Patient-Derived Xenograft Models of Breast Cancer and Their Application
Abstract
1. Introduction
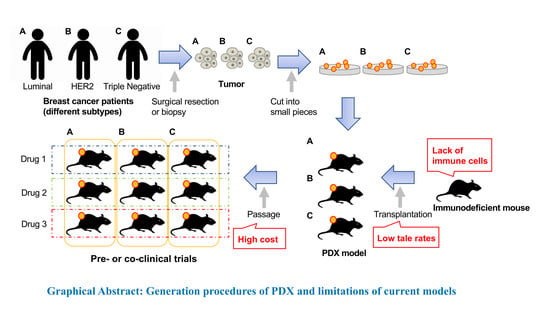
2. Generation of PDX Models
2.1. Immunodeficient Mice
2.2. Patient-Derived Tumors
3. Current Representative Line of Cancer Models
3.1. Cell line (cultured in vitro)
3.2. Cell Line Xenograft Model
3.3. Genetically Engineered Mouse Model
3.4. PDX Model
4. PDX Models of Each Breast Cancer Subtype
4.1. Luminal A and Luminal B Subtypes
4.2. HER2 Positive Subtype
4.3. Triple Negative Subtype
5. Application of PDX Models for Clinical Use
5.1. PDX Models for Drug Development
5.2. PDX Models for Precision Medicine
5.3. PDX Models for Co-Clinical Trials
6. Limitations of Current PDX Models
6.1. Lack of Immune Cells
6.2. Low Take Rates
6.3. High Cost
7. Next Generation PDX Models with Human Immune System
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Polyak, K. Heterogeneity in breast cancer. J. Clin. Investig. 2011, 121, 3786–3788. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Claerhout, S.; Prat, A.; Dobrolecki, L.E.; Petrovic, I.; Lai, Q.; Landis, M.D.; Wiechmann, L.; Schiff, R.; Giuliano, M.; et al. A renewable tissue resource of phenotypically stable, biologically and ethnically diverse, patient-derived human breast cancer xenograft models. Cancer Res. 2013, 73, 4885–4897. [Google Scholar] [CrossRef] [PubMed]
- Julien, S.; Merino-Trigo, A.; Lacroix, L.; Pocard, M.; Goere, D.; Mariani, P.; Landron, S.; Bigot, L.; Nemati, F.; Dartigues, P.; et al. Characterization of a large panel of patient-derived tumor xenografts representing the clinical heterogeneity of human colorectal cancer. Clin. Cancer Res. 2012, 18, 5314–5328. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.P.; Evans, D.B.; Wang, H.; Abbruzzese, J.L.; Fleming, J.B.; Gallick, G.E. Generation of orthotopic and heterotopic human pancreatic cancer xenografts in immunodeficient mice. Nat. Protoc. 2009, 4, 1670–1680. [Google Scholar] [CrossRef]
- Chapuy, B.; Cheng, H.; Watahiki, A.; Ducar, M.D.; Tan, Y.; Chen, L.; Roemer, M.G.; Ouyang, J.; Christie, A.L.; Zhang, L.; et al. Diffuse large B-cell lymphoma patient-derived xenograft models capture the molecular and biological heterogeneity of the disease. Blood 2016, 127, 2203–2213. [Google Scholar] [CrossRef] [PubMed]
- Dong, X.; Guan, J.; English, J.C.; Flint, J.; Yee, J.; Evans, K.; Murray, N.; Macaulay, C.; Ng, R.T.; Gout, P.W.; et al. Patient-derived first generation xenografts of non-small cell lung cancers: Promising tools for predicting drug responses for personalized chemotherapy. Clin. Cancer Res. 2010, 16, 1442–1451. [Google Scholar] [CrossRef] [PubMed]
- Weroha, S.J.; Becker, M.A.; Enderica-Gonzalez, S.; Harrington, S.C.; Oberg, A.L.; Maurer, M.J.; Perkins, S.E.; AlHilli, M.; Butler, K.A.; McKinstry, S.; et al. Tumorgrafts as in vivo surrogates for women with ovarian cancer. Clin. Cancer Res. 2014, 20, 1288–1297. [Google Scholar] [CrossRef] [PubMed]
- Okada, S.; Vaeteewoottacharn, K.; Kariya, R. Establishment of a Patient-Derived Tumor Xenograft Model and Application for Precision Cancer Medicine. Chem. Pharm. Bull. 2018, 66, 225–230. [Google Scholar] [CrossRef] [PubMed]
- Shultz, L.D.; Goodwin, N.; Ishikawa, F.; Hosur, V.; Lyons, B.L.; Greiner, D.L. Human cancer growth and therapy in immunodeficient mouse models. Cold Spring Harb. Protoc. 2014, 2014, 694–708. [Google Scholar] [CrossRef]
- Schmitt, K.; Akkina, R. Ultra-Sensitive HIV-1 Latency Viral Outgrowth Assays Using Humanized Mice. Front. Immunol. 2018, 9, 344. [Google Scholar] [CrossRef]
- Shultz, L.D.; Ishikawa, F.; Greiner, D.L. Humanized mice in translational biomedical research. Nat. Rev. Immunol. 2007, 7, 118–130. [Google Scholar] [CrossRef] [PubMed]
- Brehm, M.A.; Wiles, M.V.; Greiner, D.L.; Shultz, L.D. Generation of improved humanized mouse models for human infectious diseases. J. Immunol. Methods 2014, 410, 3–17. [Google Scholar] [CrossRef]
- Sheth, R.A.; Perkons, N.; Dondossola, E.; Subudhi, S.K.; Gade, T.P.; Tam, A.L. Patient-Derived Xenograft Tumor Models: Overview and Relevance to IR. J. Vasc. Interv. Radiol. 2018, 29, 880.e1–882.e1. [Google Scholar] [CrossRef] [PubMed]
