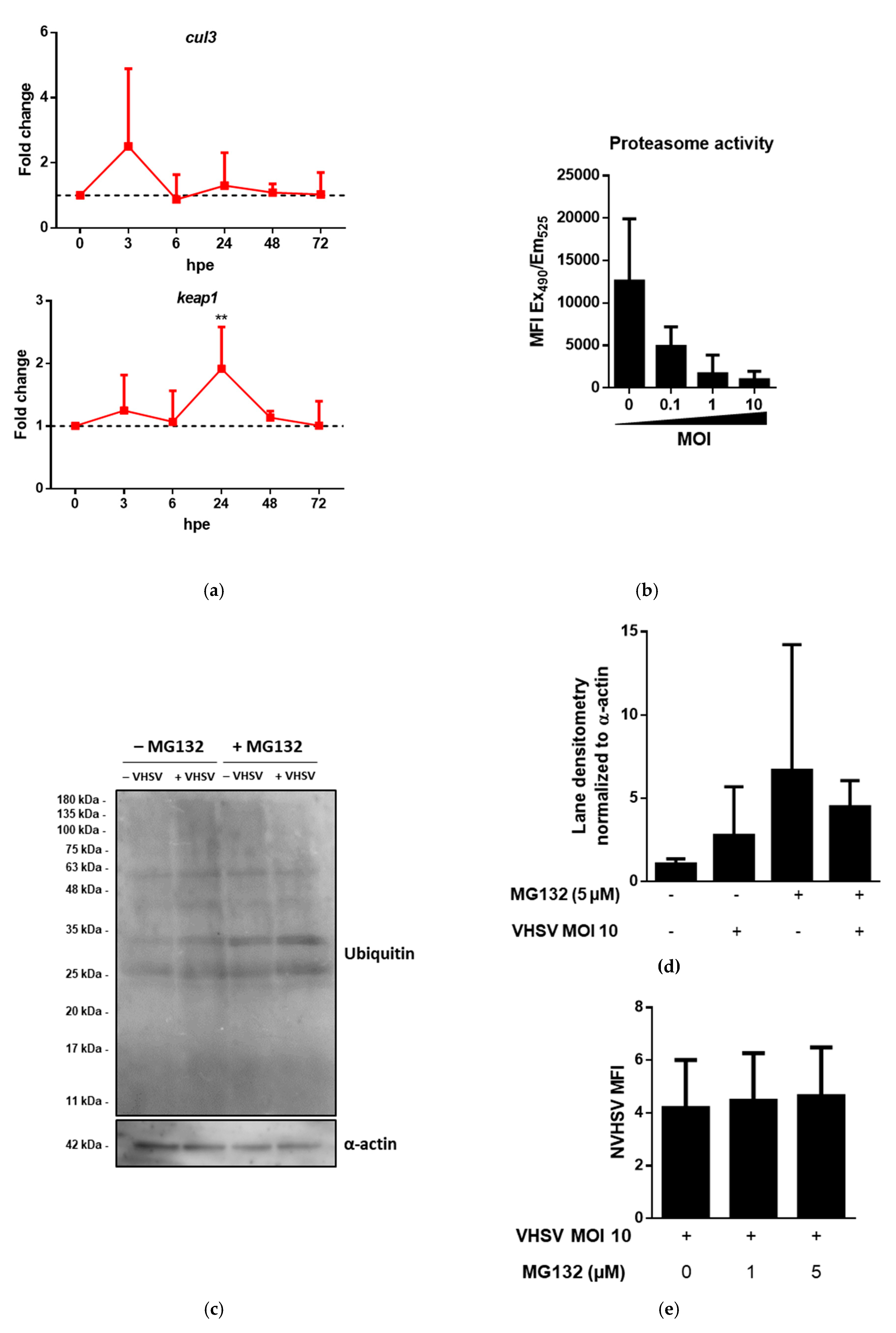
3.2. Proteomic Analysis of VHSV-Exposed RBCs Showed Proteasome Down-Regulation, Increased Ubiquitination, and Regulation of Antigen Presentation-Related Molecules at 72 hpe
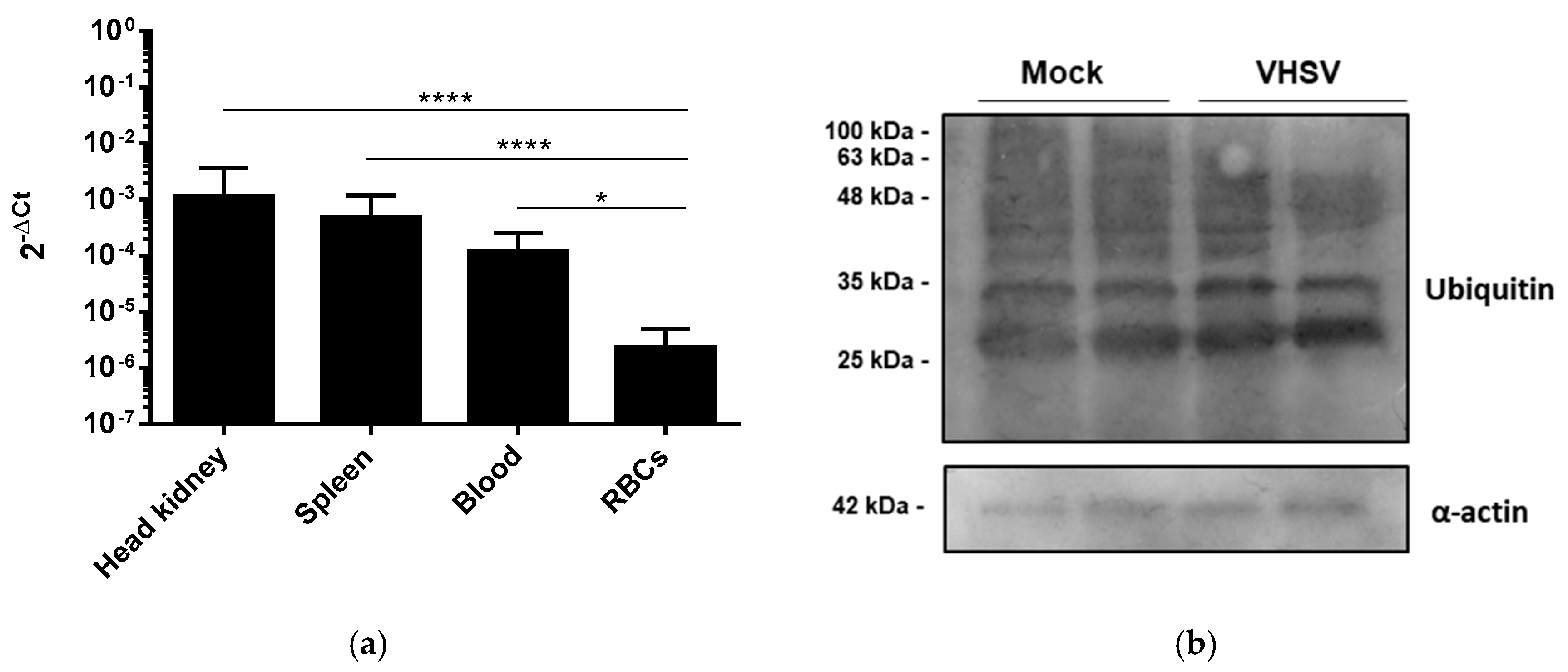
3.3. VHSV Induced Ubiquitination But Impaired Proteasome Degradation in Ex Vivo VHSV-exposed Rainbow Trout RBCs
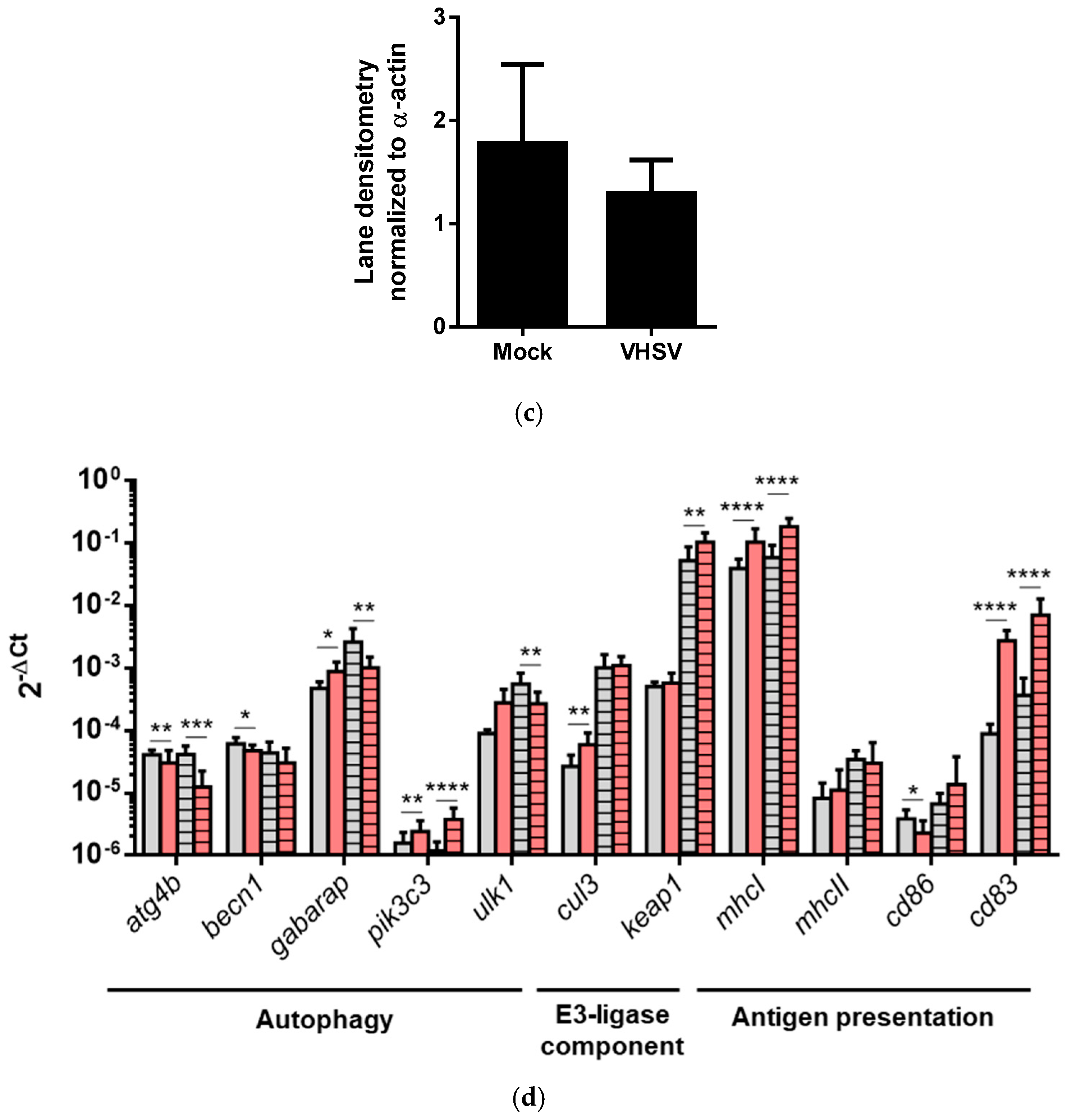
3.4. VHSV Induced Autophagy in Ex Vivo VHSV-exposed Rainbow Trout RBCs
3.5. Niclosamide Increased p62 and Intracellular VHSV Levels in Ex Vivo VHSV-exposed RBCs
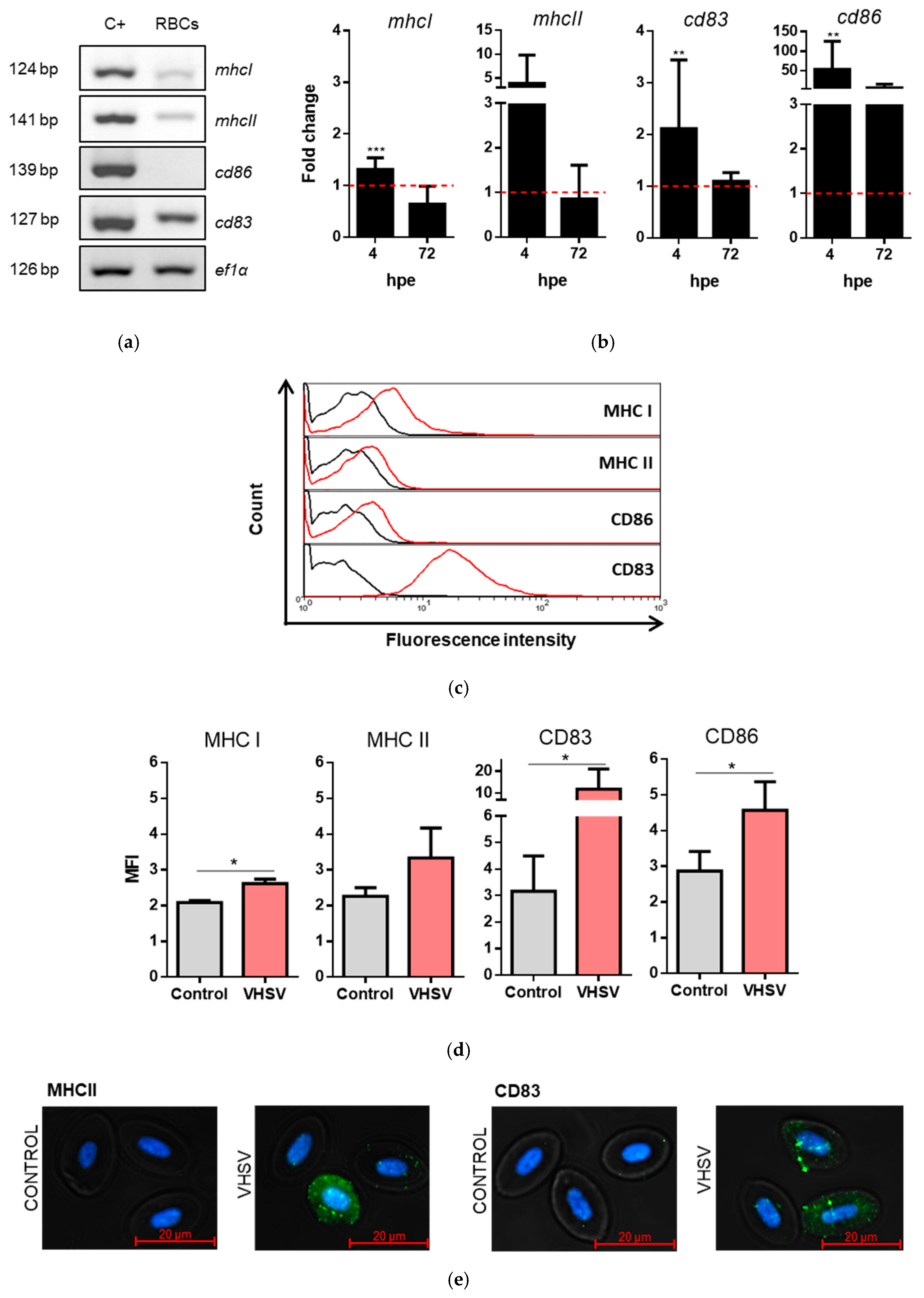
3.6. Rainbow Trout RBCs Up-Regulated MHCI, MHCII, CD86, and CD83 after VHSV Exposure
3.7. VHSV Induced Autophagy and Antigen Presentation Genes Expression in RBCs from VHSV-challenged Rainbow Trout
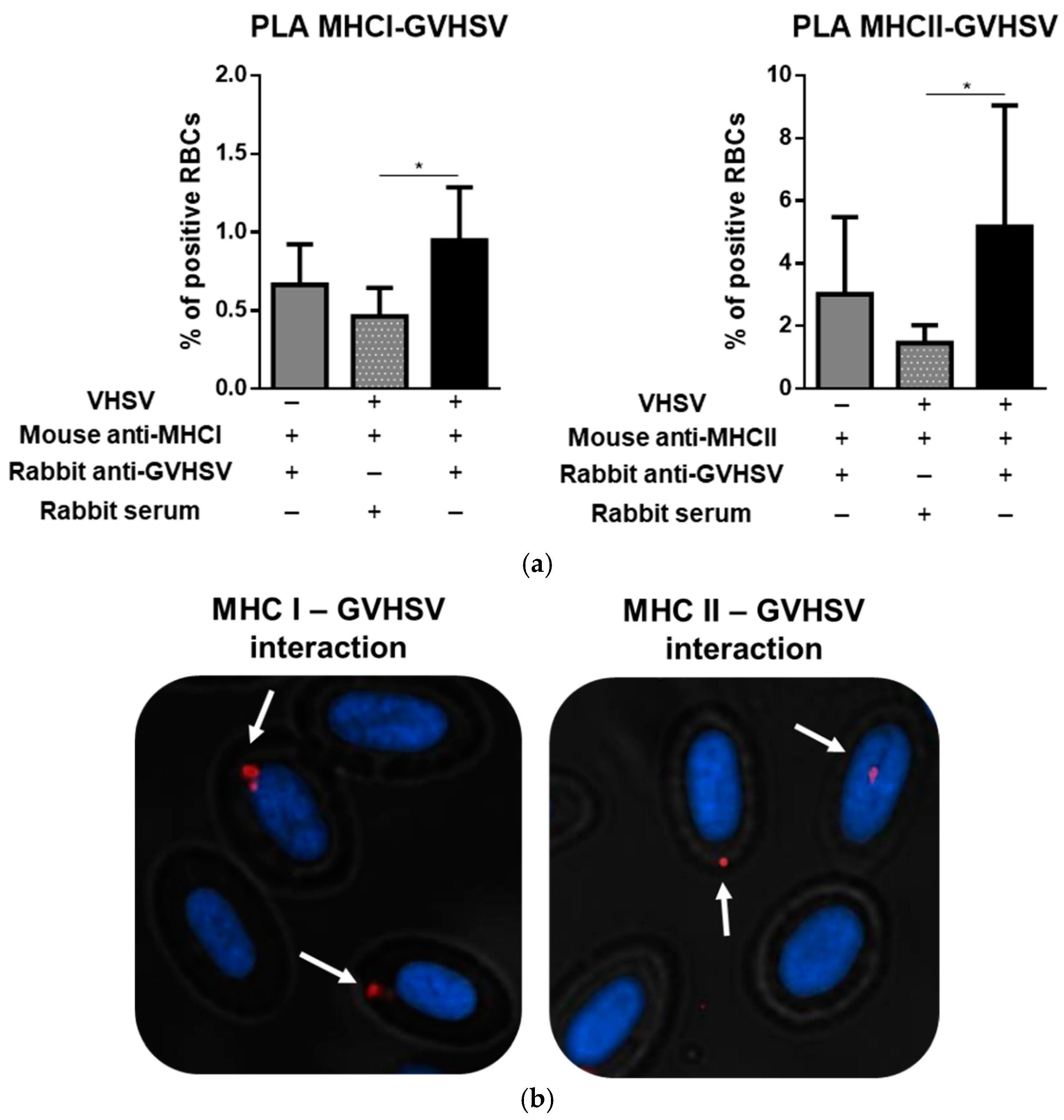
3.8. GVHSV Protein Peptides Colocalize with MHCI and MHCII in VHSV-Exposed Rainbow Trout RBCs
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Workenhe, S.T.; Kibenge, M.J.; Wright, G.M.; Wadowska, D.W.; Groman, D.B.; Kibenge, F.S. Infectious salmon anaemia virus replication and induction of alpha interferon in atlantic salmon erythrocytes. Virol. J. 2008, 5, 36. [Google Scholar] [CrossRef]
- Finstad, O.W.; Dahle, M.K.; Lindholm, T.H.; Nyman, I.B.; Lovoll, M.; Wallace, C.; Olsen, C.M.; Storset, A.K.; Rimstad, E. Piscine orthoreovirus (prv) infects atlantic salmon erythrocytes. Vet. Res. 2014, 45, 35. [Google Scholar] [CrossRef] [PubMed]
- Wessel, O.; Olsen, C.M.; Rimstad, E.; Dahle, M.K. Piscine orthoreovirus (prv) replicates in atlantic salmon (salmo salar l.) erythrocytes ex vivo. Vet. Res. 2015, 46, 26. [Google Scholar] [CrossRef]
- Wessel, O.; Krasnov, A.; Timmerhaus, G.; Rimstad, E.; Dahle, M.K. Antiviral responses and biological concequences of piscine orthoreovirus infection in salmonid erythrocytes. Front. Immunol. 2018, 9, 3182. [Google Scholar] [CrossRef] [PubMed]
