Proteomics in the World of Induced Pluripotent Stem Cells
Abstract
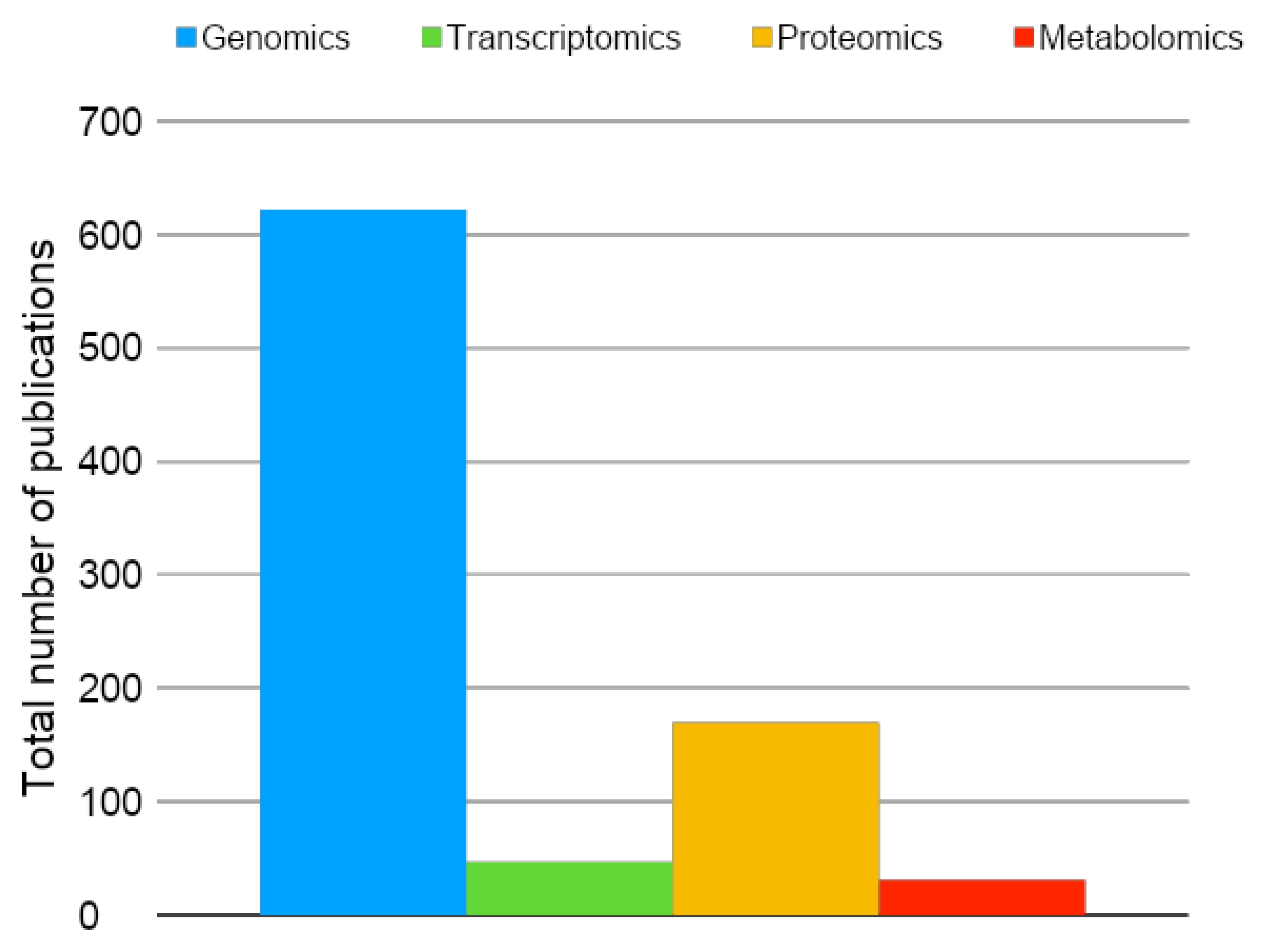
1. Introduction: A historical View and Evolution of Conceptual and Methodological Strategies in Proteomics
2. Induced Pluripotent Stem Cells (iPSC): A Historical Perspective
3. Proteomics of iPSC
4. Proteomics of iPSC-Derived Cells
4.1. iPSC Differentiation
4.2. Applications of Proteomic Approaches in the Study of Differentiated Cells Derived from iPSC
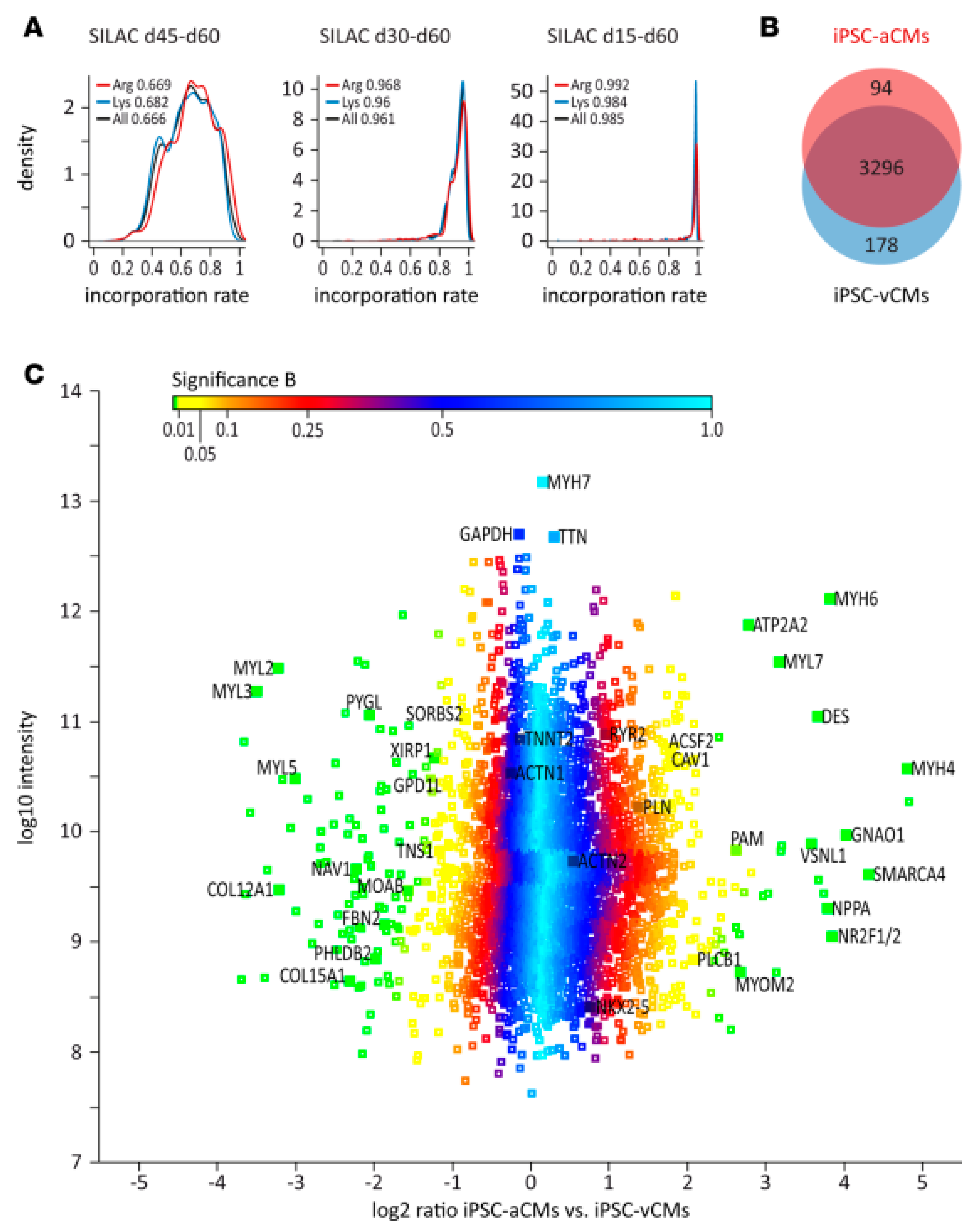
4.2.1. Cell Differentiation and Maturation
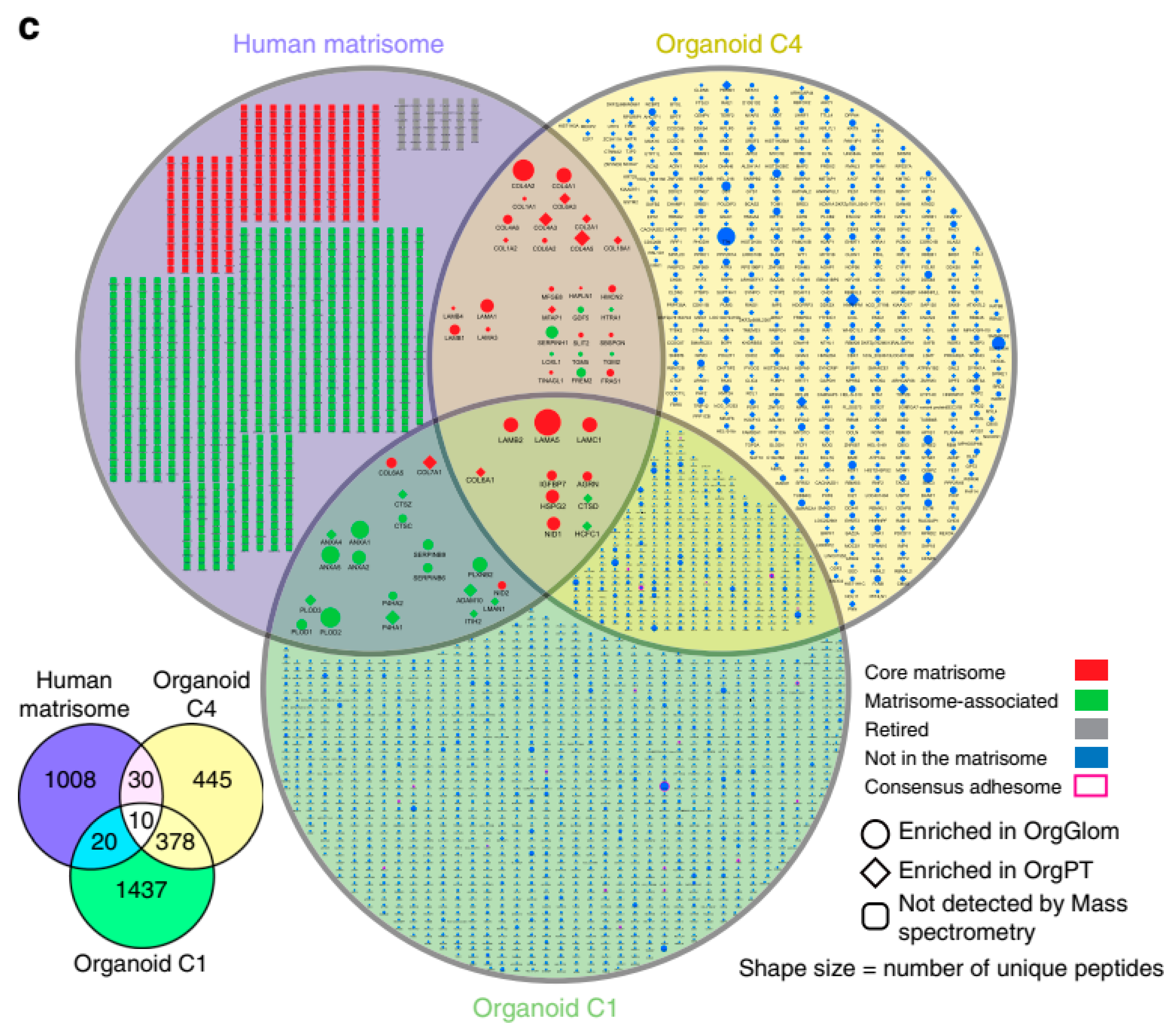
4.2.2. Proteomics in iPSC-Derived Organoids
4.2.3. Proteomics of iPSC-Derived Cells in Disease Models
4.2.4. Proteomic Approaches for the Study of Cell Interactions
4.2.5. Drug Screening Using iPSC-Derived Differentiated Cells: An Unexplored Field
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Patterson, S.D.; Aebersold, R.H. Proteomics: The first decade and beyond. Nat. Genet. 2003, 33, 311–323. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.C.; Mann, M. Mass spectrometry–based proteomics in cell biology. J. Cell Biol. 2010, 190, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Rabilloud, T.; Chevallet, M.; Luche, S.; Lelong, C. Two-dimensional gel electrophoresis in proteomics: Past, present and future. J. Proteomics 2010, 73, 2064–2077. [Google Scholar] [CrossRef] [PubMed]
- Gibson, F.; Anderson, L.; Babnigg, G.; Baker, M.; Berth, M.; Binz, P.-A.; Borthwick, A.; Cash, P.; Day, B.W.; Friedman, D.B.; et al. Guidelines for reporting the use of gel electrophoresis in proteomics. Nat. Biotechnol. 2008, 26, 863–864. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Opiteck, G.J.; Lewis, K.C.; Jorgenson, J.W.; Anderegg, R.J. Comprehensive on-line LC/LC/MS of proteins. Anal. Chem. 1997, 69, 1518–1524. [Google Scholar] [CrossRef] [PubMed]
- Yates, J.R., 3rd. The revolution and evolution of shotgun proteomics for large-scale proteome analysis. J. Am. Chem. Soc. 2013, 135, 1629–1640. [Google Scholar] [CrossRef] [PubMed]
- Perkins, D.N.; Pappin, D.J.C.; Creasy, D.M.; Cottrell, J.S. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 1999, 20, 3551–3567. [Google Scholar] [CrossRef]
- Eng, J.K.; McCormack, A.L.; Yates, J.R. An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J. Am. Soc. Mass. Spectrom. 1994, 5, 976–989. [Google Scholar] [CrossRef]
- Carvalho, P.C.; Fischer, J.S.G.; Chen, E.I.; Yates, J.R., 3rd; Barbosa, V.C. PatternLab for proteomics: A tool for differential shotgun proteomics. BMC Bioinf. 2008, 9, 316. [Google Scholar] [CrossRef]
- Cox, J.; Mann, M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008, 26, 1367–1372. [Google Scholar] [CrossRef]
- Breuza, L.; Poux, S.; Estreicher, A.; Famiglietti, M.L.; Magrane, M.; Tognolli, M.; Bridge, A.; Baratin, D.; Redaschi, N. UniProt Consortium The UniProtKB guide to the human proteome. Database 2016, bav120. [Google Scholar] [CrossRef]
