Transcriptional Regulation of Selenoprotein F by Heat Shock Factor 1 during Selenium Supplementation and Stress Response
Abstract
:1. Introduction
2. Materials and Methods
2.1. Promoter Analysis and Transcription Factor Binding Sites Prediction by Bioinformatics
2.2. Plasmids Construction and Antibodies
2.3. Cell Culture and Transfection
2.4. Cell Treatment
2.5. Dual-Glo Luciferase Assay
2.6. Quantitative Real-Time PCR (Q-PCR)
2.7. Chromatin Immunoprecipitation (ChIP) Assay
2.8. Western Blotting
2.9. Data Analysis
3. Results
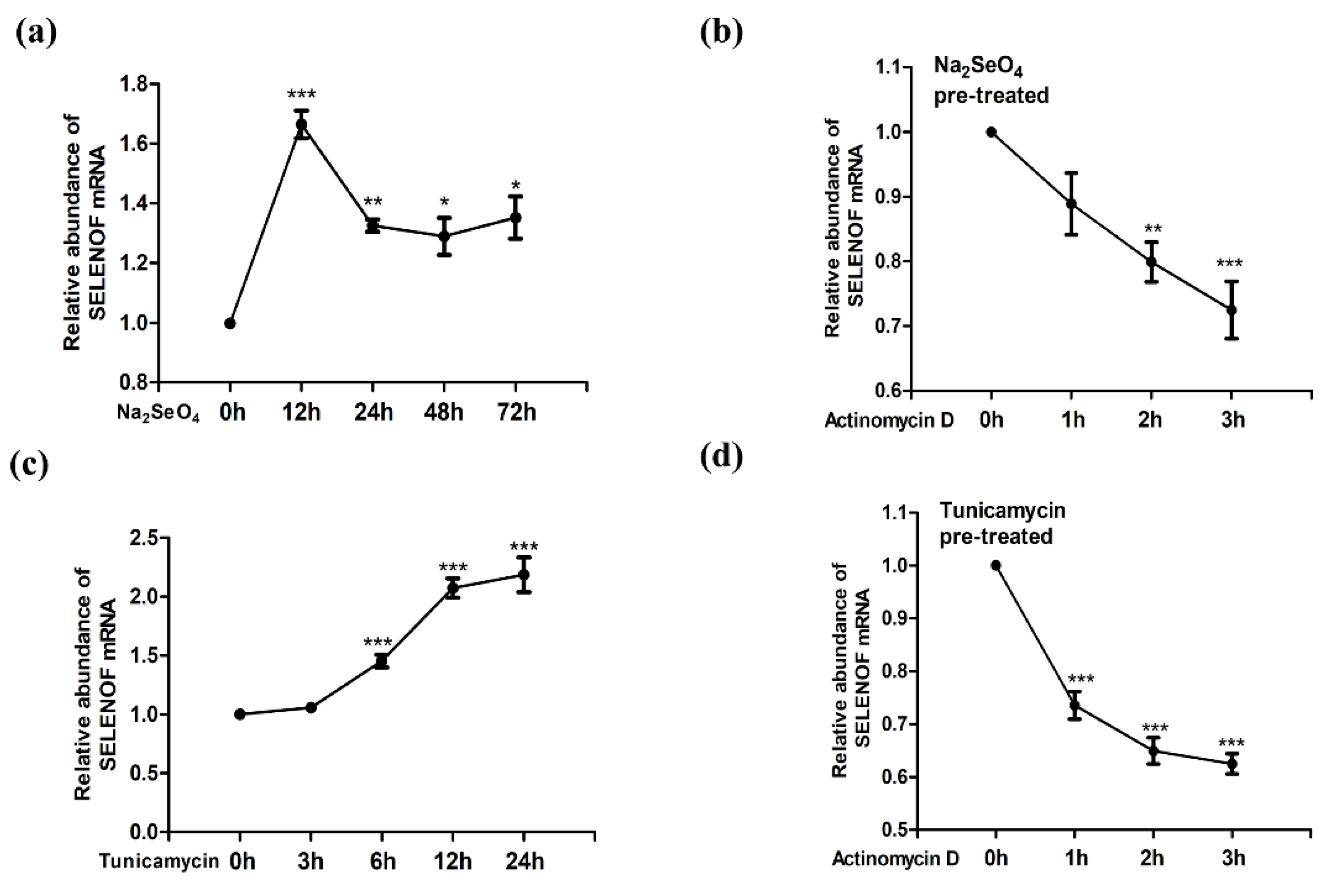
3.1. Up-Regulation of SELENOF mRNA by Selenate and Tm Treatment
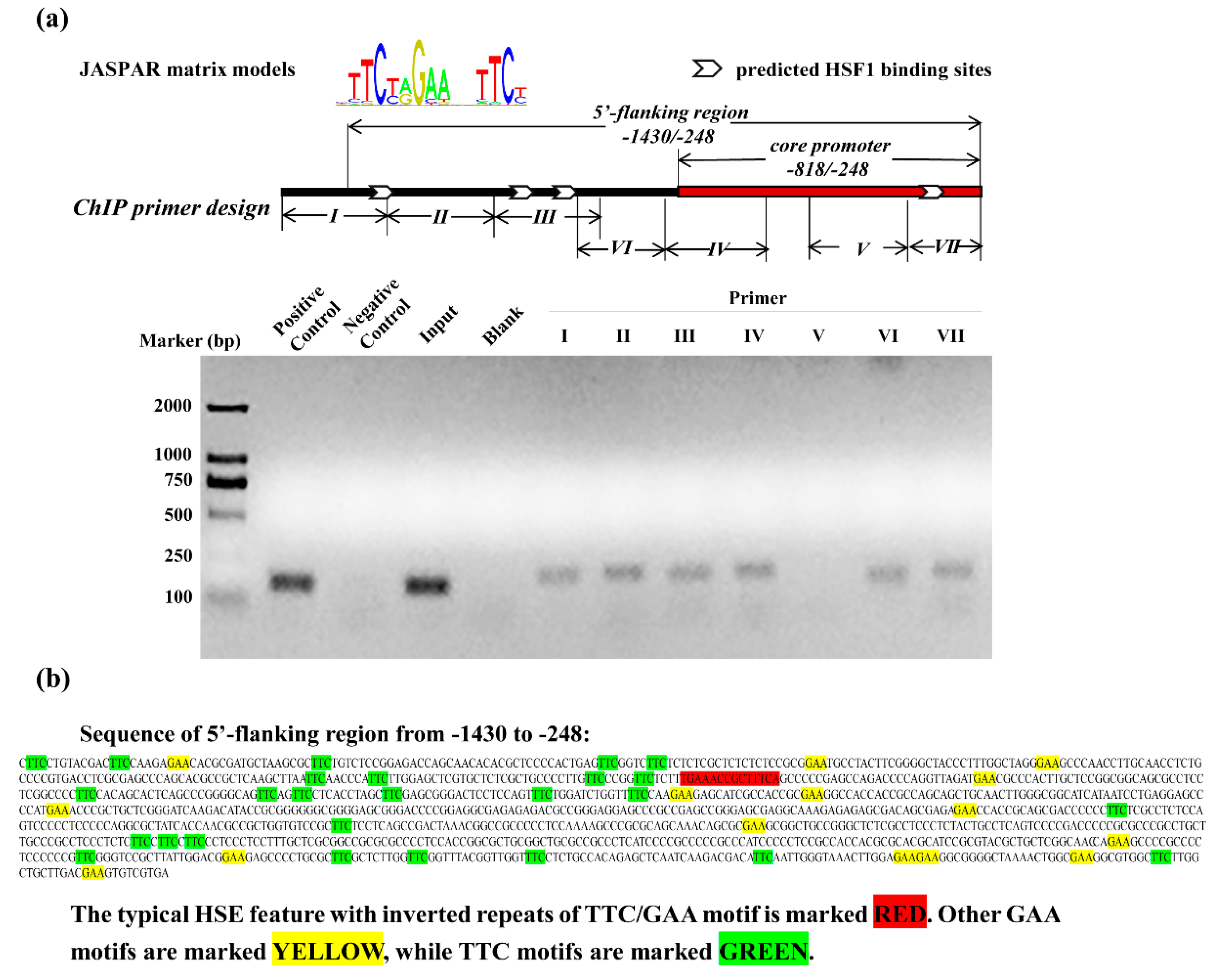
3.2. Identification of the Core Positive Regulatory Element in 5′-Flanking Region of SELENOF
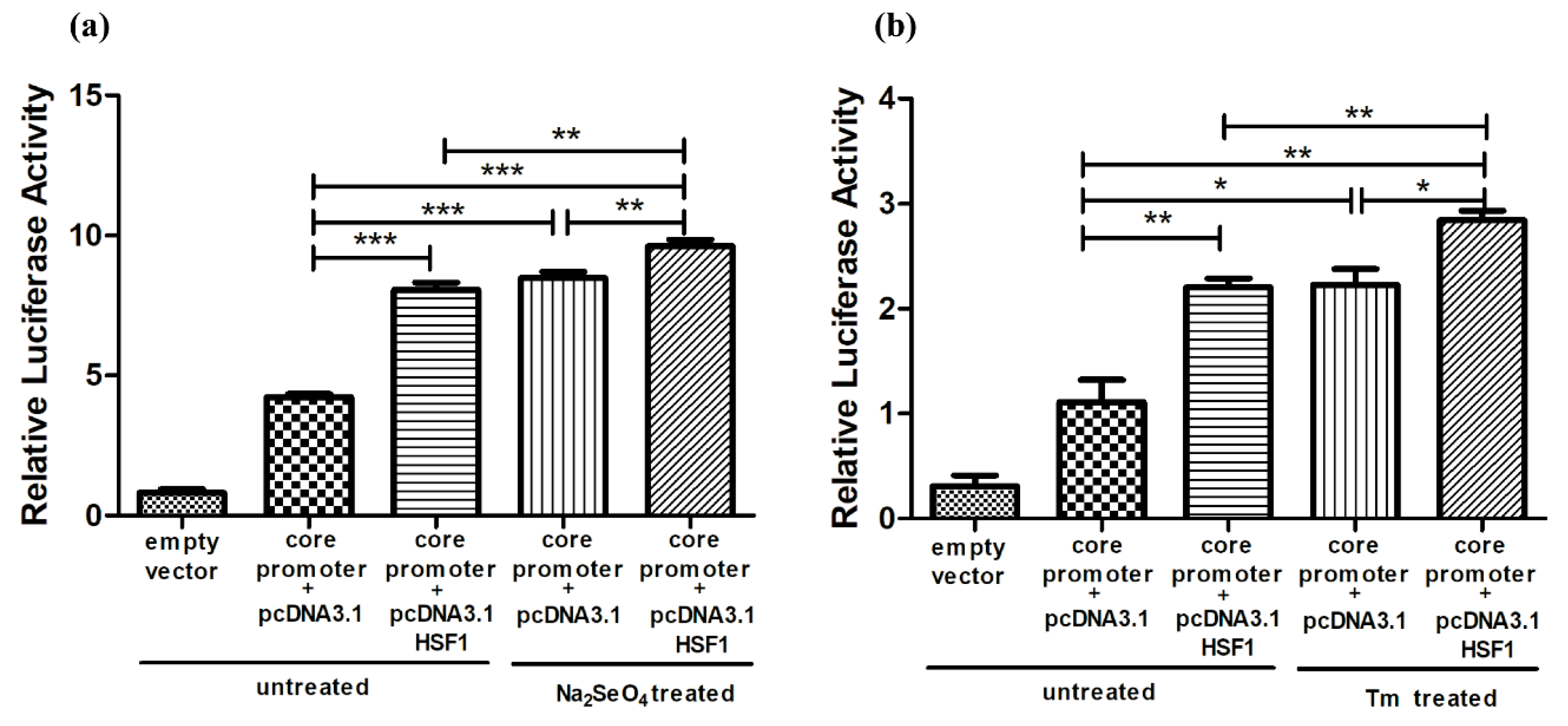
3.3. Increased Promotor Activities and Expression of SELENOF by HSF1 Over-Expression
3.4. Recruitment of HSF1 to the SELENOF 5′-Flanking Regions
3.5. Activation of HSF1 by Heat Shock Treatment Up-Regulated the Promotor Activities and Expression of SELENOF
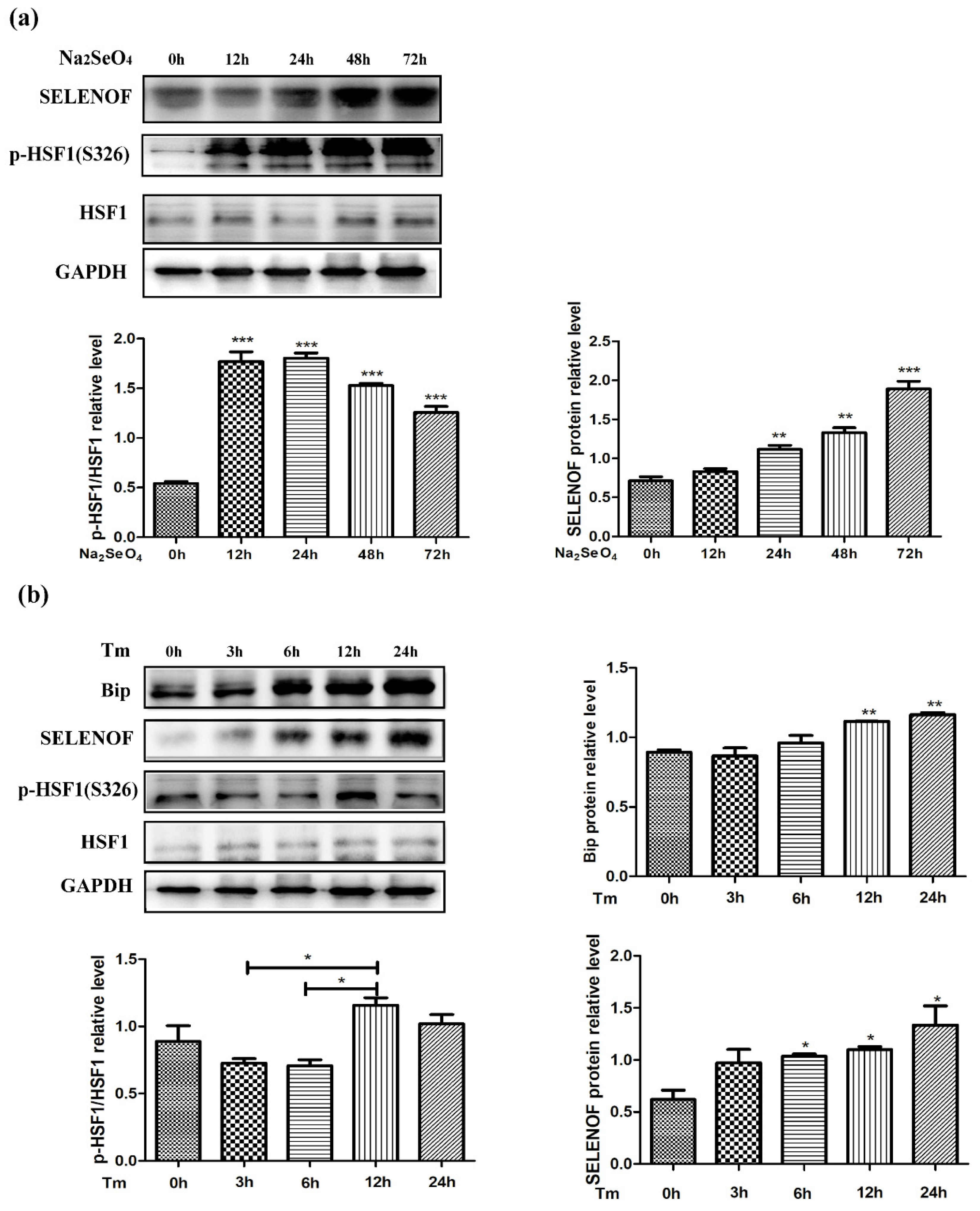
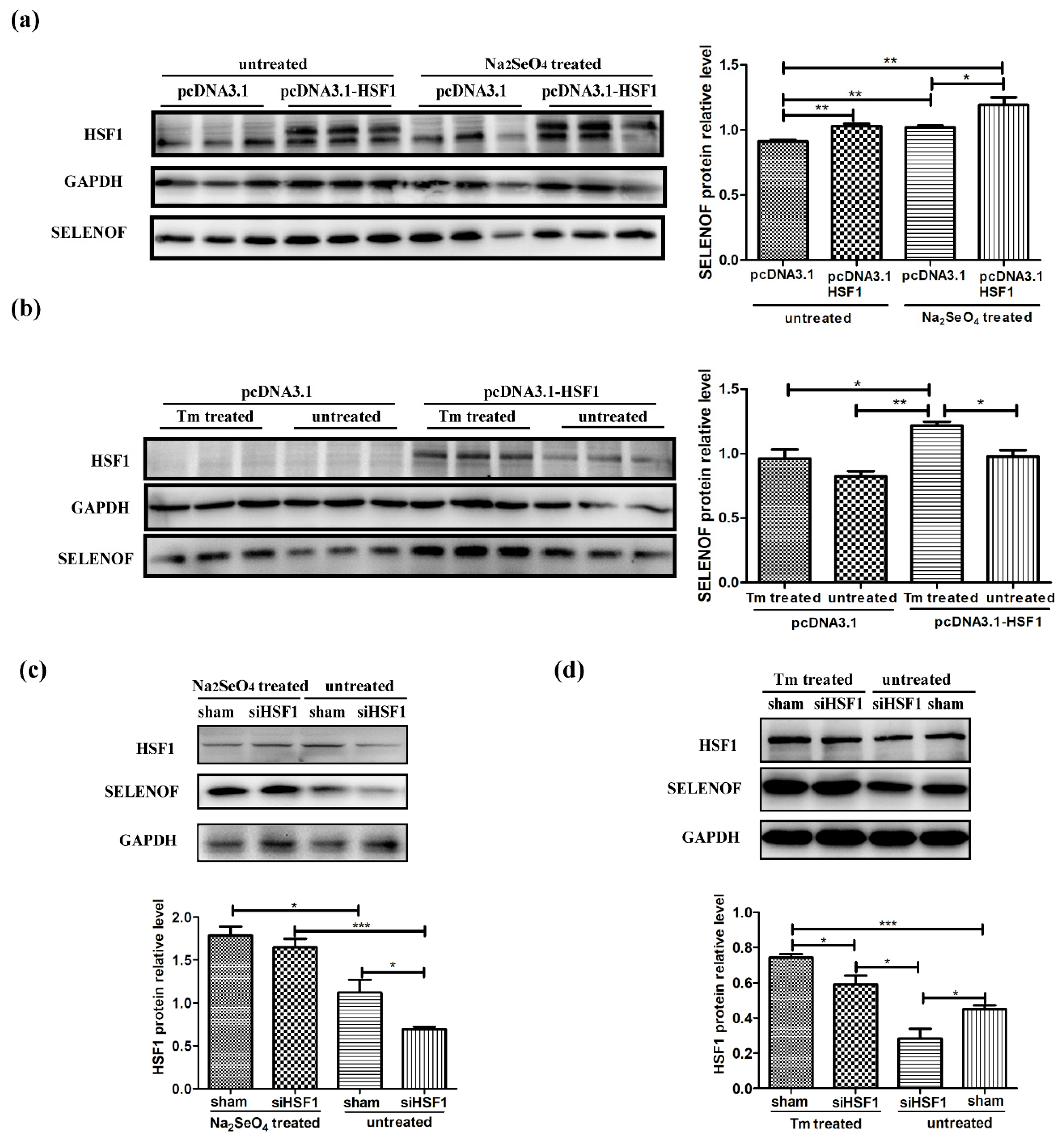
3.6. Endogenous Protein Changes of HSF1 and SELENOF During Selenate and Tm Treatment
