The Effects of Apigenin-Biosynthesized Ultra-Small Platinum Nanoparticles on the Human Monocytic THP-1 Cell Line
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
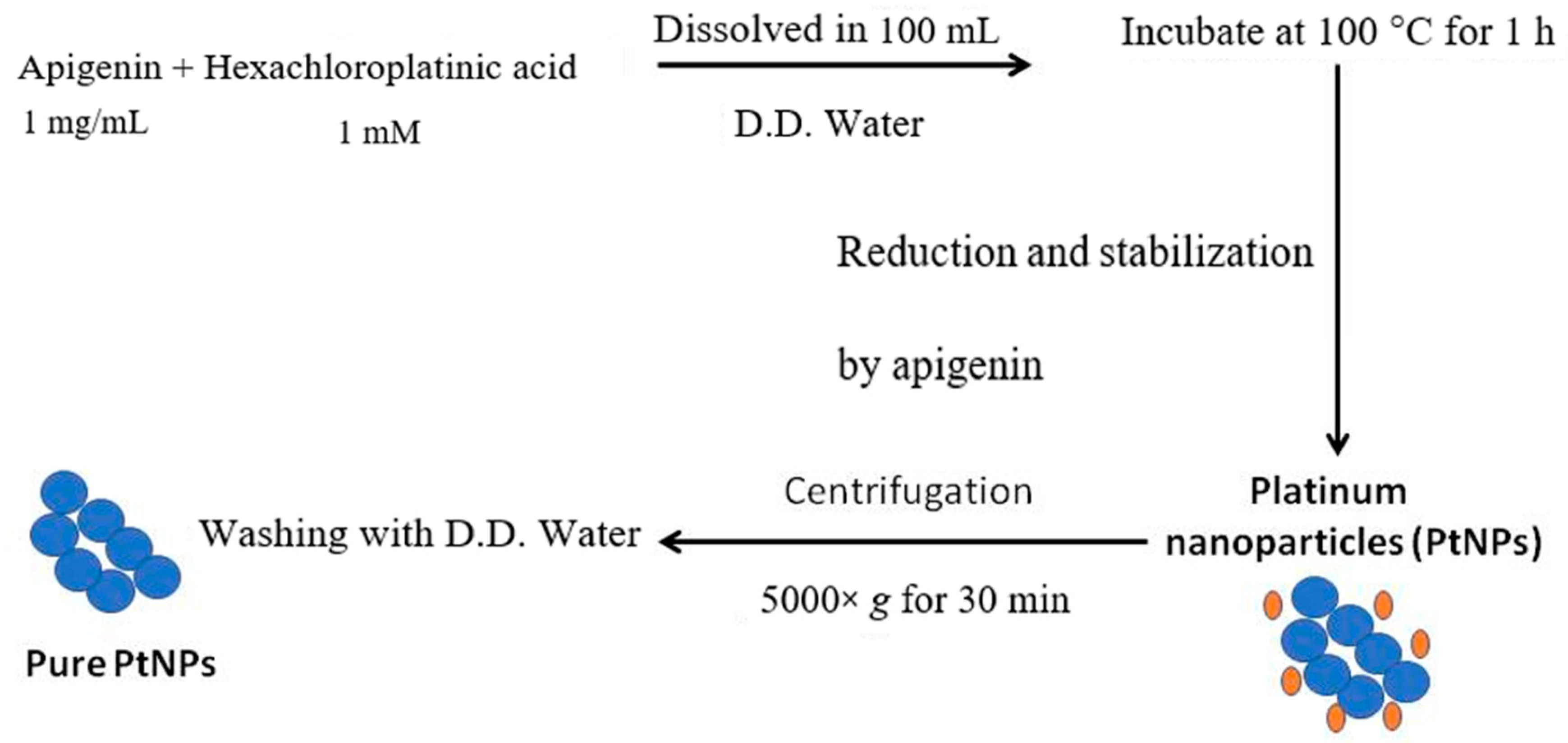
2.2. Synthesis and Characterization of PtNPs
2.3. Cell Culture Conditions and PtNP Exposures
2.4. Cell Viability Assay
2.5. BrdU Cell Proliferation Assay
2.6. Assessment of Membrane Integrity
2.7. Cell Mortality Assay
2.8. Determination of ROS, MDA, Nitric Oxide (NO), and Carbonylated Protein Levels
2.9. Measurement of Anti-Oxidative Marker Levels
2.10. Measurement of 8-Oxo-7,8-Dihydro-2′-Deoxyguanosine (8-Oxo-Dg) and 8-Oxo-G Levels
2.11. Reverse Transcription-Quantitative Polymerase Chain Reaction (RT-qPCR)
2.12. Measurement of Cytokine Levels
2.13. Statistical Analysis
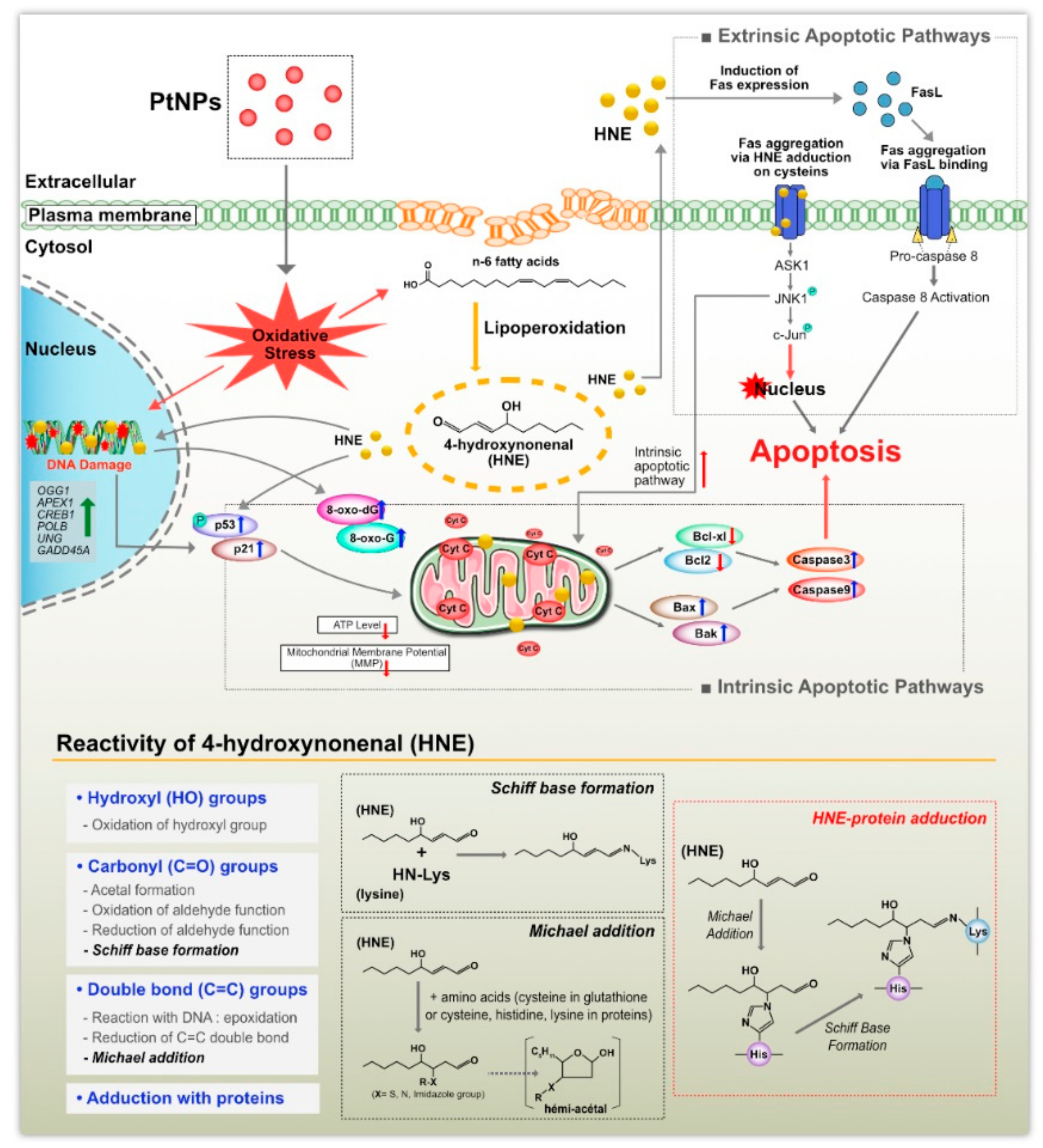
3. Results and Discussion
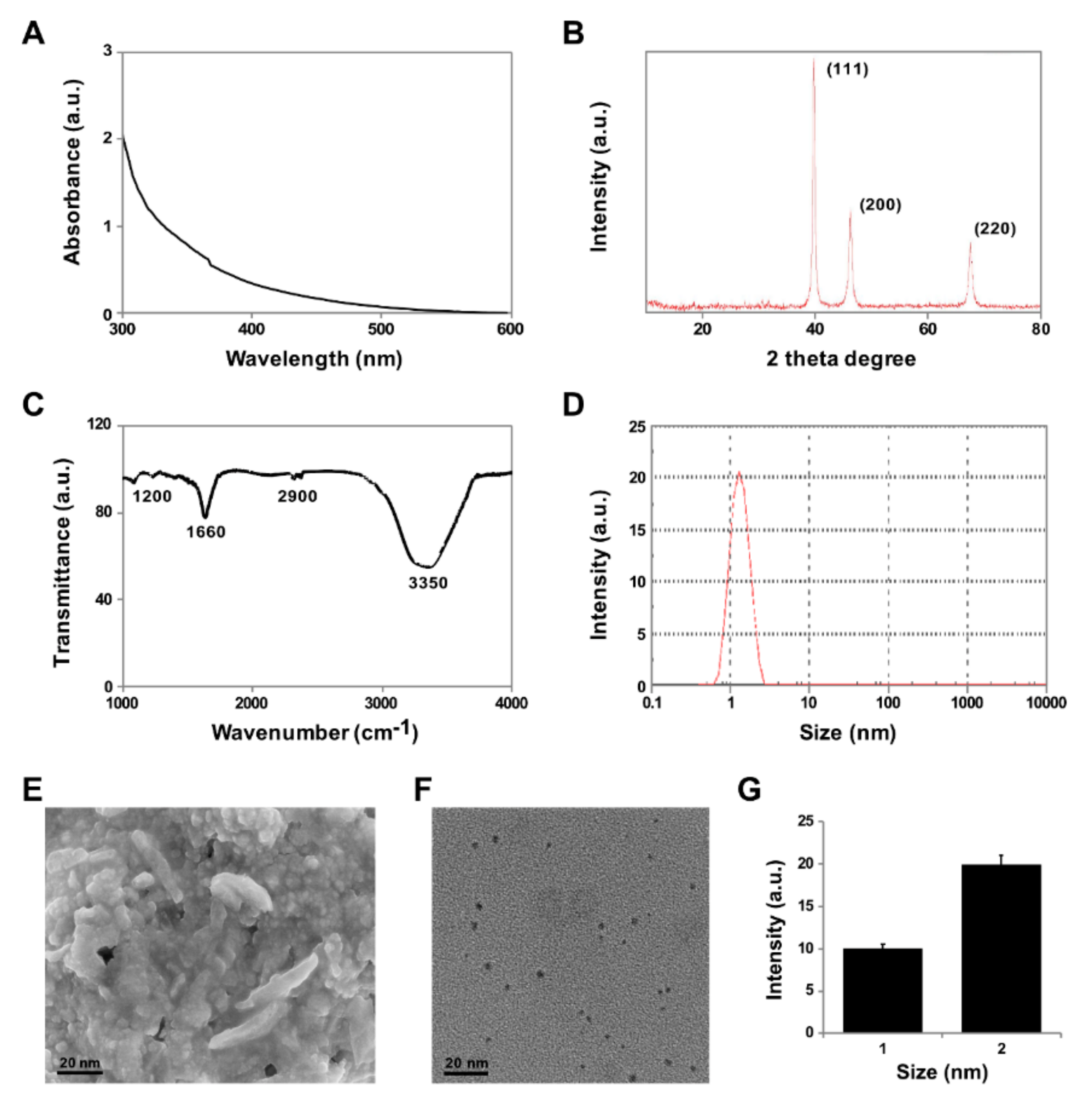
3.1. Synthesis and Characterization of PtNPs Using Apigenin
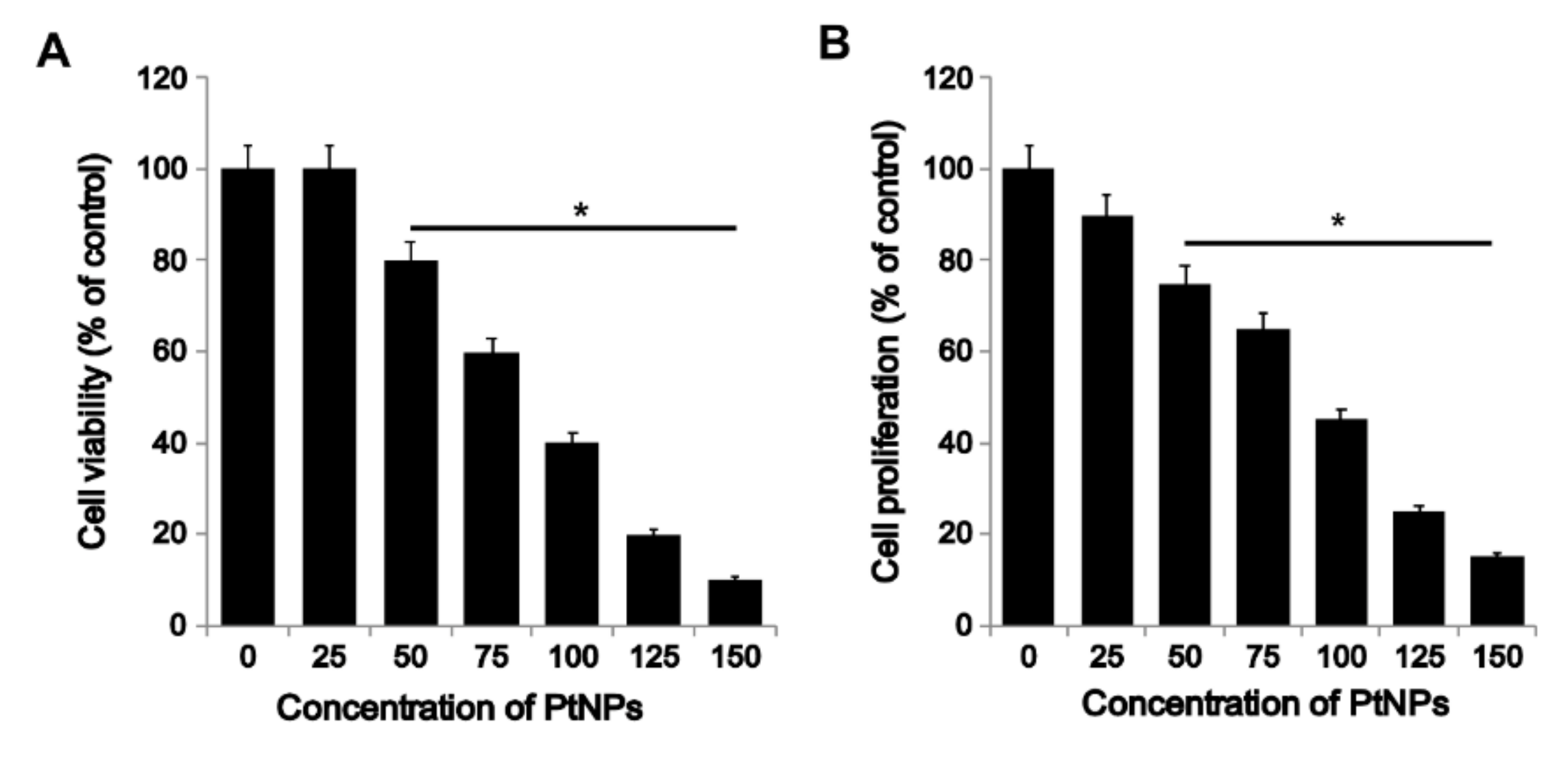
3.2. PtNPs Decreased the Survival and Proliferation of THP-1 Cells
3.3. PtNPs Altered Cell Morphology
3.4. PtNPs Induced Toxicity via Increase in LDH Leakage and Decrease in THP-1 Cell Viability
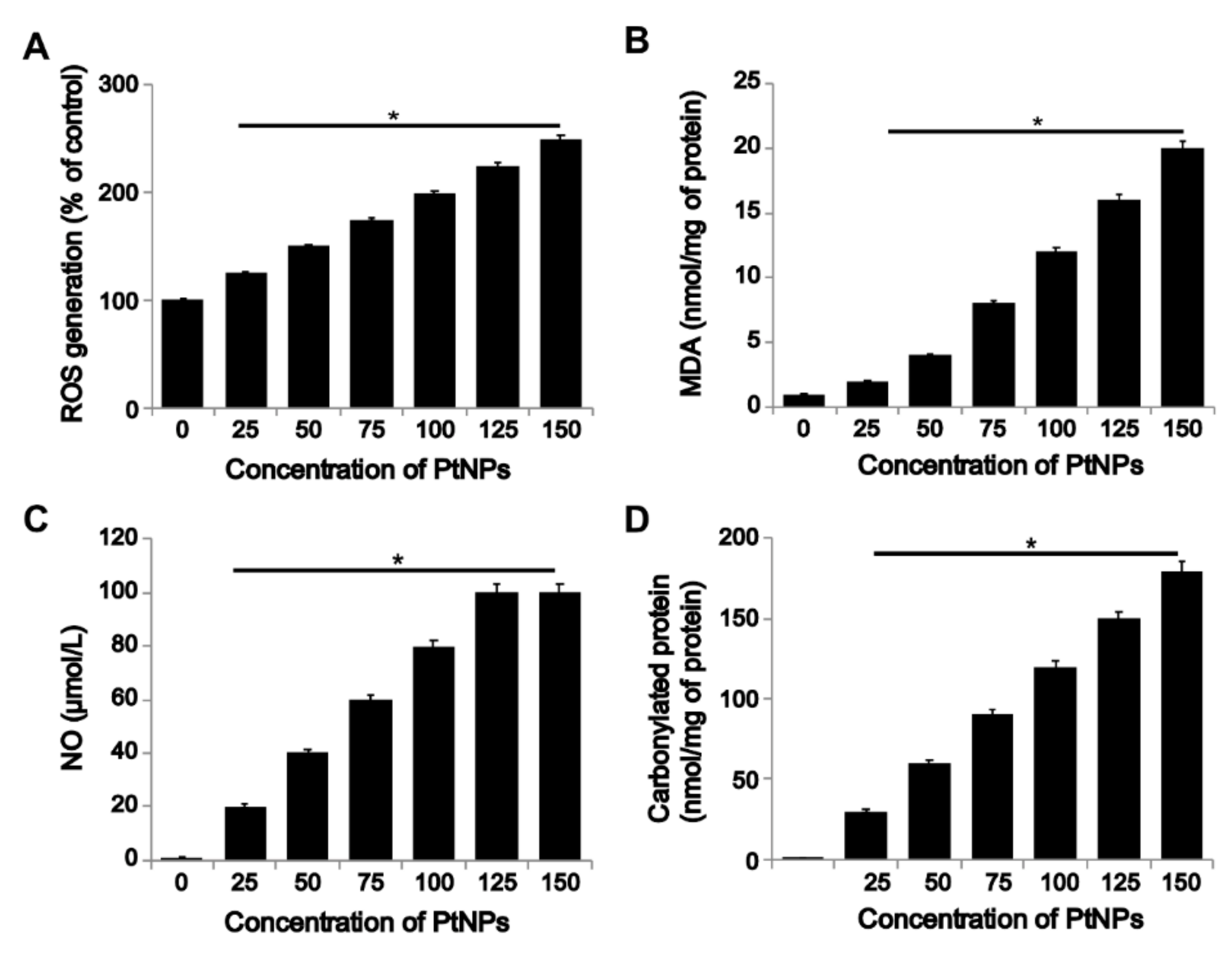
3.5. PtNPs Induced ROS Generation and Expression of Oxidative Stress Markers
3.6. Effect of PtNPs on Antioxidant Markers
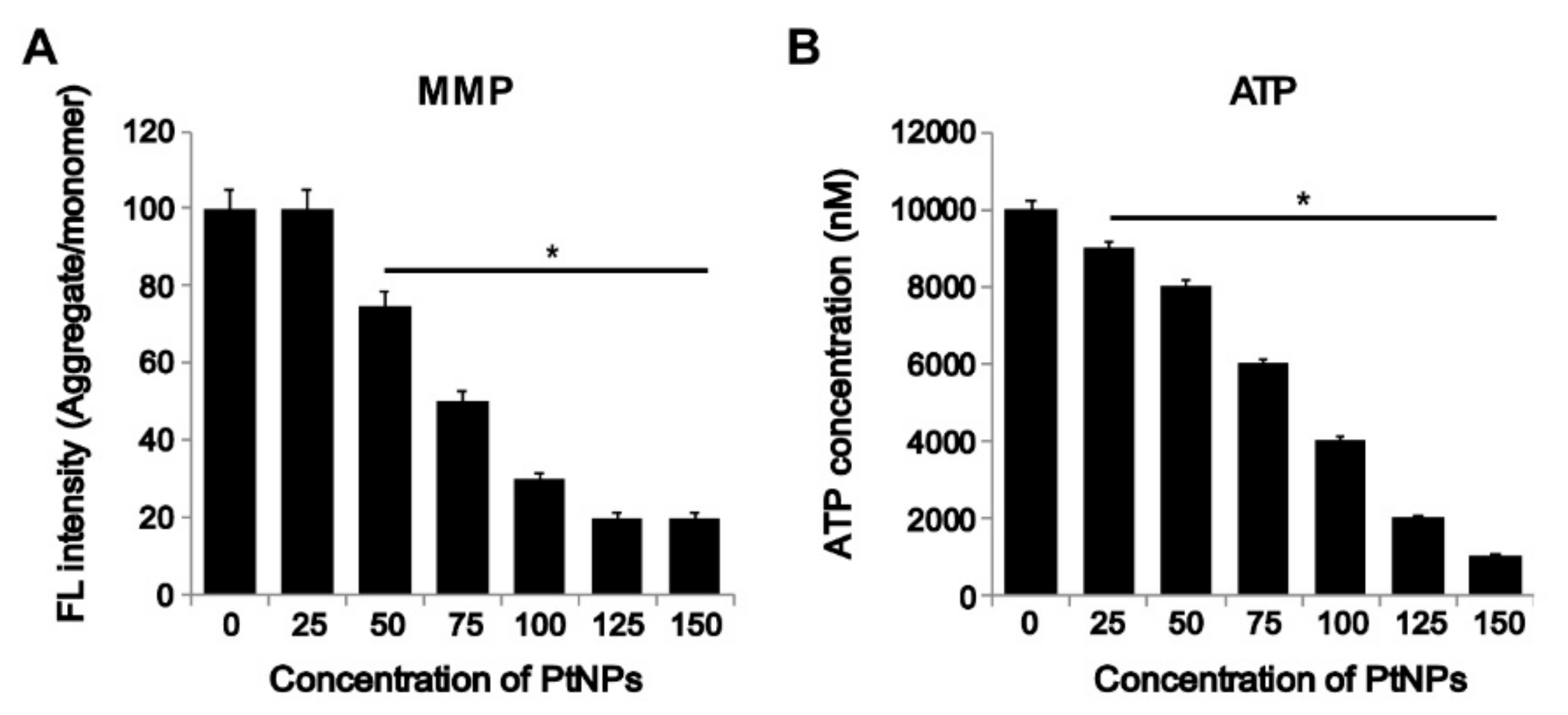
3.7. PtNPs Impaired Mitochondrial Membrane Potential (MMP) and Decreased ATP Level
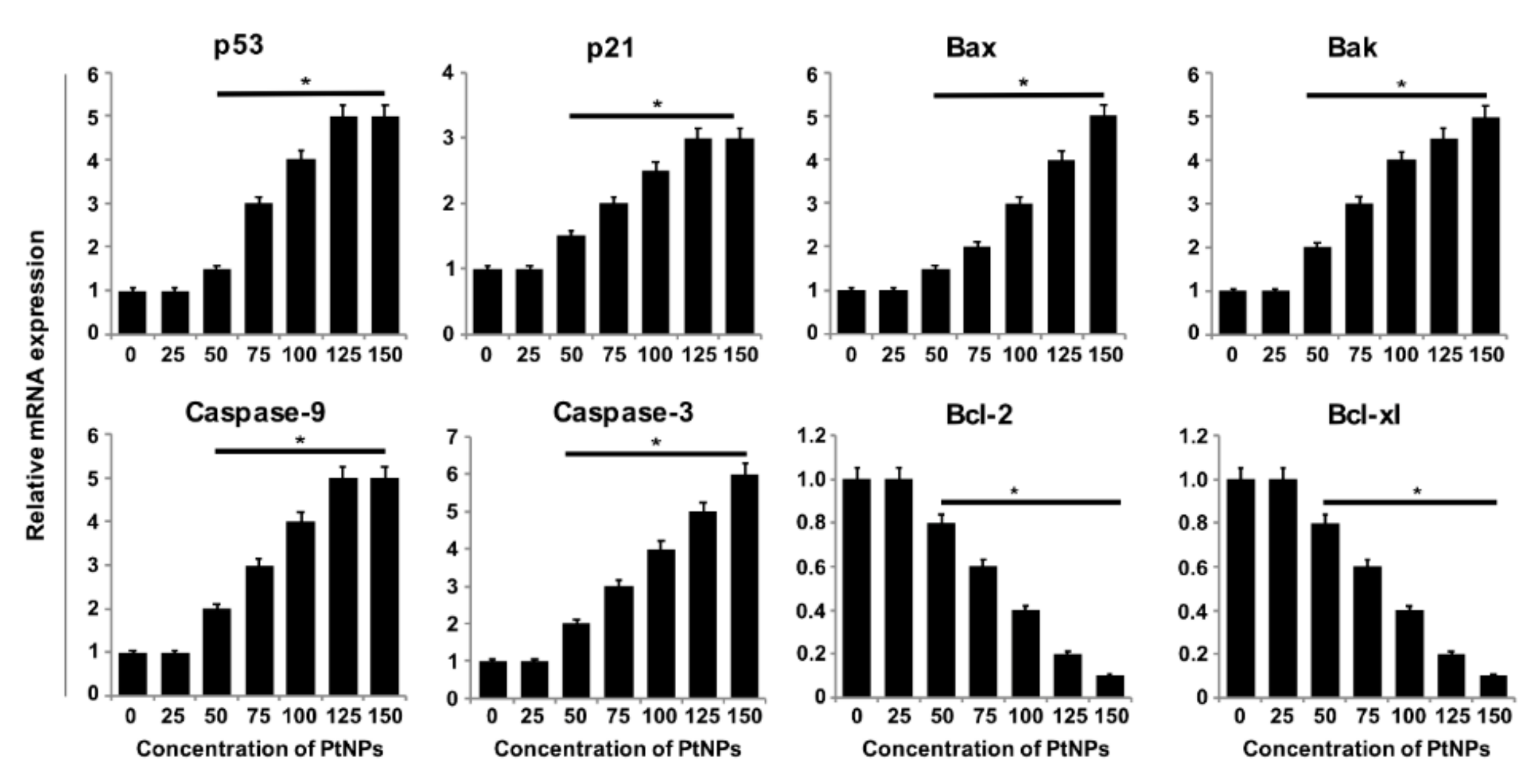
3.8. PtNPs Increased the Expression of Proapoptotic Genes and Decreased the Expression of Antiapoptotic Genes
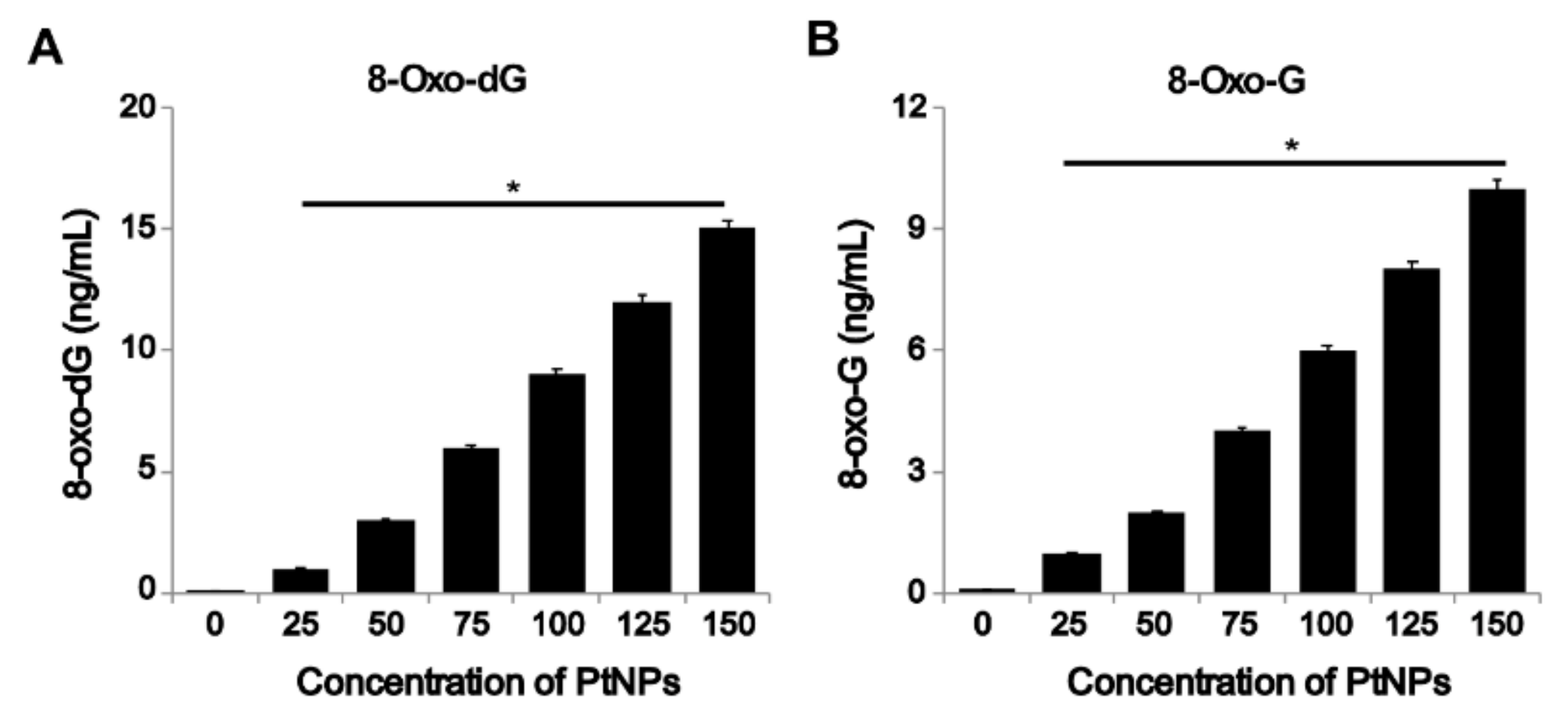
3.9. PtNPs Increased Oxidative Damage to DNA and RNA
3.10. PtNPs Regulated DNA Damage and Repair Genes
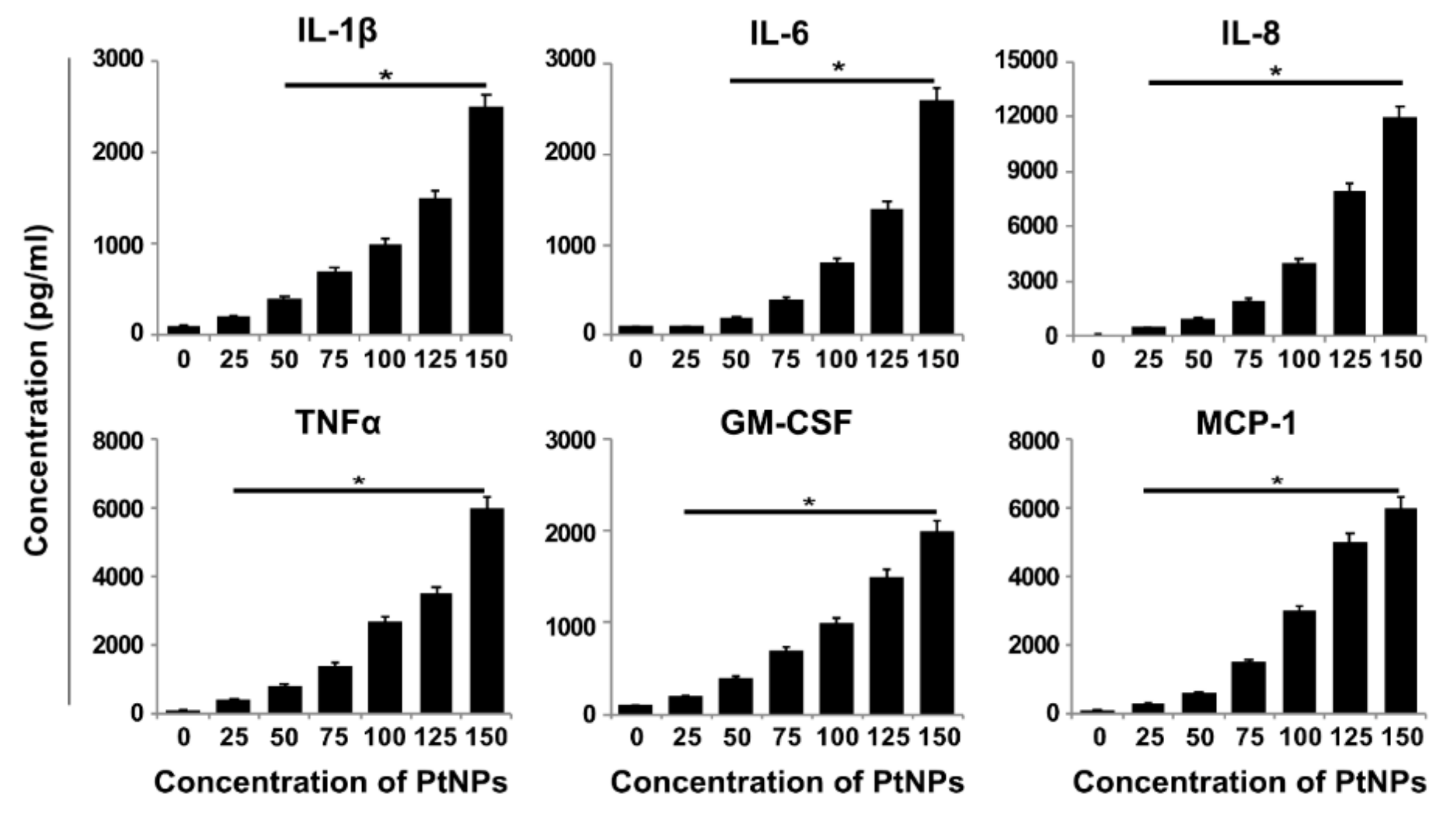
3.11. PtNPs Increased the Expression of Proinflammatory Cytokines and Chemokines
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pedone, D.; Moglianetti, M.; De Luca, E.; Bardi, G.; Pompa, P.P. Platinum nanoparticles in nanobiomedicine. Chem. Soc. Rev. 2017, 46, 4951–4975. [Google Scholar] [CrossRef]
- Shi, Y.; Lin, M.; Jiang, X.; Liang, S. Recent advances in FePt nanoparticles for biomedicine. J. Nanomater. 2015, 2015, 13. [Google Scholar] [CrossRef]
- Liu, Y.; Li, D.; Sun, S. Pt-based composite nanoparticles for magnetic, catalytic, and biomedical applications. J. Mater. Chem. 2011, 21, 12579–12587. [Google Scholar] [CrossRef]
- Jiang, K.; Zhao, D.; Guo, S.; Zhang, X.; Zhu, X.; Guo, J.; Lu, G.; Huang, X. Efficient oxygen reduction catalysis by subnanometer Pt alloy nanowires. Sci. Adv. 2017, 3, e1601705. [Google Scholar] [CrossRef] [PubMed]
- Tsung, C.-K.; Kuhn, J.N.; Huang, W.; Aliaga, C.; Hung, L.-I.; Somorjai, G.A.; Yang, P. Sub-10 nm platinum nanocrystals with size and shape control: Catalytic study for ethylene and pyrrole hydrogenation. J. Am. Chem. Soc. 2009, 131, 5816–5822. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, J.N.; Tsung, C.-K.; Huang, W.; Somorjai, G.A. Effect of organic capping layers over monodisperse platinum nanoparticles upon activity for ethylene hydrogenation and carbon monoxide oxidation. J. Catal. 2009, 265, 209–215. [Google Scholar] [CrossRef]
- Chen, A.; Holt-Hindle, P. Platinum-based nanostructured materials: synthesis, properties, and applications. Chem. Rev. 2010, 110, 3767–3804. [Google Scholar] [CrossRef] [PubMed]