- Cox, J.; Hein, M.Y.; Luber, C.A.; Paron, I.; Nagaraj, N.; Mann, M. Accurate proteome-wide label-free quantification by delayed normalization and maximal peptide ratio extraction, termed MaxLFQ. Mol. Cell. Proteomics 2014, 13, 2513–2526. [Google Scholar] [CrossRef] [PubMed]
- Wiese, S.; Reidegeld, K.A.; Meyer, H.E.; Warscheid, B. Protein labeling by iTRAQ: A new tool for quantitative mass spectrometry in proteome research. Proteomics 2007, 7, 1004. [Google Scholar] [CrossRef]
- Graumann, J.; Hubner, N.C.; Kim, J.B.; Ko, K.; Moser, M.; Kumar, C.; Cox, J.; Schöler, H.; Mann, M. Stable isotope labeling by amino acids in cell culture (SILAC) and proteome quantitation of mouse embryonic stem cells to a depth of 5,111 proteins. Mol. Cell. Proteomics 2008, 7, 672–683. [Google Scholar] [CrossRef] [PubMed]
- Lange, V.; Picotti, P.; Domon, B.; Aebersold, R. Selected reaction monitoring for quantitative proteomics: A tutorial. Mol. Syst. Biol. 2008, 4, 222. [Google Scholar] [CrossRef] [PubMed]
- Bourmaud, A.; Gallien, S.; Domon, B. Parallel reaction monitoring using quadrupole-Orbitrap mass spectrometer: Principle and applications. Proteomics 2016, 16, 2146–2159. [Google Scholar] [CrossRef] [PubMed]
- Vidova, V.; Spacil, Z. A review on mass spectrometry-based quantitative proteomics: Targeted and data independent acquisition. Anal. Chim. Acta 2017, 964, 7–23. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Jensen, O.N. Modification-specific proteomics: strategies for characterization of post-translational modifications using enrichment techniques. Proteomics 2009, 9, 4632–4641. [Google Scholar] [CrossRef] [PubMed]
- Olsen, J.V.; Mann, M. Status of large-scale analysis of post-translational modifications by mass spectrometry. Mol. Cell. Proteomics 2013, 12, 3444–3452. [Google Scholar] [CrossRef] [PubMed]
- Orre, L.M.; Vesterlund, M.; Pan, Y.; Arslan, T.; Zhu, Y.; Fernandez Woodbridge, A.; Frings, O.; Fredlund, E.; Lehtiö, J. SubCellBarCode: Proteome-wide Mapping of Protein Localization and Relocalization. Mol. Cell 2019, 73, 166–182. [Google Scholar] [CrossRef] [PubMed]
- Toby, T.K.; Fornelli, L.; Kelleher, N.L. Progress in Top-Down Proteomics and the Analysis of Proteoforms. Annu. Rev. Anal. Chem. 2016, 9, 499–519. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 2006, 126, 663–676. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, K.; Yamanaka, S. A decade of transcription factor-mediated reprogramming to pluripotency. Nat. Rev. Mol. Cell Biol. 2016, 17, 183–193. [Google Scholar] [CrossRef] [PubMed]
- Gurdon, J.B. The developmental capacity of nuclei taken from intestinal epithelium cells of feeding tadpoles. J. Embryol. Exp. Morphol. 1962, 10, 622–640. [Google Scholar] [PubMed]
- Gurdon, J.B. The egg and the nucleus: A battle for supremacy. Rambam Maimonides Med. J. 2015, 6, e0023. [Google Scholar] [CrossRef] [PubMed]
- Wilmut, I.; Schnieke, A.E.; McWhir, J.; Kind, A.J.; Campbell, K.H. Viable offspring derived from fetal and adult mammalian cells. Nature 1997, 385, 810–813. [Google Scholar] [CrossRef] [PubMed]