- Tominaga, K.; Minato, H.; Murayama, T.; Sasahara, A.; Nishimura, T.; Kiyokawa, E.; Kanauchi, H.; Shimizu, S.; Sato, A.; Nishioka, K.; et al. Semaphorin signaling via MICAL3 induces symmetric cell division to expand breast cancer stem-like cells. Proc. Natl. Acad. Sci. USA 2019, 116, 625–630. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Taftaf, R.; Kawaguchi, M.; Chang, Y.F.; Chen, W.; Entenberg, D.; Zhang, Y.; Gerratana, L.; Huang, S.; Patel, D.B.; et al. Homophilic CD44 Interactions Mediate Tumor Cell Aggregation and Polyclonal Metastasis in Patient-Derived Breast Cancer Models. Cancer Discov. 2019, 9, 96–113. [Google Scholar] [CrossRef] [PubMed]
- Hoffman, R.M. Patient-derived orthotopic xenografts: Better mimic of metastasis than subcutaneous xenografts. Nat. Rev. Cancer 2015, 15, 451–452. [Google Scholar] [CrossRef]
- Ni, J.; Ramkissoon, S.H.; Xie, S.; Goel, S.; Stover, D.G.; Guo, H.; Luu, V.; Marco, E.; Ramkissoon, L.A.; Kang, Y.J.; et al. Combination inhibition of PI3K and mTORC1 yields durable remissions in mice bearing orthotopic patient-derived xenografts of HER2-positive breast cancer brain metastases. Nat. Med. 2016, 22, 723–726. [Google Scholar] [CrossRef] [PubMed]
- Neve, R.M.; Chin, K.; Fridlyand, J.; Yeh, J.; Baehner, F.L.; Fevr, T.; Clark, L.; Bayani, N.; Coppe, J.P.; Tong, F.; et al. A collection of breast cancer cell lines for the study of functionally distinct cancer subtypes. Cancer Cell 2006, 10, 515–527. [Google Scholar] [CrossRef]
- Imamura, Y.; Mukohara, T.; Shimono, Y.; Funakoshi, Y.; Chayahara, N.; Toyoda, M.; Kiyota, N.; Takao, S.; Kono, S.; Nakatsura, T.; et al. Comparison of 2D- and 3D-culture models as drug-testing platforms in breast cancer. Oncol. Rep. 2015, 33, 1837–1843. [Google Scholar] [CrossRef]
- Hajitou, A.; Lev, D.C.; Hannay, J.A.; Korchin, B.; Staquicini, F.I.; Soghomonyan, S.; Alauddin, M.M.; Benjamin, R.S.; Pollock, R.E.; Gelovani, J.G.; et al. A preclinical model for predicting drug response in soft-tissue sarcoma with targeted AAVP molecular imaging. Proc. Natl. Acad. Sci. USA 2008, 105, 4471–4476. [Google Scholar] [CrossRef]
- Whittle, J.R.; Lewis, M.T.; Lindeman, G.J.; Visvader, J.E. Patient-derived xenograft models of breast cancer and their predictive power. Breast Cancer Res. 2015, 17, 17. [Google Scholar] [CrossRef] [PubMed]
- Khaled, W.T.; Liu, P. Cancer mouse models: Past, present and future. Semin. Cell Dev. Biol. 2014, 27, 54–60. [Google Scholar] [CrossRef] [PubMed]
- Borowsky, A.D. Genetically engineering a mouse. Comp. Med. 2003, 53, 249–250. [Google Scholar] [PubMed]
- Menezes, M.E.; Das, S.K.; Emdad, L.; Windle, J.J.; Wang, X.Y.; Sarkar, D.; Fisher, P.B. Genetically engineered mice as experimental tools to dissect the critical events in breast cancer. Adv. Cancer Res. 2014, 121, 331–382. [Google Scholar] [PubMed]
- Voncken, J.W. Genetic modification of the mouse: General technology—Pronuclear and blastocyst injection. Methods Mol. Biol. 2011, 693, 11–36. [Google Scholar] [PubMed]
- Shibata, H.; Komura, S.; Yamada, Y.; Sankoda, N.; Tanaka, A.; Ukai, T.; Kabata, M.; Sakurai, S.; Kuze, B.; Woltjen, K.; et al. In vivo reprogramming drives Kras-induced cancer development. Nat. Commun. 2018, 9, 2081. [Google Scholar] [CrossRef] [PubMed]
- Goel, S.; Wang, Q.; Watt, A.C.; Tolaney, S.M.; Dillon, D.A.; Li, W.; Ramm, S.; Palmer, A.C.; Yuzugullu, H.; Varadan, V.; et al. Overcoming Therapeutic Resistance in HER2-Positive Breast Cancers with CDK4/6 Inhibitors. Cancer Cell 2016, 29, 255–269. [Google Scholar] [CrossRef]
- Cardiff, R.D.; Rosner, A.; Hogarth, M.A.; Galvez, J.J.; Borowsky, A.D.; Gregg, J.P. Validation: The new challenge for pathology. Toxicol. Pathol. 2004, 32 (Suppl. 1), 31–39. [Google Scholar] [CrossRef]
- Eyre, R.; Alferez, D.G.; Spence, K.; Kamal, M.; Shaw, F.L.; Simoes, B.M.; Santiago-Gomez, A.; Sarmiento-Castro, A.; Bramley, M.; Absar, M.; et al. Patient-derived Mammosphere and Xenograft Tumour Initiation Correlates with Progression to Metastasis. J. Mammary Gland Biol. Neoplasia 2016, 21, 99–109. [Google Scholar] [CrossRef]
- Perou, C.M.; Sorlie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; Rees, C.A.; Pollack, J.R.; Ross, D.T.; Johnsen, H.; Akslen, L.A.; et al. Molecular portraits of human breast tumours. Nature 2000, 406, 747–752. [Google Scholar] [CrossRef]
- Sorlie, T.; Perou, C.M.; Tibshirani, R.; Aas, T.; Geisler, S.; Johnsen, H.; Hastie, T.; Eisen, M.B.; van de Rijn, M.; Jeffrey, S.S.; et al. Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc. Natl. Acad. Sci. USA 2001, 98, 10869–10874. [Google Scholar] [CrossRef] [PubMed]
- Agnoletto, C.; Minotti, L.; Brulle-Soumare, L.; Pasquali, L.; Galasso, M.; Corra, F.; Baldassari, F.; Judde, J.G.; Cairo, S.; Volinia, S. Heterogeneous expression of EPCAM in human circulating tumour cells from patient-derived xenografts. Biomark. Res. 2018, 6, 31. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef] [PubMed]