3.7. HSF1 Mediated Up-Regulation of SELENOF During Selenate and Tm Treatment
4. Discussion
4.1. Specific Expression of SELENOF
4.2. Regulation of SELENOF by Selenium Compounds at the Transcription and Translation Levels
4.3. Potential Role of SELENOF in Selenium Supplementation and Stress Response
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Oropeza-Moe, M.; Wisloff, H.; Bernhoft, A. Selenium deficiency associated porcine and human cardiomyopathies. J. Trace Elem. Med. Biol. 2015, 31, 148–156. [Google Scholar] [CrossRef]
- Iglesias, S.B.D.; Leite, H.P.; Paes, A.T.; de Oliveira, S.V.; Sarni, R.O.S. Low plasma selenium concentrations in critically ill children: The interaction effect between inflammation and selenium deficiency. Crit. Care 2014, 18, R101. [Google Scholar] [CrossRef]
- McCann, J.C.; Ames, B.N. Adaptive dysfunction of selenoproteins from the perspective of the triage theory: Why modest selenium deficiency may increase risk of diseases of aging. FASEB J. 2011, 25, 1793–1814. [Google Scholar] [CrossRef] [PubMed]
- Low, S.C.; Berry, M.J. Knowing when not to stop: Selenocysteine incorporation in eukaryotes. Trends Biochem. Sci. 1996, 21, 203–208. [Google Scholar] [CrossRef]