- Tachibana, M.; Amato, P.; Sparman, M.; Gutierrez, N.M.; Tippner-Hedges, R.; Ma, H.; Kang, E.; Fulati, A.; Lee, H.-S.; Sritanaudomchai, H.; et al. Human embryonic stem cells derived by somatic cell nuclear transfer. Cell 2013, 153, 1228–1238. [Google Scholar] [CrossRef] [PubMed]
- Evans, M.J.; Kaufman, M.H. Establishment in culture of pluripotential cells from mouse embryos. Nature 1981, 292, 154–156. [Google Scholar] [CrossRef]
- Thomson, J.A.; Itskovitz-Eldor, J.; Shapiro, S.S.; Waknitz, M.A.; Swiergiel, J.J.; Marshall, V.S.; Jones, J.M. Embryonic stem cell lines derived from human blastocysts. Science 1998, 282, 1145–1147. [Google Scholar] [CrossRef]
- Davis, R.L.; Weintraub, H.; Lassar, A.B. Expression of a single transfected cDNA converts fibroblasts to myoblasts. Cell 1987, 51, 987–1000. [Google Scholar] [CrossRef]
- Takahashi, K.; Tanabe, K.; Ohnuki, M.; Narita, M.; Ichisaka, T.; Tomoda, K.; Yamanaka, S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 2007, 131, 861–872. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Vodyanik, M.A.; Smuga-Otto, K.; Antosiewicz-Bourget, J.; Frane, J.L.; Tian, S.; Nie, J.; Jonsdottir, G.A.; Ruotti, V.; Stewart, R.; et al. Induced pluripotent stem cell lines derived from human somatic cells. Science 2007, 318, 1917–1920. [Google Scholar] [CrossRef] [PubMed]
- Yamanaka, S. Elite and stochastic models for induced pluripotent stem cell generation. Nature 2009, 460, 49–52. [Google Scholar] [CrossRef] [PubMed]
- Buganim, Y.; Faddah, D.A.; Cheng, A.W.; Itskovich, E.; Markoulaki, S.; Ganz, K.; Klemm, S.L.; van Oudenaarden, A.; Jaenisch, R. Single-cell expression analyses during cellular reprogramming reveal an early stochastic and a late hierarchic phase. Cell 2012, 150, 1209–1222. [Google Scholar] [CrossRef] [PubMed]
- Soufi, A.; Donahue, G.; Zaret, K.S. Facilitators and impediments of the pluripotency reprogramming factors’ initial engagement with the genome. Cell 2012, 151, 994–1004. [Google Scholar] [CrossRef] [PubMed]
- Shu, J.; Wu, C.; Wu, Y.; Li, Z.; Shao, S.; Zhao, W.; Tang, X.; Yang, H.; Shen, L.; Zuo, X.; et al. Induction of pluripotency in mouse somatic cells with lineage specifiers. Cell 2013, 153, 963–975. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Brown, J.; Kanarek, A.; Rajagopal, J.; Melton, D.A. In vivo reprogramming of adult pancreatic exocrine cells to beta-cells. Nature 2008, 455, 627–632. [Google Scholar] [CrossRef] [PubMed]
- Vierbuchen, T.; Ostermeier, A.; Pang, Z.P.; Kokubu, Y.; Südhof, T.C.; Wernig, M. Direct conversion of fibroblasts to functional neurons by defined factors. Nature 2010, 463, 1035–1041. [Google Scholar] [CrossRef]
- Ieda, M.; Fu, J.-D.; Delgado-Olguin, P.; Vedantham, V.; Hayashi, Y.; Bruneau, B.G.; Srivastava, D. Direct reprogramming of fibroblasts into functional cardiomyocytes by defined factors. Cell 2010, 142, 375–386. [Google Scholar] [CrossRef]
- Yu, J.; Hu, K.; Smuga-Otto, K.; Tian, S.; Stewart, R.; Slukvin, I.I.; Thomson, J.A. Human induced pluripotent stem cells free of vector and transgene sequences. Science 2009, 324, 797–801. [Google Scholar] [CrossRef]