- Leong, G.J.; Schulze, M.C.; Strand, M.B.; Maloney, D.; Frisco, S.L.; Dinh, H.N.; Pivovar, B.; Richards, R.M. Shape-directed platinum nanoparticle synthesis: Nanoscale design of novel catalysts. Appl. Organomet. Chem. 2014, 28, 1–17. [Google Scholar] [CrossRef]
- Konishi, Y.; Ohno, K.; Saitoh, N.; Nomura, T.; Nagamine, S.; Hishida, H.; Takahashi, Y.; Uruga, T. Bioreductive deposition of platinum nanoparticles on the bacterium Shewanella algae. J. Biotechnol. 2007, 128, 648–653. [Google Scholar] [CrossRef]
- Brayner, R.; Barberousse, H.; Hemadi, M.; Djedjat, C.; Yéprémian, C.; Coradin, T.; Livage, J.; Fiévet, F.; Couté, A. Cyanobacteria as bioreactors for the synthesis of Au, Ag, Pd, and Pt nanoparticles via an enzyme-mediated route. J. Nanosci. Nanotechnol. 2007, 7, 2696–2708. [Google Scholar] [CrossRef]
- Lengke, M.F.; Fleet, M.E.; Southam, G. Synthesis of platinum nanoparticles by reaction of filamentous cyanobacteria with platinum(IV)−chloride complex. Langmuir 2006, 22, 7318–7323. [Google Scholar] [CrossRef]
- Shiny, P.J.; Mukherjee, A.; Chandrasekaran, N. DNA damage and mitochondria-mediated apoptosis of A549 lung carcinoma cells induced by biosynthesised silver and platinum nanoparticles. RSC Adv. 2016, 6, 27775–27787. [Google Scholar] [CrossRef]
- Govender, Y.; Riddin, T.; Gericke, M.; Whiteley, C.G. Bioreduction of platinum salts into nanoparticles: A mechanistic perspective. Biotechnol. Lett. 2009, 31, 95–100. [Google Scholar] [CrossRef]
- Lin, X.; Wu, M.; Wu, D.; Kuga, S.; Endo, T.; Huang, Y. Platinum nanoparticles using wood nanomaterials: Eco-friendly synthesis, shape control and catalytic activity for p-nitrophenol reduction. Green Chem. 2011, 13, 283–287. [Google Scholar] [CrossRef]
- Venu, R.; Ramulu, T.S.; Anandakumar, S.; Rani, V.S.; Kim, C.G. Bio-directed synthesis of platinum nanoparticles using aqueous honey solutions and their catalytic applications. Colloids. Surf. A Physicochem. Eng. Asp. 2011, 384, 733–738. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Yan, Q.; Shen, W.; Gurunathan, S. Trichostatin a enhances the apoptotic potential of palladium nanoparticles in human cervical cancer cells. Int. J. Mol. Sci. 2016, 17, 1354. [Google Scholar] [CrossRef]
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Effects of silver nanoparticles on multiple drug-resistant strains of Staphylococcus aureus and Pseudomonas aeruginosa from mastitis-infected goats: An alternative approach for antimicrobial therapy. Int. J. Mol. Sci. 2017, 18, 569. [Google Scholar] [CrossRef]
- Yuan, Y.-G.; Peng, Q.-L.; Gurunathan, S. Silver nanoparticles enhance the apoptotic potential of gemcitabine in human ovarian cancer cells: Combination therapy for effective cancer treatment. Int. J. Nanomed. 2017, 12, 6487–6502. [Google Scholar] [CrossRef]
- Zhang, X.-F.; Huang, F.-H.; Zhang, G.-L.; Bai, D.-P.; Massimo, D.F.; Huang, Y.-F.; Gurunathan, S. Novel biomolecule lycopene-reduced graphene oxide-silver nanoparticle enhances apoptotic potential of trichostatin A in human ovarian cancer cells (SKOV3). Int. J. Nanomed. 2017, 12, 7551–7575. [Google Scholar] [CrossRef] [PubMed]
- Choi, Y.-J.; Gurunathan, S.; Kim, J.-H. Graphene oxide–silver nanocomposite enhances cytotoxic and apoptotic potential of salinomycin in human ovarian cancer stem cells (OvCSCs): A novel approach for cancer therapy. Int. J. Mol. Sci. 2018, 19, 710. [Google Scholar] [CrossRef]
- Gurunathan, S.; Kim, J.-H. Biocompatible gold nanoparticles ameliorate retinoic acid-induced cell death and induce differentiation in F9 teratocarcinoma stem cells. Nanomaterials 2018, 8, 396. [Google Scholar] [CrossRef]