- Dahle, M.K.; Jorgensen, J.B. Antiviral defense in salmonids - mission made possible? Fish. Shellfish Immunol. 2019, 87, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Dahle, M.K.; Wessel, O.; Timmerhaus, G.; Nyman, I.B.; Jorgensen, S.M.; Rimstad, E.; Krasnov, A. Transcriptome analyses of atlantic salmon (salmo salar l.) erythrocytes infected with piscine orthoreovirus (prv). Fish. Shellfish Immunol. 2015, 45, 780–790. [Google Scholar] [CrossRef] [PubMed]
- Nombela, I.; Puente-Marin, S.; Chico, V.; Villena, A.J.; Carracedo, B.; Ciordia, S.; Mena, M.C.; Mercado, L.; Perez, L.; Coll, J.; et al. Identification of diverse defense mechanisms in rainbow trout red blood cells in response to halted replication of vhs virus. F1000Research 2017, 6, 1958. [Google Scholar] [CrossRef] [PubMed]
- Nombela, I.; Carrion, A.; Puente-Marin, S.; Chico, V.; Mercado, L.; Perez, L.; Coll, J.; Ortega-Villaizan, M.d.M. Infectious pancreatic necrosis virus triggers antiviral immune response in rainbow trout red blood cells, despite not being infective. F1000Research 2017, 6, 1968. [Google Scholar] [CrossRef] [PubMed]
- Chico, V.; Salvador-Mira, M.; Nombela, I.; Puente-Marin, S.; Ciordia, S.; Mena, M.C.; Perez, L.; Coll, J.; Guzman, F.; Encinar, J.A.; et al. Ifit5 participates in the antiviral mechanisms of rainbow trout red blood cells. Front. Immunol. 2019, 10, 613. [Google Scholar] [CrossRef]
- Jackson, W.T. Viruses and the autophagy pathway. Virology 2015, 479–480, 450–456. [Google Scholar] [CrossRef]
- Chiramel, A.I.; Brady, N.R.; Bartenschlager, R. Divergent roles of autophagy in virus infection. Cells 2013, 2, 83–104. [Google Scholar] [CrossRef]
- Lee, H.K.; Lund, J.M.; Ramanathan, B.; Mizushima, N.; Iwasaki, A. Autophagy-dependent viral recognition by plasmacytoid dendritic cells. Science 2007, 315, 1398–1401. [Google Scholar] [CrossRef]
- Gorbea, C.; Makar, K.A.; Pauschinger, M.; Pratt, G.; Bersola, J.L.; Varela, J.; David, R.M.; Banks, L.; Huang, C.H.; Li, H.; et al. A role for toll-like receptor 3 variants in host susceptibility to enteroviral myocarditis and dilated cardiomyopathy. J. Biol. Chem. 2010, 285, 23208–23223. [Google Scholar] [CrossRef]