- Kim, D.; Kim, C.-H.; Moon, J.-I.; Chung, Y.-G.; Chang, M.-Y.; Han, B.-S.; Ko, S.; Yang, E.; Cha, K.Y.; Lanza, R.; et al. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell 2009, 4, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Warren, L.; Manos, P.D.; Ahfeldt, T.; Loh, Y.-H.; Li, H.; Lau, F.; Ebina, W.; Mandal, P.K.; Smith, Z.D.; Meissner, A.; et al. Highly efficient reprogramming to pluripotency and directed differentiation of human cells with synthetic modified mRNA. Cell Stem Cell 2010, 7, 618–630. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, K.; Sano, M.; Ohtaka, M.; Furuta, B.; Umemura, Y.; Nakajima, Y.; Ikehara, Y.; Kobayashi, T.; Segawa, H.; Takayasu, S.; et al. Development of defective and persistent Sendai virus vector: A unique gene delivery/expression system ideal for cell reprogramming. J. Biol. Chem. 2011, 286, 4760–4771. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.J.; Suhr, S.T.; Rodriguez, R.M.; Chang, E.-A.; Wang, K.; Siripattarapravat, K.; Ko, T.; Cibelli, J.B. Human-induced pluripotent stem cells produced under xeno-free conditions. Stem Cells Dev. 2010, 19, 1221–1229. [Google Scholar] [CrossRef] [PubMed]
- Bergström, R.; Ström, S.; Holm, F.; Feki, A.; Hovatta, O. Xeno-free culture of human pluripotent stem cells. Methods Mol. Biol. 2011, 767, 125–136. [Google Scholar] [PubMed]
- Mandai, M.; Watanabe, A.; Kurimoto, Y.; Hirami, Y.; Morinaga, C.; Daimon, T.; Fujihara, M.; Akimaru, H.; Sakai, N.; Shibata, Y.; et al. Autologous induced stem-cell-derived retinal cells for macular degeneration. N. Engl. J. Med. 2017, 376, 1038–1046. [Google Scholar] [CrossRef] [PubMed]
- Monteiro, J. iPS Cells 10 Years Later. Cell 2016, 166, 1356–1359. [Google Scholar]
- Normile, D. iPS cell therapy reported safe. Science 2017, 355, 1109–1110. [Google Scholar] [CrossRef]
- Carvajal-Vergara, X.; Sevilla, A.; D’Souza, S.L.; Ang, Y.-S.; Schaniel, C.; Lee, D.-F.; Yang, L.; Kaplan, A.D.; Adler, E.D.; Rozov, R.; et al. Patient-specific induced pluripotent stem-cell-derived models of LEOPARD syndrome. Nature 2010, 465, 808–812. [Google Scholar] [CrossRef]
- Moretti, A.; Bellin, M.; Welling, A.; Jung, C.B.; Lam, J.T.; Bott-Flügel, L.; Dorn, T.; Goedel, A.; Höhnke, C.; Hofmann, F.; et al. Patient-specific induced pluripotent stem-cell models for long-QT syndrome. N. Engl. J. Med. 2010, 363, 1397–1409. [Google Scholar] [CrossRef]
- Itzhaki, I.; Maizels, L.; Huber, I.; Zwi-Dantsis, L.; Caspi, O.; Winterstern, A.; Feldman, O.; Gepstein, A.; Arbel, G.; Hammerman, H.; et al. Modelling the long QT syndrome with induced pluripotent stem cells. Nature 2011, 471, 225–229. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Disatnik, M.-H.; Monbureau, M.; Shamloo, M.; Mochly-Rosen, D.; Qi, X. Inhibition of mitochondrial fragmentation diminishes Huntington’s disease-associated neurodegeneration. J. Clin. Invest. 2013, 123, 5371–5388. [Google Scholar] [CrossRef] [PubMed]