- Horie, M.; Kato, H.; Endoh, S.; Fujita, K.; Nishio, K.; Komaba, L.K.; Fukui, H.; Nakamura, A.; Miyauchi, A.; Nakazato, T.; et al. Evaluation of cellular influences of platinum nanoparticles by stable medium dispersion. Metallomics 2011, 3, 1244–1252. [Google Scholar] [CrossRef]
- Moglianetti, M.; De Luca, E.; Pedone, D.; Marotta, R.; Catelani, T.; Sartori, B.; Amenitsch, H.; Retta, S.F.; Pompa, P.P. Platinum nanozymes recover cellular ROS homeostasis in an oxidative stress-mediated disease model. Nanoscale 2016, 8, 3739–3752. [Google Scholar] [CrossRef] [PubMed]
- Loos, C.; Syrovets, T.; Musyanovych, A.; Mailänder, V.; Landfester, K.; Nienhaus, G.U.; Simmet, T. Functionalized polystyrene nanoparticles as a platform for studying bio-nano interactions. Beilstein J. Nanotechnol. 2014, 5, 2403–2412. [Google Scholar] [CrossRef] [PubMed]
- Conti, M.E.; Pino, A.; Botrè, F.; Bocca, B.; Alimonti, A. Lichen Usnea barbata as biomonitor of airborne elements deposition in the Province of Tierra del Fuego (southern Patagonia, Argentina). Ecotoxicol. Environ. Saf. 2009, 72, 1082–1089. [Google Scholar] [CrossRef] [PubMed]
- Bendale, Y.; Bendale, V.; Paul, S. Evaluation of cytotoxic activity of platinum nanoparticles against normal and cancer cells and its anticancer potential through induction of apoptosis. Integr. Med. Res. 2017, 6, 141–148. [Google Scholar] [CrossRef] [PubMed]
- Brown, A.L.; Kai, M.P.; DuRoss, A.N.; Sahay, G.; Sun, C. Biodistribution and toxicity of micellar platinum nanoparticles in mice via intravenous administration. Nanomaterials 2018, 8, 410. [Google Scholar] [CrossRef]
- Gatto, F.; Moglianetti, M.; Pompa, P.P.; Bardi, G. Platinum nanoparticles decrease reactive oxygen species and modulate gene expression without alteration of immune responses in THP-1 monocytes. Nanomaterials 2018, 8, 392. [Google Scholar] [CrossRef]
- Katsumi, H.; Fukui, K.; Sato, K.; Maruyama, S.; Yamashita, S.; Mizumoto, E.; Kusamori, K.; Oyama, M.; Sano, M.; Sakane, T.; et al. Pharmacokinetics and preventive effects of platinum nanoparticles as reactive oxygen species scavengers on hepatic ischemia/reperfusion injury in mice. Metallomics 2014, 6, 1050–1056. [Google Scholar] [CrossRef] [PubMed]
- Konieczny, P.; Goralczyk, A.G.; Szmyd, R.; Skalniak, L.; Koziel, J.; Filon, F.L.; Crosera, M.; Cierniak, A.; Zuba-Surma, E.K.; Borowczyk, J.; et al. Effects triggered by platinum nanoparticles on primary keratinocytes. Int. J. Nanomed. 2013, 8, 3963–3975. [Google Scholar] [CrossRef]
- Hashimoto, M.; Kawai, K.; Kawakami, H.; Imazato, S. Matrix metalloproteases inhibition and biocompatibility of gold and platinum nanoparticles. J. Biomed. Mater. Res. A 2016, 104, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Nejdl, L.; Kudr, J.; Moulick, A.; Hegerova, D.; Ruttkay-Nedecky, B.; Gumulec, J.; Cihalova, K.; Smerkova, K.; Dostalova, S.; Krizkova, S.; et al. Platinum nanoparticles induce damage to DNA and inhibit DNA replication. PLoS ONE 2017, 12, e0180798. [Google Scholar] [CrossRef]
- Asharani, P.V.; Lianwu, Y.; Gong, Z.; Valiyaveettil, S. Comparison of the toxicity of silver, gold and platinum nanoparticles in developing zebrafish embryos. Nanotoxicology 2011, 5, 43–54. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Eppakayala, V.; Kim, J.H. Green synthesis of graphene and its cytotoxic effects in human breast cancer cells. Int. J. Nanomed. 2013, 8, 1015–1027. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Qasim, M.; Park, C.; Yoo, H.; Choi, D.Y.; Song, H.; Park, C.; Kim, J.H.; Hong, K. Cytotoxicity and transcriptomic analysis of silver nanoparticles in mouse embryonic fibroblast cells. Int. J. Mol. Sci. 2018, 19, 618. [Google Scholar] [CrossRef]