- Garcia-Valtanen, P.; Ortega-Villaizan Mdel, M.; Martinez-Lopez, A.; Medina-Gali, R.; Perez, L.; Mackenzie, S.; Figueras, A.; Coll, J.M.; Estepa, A. Autophagy-inducing peptides from mammalian vsv and fish vhsv rhabdoviral g glycoproteins (g) as models for the development of new therapeutic molecules. Autophagy 2014, 10, 1666–1680. [Google Scholar] [CrossRef]
- Ortega-Villaizan, M.; Chico, V.; Martinez-Lopez, A.; Garcia-Valtanen, P.; Coll, J.M.; Estepa, A. Development of new therapeutical/adjuvant molecules by pepscan mapping of autophagy and ifn inducing determinants of rhabdoviral g proteins. Mol. Immunol. 2016, 70, 118–124. [Google Scholar] [CrossRef] [PubMed]
- Pereiro, P.; Romero, A.; Diaz-Rosales, P.; Estepa, A.; Figueras, A.; Novoa, B. Nucleated teleost erythrocytes play an nk-lysin- and autophagy-dependent role in antiviral immunity. Front. Immunol. 2017, 8, 1458. [Google Scholar] [CrossRef] [PubMed]
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2013, 20, 21–30. [Google Scholar] [CrossRef]
- Deretic, V. Autophagy: An emerging immunological paradigm. J. Immunol. 2012, 189, 15–20. [Google Scholar] [CrossRef] [PubMed]
- Lin, X.; Li, S.; Zhao, Y.; Ma, X.; Zhang, K.; He, X.; Wang, Z. Interaction domains of p62: A bridge between p62 and selective autophagy. DNA Cell Biol. 2013, 32, 220–227. [Google Scholar] [CrossRef] [PubMed]
- Weissman, A.M. Themes and variations on ubiquitylation. Nat. Rev. Mol. Cell Biol. 2001, 2, 169–178. [Google Scholar] [CrossRef] [PubMed]
- Neelam, S.; Kakhniashvili, D.G.; Wilkens, S.; Levene, S.D.; Goodman, S.R. Functional 20s proteasomes in mature human red blood cells. Exp. Biol. Med. (Maywood) 2011, 236, 580–591. [Google Scholar] [CrossRef]
- Luo, H. Interplay between the virus and the ubiquitin-proteasome system: Molecular mechanism of viral pathogenesis. Curr. Opin. Virol. 2016, 17, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Gaczynska, M.; Rock, K.L.; Goldberg, A.L. Role of proteasomes in antigen presentation. Enzyme Protein 1993, 47, 354–369. [Google Scholar] [CrossRef] [PubMed]