- Kondo, T.; Asai, M.; Tsukita, K.; Kutoku, Y.; Ohsawa, Y.; Sunada, Y.; Imamura, K.; Egawa, N.; Yahata, N.; Okita, K.; et al. Modeling Alzheimer’s disease with iPSCs reveals stress phenotypes associated with intracellular Aβ and differential drug responsiveness. Cell Stem Cell 2013, 12, 487–496. [Google Scholar] [CrossRef] [PubMed]
- Kiskinis, E.; Sandoe, J.; Williams, L.A.; Boulting, G.L.; Moccia, R.; Wainger, B.J.; Han, S.; Peng, T.; Thams, S.; Mikkilineni, S.; et al. Pathways disrupted in human ALS motor neurons identified through genetic correction of mutant SOD1. Cell Stem Cell 2014, 14, 781–795. [Google Scholar] [CrossRef] [PubMed]
- Tucker, B.A.; Solivan-Timpe, F.; Roos, B.R.; Anfinson, K.R.; Robin, A.L.; Wiley, L.A.; Mullins, R.F.; Fingert, J.H. Duplication of TBK1 stimulates autophagy in iPSC-derived retinal cells from a patient with normal tension glaucoma. J. Stem Cell Res. Ther. 2014, 3, 161. [Google Scholar] [CrossRef]
- Jiang, W.; Lan, F.; Zhang, H. Human Induced Pluripotent Stem Cells for Inherited Cardiovascular Diseases Modeling. Curr. Stem Cell Res. Ther. 2016, 11, 533–541. [Google Scholar] [CrossRef]
- Qian, X.; Nguyen, H.N.; Song, M.M.; Hadiono, C.; Ogden, S.C.; Hammack, C.; Yao, B.; Hamersky, G.R.; Jacob, F.; Zhong, C.; et al. Brain-Region-Specific Organoids Using Mini-bioreactors for Modeling ZIKV Exposure. Cell 2016, 165, 1238–1254. [Google Scholar] [CrossRef]
- Chin, M.H.; Mason, M.J.; Xie, W.; Volinia, S.; Singer, M.; Peterson, C.; Ambartsumyan, G.; Aimiuwu, O.; Richter, L.; Zhang, J.; et al. Induced pluripotent stem cells and embryonic stem cells are distinguished by gene expression signatures. Cell Stem Cell 2009, 5, 111–123. [Google Scholar] [CrossRef]
- Bar, S.; Benvenisty, N. Epigenetic aberrations in human pluripotent stem cells. EMBO J. 2019, 38, e101033. [Google Scholar] [CrossRef]
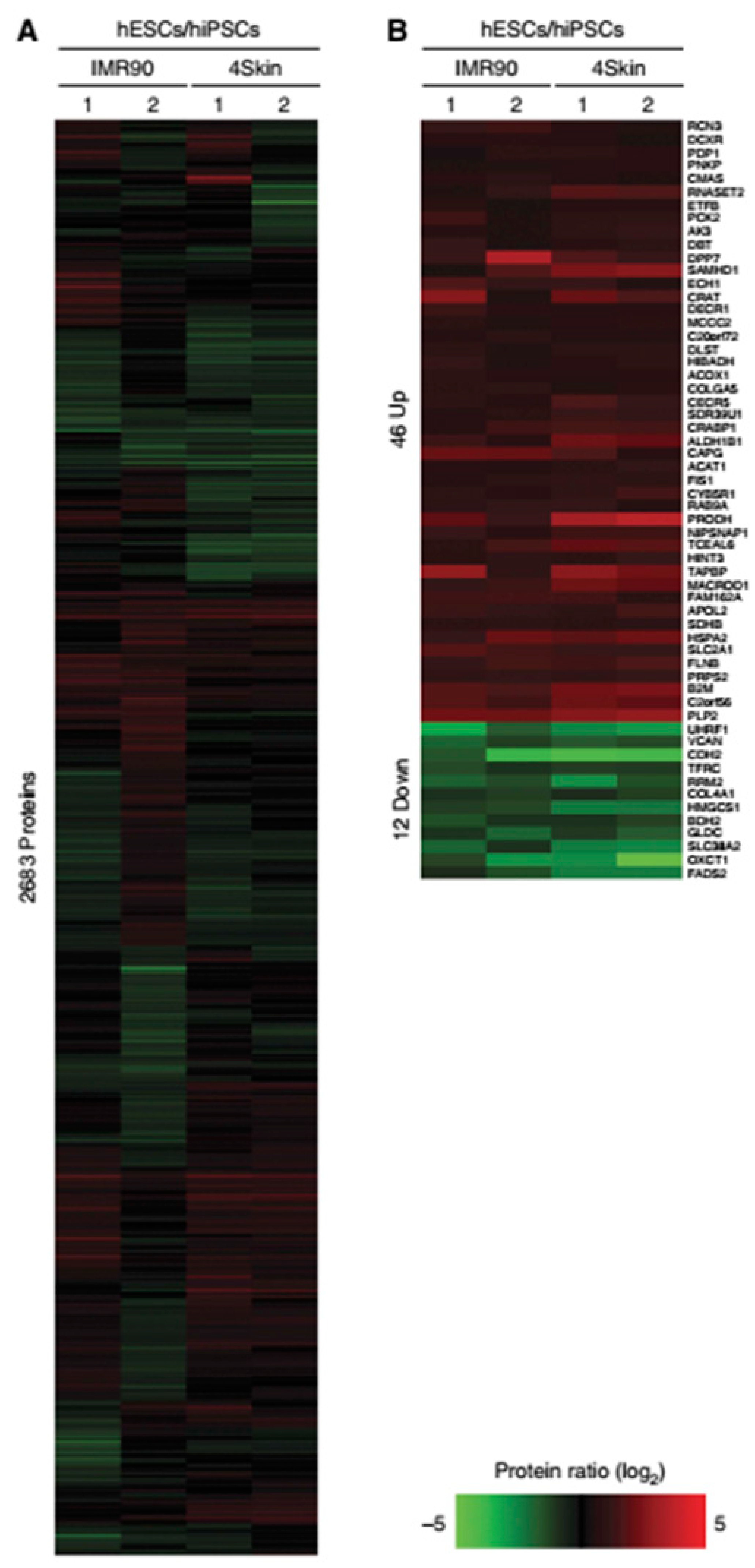
- Munoz, J.; Low, T.Y.; Kok, Y.J.; Chin, A.; Frese, C.K.; Ding, V.; Choo, A.; Heck, A.J.R. The quantitative proteomes of human-induced pluripotent stem cells and embryonic stem cells. Mol. Syst. Biol. 2011, 7, 550. [Google Scholar] [CrossRef]
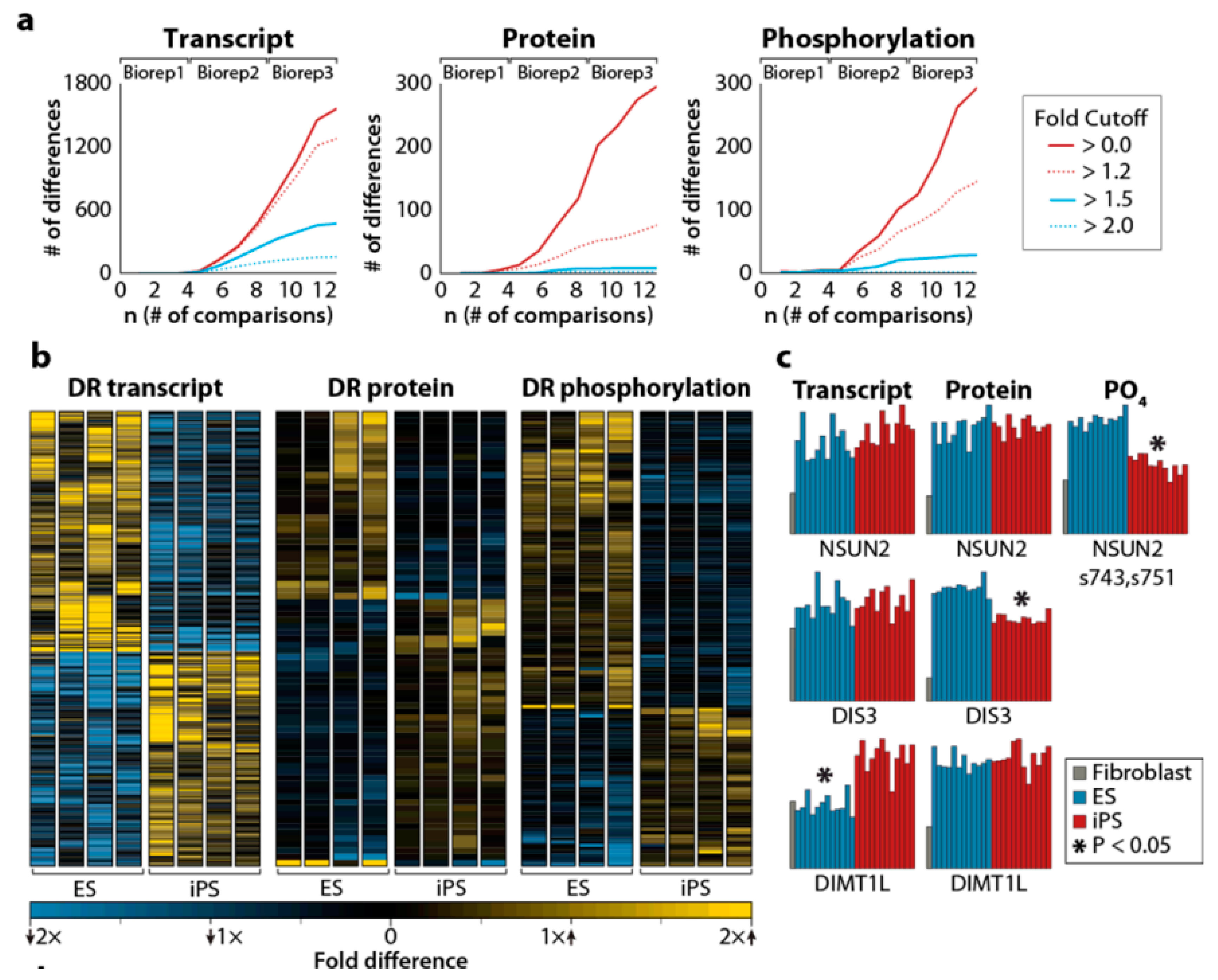
- Phanstiel, D.H.; Brumbaugh, J.; Wenger, C.D.; Tian, S.; Probasco, M.D.; Bailey, D.J.; Swaney, D.L.; Tervo, M.A.; Bolin, J.M.; Ruotti, V.; et al. Proteomic and phosphoproteomic comparison of human ES and iPS cells. Nat. Methods 2011, 8, 821–827. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.Y.; Kim, M.-J.; Jung, H.; Kim, W.K.; Kwon, S.O.; Son, M.J.; Jang, I.-S.; Choi, J.-S.; Park, S.G.; Park, B.C.; et al. Comparative proteomic analysis of human somatic cells, induced pluripotent stem cells, and embryonic stem cells. Stem Cells Dev. 2012, 21, 1272–1286. [Google Scholar] [CrossRef]
- Yamana, R.; Iwasaki, M.; Wakabayashi, M.; Nakagawa, M.; Yamanaka, S.; Ishihama, Y. Rapid and deep profiling of human induced pluripotent stem cell proteome by one-shot NanoLC-MS/MS analysis with meter-scale monolithic silica columns. J. Proteome Res. 2013, 12, 214–221. [Google Scholar] [CrossRef] [PubMed]