- Arango, N.P.; Yuca, E.; Zhao, M.; Evans, K.W.; Scott, S.; Kim, C.; Gonzalez-Angulo, A.M.; Janku, F.; Ueno, N.T.; Tripathy, D.; et al. Selinexor (KPT-330) demonstrates anti-tumor efficacy in preclinical models of triple-negative breast cancer. Breast Cancer Res. 2017, 19, 93. [Google Scholar] [CrossRef]
- Bruna, A.; Rueda, O.M.; Greenwood, W.; Batra, A.S.; Callari, M.; Batra, R.N.; Pogrebniak, K.; Sandoval, J.; Cassidy, J.W.; Tufegdzic-Vidakovic, A.; et al. A Biobank of Breast Cancer Explants with Preserved Intra-tumor Heterogeneity to Screen Anticancer Compounds. Cell 2016, 167, 260.e22–274.e22. [Google Scholar] [CrossRef] [PubMed]
- Castroviejo-Bermejo, M.; Cruz, C.; Llop-Guevara, A.; Gutierrez-Enriquez, S.; Ducy, M.; Ibrahim, Y.H.; Gris-Oliver, A.; Pellegrino, B.; Bruna, A.; Guzman, M.; et al. A RAD51 assay feasible in routine tumor samples calls PARP inhibitor response beyond BRCA mutation. EMBO Mol. Med. 2018, 10. [Google Scholar] [CrossRef]
- Contreras-Zarate, M.J.; Ormond, D.R.; Gillen, A.E.; Hanna, C.; Day, N.L.; Serkova, N.J.; Jacobsen, B.M.; Edgerton, S.M.; Thor, A.D.; Borges, V.F.; et al. Development of Novel Patient-Derived Xenografts from Breast Cancer Brain Metastases. Front. Oncol. 2017, 7, 252. [Google Scholar] [CrossRef]
- Coussy, F.; de Koning, L.; Lavigne, M.; Bernard, V.; Ouine, B.; Boulai, A.; El Botty, R.; Dahmani, A.; Montaudon, E.; Assayag, F.; et al. A large collection of integrated genomically characterized patient-derived xenografts highlighting the heterogeneity of triple-negative breast cancer. Int. J. Cancer 2019. [Google Scholar] [CrossRef]
- Cruz, C.; Castroviejo-Bermejo, M.; Gutierrez-Enriquez, S.; Llop-Guevara, A.; Ibrahim, Y.H.; Gris-Oliver, A.; Bonache, S.; Morancho, B.; Bruna, A.; Rueda, O.M.; et al. RAD51 foci as a functional biomarker of homologous recombination repair and PARP inhibitor resistance in germline BRCA-mutated breast cancer. Ann. Oncol. 2018, 29, 1203–1210. [Google Scholar] [CrossRef]
- Davila-Gonzalez, D.; Choi, D.S.; Rosato, R.R.; Granados-Principal, S.M.; Kuhn, J.G.; Li, W.F.; Qian, W.; Chen, W.; Kozielski, A.J.; Wong, H.; et al. Pharmacological Inhibition of NOS Activates ASK1/JNK Pathway Augmenting Docetaxel-Mediated Apoptosis in Triple-Negative Breast Cancer. Clin. Cancer Res. 2018, 24, 1152–1162. [Google Scholar] [CrossRef]
- DeRose, Y.S.; Wang, G.; Lin, Y.C.; Bernard, P.S.; Buys, S.S.; Ebbert, M.T.; Factor, R.; Matsen, C.; Milash, B.A.; Nelson, E.; et al. Tumor grafts derived from women with breast cancer authentically reflect tumor pathology, growth, metastasis and disease outcomes. Nat. Med. 2011, 17, 1514–1520. [Google Scholar] [CrossRef] [PubMed]
- Evans, K.W.; Yuca, E.; Akcakanat, A.; Scott, S.M.; Arango, N.P.; Zheng, X.; Chen, K.; Tapia, C.; Tarco, E.; Eterovic, A.K.; et al. A Population of Heterogeneous Breast Cancer Patient-Derived Xenografts Demonstrate Broad Activity of PARP Inhibitor in BRCA1/2 Wild-Type Tumors. Clin. Cancer Res. 2017, 23, 6468–6477. [Google Scholar] [CrossRef] [PubMed]
- Fatima, I.; El-Ayachi, I.; Taotao, L.; Lillo, M.A.; Krutilina, R.I.; Seagroves, T.N.; Radaszkiewicz, T.W.; Hutnan, M.; Bryja, V.; Krum, S.A.; et al. The natural compound Jatrophone interferes with Wnt/beta-catenin signaling and inhibits proliferation and EMT in human triple-negative breast cancer. PLoS ONE 2017, 12, e0189864. [Google Scholar] [CrossRef] [PubMed]
- Fleming, J.M.; Miller, T.C.; Meyer, M.J.; Ginsburg, E.; Vonderhaar, B.K. Local regulation of human breast xenograft models. J. Cell. Physiol. 2010, 224, 795–806. [Google Scholar] [CrossRef] [PubMed]
- Formisano, L.; Stauffer, K.M.; Young, C.D.; Bhola, N.E.; Guerrero-Zotano, A.L.; Jansen, V.M.; Estrada, M.M.; Hutchinson, K.E.; Giltnane, J.M.; Schwarz, L.J.; et al. Association of FGFR1 with ERalpha Maintains Ligand-Independent ER Transcription and Mediates Resistance to Estrogen Deprivation in ER(+) Breast Cancer. Clin. Cancer Res. 2017, 23, 6138–6150. [Google Scholar] [CrossRef]
- Gonzalez-Gonzalez, A.; Munoz-Muela, E.; Marchal, J.A.; Cara, F.E.; Molina, M.P.; Cruz-Lozano, M.; Jimenez, G.; Verma, A.; Ramirez, A.; Qian, W.; et al. Activating Transcription Factor 4 Modulates TGFbeta-Induced Aggressiveness in Triple-Negative Breast Cancer via SMAD2/3/4 and mTORC2 Signaling. Clin. Cancer Res. 2018, 24, 5697–5709. [Google Scholar] [CrossRef] [PubMed]