- Gannage, M.; Munz, C. MHC presentation via autophagy and how viruses escape from it. Semin. Immunopathol. 2010, 32, 373–381. [Google Scholar] [CrossRef] [PubMed]
- English, L.; Chemali, M.; Duron, J.; Rondeau, C.; Laplante, A.; Gingras, D.; Alexander, D.; Leib, D.; Norbury, C.; Lippe, R.; et al. Autophagy enhances the presentation of endogenous viral antigens on mhc class i molecules during hsv-1 infection. Nat. Immunol. 2009, 10, 480–487. [Google Scholar] [CrossRef]
- Paludan, C.; Schmid, D.; Landthaler, M.; Vockerodt, M.; Kube, D.; Tuschl, T.; Munz, C. Endogenous mhc class ii processing of a viral nuclear antigen after autophagy. Science 2005, 307, 593–596. [Google Scholar] [CrossRef] [PubMed]
- Lugo-Villarino, G.; Balla, K.M.; Stachura, D.L.; Banuelos, K.; Werneck, M.B.; Traver, D. Identification of dendritic antigen-presenting cells in the zebrafish. Proc. Natl. Acad. Sci. USA 2010, 107, 15850–15855. [Google Scholar] [CrossRef] [PubMed]
- Bassity, E.; Clark, T.G. Functional identification of dendritic cells in the teleost model, rainbow trout (oncorhynchus mykiss). PLoS ONE 2012, 7, e33196. [Google Scholar] [CrossRef] [PubMed]
- Sarder, M.R.; Fischer, U.; Dijkstra, J.M.; Kiryu, I.; Yoshiura, Y.; Azuma, T.; Kollner, B.; Ototake, M. The mhc class i linkage group is a major determinant in the in vivo rejection of allogeneic erythrocytes in rainbow trout (oncorhynchus mykiss). Immunogenetics 2003, 55, 315–324. [Google Scholar] [CrossRef][Green Version]
- Dijkstra, J.M.; Kollner, B.; Aoyagi, K.; Sawamoto, Y.; Kuroda, A.; Ototake, M.; Nakanishi, T.; Fischer, U. The rainbow trout classical mhc class i molecule onmy-uba*501 is expressed in similar cell types as mammalian classical mhc class i molecules. Fish. Shellfish Immunol. 2003, 14, 1–23. [Google Scholar] [CrossRef] [PubMed]