- Rayman, M.P. The importance of selenium to human health. Lancet 2000, 356, 233–241. [Google Scholar] [CrossRef] [Green Version]
- Duntas, L.H.; Benvenga, S. Selenium: An element for life. Endocrine 2015, 48, 756–775. [Google Scholar] [CrossRef] [PubMed]
- Kumar, N.; Krishnani, K.K.; Gupta, S.K.; Singh, N.P. Selenium nanoparticles enhanced thermal tolerance and maintain cellular stress protection of pangasius hypophthalmus reared under lead and high temperature. Respir. Physiol. Neurobiol. 2017, 246, 107–116. [Google Scholar] [CrossRef] [PubMed]
- Gladyshev, V.N.; Arner, E.S.; Berry, M.J.; Brigelius-Flohe, R.; Bruford, E.A.; Burk, R.F.; Carlson, B.A.; Castellano, S.; Chavatte, L.; Conrad, M.; et al. Selenoprotein gene nomenclature. J. Biol. Chem. 2016, 291, 24036–24040. [Google Scholar] [CrossRef]
- Gladyshev, V.N.; Jeang, K.T.; Wootton, J.C.; Hatfield, D.L. A new human selenium-containing protein. Purification, characterization, and cdna sequence. J. Biol. Chem. 1998, 273, 8910–8915. [Google Scholar] [CrossRef]
- Wu, H.J.; Lin, C.; Zha, Y.Y.; Yang, J.G.; Zhang, M.C.; Zhang, X.Y.; Liang, X.; Fu, M.; Wu, M. Redox reactions of sep15 and its relationship with tumor development. Chin. J. Cancer 2003, 22, 119–122. [Google Scholar]