- Chen, F.; Kuhn, D.C.; Sun, S.-C.; Gaydos, L.J.; Demers, L.M. Dependence and reversal of nitric oxide production on NF-κ-B in silica and lipopolysaccharide induced macrophages. Biochem. Biophys. Res. Commun. 1995, 214, 839–846. [Google Scholar] [CrossRef]
- Maisonneuve, E.; Fraysse, L.; Lignon, S.; Capron, L.; Dukan, S. Carbonylated proteins are detectable only in a degradation-resistant aggregate state in Escherichia coli. J. Bacteriol. 2008, 190, 6609–6614. [Google Scholar] [CrossRef] [PubMed]
- Gurunathan, S.; Kang, M.-H.; Jeyaraj, M.; Kim, J.-H. Differential immunomodulatory effect of graphene oxide and vanillin-functionalized graphene oxide nanoparticles in human acute monocytic leukemia cell line (THP-1). Int. J. Mol. Sci. 2019, 20, 247. [Google Scholar] [CrossRef] [PubMed]
- Sliwinska, A.; Kwiatkowski, D.; Czarny, P.; Toma, M.; Wigner, P.; Drzewoski, J.; Fabianowska-Majewska, K.; Szemraj, J.; Maes, M.; Galecki, P. The levels of 7, 8-dihydrodeoxyguanosine (8-oxoG) and 8-oxoguanine DNA glycosylase 1 (OGG1)–A potential diagnostic biomarkers of Alzheimer’s disease. J. Neurol. Sci. 2016, 368, 155–159. [Google Scholar] [CrossRef] [PubMed]
- Karthik, R.; Sasikumar, R.; Chen, S.-M.; Govindasamy, M.; Kumar, J.V.; Muthuraj, V. Green synthesis of platinum nanoparticles using quercus glauca extract and its electrochemical oxidation of hydrazine in water samples. Int. J. Electrochem. Sci. 2016, 11, 8245–8255. [Google Scholar] [CrossRef]
- Ghosh, S.; Nitnavare, R.; Dewle, A.; Tomar, G.B.; Chippalkatti, R.; More, P.; Kitture, R.; Kale, S.; Bellare, J.; Chopade, B.A. Novel platinum-palladium bimetallic nanoparticles synthesized by Dioscorea bulbifera: Anticancer and antioxidant activities. Int. J. Nanomed. 2015, 10, 7477–7490. [Google Scholar] [CrossRef]
- Labrador-Rached, C.J.; Browning, R.T.; Braydich-Stolle, L.K.; Comfort, K.K. Toxicological implications of platinum nanoparticle exposure: Stimulation of intracellular stress, inflammatory response, and akt signaling in vitro. J. Toxicol. 2018, 2018, 8. [Google Scholar] [CrossRef]
- Chekin, F.; Ghasemi, S. Silver nanoparticles prepared in presence of ascorbic acid and gelatin, and their electrocatalytic application. Bull. Mater. Sci. 2014, 37, 1433–1437. [Google Scholar] [CrossRef]
- Liu, Y.; Zheng, Z.; Zara, J.N.; Hsu, C.; Soofer, D.E.; Lee, K.S.; Siu, R.K.; Miller, L.S.; Zhang, X.; Carpenter, D.; et al. The antimicrobial and osteoinductive properties of silver nanoparticle/poly (dl-lactic-co-glycolic acid)-coated stainless steel. Biomaterials 2012, 33, 8745–8756. [Google Scholar] [CrossRef]
- Shah, A.; Khan, S.B.; Asiri, A.M.; Hussain, H.; Han, C.; Qureshi, R.; Ashiq, M.N.; Zia, M.A.; Ishaq, M.; Kraatz, H.-B. Synthesis, characterization, and application of Au–Ag alloy nanoparticles for the sensing of an environmental toxin, pyrene. J. App. Electrochem. 2015, 45, 463–472. [Google Scholar] [CrossRef]
- Al-Radadi, N.S. Green synthesis of platinum nanoparticles using Saudi’s Dates extract and their usage on the cancer cell treatment. Arab. J. Chem. 2019, 12, 330–349. [Google Scholar] [CrossRef]
- Han, J.W.; Gurunathan, S.; Jeong, J.K.; Choi, Y.J.; Kwon, D.N.; Park, J.K.; Kim, J.H. Oxidative stress mediated cytotoxicity of biologically synthesized silver nanoparticles in human lung epithelial adenocarcinoma cell line. Nanoscale Res. Lett. 2014, 9, 459. [Google Scholar] [CrossRef] [PubMed]