- Hsu, P.Y.; Wu, V.S.; Kanaya, N.; Petrossian, K.; Hsu, H.K.; Nguyen, D.; Schmolze, D.; Kai, M.; Liu, C.Y.; Lu, H.; et al. Dual mTOR Kinase Inhibitor MLN0128 Sensitizes HR(+)/HER2(+) Breast Cancer Patient-Derived Xenografts to Trastuzumab or Fulvestrant. Clin. Cancer Res. 2018, 24, 395–406. [Google Scholar] [CrossRef] [PubMed]
- Hu, Y.; Yague, E.; Zhao, J.; Wang, L.; Bai, J.; Yang, Q.; Pan, T.; Zhao, H.; Liu, J.; Zhang, J. Sabutoclax, pan-active BCL-2 protein family antagonist, overcomes drug resistance and eliminates cancer stem cells in breast cancer. Cancer Lett. 2018, 423, 47–59. [Google Scholar] [CrossRef]
- Jung, J.; Jang, K.; Ju, J.M.; Lee, E.; Lee, J.W.; Kim, H.J.; Kim, J.; Lee, S.B.; Ko, B.S.; Son, B.H.; et al. Novel cancer gene variants and gene fusions of triple-negative breast cancers (TNBCs) reveal their molecular diversity conserved in the patient-derived xenograft (PDX) model. Cancer Lett. 2018, 428, 127–138. [Google Scholar] [CrossRef]
- Kabos, P.; Finlay-Schultz, J.; Li, C.; Kline, E.; Finlayson, C.; Wisell, J.; Manuel, C.A.; Edgerton, S.M.; Harrell, J.C.; Elias, A.; et al. Patient-derived luminal breast cancer xenografts retain hormone receptor heterogeneity and help define unique estrogen-dependent gene signatures. Breast Cancer Res. Treat. 2012, 135, 415–432. [Google Scholar] [CrossRef]
- Kanaya, N.; Somlo, G.; Wu, J.; Frankel, P.; Kai, M.; Liu, X.; Wu, S.V.; Nguyen, D.; Chan, N.; Hsieh, M.Y.; et al. Characterization of patient-derived tumor xenografts (PDXs) as models for estrogen receptor positive (ER+HER2- and ER+HER2+) breast cancers. J. Steroid Biochem. Mol. Biol. 2017, 170, 65–74. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Shen, D.; Shao, J.; Crowder, R.; Liu, W.; Prat, A.; He, X.; Liu, S.; Hoog, J.; Lu, C.; et al. Endocrine-therapy-resistant ESR1 variants revealed by genomic characterization of breast-cancer-derived xenografts. Cell Rep. 2013, 4, 1116–1130. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Patel, M.R.; Prescher, J.A.; Patsialou, A.; Qian, D.; Lin, J.; Wen, S.; Chang, Y.F.; Bachmann, M.H.; Shimono, Y.; et al. Cancer stem cells from human breast tumors are involved in spontaneous metastases in orthotopic mouse models. Proc. Natl. Acad. Sci. USA 2010, 107, 18115–18120. [Google Scholar] [CrossRef] [PubMed]
- Ma, H.Y.; Liu, X.Z.; Liang, C.M. Inflammatory microenvironment contributes to epithelial-mesenchymal transition in gastric cancer. World J. Gastroenterol. 2016, 22, 6619–6628. [Google Scholar] [CrossRef] [PubMed]
- Marangoni, E.; Vincent-Salomon, A.; Auger, N.; Degeorges, A.; Assayag, F.; de Cremoux, P.; de Plater, L.; Guyader, C.; De Pinieux, G.; Judde, J.G.; et al. A new model of patient tumor-derived breast cancer xenografts for preclinical assays. Clin. Cancer Res. 2007, 13, 3989–3998. [Google Scholar] [CrossRef]
- Matossian, M.D.; Burks, H.E.; Elliott, S.; Hoang, V.T.; Bowles, A.C.; Sabol, R.A.; Wahba, B.; Anbalagan, M.; Rowan, B.; Abazeed, M.E.; et al. Drug resistance profiling of a new triple negative breast cancer patient-derived xenograft model. BMC Cancer 2019, 19, 205. [Google Scholar] [CrossRef] [PubMed]
- Mendez-Pertuz, M.; Martinez, P.; Blanco-Aparicio, C.; Gomez-Casero, E.; Belen Garcia, A.; Martinez-Torrecuadrada, J.; Palafox, M.; Cortes, J.; Serra, V.; Pastor, J.; et al. Modulation of telomere protection by the PI3K/AKT pathway. Nat. Commun. 2017, 8, 1278. [Google Scholar] [CrossRef]
- Merino, D.; Weber, T.S.; Serrano, A.; Vaillant, F.; Liu, K.; Pal, B.; Di Stefano, L.; Schreuder, J.; Lin, D.; Chen, Y.; et al. Barcoding reveals complex clonal behavior in patient-derived xenografts of metastatic triple negative breast cancer. Nat. Commun. 2019, 10, 766. [Google Scholar] [CrossRef]
- Pillai, S.G.; Li, S.; Siddappa, C.M.; Ellis, M.J.; Watson, M.A.; Aft, R. Identifying biomarkers of breast cancer micrometastatic disease in bone marrow using a patient-derived xenograft mouse model. Breast Cancer Res. 2018, 20, 2. [Google Scholar] [CrossRef]
- Rather, G.M.; Lin, S.Y.; Lin, H.; Banach-Petrosky, W.; Hirshfield, K.M.; Lin, C.Y.; Johnson, M.D.; Szekely, Z.; Bertino, J.R. Activated matriptase as a target to treat breast cancer with a drug conjugate. Oncotarget 2018, 9, 25983–25992. [Google Scholar] [CrossRef]
- Rosato, R.R.; Davila-Gonzalez, D.; Choi, D.S.; Qian, W.; Chen, W.; Kozielski, A.J.; Wong, H.; Dave, B.; Chang, J.C. Evaluation of anti-PD-1-based therapy against triple-negative breast cancer patient-derived xenograft tumors engrafted in humanized mouse models. Breast Cancer Res. 2018, 20, 108. [Google Scholar] [CrossRef] [PubMed]