- Mohammaddoust, S.; Salehi, Z.; Saedi, H. Sepp1 and sep15 gene polymorphisms and susceptibility to breast cancer. Br. J. Biomed. Sci. 2018, 75, 36–39. [Google Scholar] [CrossRef]
- Chen, Q.C.; Ding, X.L.; Zhu, S.F.; Su, L.; Cai, D.M.; Chen, L.; He, W. Common SEP15 polymorphisms and susceptibility to cancer: A systematic review and meta-analysis. Transl. Cancer Res. 2017, 6, 886. [Google Scholar] [CrossRef]
- Hatfield, D.L.; Yoo, M.H.; Carlson, B.A.; Gladyshev, V.N. Selenoproteins that function in cancer prevention and promotion. Biochim. Biophys. Acta 2009, 1790, 1541–1545. [Google Scholar] [CrossRef] [Green Version]
- Korotkov, K.V.; Kumaraswamy, E.; Zhou, Y.; Hatfield, D.L.; Gladyshev, V.N. Association between the 15-kda selenoprotein and udp-glucose: Glycoprotein glucosyltransferase in the endoplasmic reticulum of mammalian cells. J. Biol. Chem. 2001, 276, 15330–15336. [Google Scholar] [CrossRef] [PubMed]
- Labunskyy, V.M.; Hatfield, D.L.; Gladyshev, V.N. The sep15 protein family: Roles in disulfide bond formation and quality control in the endoplasmic reticulum. IUBMB Life 2007, 59, 1–5. [Google Scholar] [CrossRef] [PubMed]