- Bendale, Y.; Bendale, V.; Natu, R.; Paul, S. Biosynthesized platinum nanoparticles inhibit the proliferation of human lung-cancer cells in vitro and delay the growth of a human lung-tumor xenograft in vivo: In vitro and in vivo anticancer activity of bio-Pt NPs. J. Pharmacopunct. 2016, 19, 114–121. [Google Scholar] [CrossRef]
- Kutwin, M.; Sawosz, E.; Jaworski, S.; Wierzbicki, M.; Strojny, B.; Grodzik, M.; Chwalibog, A. Assessment of the proliferation status of glioblastoma cell and tumour tissue after nanoplatinum treatment. PLoS ONE 2017, 12, e0178277. [Google Scholar] [CrossRef]
- Antognelli, C.; Gambelunghe, A.; Muzi, G.; Talesa, V.N. Glyoxalase I drives epithelial-to-mesenchymal transition via argpyrimidine-modified Hsp70, miR-21 and SMAD signalling in human bronchial cells BEAS-2B chronically exposed to crystalline silica Min-U-Sil 5: Transformation into a neoplastic-like phenotype. Free Radic. Biol. Med. 2016, 92, 110–125. [Google Scholar] [CrossRef]
- Hussain, S.M.; Hess, K.L.; Gearhart, J.M.; Geiss, K.T.; Schlager, J.J. In vitro toxicity of nanoparticles in BRL 3A rat liver cells. Toxicol. In Vitro 2005, 19, 975–983. [Google Scholar] [CrossRef]
- Park, J.H.; Gurunathan, S.; Choi, Y.-J.; Han, J.W.; Song, H.; Kim, J.-H. Silver nanoparticles suppresses brain-derived neurotrophic factor-induced cell survival in the human neuroblastoma cell line SH-SY5Y. J. Ind. Eng. Chem. 2017, 47, 62–73. [Google Scholar] [CrossRef]
- Gurunathan, S.; Han, J.W.; Park, J.H.; Kim, E.; Choi, Y.J.; Kwon, D.N.; Kim, J.H. Reduced graphene oxide-silver nanoparticle nanocomposite: A potential anticancer nanotherapy. Int. J. Nanomed. 2015, 10, 6257–6276. [Google Scholar] [CrossRef]
- Wright, P.C.; Qin, H.; Choi, M.M.; Chiu, N.H.; Jia, Z. Carbon nanodots interference with lactate dehydrogenase assay in human monocyte THP-1 cells. SpringerPlus 2014, 3, 615–615. [Google Scholar] [CrossRef][Green Version]
- Athinarayanan, J.; Periasamy, V.S.A.; Alshatwi, A. Eco-friendly synthesis and characterization of platinum-copper alloy nanoparticles induce cell death in human cervical cancer cells. Process. Biochem. 2016, 51, 925–932. [Google Scholar] [CrossRef]
- Poli, G.; Leonarduzzi, G.; Biasi, F.; Chiarpotto, E. Oxidative stress and cell signalling. Curr. Med. Chem. 2004, 11, 1163–1182. [Google Scholar] [CrossRef]
- Kim, W.-K.; Kim, J.-C.; Park, H.-J.; Sul, O.-J.; Lee, M.-H.; Kim, J.-S.; Choi, H.-S. Platinum nanoparticles reduce ovariectomy-induced bone loss by decreasing osteoclastogenesis. Exp. Mol. Med. 2012, 44, 432–439. [Google Scholar] [CrossRef] [PubMed]
- Gurr, J.R.; Wang, A.S.; Chen, C.H.; Jan, K.Y. Ultrafine titanium dioxide particles in the absence of photoactivation can induce oxidative damage to human bronchial epithelial cells. Toxicology 2005, 213, 66–73. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, A.; Weiss, D.G.; Jonas, L.; Kriehuber, R. Oxidative stress-induced cytotoxic and genotoxic effects of nano-sized titanium dioxide particles in human HaCaT keratinocytes. Toxicology 2012, 296, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Xu, L.; Zhang, T.; Ren, G.; Yang, Z. Oxidative stress and apoptosis induced by nanosized titanium dioxide in PC12 cells. Toxicology 2010, 267, 172–177. [Google Scholar] [CrossRef]
- Xue, C.; Wu, J.; Lan, F.; Liu, W.; Yang, X.; Zeng, F.; Xu, H. Nano titanium dioxide induces the generation of ROS and potential damage in HaCaT cells under UVA irradiation. J. Nanosci. Nanotechnol. 2010, 10, 8500–8507. [Google Scholar] [CrossRef]