- Ruiz de Garibay, G.; Mateo, F.; Stradella, A.; Valdes-Mas, R.; Palomero, L.; Serra-Musach, J.; Puente, D.A.; Diaz-Navarro, A.; Vargas-Parra, G.; Tornero, E.; et al. Tumor xenograft modeling identifies an association between TCF4 loss and breast cancer chemoresistance. Dis. Model. Mech. 2018, 11. [Google Scholar] [CrossRef] [PubMed]
- Ryu, J.S.; Sim, S.H.; Park, I.H.; Lee, E.G.; Lee, E.S.; Kim, Y.H.; Kwon, Y.; Kong, S.Y.; Lee, K.S. Integrative In Vivo Drug Testing Using Gene Expression Signature and Patient-Derived Xenografts from Treatment-Refractory HER2 Positive and Triple-Negative Subtypes of Breast Cancer. Cancers 2019, 11, 574. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Yao, L.C.; Cheng, M.; Cai, D.; Martinek, J.; Pan, C.X.; Shi, W.; Ma, A.H.; De Vere White, R.W.; Airhart, S.; et al. Humanized mice in studying efficacy and mechanisms of PD-1-targeted cancer immunotherapy. FASEB J. 2018, 32, 1537–1549. [Google Scholar] [CrossRef] [PubMed]
- Wang, D.; Xu, J.; Liu, B.; He, X.; Zhou, L.; Hu, X.; Qiao, F.; Zhang, A.; Xu, X.; Zhang, H.; et al. IL6 blockade potentiates the anti-tumor effects of gamma-secretase inhibitors in Notch3-expressing breast cancer. Cell Death Differ. 2018, 25, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Cohen, A.L.; Krishnakumar, S.; Wapnir, I.L.; Veeriah, S.; Deng, G.; Coram, M.A.; Piskun, C.M.; Longacre, T.A.; Herrler, M.; et al. Patient-derived xenografts of triple-negative breast cancer reproduce molecular features of patient tumors and respond to mTOR inhibition. Breast Cancer Res. 2014, 16, R36. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Zhang, H.; Ghia, E.M.; Huang, J.; Wu, L.; Zhang, J.; Lam, S.; Lei, Y.; He, J.; Cui, B.; et al. Inhibition of chemotherapy resistant breast cancer stem cells by a ROR1 specific antibody. Proc. Natl. Acad. Sci. USA 2019, 116, 1370–1377. [Google Scholar] [CrossRef] [PubMed]
- Cheang, M.C.; Chia, S.K.; Voduc, D.; Gao, D.; Leung, S.; Snider, J.; Watson, M.; Davies, S.; Bernard, P.S.; Parker, J.S.; et al. Ki67 index, HER2 status, and prognosis of patients with luminal B breast cancer. J. Natl. Cancer Inst. 2009, 101, 736–750. [Google Scholar] [CrossRef]
- Goetz, M.P.; Kalari, K.R.; Suman, V.J.; Moyer, A.M.; Yu, J.; Visscher, D.W.; Dockter, T.J.; Vedell, P.T.; Sinnwell, J.P.; Tang, X.; et al. Tumor Sequencing and Patient-Derived Xenografts in the Neoadjuvant Treatment of Breast Cancer. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef]
- Vaillant, F.; Merino, D.; Lee, L.; Breslin, K.; Pal, B.; Ritchie, M.E.; Smyth, G.K.; Christie, M.; Phillipson, L.J.; Burns, C.J.; et al. Targeting BCL-2 with the BH3 mimetic ABT-199 in estrogen receptor-positive breast cancer. Cancer Cell 2013, 24, 120–129. [Google Scholar] [CrossRef]
- Kang, S.A.; Guan, J.S.; Tan, H.J.; Chu, T.; Thike, A.A.; Bernado Morales, C.; Arribas, J.; Wong, C.Y.; Tan, P.H.; Gudi, M.; et al. Elevated WBP2 expression in HER2-positive breast cancers correlates with sensitivity to trastuzumab-based neo-adjuvant therapy:A Retrospective and Multicentric Study. Clin. Cancer Res. 2018. [Google Scholar] [CrossRef]
- Cleator, S.; Heller, W.; Coombes, R.C. Triple-negative breast cancer: Therapeutic options. Lancet Oncol. 2007, 8, 235–244. [Google Scholar] [CrossRef]
- Foulkes, W.D.; Smith, I.E.; Reis-Filho, J.S. Triple-negative breast cancer. N. Engl. J. Med. 2010, 363, 1938–1948. [Google Scholar] [CrossRef] [PubMed]
- Leconet, W.; Chentouf, M.; du Manoir, S.; Chevalier, C.; Sirvent, A.; Ait-Arsa, I.; Busson, M.; Jarlier, M.; Radosevic-Robin, N.; Theillet, C.; et al. Therapeutic Activity of Anti-AXL Antibody against Triple-Negative Breast Cancer Patient-Derived Xenografts and Metastasis. Clin. Cancer Res. 2017, 23, 2806–2816. [Google Scholar] [CrossRef] [PubMed]
- Byrd, T.T.; Fousek, K.; Pignata, A.; Szot, C.; Samaha, H.; Seaman, S.; Dobrolecki, L.; Salsman, V.S.; Oo, H.Z.; Bielamowicz, K.; et al. TEM8/ANTXR1-Specific CAR T Cells as a Targeted Therapy for Triple-Negative Breast Cancer. Cancer Res. 2018, 78, 489–500. [Google Scholar] [CrossRef] [PubMed]