- Yim, S.H.; Everley, R.A.; Schildberg, F.A.; Lee, S.G.; Orsi, A.; Barbati, Z.R.; Karatepe, K.; Fomenko, D.E.; Tsuji, P.A.; Luo, H.R.; et al. Role of selenof as a gatekeeper of secreted disulfide-rich glycoproteins. Cell Rep. 2018, 23, 1387–1398. [Google Scholar] [CrossRef]
- Guariniello, S.; Di Bernardo, G.; Colonna, G.; Cammarota, M.; Castello, G.; Costantini, S. Evaluation of the selenotranscriptome expression in two hepatocellular carcinoma cell lines. Anal. Cell. Pathol. 2015, 2015, 419561. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ye, Y.L.; Zhu, H.; Sun, S.N.; Zheng, J.; Fan, H.H.; Wu, H.M.; Chen, S.F.; Cheng, W.H.; Zhu, J.H. Selenotranscriptomic analyses identify signature selenoproteins in brain regions in a mouse model of parkinson’s disease. PLoS ONE 2016, 11, e0163372. [Google Scholar] [CrossRef]
- Stoytcheva, Z.R.; Berry, M.J. Transcriptional regulation of mammalian selenoprotein expression. Biochim. Biophys. Acta 2009, 1790, 1429–1440. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.L.; Xu, B.; Huang, X.D.; Gao, Y.H.; Chen, Y.; Shan, A.S. Selenium deficiency affects the mrna expression of inflammatory factors and selenoprotein genes in the kidneys of broiler chicks. Biol. Trace Elem. Res. 2016, 171, 201–207. [Google Scholar] [CrossRef]