- Valko, M.; Rhodes, C.J.; Moncol, J.; Izakovic, M.; Mazur, M. Free radicals, metals and antioxidants in oxidative stress-induced cancer. Chem. Biol. Interact. 2006, 160, 1–40. [Google Scholar] [CrossRef]
- Radu, M.; Munteanu, M.C.; Petrache, S.; Serban, A.I.; Dinu, D.; Hermenean, A.; Sima, C.; Dinischiotu, A. Depletion of intracellular glutathione and increased lipid peroxidation mediate cytotoxicity of hematite nanoparticles in MRC-5 cells. Acta Biochim. Pol. 2010, 57, 355–360. [Google Scholar] [CrossRef] [PubMed]
- Brown, G.C. Nitric oxide and neuronal death. Nitric Oxide 2010, 23, 153–165. [Google Scholar] [CrossRef] [PubMed]
- Wink, D.A.; Mitchell, J.B. Chemical biology of nitric oxide: Insights into regulatory, cytotoxic, and cytoprotective mechanisms of nitric oxide. Free Radic. Biol. Med. 1998, 25, 434–456. [Google Scholar] [CrossRef]
- Antognelli, C.; Gambelunghe, A.; Muzi, G.; Talesa, V.N. Peroxynitrite-mediated glyoxalase I epigenetic inhibition drives apoptosis in airway epithelial cells exposed to crystalline silica via a novel mechanism involving argpyrimidine-modified Hsp70, JNK, and NF-kappaB. Free Radic. Biol. Med. 2015, 84, 128–141. [Google Scholar] [CrossRef]
- Curtis, J.M.; Hahn, W.S.; Long, E.K.; Burrill, J.S.; Arriaga, E.A.; Bernlohr, D.A. Protein carbonylation and metabolic control systems. TEM 2012, 23, 399–406. [Google Scholar] [CrossRef] [PubMed]
- Antognelli, C.; Gambelunghe, A.; Del Buono, C.; Murgia, N.; Talesa, V.N.; Muzi, G. Crystalline silica Min-U-Sil 5 induces oxidative stress in human bronchial epithelial cells BEAS-2B by reducing the efficiency of antiglycation and antioxidant enzymatic defenses. Chem. Biol. Interact. 2009, 182, 13–21. [Google Scholar] [CrossRef]
- Haase, A.; Tentschert, J.; Jungnickel, H.; Graf, P.; Mantion, A.; Draude, F.; Plendl, J.; Goetz, M.E.; Galla, S.; Mašić, A.; et al. Toxicity of silver nanoparticles in human macrophages: Uptake, intracellular distribution and cellular responses. J. Phys. Conf. Ser. 2011, 304, 012030. [Google Scholar] [CrossRef]
- Dayem, A.A.; Kim, B.; Gurunathan, S.; Choi, H.Y.; Yang, G.; Saha, S.K.; Han, D.; Han, J.; Kim, K.; Kim, J.-H.; et al. Biologically synthesized silver nanoparticles induce neuronal differentiation of SH-SY5Y cells via modulation of reactive oxygen species, phosphatases, and kinase signaling pathways. Biotechnol. J. 2014, 9, 934–943. [Google Scholar] [CrossRef]
- Yusof, F.; Ismail, N.A.S. Antioxidants effects of platinum nanoparticles: A potential alternative treatment to lung diseases. J. App. Pharm. Sci. 2015, 5, 140–145. [Google Scholar] [CrossRef]
- Fu, P.P.; Xia, Q.; Hwang, H.-M.; Ray, P.C.; Yu, H. Mechanisms of nanotoxicity: Generation of reactive oxygen species. J. Food Drug Anal. 2014, 22, 64–75. [Google Scholar] [CrossRef] [PubMed]
- Nel, A.; Xia, T.; Mädler, L.; Li, N. Toxic potential of materials at the nanolevel. Science 2006, 311, 622. [Google Scholar] [CrossRef] [PubMed]
- Lushchak, O.; Zayachkivska, A.; Vaiserman, A. Metallic nanoantioxidants as potential therapeutics for type 2 diabetes: A hypothetical background and translational perspectives. Oxid. Med. Cell Longev. 2018, 2018, 9. [Google Scholar] [CrossRef] [PubMed]
- Almeer, R.S.; Ali, D.; Alarifi, S.; Alkahtani, S.; Almansour, M. Green platinum nanoparticles interaction with HEK293 cells: Cellular toxicity, apoptosis, and genetic damage. Dose Response 2018, 16. [Google Scholar] [CrossRef]