- Tentler, J.J.; Tan, A.C.; Weekes, C.D.; Jimeno, A.; Leong, S.; Pitts, T.M.; Arcaroli, J.J.; Messersmith, W.A.; Eckhardt, S.G. Patient-derived tumour xenografts as models for oncology drug development. Nat. Rev. Clin. Oncol. 2012, 9, 338–350. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Qin, B.; Moyer, A.M.; Sinnwell, J.P.; Thompson, K.J.; Copland, J.A., 3rd; Marlow, L.A.; Miller, J.L.; Yin, P.; Gao, B.; et al. Establishing and characterizing patient-derived xenografts using pre-chemotherapy percutaneous biopsy and post-chemotherapy surgical samples from a prospective neoadjuvant breast cancer study. Breast Cancer Res. 2017, 19, 130. [Google Scholar] [CrossRef]
- El Ayachi, I.; Fatima, I.; Wend, P.; Alva-Ornelas, J.A.; Runke, S.; Kuenzinger, W.L.; Silva, J.; Silva, W.; Gray, J.K.; Lehr, S.; et al. The WNT10B network is associated with survival and metastases in chemoresistant triple-negative breast cancer. Cancer Res. 2018. [Google Scholar] [CrossRef]
- Qiu, M.; Peng, Q.; Jiang, I.; Carroll, C.; Han, G.; Rymer, I.; Lippincott, J.; Zachwieja, J.; Gajiwala, K.; Kraynov, E.; et al. Specific inhibition of Notch1 signaling enhances the antitumor efficacy of chemotherapy in triple negative breast cancer through reduction of cancer stem cells. Cancer Lett. 2013, 328, 261–270. [Google Scholar] [CrossRef]
- Chen, X.; Low, K.H.; Alexander, A.; Jiang, Y.; Karakas, C.; Hess, K.R.; Carey, J.P.W.; Bui, T.N.; Vijayaraghavan, S.; Evans, K.W.; et al. Cyclin E Overexpression Sensitizes Triple-Negative Breast Cancer to Wee1 Kinase Inhibition. Clin. Cancer Res. 2018, 24, 6594–6610. [Google Scholar] [CrossRef]
- Morgan, K.M.; Riedlinger, G.M.; Rosenfeld, J.; Ganesan, S.; Pine, S.R. Patient-Derived Xenograft Models of Non-Small Cell Lung Cancer and Their Potential Utility in Personalized Medicine. Front. Oncol. 2017, 7, 2. [Google Scholar] [CrossRef] [PubMed]
- Scott, C.L.; Becker, M.A.; Haluska, P.; Samimi, G. Patient-derived xenograft models to improve targeted therapy in epithelial ovarian cancer treatment. Front. Oncol. 2013, 3, 295. [Google Scholar] [CrossRef] [PubMed]
- Annibali, D.; Leucci, E.; Hermans, E.; Amant, F. Development of Patient-Derived Tumor Xenograft Models. Methods Mol. Biol. 2019, 1862, 217–225. [Google Scholar] [PubMed]
- Moro, M.; Bertolini, G.; Caserini, R.; Borzi, C.; Boeri, M.; Fabbri, A.; Leone, G.; Gasparini, P.; Galeone, C.; Pelosi, G.; et al. Establishment of patient derived xenografts as functional testing of lung cancer aggressiveness. Sci. Rep. 2017, 7, 6689. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, T.; Kobori, G.; Goto, T.; Akamatsu, S.; Terada, N.; Kobayashi, T.; Tanaka, Y.; Jung, G.; Kamba, T.; Ogawa, O.; et al. An original patient-derived xenograft of prostate cancer with cyst formation. Prostate 2016, 76, 994–1003. [Google Scholar] [CrossRef] [PubMed]
- Bernardo, C.; Costa, C.; Sousa, N.; Amado, F.; Santos, L. Patient-derived bladder cancer xenografts: A systematic review. Transl. Res. 2015, 166, 324–331. [Google Scholar] [CrossRef] [PubMed]
- Li, H.; Wheeler, S.; Park, Y.; Ju, Z.; Thomas, S.M.; Fichera, M.; Egloff, A.M.; Lui, V.W.; Duvvuri, U.; Bauman, J.E.; et al. Proteomic Characterization of Head and Neck Cancer Patient-Derived Xenografts. Mol. Cancer Res. 2016, 14, 278–286. [Google Scholar] [CrossRef]
- Gao, H.; Korn, J.M.; Ferretti, S.; Monahan, J.E.; Wang, Y.; Singh, M.; Zhang, C.; Schnell, C.; Yang, G.; Zhang, Y.; et al. High-throughput screening using patient-derived tumor xenografts to predict clinical trial drug response. Nat. Med. 2015, 21, 1318–1325. [Google Scholar] [CrossRef]
- Karamboulas, C.; Bruce, J.P.; Hope, A.J.; Meens, J.; Huang, S.H.; Erdmann, N.; Hyatt, E.; Pereira, K.; Goldstein, D.P.; Weinreb, I.; et al. Patient-Derived Xenografts for Prognostication and Personalized Treatment for Head and Neck Squamous Cell Carcinoma. Cell Rep. 2018, 25, 1318.e4–1331.e4. [Google Scholar] [CrossRef]
- Karamboulas, C.; Ailles, L. Patient-derived xenografts: A promising resource for preclinical cancer research. Mol. Cell. Oncol. 2019, 6, 1558684. [Google Scholar] [CrossRef]
- Grunewald, S.; Politz, O.; Bender, S.; Heroult, M.; Lustig, K.; Thuss, U.; Kneip, C.; Kopitz, C.; Zopf, D.; Collin, M.P.; et al. Rogaratinib: A potent and selective pan-FGFR inhibitor with broad antitumor activity in FGFR-overexpressing preclinical cancer models. Int. J. Cancer 2019. [Google Scholar] [CrossRef] [PubMed]
- Engebraaten, O.; Vollan, H.K.M.; Borresen-Dale, A.L. Triple-negative breast cancer and the need for new therapeutic targets. Am. J. Pathol. 2013, 183, 1064–1074. [Google Scholar] [CrossRef] [PubMed]
- Abramson, V.G.; Lehmann, B.D.; Ballinger, T.J.; Pietenpol, J.A. Subtyping of triple-negative breast cancer: Implications for therapy. Cancer 2015, 121, 8–16. [Google Scholar] [CrossRef] [PubMed]