- Mallonee, D.H.; Crowdus, C.A.; Barger, J.L.; Dawson, K.A.; Power, R.F. Use of stringent selection parameters for the identification of possible selenium-responsive marker genes in mouse liver and gastrocnemius. Biol. Trace Elem. Res. 2011, 143, 992–1006. [Google Scholar] [CrossRef] [PubMed]
- Labunskyy, V.M.; Yoo, M.H.; Hatfield, D.L.; Gladyshev, V.N. Sep15, a thioredoxin-like selenoprotein, is involved in the unfolded protein response and differentially regulated by adaptive and acute er stresses. Biochemistry 2009, 48, 8458–8465. [Google Scholar] [CrossRef]
- Suzuki, Y.; Yamashita, R.; Sugano, S.; Nakai, K. Dbtss, database of transcriptional start sites: Progress report 2004. Nucleic Acids Res. 2004, 32, D78–D81. [Google Scholar] [CrossRef]
- Database of Transcriptional Start Sites. Release 9.0 ed. Available online: http://dbtss.hgc.jp (accessed on 9 November 2016).
- Khan, A.; Fornes, O.; Stigliani, A.; Gheorghe, M.; Castro-Mondragon, J.A.; van der Lee, R.; Bessy, A.; Cheneby, J.; Kulkarni, S.R.; Tan, G.; et al. Jaspar 2018: Update of the open-access database of transcription factor binding profiles and its web framework. Nucleic Acids Res. 2018, 46, D260–d266. [Google Scholar] [CrossRef]
- The Jaspar Database. Version: 5.0_ALPHA ed. Available online: http://jaspardev.genereg.net/ (accessed on 2 December 2016).