- Gong, Y.; Li, X.; Liao, G.; Ding, Y.; Li, J.; Cao, Y. Cytotoxicity and ER stress–apoptosis gene expression in ZnO nanoparticle exposed THP-1 macrophages: Influence of pre-incubation with BSA or palmitic acids complexed to BSA. RSC Adv. 2018, 8, 15380–15388. [Google Scholar] [CrossRef]
- Eustaquio, T.; Leary, J.F. Single-cell nanotoxicity assays of superparamagnetic iron oxide nanoparticles. Methods Mol. Biol. 2012, 926, 69–85. [Google Scholar] [CrossRef] [PubMed]
- Guo, C.; Ding, P.; Xie, C.; Ye, C.; Ye, M.; Pan, C.; Cao, X.; Zhang, S.; Zheng, S. Potential application of the oxidative nucleic acid damage biomarkers in detection of diseases. Oncotarget 2017, 8, 75767. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Zhou, J.; Wallace, S.S.; Burrows, C.J. A Role for the fifth G-Track in G-Quadruplex forming oncogene promoter sequences during oxidative stress: Do these “Spare Tires” have an evolved function? ACS Central Sci. 2015, 1, 226–233. [Google Scholar] [CrossRef]
- Guo, C.; Li, X.; Wang, R.; Yu, J.; Ye, M.; Mao, L.; Zhang, S.; Zheng, S. Association between oxidative DNA damage and risk of colorectal cancer: sensitive determination of urinary 8-Hydroxy-2′-deoxyguanosine by UPLC-MS/MS Analysis. Sci. Rep. 2016, 6, 32581. [Google Scholar] [CrossRef]
- Valko, M.; Izakovic, M.; Mazur, M.; Rhodes, C.J.; Telser, J. Role of oxygen radicals in DNA damage and cancer incidence. Mol. Cell. Biochem. 2004, 266, 37–56. [Google Scholar] [CrossRef]
- Nunomura, A.; Moreira, P.I.; Takeda, A.; Smith, M.A.; Perry, G. Oxidative RNA Damage and Neurodegeneration. Curr. Med. Chem. 2007, 14, 2968–2975. [Google Scholar] [CrossRef] [PubMed]
- Tsoli, M.; Kuhn, H.; Brandau, W.; Esche, H.; Schmid, G. Cellular uptake and toxicity of Au55 clusters. Small 2005, 1, 841–844. [Google Scholar] [CrossRef] [PubMed]
- Fleming, A.M.; Ding, Y.; Burrows, C.J. Oxidative DNA damage is epigenetic by regulating gene transcription via base excision repair. Proc. Nat. Acad. Sci. USA 2017, 114, 2604–2609. [Google Scholar] [CrossRef] [PubMed]
- Prasad, R.Y.; Chastain, P.D.; Nikolaishvili-Feinberg, N.; Smeester, L.; Kaufmann, W.K.; Fry, R.C. Titanium dioxide nanoparticles activate the ATM-Chk2 DNA damage response in human dermal fibroblasts. Nanotoxicology 2013, 7, 1111–1119. [Google Scholar] [CrossRef] [PubMed][Green Version]
- El-Said, K.S.; Ali, E.M.; Kanehira, K.; Taniguchi, A. Molecular mechanism of DNA damage induced by titanium dioxide nanoparticles in toll-like receptor 3 or 4 expressing human hepatocarcinoma cell lines. J. Nanobiotechnol. 2014, 12, 48. [Google Scholar] [CrossRef] [PubMed]
- Singh, N.; Jenkins, G.J.S.; Nelson, B.C.; Marquis, B.J.; Maffeis, T.G.G.; Brown, A.P.; Williams, P.M.; Wright, C.J.; Doak, S.H. The role of iron redox state in the genotoxicity of ultrafine superparamagnetic iron oxide nanoparticles. Biomaterials 2012, 33, 163–170. [Google Scholar] [CrossRef] [PubMed]
- Broustas, C.G.; Lieberman, H.B. BIOONE 2014, 181, 111–130.
- Evans, S.J.; Clift, M.J.D.; Singh, N.; de Oliveira Mallia, J.; Burgum, M.; Wills, J.W.; Wilkinson, T.S.; Jenkins, G.J.S.; Doak, S.H. Critical review of the current and future challenges associated with advanced in vitro systems towards the study of nanoparticle (secondary) genotoxicity. Mutagenesis 2017, 32, 233–241. [Google Scholar] [CrossRef] [PubMed]
- Biola-Clier, M.; Beal, D.; Caillat, S.; Libert, S.; Armand, L.; Herlin-Boime, N.; Sauvaigo, S.; Douki, T.; Carriere, M. Comparison of the DNA damage response in BEAS-2B and A549 cells exposed to titanium dioxide nanoparticles. Mutagenesis 2017, 32, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Radak, Z.; Boldogh, I. 8-Oxo-7,8-dihydroguanine: Links to gene expression, aging, and defense against oxidative stress. Free Radic. Biol. Med. 2010, 49, 587–596. [Google Scholar] [CrossRef]