- Lehmann, B.D.; Jovanovic, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef] [PubMed]
- Clohessy, J.G.; Pandolfi, P.P. Mouse hospital and co-clinical trial project--from bench to bedside. Nat. Rev. Clin. Oncol. 2015, 12, 491–498. [Google Scholar] [CrossRef]
- Byrne, A.T.; Alferez, D.G.; Amant, F.; Annibali, D.; Arribas, J.; Biankin, A.V.; Bruna, A.; Budinska, E.; Caldas, C.; Chang, D.K.; et al. Interrogating open issues in cancer precision medicine with patient-derived xenografts. Nat. Rev. Cancer 2017, 17, 254–268. [Google Scholar] [CrossRef] [PubMed]
- Gopinathan, A.; Morton, J.P.; Jodrell, D.I.; Sansom, O.J. GEMMs as preclinical models for testing pancreatic cancer therapies. Dis. Model. Mech. 2015, 8, 1185–1200. [Google Scholar] [CrossRef]
- Lunardi, A.; Pandolfi, P.P. A co-clinical platform to accelerate cancer treatment optimization. Trends Mol. Med. 2015, 21, 1–5. [Google Scholar] [CrossRef]
- Dutta, A.; Panja, S.; Virk, R.K.; Kim, J.Y.; Zott, R.; Cremers, S.; Golombos, D.M.; Liu, D.; Mosquera, J.M.; Mostaghel, E.A.; et al. Co-clinical Analysis of a Genetically Engineered Mouse Model and Human Prostate Cancer Reveals Significance of NKX3.1 Expression for Response to 5alpha-reductase Inhibition. Eur. Urol. 2017, 72, 499–506. [Google Scholar] [CrossRef]
- Mantovani, A.; Marchesi, F.; Malesci, A.; Laghi, L.; Allavena, P. Tumour-associated macrophages as treatment targets in oncology. Nat. Rev. Clin. Oncol. 2017, 14, 399–416. [Google Scholar] [CrossRef]
- Choi, J.; Gyamfi, J.; Jang, H.; Koo, J.S. The role of tumor-associated macrophage in breast cancer biology. Histol. Histopathol. 2018, 33, 133–145. [Google Scholar] [PubMed]
- Inoue, T.; Terada, N.; Kobayashi, T.; Ogawa, O. Patient-derived xenografts as in vivo models for research in urological malignancies. Nat. Rev. Urol. 2017, 14, 267–283. [Google Scholar] [CrossRef] [PubMed]
- Bartucci, M.; Ferrari, A.C.; Kim, I.Y.; Ploss, A.; Yarmush, M.; Sabaawy, H.E. Personalized Medicine Approaches in Prostate Cancer Employing Patient Derived 3D Organoids and Humanized Mice. Front. Cell Dev. Biol. 2016, 4, 64. [Google Scholar] [PubMed]
- Hui, L.; Chen, Y. Tumor microenvironment: Sanctuary of the devil. Cancer Lett. 2015, 368, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Joyce, J.A.; Pollard, J.W. Microenvironmental regulation of metastasis. Nat. Rev. Cancer 2009, 9, 239–252. [Google Scholar] [CrossRef] [PubMed]
- Cassidy, J.W.; Caldas, C.; Bruna, A. Maintaining Tumor Heterogeneity in Patient-Derived Tumor Xenografts. Cancer Res. 2015, 75, 2963–2968. [Google Scholar] [CrossRef]
- Morton, J.J.; Bird, G.; Refaeli, Y.; Jimeno, A. Humanized Mouse Xenograft Models: Narrowing the Tumor-Microenvironment Gap. Cancer Res. 2016, 76, 6153–6158. [Google Scholar] [CrossRef]
- Pearson, T.; Greiner, D.L.; Shultz, L.D. Creation of “humanized” mice to study human immunity. Curr. Protoc. Immunol. 2008, 81, 15–21. [Google Scholar]

| Advantages | Limitations | Recommendations to Overcome Limitations | |
|---|---|---|---|
| Cell line (cultured in vitro) | ·Maintained inexpensively ·Treated very easily ·Grow infinitely | ·Completely lack the tumor microenvironment ·Can’t maintain original cell properties ⇒Very low predictive value | ·Should be used in basic studies and very early stages of drug development ·Co-culture with cancer associated fibroblasts (CAFs) or immune cells will improve the predictive value |
| Cell line xenograft | ·High take rates ·Slightly recapitulate tumor microenvironment ·Take short time to be established | ·Can’t reproduce heterogeneity ·Can’t maintain the original cell properties ⇒Low predictive value | ·Should be used in the relatively early stages of drug development with a large number of mice, which can reflect the inter-tumor heterogeneity |
| Genetically engineered mouse | ·Recapitulate tumor initiation and early development process ·Gene of interest can be studied in detail ·Can be increased easily after establishment | ·Can’t reproduce heterogeneity of human tumor ⇒Low predictive value ·Take long time to be established | ·Should be used when investigating how a specific gene of interest could contribute to tumor initiation and relapse |
| PDX | ·Partly recapitulate tumor microenvironment ·Maintain histologic and genetic features of origin ⇒High predictive value ·Can be used for metastatic model | ·Low take rate ·Very expensive ·Take long time to be established | ·Development of new immunodeficient mice and/or better methods of tumor transplantation will improve the take rates and the cost |