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2−δδct method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef]
- Dai, J.; Zhou, J.; Liu, H.; Huang, K. Selenite and ebselen supplementation attenuates d-galactose-induced oxidative stress and increases expression of selr and sep15 in rat lens. J. Biol. Inorg. Chem. 2016, 21, 1037–1046. [Google Scholar] [CrossRef] [PubMed]
- Juszczuk-Kubiak, E.; Bujko, K.; Cymer, M.; Wicinska, K.; Gabryszuk, M.; Pierzchala, M. Effect of inorganic dietary selenium supplementation on selenoprotein and lipid metabolism gene expression patterns in liver and loin muscle of growing lambs. Biol. Trace Elem. Res. 2016, 172, 336–345. [Google Scholar] [CrossRef] [PubMed]
- Misra, S.; Boylan, M.; Selvam, A.; Spallholz, J.E.; Björnstedt, M. Redox-active selenium compounds—From toxicity and cell death to cancer treatment. Nutrients 2015, 7, 3536–3556. [Google Scholar] [CrossRef]
- Malhotra, J.D.; Kaufman, R.J. Endoplasmic reticulum stress and oxidative stress: A vicious cycle or a double-edged sword? Antioxid. Redox Signal. 2007, 9, 2277–2293. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, N.M.; Martin, D.; Hutter-Paier, B.; Windisch, M.; Nguyen, T.; Nheu, L.; Sundstrom, L.E.; Costello, A.J.; Hovens, C.M. Sodium selenate specifically activates pp2a phosphatase, dephosphorylates tau and reverses memory deficits in an alzheimer’s disease model. J. Clin. Neurosci. 2010, 17, 1025–1033. [Google Scholar] [CrossRef]
- Kipp, A.P.; Frombach, J.; Deubel, S.; Brigelius-Flohe, R. Selenoprotein was biomarker for the efficacy of selenium compounds to act as source for selenoprotein biosynthesis. In Hydrogen Peroxide and Cell Signaling, Part B; Cadenas, E., Packer, L., Eds.; Elsevier Academic Press Inc.: San Diego, CA, USA, 2013; Volume 527, pp. 87–112. [Google Scholar]
- Ren, B.; Wang, Y.; Wang, H.; Wu, Y.; Li, J.; Tian, J. Comparative proteomics reveals the neurotoxicity mechanism of er stressors tunicamycin and dithiothreitol. NeuroToxicology 2018, 68, 25–37. [Google Scholar] [CrossRef] [PubMed]
- Deaton, A.M.; Bird, A. Cpg islands and the regulation of transcription. Genes Dev. 2011, 25, 1010–1022. [Google Scholar] [CrossRef] [PubMed]
- Jiang, S.; Zhang, E.; Zhang, R.; Li, X. Altered activity patterns of transcription factors induced by endoplasmic reticulum stress. BMC Biochem. 2016, 17, 8. [Google Scholar] [CrossRef]
- Luan, Q.; Jin, L.; Jiang, C.C.; Tay, K.H.; Lai, F.; Liu, X.Y.; Liu, Y.L.; Guo, S.T.; Li, C.Y.; Yan, X.G.; et al. Ripk1 regulates survival of human melanoma cells upon endoplasmic reticulum stress through autophagy. Autophagy 2015, 11, 975–994. [Google Scholar] [CrossRef]
- Kim, E.; Sakata, K.; Liao, F.-F. Bidirectional interplay of hsf1 degradation and upr activation promotes tau hyperphosphorylation. PLoS Genet. 2017, 13, e1006849. [Google Scholar] [CrossRef] [PubMed]
- Zou, C.; Z, J.; Li, J.; Tian, J. Human Gene of 15-kda Selenoprotein is Regulated by nf κb in Selenium in the Environment and Human Health; Gary, S., Banuelos, Z.-Q.L., Yin, X., Eds.; CRC Press: Boca Raton, FL, USA, 2013; pp. 172–173. [Google Scholar]
- Song, M.; Pinsky, M.R.; Kellum, J.A. Heat shock factor 1 inhibits nuclear factor–κb nuclear binding activity during endotoxin tolerance and heat shock. J. Crit. Care 2008, 23, 406–415. [Google Scholar] [CrossRef] [PubMed]
- Pan, X.-Y.; Zhao, W.; Zeng, X.-Y.; Lin, J.; Li, M.-M.; Shen, X.-T.; Liu, S.-W. Heat shock factor 1 mediates latent HIV reactivation. Sci. Rep. 2016, 6, 26294. [Google Scholar] [CrossRef]
- Hahn, J.S.; Hu, Z.; Thiele, D.J.; Iyer, V.R. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol. 2004, 24, 5249–5256. [Google Scholar] [CrossRef]
- Vihervaara, A.; Sistonen, L. Hsf1 at a glance. J. Cell Sci. 2014, 127, 261–266. [Google Scholar] [CrossRef]