- Beal, M.F. Oxidatively modified proteins in aging and disease1, 21Guest Editor: Earl Stadtman2This article is part of a series of reviews on “Oxidatively Modified Proteins in Aging and Disease.” The full list of papers may be found on the homepage of the journal. Free Radic. Biol. Med. 2002, 32, 797–803. [Google Scholar] [CrossRef]
- Jalava, P.I.; Salonen, R.O.; Pennanen, A.S.; Sillanpää, M.; Hälinen, A.I.; Happo, M.S.; Hillamo, R.; Brunekreef, B.; Katsouyanni, K.; Sunyer, J.; et al. Heterogeneities in inflammatory and cytotoxic responses of RAW 264.7 macrophage cell line to urban air coarse, fine, and ultrafine particles from six european sampling campaigns. Inhal. Toxicol. 2007, 19, 213–225. [Google Scholar] [CrossRef] [PubMed]
- Corsini, E.; Budello, S.; Marabini, L.; Galbiati, V.; Piazzalunga, A.; Barbieri, P.; Cozzutto, S.; Marinovich, M.; Pitea, D.; Galli, C.L. Comparison of wood smoke PM2.5 obtained from the combustion of FIR and beech pellets on inflammation and DNA damage in A549 and THP-1 human cell lines. Arch. Toxicol. 2013, 87, 2187–2199. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Wang, S.; Zhu, J.; Li, C.; Zhang, T.; Liu, H.; Xu, Q.; Ye, X.; Zhou, L.; Ye, L. Effect of Atmospheric PM2.5 on expression levels of NF-kappaB genes and inflammatory cytokines regulated by nf-kappab in human macrophage. Inflammation 2018, 41, 784–794. [Google Scholar] [CrossRef]
- Kusaka, T.; Nakayama, M.; Nakamura, K.; Ishimiya, M.; Furusawa, E.; Ogasawara, K. Effect of silica particle size on macrophage inflammatory responses. PLoS ONE 2014, 9, e92634. [Google Scholar] [CrossRef] [PubMed]
- Murphy, A.; Casey, A.; Byrne, G.; Chambers, G.; Howe, O. Silver nanoparticles induce pro-inflammatory gene expression and inflammasome activation in human monocytes. J. App. Toxicol. 2016, 36, 1311–1320. [Google Scholar] [CrossRef]
- Khan, H.A.; Alamery, S.; Ibrahim, K.E.; El-Nagar, D.M.; Al-Harbi, N.; Rusop, M.; Alrokayan, S.H. Size and time-dependent induction of proinflammatory cytokines expression in brains of mice treated with gold nanoparticles. Saudi J. Biol. Sci. 2019, 26, 625–631. [Google Scholar] [CrossRef]
- Gómez, D.M.; Urcuqui-Inchima, S.; Hernandez, J.C. Silica nanoparticles induce NLRP3 inflammasome activation in human primary immune cells. Innate Immun. 2017, 23, 697–708. [Google Scholar] [CrossRef] [PubMed]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gurunathan, S.; Jeyaraj, M.; Kang, M.-H.; Kim, J.-H. The Effects of Apigenin-Biosynthesized Ultra-Small Platinum Nanoparticles on the Human Monocytic THP-1 Cell Line. Cells 2019, 8, 444. https://doi.org/10.3390/cells8050444
Gurunathan S, Jeyaraj M, Kang M-H, Kim J-H. The Effects of Apigenin-Biosynthesized Ultra-Small Platinum Nanoparticles on the Human Monocytic THP-1 Cell Line. Cells. 2019; 8(5):444. https://doi.org/10.3390/cells8050444
Chicago/Turabian StyleGurunathan, Sangiliyandi, Muniyandi Jeyaraj, Min-Hee Kang, and Jin-Hoi Kim. 2019. "The Effects of Apigenin-Biosynthesized Ultra-Small Platinum Nanoparticles on the Human Monocytic THP-1 Cell Line" Cells 8, no. 5: 444. https://doi.org/10.3390/cells8050444
APA StyleGurunathan, S., Jeyaraj, M., Kang, M.-H., & Kim, J.-H. (2019). The Effects of Apigenin-Biosynthesized Ultra-Small Platinum Nanoparticles on the Human Monocytic THP-1 Cell Line. Cells, 8(5), 444. https://doi.org/10.3390/cells8050444