| Authors | Host Mouse | Tissue Source | Subtypes of PDX (Number) | Method | Reference |
|---|---|---|---|---|---|
| Agnoletto and collegues | nude | primary, metastasis | triple negative (7), HER2+ (2), luminal (2) | interscapular | [31] |
| Al-Hajj and collegues | NOD/SCID | primary | triple negative (4) | orthotopic | [32] |
| Arango and collegues | nude | primary | triple negative (5) | orthotopic | [33] |
| Bruna and collegues | NSG | primary, biopsy, plueral effusion | triple negative (24), HER2+ (6), luminal (52) | orthotopic | [34] |
| Castroviejo-Bermejo and collegues | NSG | primary, biopsy | triple negative (8), luminal (5) | orthotopic | [35] |
| Contreras-Zárate and collegues | NSG | metastasis | triple negative (3), HER2+ (5), luminal (1) | orthotopic | [36] |
| Coussy and collegues | nude | primary | triple negative (61) | orthotopic | [37] |
| Cruz and colleagues | nude | primary | triple negative (9), luminal (1) | orthotopic | [38] |
| Dávila-González and collegues | SCID/Bg | primary | triple negative (5) | orthotopic | [39] |
| DeRose and collegues | NOD/SCID | primary, pleural effusion | triple negative (5), HER2+ (2), luminal (5) | orthotopic | [40] |
| Evans and collegues | NOD/SCID, nude | primary | triple negative (24) | orthotopic | [41] |
| Fatima and collegues | NSG | primary | triple negative (2) | orthotopic | [42] |
| Fleming and collegues | NOD/SCID | pleural effusion | - | orthotopic | [43] |
| Formisano and collegues | SCID/Bg | primary | luminal (2) | orthotopic | [44] |
| González-González and collegues | NSG | primary | triple negative (2) | orthotopic | [45] |
| Hsu and collegues | NSG | primary | luminal (2) | orthotopic | [46] |
| Hu and collegues | NOD/SCID | primary | - | orthotopic | [47] |
| Jung and collegues | NOD/SCID | primary | triple negative (24) | orthotopic | [48] |
| Kabos and collegues | NOD/SCID, NSG | primary, metastasis | triple negative (2), luminal (8) | orthotopic | [49] |
| Kanaya and collegues | NSG | primary | luminal (9) | orthotopic | [50] |
| Li and collegues | NOD/SCID | primary, metastasis | triple negative (12), HER2+ (2), luminal (8) | orthotopic | [51] |
| Liu and collegues | NSG | pleural effusion | HER2+ (2), luminal (2) | orthotopic | [52] |
| Ma and collegues | NOD/SCID | primary, metastasis | triple negative (3) | orthotopic | [53] |
| Marangoni and collegues | nude | primary | triple negative (15), HER2+ (2), luminal (1) | orthotopic | [54] |
| Matossian and collegues | SCID/Bg | primary | triple negative (1) | orthotopic | [55] |
| Méndez-Pertuz and collegues | nude | primary, metastasis | luminal (7) | orthotopic, lower flank | [56] |
| Merino and collegues | NSG | primary | triple negative (2) | orthotopic | [57] |
| Pillai and collegues | NOD/SCID | primary | triple negative (3), luminal (2) | orthotopic | [58] |
| Rather and collegues | NSG | primary | triple negative (1) | s.c. in the right flank | [59] |
| Rosato and collegues | NSG | triple negative (5) | orthotopic | [60] | |
| Ruiz de Garibay and collegues | nude | primary | triple negative (1) | orthotopic | [61] |
| Ryu and collegues | NSG, NOG | primary | triple negative (9), HER2+ (7), luminal (4) | orthotopic | [62] |
| Wang and collegues | SCID/Bg | primary | triple negative (2) | orthotopic | [63] |
| Wang and collegues | nude | primary | luminal (1) | orthotopic | [64] |
| Zhang and collegues | SCID/Bg, NSG | primary, pleural effusion | triple negative (12), HER2+ (3), luminal (2) | orthotopic | [2] |
| Zhang and collegues | NOD/SCID | primary, metastasis | triple negative (7) | orthotopic | [65] |
| Zhang and collegues | NSG | biopsy | - | orthotopic | [66] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Murayama, T.; Gotoh, N. Patient-Derived Xenograft Models of Breast Cancer and Their Application. Cells 2019, 8, 621. https://doi.org/10.3390/cells8060621
Murayama T, Gotoh N. Patient-Derived Xenograft Models of Breast Cancer and Their Application. Cells. 2019; 8(6):621. https://doi.org/10.3390/cells8060621
Chicago/Turabian StyleMurayama, Takahiko, and Noriko Gotoh. 2019. "Patient-Derived Xenograft Models of Breast Cancer and Their Application" Cells 8, no. 6: 621. https://doi.org/10.3390/cells8060621
APA StyleMurayama, T., & Gotoh, N. (2019). Patient-Derived Xenograft Models of Breast Cancer and Their Application. Cells, 8(6), 621. https://doi.org/10.3390/cells8060621