- Hu, Y.J.; Korotkov, K.V.; Mehta, R.; Hatfield, D.L.; Rotimi, C.N.; Luke, A.; Prewitt, T.E.; Cooper, R.S.; Stock, W.; Vokes, E.E.; et al. Distribution and functional consequences of nucleotide polymorphisms in the 3′-untranslated region of the human sep15 gene. Cancer Res. 2001, 61, 2307–2310. [Google Scholar]
- Kasaikina, M.V.; Fomenko, D.E.; Labunskyy, V.M.; Lachke, S.A.; Qiu, W.; Moncaster, J.A.; Zhang, J.; Wojnarowicz, M.W.; Natarajan, S.K.; Malinouski, M.; et al. Roles of the 15-kda selenoprotein (sep15) in redox homeostasis and cataract development revealed by the analysis of sep 15 knockout mice. J. Biol. Chem. 2011, 286, 33203–33212. [Google Scholar] [CrossRef] [PubMed]
- Touat-Hamici, Z.; Bulteau, A.L.; Bianga, J.; Jean-Jacques, H.; Szpunar, J.; Lobinski, R.; Chavatte, L. Selenium-regulated hierarchy of human selenoproteome in cancerous and immortalized cells lines. Biochim. Biophys. Acta 2018. [Google Scholar] [CrossRef]
- Maniatis, T.; Goodbourn, S.; Fischer, J.A. Regulation of inducible and tissue-specific gene-expression. Science 1987, 236, 1237–1245. [Google Scholar] [CrossRef]
- Berry, M.J. Insights into the hierarchy of selenium incorporation. Nat. Genet. 2005, 37, 1162. [Google Scholar] [CrossRef]
- Bermano, G.; Arthur, J.R.; Hesketh, J.E. Selective control of cytosolic glutathione peroxidase and phospholipid hydroperoxide glutathione peroxidase mrna stability by selenium supply. FEBS Lett. 1996, 387, 157–160. [Google Scholar] [CrossRef]
- Maquat, L.E. Evidence that selenium deficiency results in the cytoplasmic decay of gpx1 mrna dependent on pre-mrna splicing proteins bound to the mRNA exon-exon junction. BioFactors 2001, 14, 37–42. [Google Scholar] [CrossRef]
- Yuan, B.; Webster, T.J.; Roy, A.K. Cytoprotective effects of cerium and selenium nanoparticles on heat-shocked human dermal fibroblasts: An in vitro evaluation. Int. J. Nanomed. 2016, 11, 1427–1433. [Google Scholar]
- Kaur, S.; Bansal, M.P. Protective role of dietary-supplemented selenium and vitamin e in heat-induced apoptosis and oxidative stress in mice testes. Andrologia 2015, 47, 1109–1119. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; An, Y.; Jiao, W.; Zhang, Z.; Han, H.; Gu, X.; Teng, X. Selenium protects against lead-induced apoptosis via endoplasmic reticulum stress in chicken kidneys. Biol. Trace Elem. Res. 2018, 182, 354–363. [Google Scholar] [CrossRef]
- De Maio, A. Heat shock proteins: Facts, thoughts, and dreams. Shock 1999, 11, 1–12. [Google Scholar] [CrossRef]
- Griesemer, M.; Young, C.; Robinson, A.S.; Petzold, L. Bip clustering facilitates protein folding in the endoplasmic reticulum. PLoS Comput. Biol. 2014, 10, e1003675. [Google Scholar] [CrossRef] [PubMed]



© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ren, B.; Huang, Y.; Zou, C.; Wu, Y.; Huang, Y.; Ni, J.; Tian, J. Transcriptional Regulation of Selenoprotein F by Heat Shock Factor 1 during Selenium Supplementation and Stress Response. Cells 2019, 8, 479. https://doi.org/10.3390/cells8050479
Ren B, Huang Y, Zou C, Wu Y, Huang Y, Ni J, Tian J. Transcriptional Regulation of Selenoprotein F by Heat Shock Factor 1 during Selenium Supplementation and Stress Response. Cells. 2019; 8(5):479. https://doi.org/10.3390/cells8050479
Chicago/Turabian StyleRen, Bingyu, Yanmei Huang, Chen Zou, Yingying Wu, Yuru Huang, Jiazuan Ni, and Jing Tian. 2019. "Transcriptional Regulation of Selenoprotein F by Heat Shock Factor 1 during Selenium Supplementation and Stress Response" Cells 8, no. 5: 479. https://doi.org/10.3390/cells8050479
APA StyleRen, B., Huang, Y., Zou, C., Wu, Y., Huang, Y., Ni, J., & Tian, J. (2019). Transcriptional Regulation of Selenoprotein F by Heat Shock Factor 1 during Selenium Supplementation and Stress Response. Cells, 8(5), 479. https://doi.org/10.3390/cells8050479