- Tien, W.-S.; Chen, P.-M.; Chuang, C.-Y.; Lui, S.-M.; Kuo, H.-C.; Chen, Y.-J.; Wu, K.-P. Combining membrane proteomics and computational three-way pathway analysis revealed signalling pathways preferentially regulated in human iPSCs and human ESCs. Sci. Rep. 2017, 7, 15055. [Google Scholar] [CrossRef] [PubMed]
- Marchetto, M.C.N.; Yeo, G.W.; Kainohana, O.; Marsala, M.; Gage, F.H.; Muotri, A.R. Transcriptional signature and memory retention of human-induced pluripotent stem cells. PLoS ONE 2009, 4, e7076. [Google Scholar] [CrossRef] [PubMed]
- Ohi, Y.; Qin, H.; Hong, C.; Blouin, L.; Polo, J.M.; Guo, T.; Qi, Z.; Downey, S.L.; Manos, P.D.; Rossi, D.J.; et al. Incomplete DNA methylation underlies a transcriptional memory of somatic cells in human iPS cells. Nat. Cell Biol. 2011, 13, 541–549. [Google Scholar] [CrossRef]
- Benevento, M.; Munoz, J. Role of mass spectrometry-based proteomics in the study of cellular reprogramming and induced pluripotent stem cells. Expert Rev. Proteomics 2012, 9, 379–399. [Google Scholar] [CrossRef]
- Pripuzova, N.S.; Getie-Kebtie, M.; Grunseich, C.; Sweeney, C.; Malech, H.; Alterman, M.A. Development of a protein marker panel for characterization of human induced pluripotent stem cells (hiPSCs) using global quantitative proteome analysis. Stem Cell Res. 2015, 14, 323–338. [Google Scholar] [CrossRef]
- Faradonbeh, M.Z.; Gharechahi, J.; Mollamohammadi, S.; Pakzad, M.; Taei, A.; Rassouli, H.; Baharvand, H.; Salekdeh, G.H. An orthogonal comparison of the proteome of human embryonic stem cells with that of human induced pluripotent stem cells of different genetic background. Mol. Biosyst. 2012, 8, 1833–1840. [Google Scholar] [CrossRef]
- Brenes, A.; Bensaddek, D.; Hukelmann, J.; Afzal, V.; Lamond, A.I. The iPSC proteomic compendium. bioRxiv 2018. [Google Scholar] [CrossRef]
- Baud, A.; Wessely, F.; Mazzacuva, F.; McCormick, J.; Camuzeaux, S.; Heywood, W.E.; Little, D.; Vowles, J.; Tuefferd, M.; Mosaku, O.; et al. Multiplex High-Throughput Targeted Proteomic Assay To Identify Induced Pluripotent Stem Cells. Anal. Chem. 2017, 89, 2440–2448. [Google Scholar] [CrossRef] [PubMed]
- Baud, A.; Heywood, W.E.; Little, D.; Gissen, P.; Mills, K. Preparation of iPSCs for Targeted Proteomic Analysis. Methods Mol. Biol. 2019, 1994, 131–139. [Google Scholar] [PubMed]
- Miyagawa, S.; Domae, K.; Yoshikawa, Y.; Fukushima, S.; Nakamura, T.; Saito, A.; Sakata, Y.; Hamada, S.; Toda, K.; Pak, K.; et al. Phase I Clinical trial of autologous stem cell-sheet transplantation therapy for treating cardiomyopathy. J. Am. Heart Assoc. 2017, 6, e003918. [Google Scholar] [CrossRef] [PubMed]
- Lee, G.; Papapetrou, E.P.; Kim, H.; Chambers, S.M.; Tomishima, M.J.; Fasano, C.A.; Ganat, Y.M.; Menon, J.; Shimizu, F.; Viale, A.; et al. Modelling pathogenesis and treatment of familial dysautonomia using patient-specific iPSCs. Nature 2009, 461, 402–406. [Google Scholar] [CrossRef] [PubMed]