- Puente-Marin, S.; Nombela, I.; Ciordia, S.; Mena, M.C.; Chico, V.; Coll, J.; Ortega-Villaizan, M.D.M. In silico functional networks identified in fish nucleated red blood cells by means of transcriptomic and proteomic profiling. Genes (Basel) 2018, 9, 202. [Google Scholar] [CrossRef]
- Puente-Marin, S.; Nombela, I.; Chico, V.; Ciordia, S.; Mena, M.C.; Coll, J.; Mercado, L.; Ortega-Villaizan, M.D.M. Rainbow trout erythrocytes ex vivo transfection with a DNA vaccine encoding vhsv glycoprotein g induces an antiviral immune response. Front. Immunol. 2018, 9, 2477. [Google Scholar] [CrossRef] [PubMed]
- St Paul, M.; Paolucci, S.; Barjesteh, N.; Wood, R.D.; Sharif, S. Chicken erythrocytes respond to toll-like receptor ligands by up-regulating cytokine transcripts. Res. Vet. Sci. 2013, 95, 87–91. [Google Scholar] [CrossRef]
- Kambayashi, T.; Laufer, T.M. Atypical mhc class ii-expressing antigen-presenting cells: Can anything replace a dendritic cell? Nat. Rev. Immunol. 2014, 14, 719–730. [Google Scholar] [CrossRef] [PubMed]
- Dasari, V.; Rehan, S.; Tey, S.K.; Smyth, M.J.; Smith, C.; Khanna, R. Autophagy and proteasome interconnect to coordinate cross-presentation through mhc class i pathway in b cells. Immunol. Cell Biol. 2016, 94, 964–974. [Google Scholar] [CrossRef] [PubMed]
- LeBerre, M.; De Kinkelin, P.; Metzger, A. Identification sérologique des rhabdovirus des salmonidés. Bull. Off. Int Epizoot 1977, 87, 391–393. [Google Scholar]
- Basurco, B.; Coll, J. Spanish isolates and reference strains of viral haemorrhagic septicaemia virus shown similar protein size patterns. Bull. Eur. Ass. Fish. Pathol. 1989, 9, 92–95. [Google Scholar]
- Sanz, F.; Basurco, B.; Babin, M.; Dominguez, J.; Coll, J. Monoclonal antibodies against the structural proteins of viral haemorrhagic septicaemia virus isolates. J. Fish. Dis. 1993, 16, 53–63. [Google Scholar] [CrossRef]
- Morales-Lange, B.; Ramirez, F.; Guzman, F.; Wong, V.; Imarai, M.; Lagos, L.; Øverland, M.; Boltana, S.; Mercado, L. Interferon gamma induces the overexpression of antigen presenting cell surface markers in spleen leukocytes from salmo salar. Unpublished manuscript.