- Cayo, M.A.; Cai, J.; DeLaForest, A.; Noto, F.K.; Nagaoka, M.; Clark, B.S.; Collery, R.F.; Si-Tayeb, K.; Duncan, S.A. JD induced pluripotent stem cell-derived hepatocytes faithfully recapitulate the pathophysiology of familial hypercholesterolemia. Hepatology 2012, 56, 2163–2171. [Google Scholar] [CrossRef] [PubMed]
- Juopperi, T.A.; Kim, W.; Chiang, C.-H.; Yu, H.; Margolis, R.L.; Ross, C.A.; Ming, G.-L.; Song, H. Astrocytes generated from patient induced pluripotent stem cells recapitulate features of Huntington’s disease patient cells. Mol. Brain 2012, 5, 17. [Google Scholar] [CrossRef]
- Imaizumi, Y.; Okada, Y.; Akamatsu, W.; Koike, M.; Kuzumaki, N.; Hayakawa, H.; Nihira, T.; Kobayashi, T.; Ohyama, M.; Sato, S.; et al. Mitochondrial dysfunction associated with increased oxidative stress and α-synuclein accumulation in PARK2 iPSC-derived neurons and postmortem brain tissue. Mol. Brain 2012, 5, 35. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Danés, A.; Richaud-Patin, Y.; Carballo-Carbajal, I.; Jiménez-Delgado, S.; Caig, C.; Mora, S.; Di Guglielmo, C.; Ezquerra, M.; Patel, B.; Giralt, A.; et al. Disease-specific phenotypes in dopamine neurons from human iPS-based models of genetic and sporadic Parkinson’s disease. EMBO Mol. Med. 2012, 4, 380–395. [Google Scholar] [CrossRef] [PubMed]
- Trounson, A.; McDonald, C. Stem Cell Therapies in Clinical Trials: Progress and Challenges. Cell Stem Cell 2015, 17, 11–22. [Google Scholar] [CrossRef]
- Huang, P.; He, Z.; Ji, S.; Sun, H.; Xiang, D.; Liu, C.; Hu, Y.; Wang, X.; Hui, L. Induction of functional hepatocyte-like cells from mouse fibroblasts by defined factors. Nature 2011, 475, 386–389. [Google Scholar] [CrossRef]
- Xu, J.; Du, Y.; Deng, H. Direct lineage reprogramming: Strategies, mechanisms, and applications. Cell Stem Cell 2015, 16, 119–134. [Google Scholar] [CrossRef] [PubMed]
- Takeuchi, J.K.; Bruneau, B.G. Directed transdifferentiation of mouse mesoderm to heart tissue by defined factors. Nature 2009, 459, 708–711. [Google Scholar] [CrossRef] [PubMed]
- Yoo, A.S.; Sun, A.X.; Li, L.; Shcheglovitov, A.; Portmann, T.; Li, Y.; Lee-Messer, C.; Dolmetsch, R.E.; Tsien, R.W.; Crabtree, G.R. MicroRNA-mediated conversion of human fibroblasts to neurons. Nature 2011, 476, 228–231. [Google Scholar] [CrossRef] [PubMed]
- Jayawardena, T.M.; Egemnazarov, B.; Finch, E.A.; Zhang, L.; Payne, J.A.; Pandya, K.; Zhang, Z.; Rosenberg, P.; Mirotsou, M.; Dzau, V.J. MicroRNA-mediated in vitro and in vivo direct reprogramming of cardiac fibroblasts to cardiomyocytes. Circ. Res. 2012, 110, 1465–1473. [Google Scholar] [CrossRef] [PubMed]
- Bartel, D.P. MicroRNAs: Target recognition and regulatory functions. Cell 2009, 136, 215–233. [Google Scholar] [CrossRef] [PubMed]
- Theunissen, T.W.; Jaenisch, R. Molecular Control of Induced Pluripotency. Cell Stem Cell 2014, 14, 720–734. [Google Scholar] [CrossRef]
- Li, W.; Li, K.; Wei, W.; Ding, S. Chemical Approaches to Stem Cell Biology and Therapeutics. Cell Stem Cell 2013, 13, 270–283. [Google Scholar] [CrossRef]