- Lorenzen, N.; Lapatra, S.E. Immunity to rhabdoviruses in rainbow trout: The antibody response. Fish. Shellfish Immunol. 1999, 9, 345–360. [Google Scholar] [CrossRef]
- Chico, V.; Gomez, N.; Estepa, A.; Perez, L. Rapid detection and quantitation of viral hemorrhagic septicemia virus in experimentally challenged rainbow trout by real-time rt-pcr. J. Virol. Methods 2006, 132, 154–159. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Mlecnik, B.; Hackl, H.; Charoentong, P.; Tosolini, M.; Kirilovsky, A.; Fridman, W.H.; Pages, F.; Trajanoski, Z.; Galon, J. Cluego: A cytoscape plug-in to decipher functionally grouped gene ontology and pathway annotation networks. Bioinformatics 2009, 25, 1091–1093. [Google Scholar] [CrossRef] [PubMed]
- Bindea, G.; Galon, J.; Mlecnik, B. Cluepedia cytoscape plugin: Pathway insights using integrated experimental and in silico data. Bioinformatics 2013, 29, 661–663. [Google Scholar] [CrossRef]
- Shannon, P.; Markiel, A.; Ozier, O.; Baliga, N.S.; Wang, J.T.; Ramage, D.; Amin, N.; Schwikowski, B.; Ideker, T. Cytoscape: A software environment for integrated models of biomolecular interaction networks. Genome Res. 2003, 13, 2498–2504. [Google Scholar] [CrossRef] [PubMed]
- Gotz, S.; Garcia-Gomez, J.M.; Terol, J.; Williams, T.D.; Nagaraj, S.H.; Nueda, M.J.; Robles, M.; Talon, M.; Dopazo, J.; Conesa, A. High-throughput functional annotation and data mining with the blast2go suite. Nucleic Acids Res. 2008, 36, 3420–3435. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, C.A.; Ramirez-Cepeda, F.; Santana, P.; Torres, E.; Cortes, J.; Guzman, F.; Schmitt, P.; Mercado, L. Insights into the diversity of nod-like receptors: Identification and expression analysis of nlrc3, nlrc5 and nlrx1 in rainbow trout. Mol. Immunol. 2017, 87, 102–113. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(-delta delta c(t)) method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Chaves-Pozo, E.; Montero, J.; Cuesta, A.; Tafalla, C. Viral hemorrhagic septicemia and infectious pancreatic necrosis viruses replicate differently in rainbow trout gonad and induce different chemokine transcription profiles. Dev. Comp. Immunol. 2010, 34, 648–658. [Google Scholar] [CrossRef]
- Jorgensen, T.R.; Raida, M.K.; Kania, P.W.; Buchmann, K. Response of rainbow trout (oncorhynchus mykiss) in skin and fin tissue during infection with a variant of gyrodactylus salaris (monogenea: Gyrodactylidae). Folia Parasitol. (Praha) 2009, 56, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Ortega-Villaizan, M.; Martinez-Lopez, A.; Garcia-Valtanen, P.; Chico, V.; Perez, L.; Coll, J.M.; Estepa, A. Ex vivo transfection of trout pronephros leukocytes, a model for cell culture screening of fish DNA vaccine candidates. Vaccine 2012, 30, 5983–5990. [Google Scholar] [CrossRef]
- Klionsky, D.J.; Abdelmohsen, K.; Abe, A.; Abedin, M.J.; Abeliovich, H.; Acevedo Arozena, A.; Adachi, H.; Adams, C.M.; Adams, P.D.; Adeli, K.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy (3rd edition). Autophagy 2016, 12, 1–222. [Google Scholar] [CrossRef]
- Bjorkoy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Overvatn, A.; Stenmark, H.; Johansen, T. P62/sqstm1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Khambu, B.; Zhang, H.; Kang, J.H.; Chen, X.; Chen, D.; Vollmer, L.; Liu, P.Q.; Vogt, A.; Yin, X.M. Suppression of lysosome function induces autophagy via a feedback down-regulation of mtor complex 1 (mtorc1) activity. J. Biol. Chem. 2013, 288, 35769–35780. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Liang, A.; Kang, J.-H.; Liu, P.; Yin, X.-m.; Li, M. Niclosamide blocks autophagy via lysosomal dysfunction (663.18). FASEB J. 2014, 28, 663.18. [Google Scholar]
- Kao, J.C.; HuangFu, W.C.; Tsai, T.T.; Ho, M.R.; Jhan, M.K.; Shen, T.J.; Tseng, P.C.; Wang, Y.T.; Lin, C.F. The antiparasitic drug niclosamide inhibits dengue virus infection by interfering with endosomal acidification independent of mtor. PLoS Negl. Trop. Dis. 2018, 12, e0006715. [Google Scholar] [CrossRef] [PubMed]
- Nombela, I.; Ortega-Villaizan, M.D.M. Nucleated red blood cells: Immune cell mediators of the antiviral response. PLoS Pathog. 2018, 14, e1006910. [Google Scholar] [CrossRef]
- Cullinan, S.B.; Gordan, J.D.; Jin, J.; Harper, J.W.; Diehl, J.A. The keap1-btb protein is an adaptor that bridges nrf2 to a cul3-based e3 ligase: Oxidative stress sensing by a cul3-keap1 ligase. Mol. Cell. Biol. 2004, 24, 8477–8486. [Google Scholar] [CrossRef] [PubMed]
- Diedrich, G.; Bangia, N.; Pan, M.; Cresswell, P. A role for calnexin in the assembly of the mhc class i loading complex in the endoplasmic reticulum. J. Immunol. 2001, 166, 1703–1709. [Google Scholar] [CrossRef]
- Ko, A.; Lee, E.W.; Yeh, J.Y.; Yang, M.R.; Oh, W.; Moon, J.S.; Song, J. Mkrn1 induces degradation of west nile virus capsid protein by functioning as an e3 ligase. J. Virol. 2010, 84, 426–436. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Moriishi, K.; Fukuda, K.; Shirakura, M.; Ishii, K.; Shoji, I.; Wakita, T.; Miyamura, T.; Matsuura, Y.; Suzuki, T. Proteasomal turnover of hepatitis c virus core protein is regulated by two distinct mechanisms: A ubiquitin-dependent mechanism and a ubiquitin-independent but pa28gamma-dependent mechanism. J. Virol. 2009, 83, 2389–2392. [Google Scholar] [CrossRef]
- Drugeon, G.; Jupin, I. Stability in vitro of the 69k movement protein of turnip yellow mosaic virus is regulated by the ubiquitin-mediated proteasome pathway. J. Gen. Virol. 2002, 83, 3187–3197. [Google Scholar] [CrossRef] [PubMed]
- Gilfoy, F.; Fayzulin, R.; Mason, P.W. West nile virus genome amplification requires the functional activities of the proteasome. Virology 2009, 385, 74–84. [Google Scholar] [CrossRef] [PubMed]
- Kaspari, M.; Tavalai, N.; Stamminger, T.; Zimmermann, A.; Schilf, R.; Bogner, E. Proteasome inhibitor mg132 blocks viral DNA replication and assembly of human cytomegalovirus. FEBS Lett. 2008, 582, 666–672. [Google Scholar] [CrossRef]
- Reichel, C.; Beachy, R.N. Degradation of tobacco mosaic virus movement protein by the 26s proteasome. J. Virol. 2000, 74, 3330–3337. [Google Scholar] [CrossRef] [PubMed]
- Datan, E.; Roy, S.G.; Germain, G.; Zali, N.; McLean, J.E.; Golshan, G.; Harbajan, S.; Lockshin, R.A.; Zakeri, Z. Dengue-induced autophagy, virus replication and protection from cell death require er stress (perk) pathway activation. Cell Death Dis. 2016, 7, e2127. [Google Scholar] [CrossRef]
- Orvedahl, A.; MacPherson, S.; Sumpter, R., Jr.; Talloczy, Z.; Zou, Z.; Levine, B. Autophagy protects against sindbis virus infection of the central nervous system. Cell Host Microbe 2010, 7, 115–127. [Google Scholar] [CrossRef]
- Shelly, S.; Lukinova, N.; Bambina, S.; Berman, A.; Cherry, S. Autophagy is an essential component of drosophila immunity against vesicular stomatitis virus. Immunity 2009, 30, 588–598. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Kaplan, V.; Ciechanover, A.; Livneh, I. P62 at the crossroad of the ubiquitin-proteasome system and autophagy. Oncotarget 2016, 7, 83833–83834. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.J.; Ye, L.; Huang, W.F.; Guo, L.J.; Xu, Z.G.; Wu, H.L.; Yang, C.; Liu, H.F. P62 links the autophagy pathway and the ubiqutin-proteasome system upon ubiquitinated protein degradation. Cell Mol. Biol. Lett 2016, 21, 29. [Google Scholar] [CrossRef] [PubMed]
- Zaffagnini, G.; Savova, A.; Danieli, A.; Romanov, J.; Tremel, S.; Ebner, M.; Peterbauer, T.; Sztacho, M.; Trapannone, R.; Tarafder, A.K.; et al. P62 filaments capture and present ubiquitinated cargos for autophagy. EMBO J. 2018, 37. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.T.; Shahnazari, S.; Brech, A.; Lamark, T.; Johansen, T.; Brumell, J.H. The adaptor protein p62/sqstm1 targets invading bacteria to the autophagy pathway. J. Immunol. 2009, 183, 5909–5916. [Google Scholar] [CrossRef] [PubMed]
- Sahani, M.H.; Itakura, E.; Mizushima, N. Expression of the autophagy substrate sqstm1/p62 is restored during prolonged starvation depending on transcriptional upregulation and autophagy-derived amino acids. Autophagy 2014, 10, 431–441. [Google Scholar] [CrossRef] [PubMed]
- Lippai, M.; Low, P. The role of the selective adaptor p62 and ubiquitin-like proteins in autophagy. Biomed. Res. Int. 2014, 2014, 832704. [Google Scholar] [CrossRef] [PubMed]
- Berchtold, S.; Muhl-Zurbes, P.; Heufler, C.; Winklehner, P.; Schuler, G.; Steinkasserer, A. Cloning, recombinant expression and biochemical characterization of the murine cd83 molecule which is specifically upregulated during dendritic cell maturation. FEBS Lett. 1999, 461, 211–216. [Google Scholar] [CrossRef]
- Breloer, M.; Fleischer, B. Cd83 regulates lymphocyte maturation, activation and homeostasis. Trends Immunol. 2008, 29, 186–194. [Google Scholar] [CrossRef]
- Ferm, M.T.; Gronberg, A. Human mhc class i antigens are associated with a 90-kda cell surface protein. Scand J. Immunol. 1991, 34, 221–227. [Google Scholar] [CrossRef] [PubMed]
- Van Kaer, L.; Parekh, V.V.; Postoak, J.L.; Wu, L. Role of autophagy in mhc class i-restricted antigen presentation. Mol. Immunol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Dengjel, J.; Schoor, O.; Fischer, R.; Reich, M.; Kraus, M.; Muller, M.; Kreymborg, K.; Altenberend, F.; Brandenburg, J.; Kalbacher, H.; et al. Autophagy promotes mhc class ii presentation of peptides from intracellular source proteins. Proc. Natl. Acad. Sci. USA 2005, 102, 7922–7927. [Google Scholar] [CrossRef] [PubMed]
- Cassatella, M.A. Human mature neutrophils as atypical apc. Blood 2017, 129, 1895–1896. [Google Scholar] [CrossRef]
- Hirosue, S.; Dubrot, J. Modes of antigen presentation by lymph node stromal cells and their immunological implications. Front. Immunol. 2015, 6, 446. [Google Scholar] [CrossRef] [PubMed]






| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Reference or Accession Number |
|---|---|---|---|
| mhcI | CCAGAGGATGTATGGTTGTGAG | TGGAGCGATCCATGTCTTTGTC | AF287490.1 |
| mhcII | GTACTCCAGGTGGGAGTGGA | TGCAGCGCCTATGACTTCTA | AY273808.1 |
| cd86 | ATGTAACAGTGGCCTGTGA | CCACCCACTGCTGTTCACTA | FJ607781.1 |
| cd83 | GGAGCGTGAAGTGAACTTT | TCCTGGTTCTGCTCTCCTACA | AY263797.1 |
| ef1α | TGGAGACTGGCACCCTGAAG | CCAACATTGTCACCAGGCATGG | [46] |
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) | Probe (5′–3′) | Reference or Accession Number |
|---|---|---|---|---|
| atg4b | GATCCTGTCCCTGTGATGATGA | CCCCTATTGGCTTCCCTTCT | ACCCCCCCCGGCGATTCTTC | XR_002473879 |
| ulk1 | CTTCTGCTGCTGGGTCTTCTG | GGTGACGGAAGAACTCCTCAAA | CGAAACCACAAGGACCGCATGGA | XR_002473462 |
| becn1 | GCGTGGGTGTCGTCTCAGTT | CAGGGAAGCAAGGAGAGCAT | ACCCTGGGTGTGCCCCTTGACC | NM_001124429 |
| gabarap | CCTCATCCATCCATTTTTACCTCTT | ATTCAACCGAAATCCCCATCT | TCTGAATTTTATTTGCCTCCGGGTCTCC | NM_001165091 |
| pik3c3 | AGGCCAGCTGTGTGTGTTTCTA | GTTGCACATAGCGTTCCTGTTTA | TTTGCCCCCCCGGATGATTGA | XM_021577851 |
| cul3 | GCAGCTTACGTTACAGCATCACA | TGGTGTTGGAGCCTGTTACCT | AACGCCACCTTCTACGGCCCAATC | XM_021587294.1 |
| ikbkb | TGTTCCTGTTTGACCGTTCCT | CCGTCTGGACAAAGCGTATGT | CCTACGAGCCCCAGTTCACCCCC | XM_021621802.1 |
| keap1 | CCTCCACAAGCCCACCAA | AAGTATCCCCCTGCCGTGTA | CACGCCCAAAGTGCCCCAGC | XM_021556738.1 |
| rab7 | GTTGCGTGCTGGTGTTTGAC | ACTCGTCCCTCCAGCTGTCTAG | TGACCGCCCCCAACACCTTCAA | XM_021609589.1 |
| sec13 | GCAGTGATCCAGGCACAGAA | CTGGGACTAGGATAGATGGTAGAAGTG | ATTCCACTCCTCCTCCTACCCCCACA | XM_021610740.1 |
| traf6 | AGGACGCGGTGTGGAAGAG | CATGAATCTTGCTGTCCTCGTAAA | AGATGCACCAAAGCCAACACTGCCA | XM_021586866.1 |
| mhcI | GACAGTCCGTCCCTCAGTGT | CTGGAAGGTTCCATCATCGT | [48] | |
| mhcII | TGCCATGCTGATGTGCAG | GTCCCTCAGCCAGGTCACT | CGCCTATGACTTCTACCCCAAACAAAT | [49] |
| cd86 | GGTCTGTGACCCTCCCCTGTA | CCCTCGTCTTATGGTAGCCATT | XR_002470439.1 | |
| cd83 | TTGGCTGATGATTCTTTCGATATC | TGCTGCCAGGAGACACTTGT | TCCTGCCCAATGTAACGGCTGTTGA | [50] |
| NVHSV | GACTCAACGGGACAGGAATGA | GGGCAATGCCCAAGTTGTT | TGGGTTGTTCACCCAGGCCGC | [41] |
| Antigen Processing: Ubiquitination and Proteasome Degradation | Class I MHC-Mediated Antigen Processing and Presentation | ||
|---|---|---|---|
| Gene Symbol | Log2 Fold | Gene Symbol | Log2 Fold |
| cul3 | 4.77 | canx | 4.31 |
| keap1 | 7.56 | sec13 | 5.35 |
| psma6 | 5.02 | ikbkb | 5.69 |
| psmb5 | 3.72 | klhl13 | 5.36 |
| Autophagy-Related Genes | |
|---|---|
| Gene Symbol | Log2 Fold |
| ulk1 | 3.46 |
| becn1 | 5.55 |
| atg9a | 3.69 |
| Antigen Processing and Presentation of Peptide Antigen via MHC Class II | Proteasome-mediated Ubiquitin-Dependent Protein Catabolic Process | Proteasome | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Upr. Protein | Log2 Fold | Downr. Protein | Log2 Fold | Upr. Protein | Log2 Fold | Downr. Protein | Log2 Fold | Upr. Protein | Log2 Fold | Downr. Protein | Log2 Fold |
| ACTR1B | 3.37 | CAPZB | −2.68 | CD2AP | 7.50 | HSPA5 | −6.23 | PSMB3 | 4.44 | PSMA1 | −5.33 |
| AP2S1 | 5.75 | CLTC | −3.69 | DDB1 | 3.32 | PSMA1 | −5.33 | PSMB6 | 3.73 | PSMA2 | −5.43 |
| CLTA | 4.51 | RAB7A | −4.69 | GCLC | 4.63 | PSMA2 | −5.43 | PSMC2 | 2.98 | PSMA3 | −3.31 |
| DNM2 | 5.32 | HSPA1A | 4.74 | PSMA3 | −3.31 | PSMD13 | 2.26 | PSMA4 | −4.78 | ||
| DYNC1H1 | 5.43 | NPLOC4 | 1.68 | PSMA4 | −4.78 | PSMD2 | 3.98 | PSMA5 | −6.15 | ||
| KIF15 | 3.89 | PLAA | 5.08 | PSMA5 | −6.15 | PSMD4 | 5.83 | PSMA6 | −6.49 | ||
| PYCARD | 3.28 | PSMB3 | 4.44 | PSMA6 | −6.49 | PSME1 | 5.73 | PSMA8 | −5.50 | ||
| PSMB6 | 3.73 | PSMA8 | −5.50 | PSMB1 | −4.45 | ||||||
| PSMC2 | 2.98 | PSMB1 | −4.45 | PSMB2 | −5.29 | ||||||
| PSMD13 | 2.26 | PSMB2 | −5.29 | PSMB4 | −3.69 | ||||||
| PSMD2 | 3.98 | PSMB4 | −3.69 | PSME2 | −3.44 | ||||||
| PSMD4 | 5.83 | PSME2 | −3.44 | ||||||||
| PSME1 | 5.73 | RAD23B | −3.77 | ||||||||
| RACK1 | 3.95 | UBC | −5.19 | ||||||||
| RAD23A | 4.85 | UBR2 | −11.48 | ||||||||
| RPS27A | 5.03 | VCP | −2.86 | ||||||||
| UBB | 4.39 | WFS1 | −10.33 | ||||||||
| USP19 | 8.29 | ||||||||||
| YOD1 | 2.95 | ||||||||||
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nombela, I.; Requena-Platek, R.; Morales-Lange, B.; Chico, V.; Puente-Marin, S.; Ciordia, S.; Mena, M.C.; Coll, J.; Perez, L.; Mercado, L.; et al. Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-presenting Cell Markers. Cells 2019, 8, 386. https://doi.org/10.3390/cells8050386
Nombela I, Requena-Platek R, Morales-Lange B, Chico V, Puente-Marin S, Ciordia S, Mena MC, Coll J, Perez L, Mercado L, et al. Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-presenting Cell Markers. Cells. 2019; 8(5):386. https://doi.org/10.3390/cells8050386
Chicago/Turabian StyleNombela, Ivan, Ricardo Requena-Platek, Byron Morales-Lange, Veronica Chico, Sara Puente-Marin, Sergio Ciordia, Maria Carmen Mena, Julio Coll, Luis Perez, Luis Mercado, and et al. 2019. "Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-presenting Cell Markers" Cells 8, no. 5: 386. https://doi.org/10.3390/cells8050386
APA StyleNombela, I., Requena-Platek, R., Morales-Lange, B., Chico, V., Puente-Marin, S., Ciordia, S., Mena, M. C., Coll, J., Perez, L., Mercado, L., & Ortega-Villaizan, M. d. M. (2019). Rainbow Trout Red Blood Cells Exposed to Viral Hemorrhagic Septicemia Virus Up-Regulate Antigen-Processing Mechanisms and MHC I&II, CD86, and CD83 Antigen-presenting Cell Markers. Cells, 8(5), 386. https://doi.org/10.3390/cells8050386