- Ladewig, J.; Mertens, J.; Kesavan, J.; Doerr, J.; Poppe, D.; Glaue, F.; Herms, S.; Wernet, P.; Kögler, G.; Müller, F.-J.; et al. Small molecules enable highly efficient neuronal conversion of human fibroblasts. Nat. Methods 2012, 9, 575–578. [Google Scholar] [CrossRef]
- Cheng, L.; Hu, W.; Qiu, B.; Zhao, J.; Yu, Y.; Guan, W.; Wang, M.; Yang, W.; Pei, G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2015, 25, 645–646. [Google Scholar] [CrossRef]
- Zhu, S.; Ambasudhan, R.; Sun, W.; Kim, H.J.; Talantova, M.; Wang, X.; Zhang, M.; Zhang, Y.; Laurent, T.; Parker, J.; et al. Small molecules enable OCT4-mediated direct reprogramming into expandable human neural stem cells. Cell Res. 2014, 24, 126–129. [Google Scholar] [CrossRef]
- Cahan, P.; Li, H.; Morris, S.A.; Lummertz da Rocha, E.; Daley, G.Q.; Collins, J.J. CellNet: network biology applied to stem cell engineering. Cell 2014, 158, 903–915. [Google Scholar] [CrossRef] [PubMed]
- Du, Y.; Wang, J.; Jia, J.; Song, N.; Xiang, C.; Xu, J.; Hou, Z.; Su, X.; Liu, B.; Jiang, T.; et al. Human hepatocytes with drug metabolic function induced from fibroblasts by lineage reprogramming. Cell Stem Cell 2014, 14, 394–403. [Google Scholar] [CrossRef] [PubMed]
- Viiri, L.E.; Rantapero, T.; Kiamehr, M.; Alexanova, A.; Oittinen, M.; Viiri, K.; Niskanen, H.; Nykter, M.; Kaikkonen, M.U.; Aalto-Setälä, K. Extensive reprogramming of the nascent transcriptome during iPSC to hepatocyte differentiation. Sci. Rep. 2019, 9, 3562. [Google Scholar] [CrossRef]
- Camp, J.G.; Sekine, K.; Gerber, T.; Loeffler-Wirth, H.; Binder, H.; Gac, M.; Kanton, S.; Kageyama, J.; Damm, G.; Seehofer, D.; et al. Multilineage communication regulates human liver bud development from pluripotency. Nature 2017, 546, 533–538. [Google Scholar] [CrossRef] [PubMed]
- Lemoine, M.D.; Mannhardt, I.; Breckwoldt, K.; Prondzynski, M.; Flenner, F.; Ulmer, B.; Hirt, M.N.; Neuber, C.; Horváth, A.; Kloth, B.; et al. Human iPSC-derived cardiomyocytes cultured in 3D engineered heart tissue show physiological upstroke velocity and sodium current density. Sci. Rep. 2017, 7, 5464. [Google Scholar] [CrossRef] [PubMed]
- Clevers, H. Modeling Development and Disease with Organoids. Cell 2016, 165, 1586–1597. [Google Scholar] [CrossRef] [PubMed]
- Sato, T.; Vries, R.G.; Snippert, H.J.; van de Wetering, M.; Barker, N.; Stange, D.E.; van Es, J.H.; Abo, A.; Kujala, P.; Peters, P.J.; et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 2009, 459, 262–265. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Renner, M.; Martin, C.-A.; Wenzel, D.; Bicknell, L.S.; Hurles, M.E.; Homfray, T.; Penninger, J.M.; Jackson, A.P.; Knoblich, J.A. Cerebral organoids model human brain development and microcephaly. Nature 2013, 501, 373–379. [Google Scholar] [CrossRef]
- Jha, R.; Wu, Q.; Singh, M.; Preininger, M.K.; Han, P.; Ding, G.; Cho, H.C.; Jo, H.; Maher, K.O.; Wagner, M.B.; et al. Simulated microgravity and 3D culture enhance induction, viability, proliferation and differentiation of cardiac progenitors from human pluripotent stem cells. Sci. Rep. 2016, 6, 30956. [Google Scholar] [CrossRef]
- Wimmer, R.A.; Leopoldi, A.; Aichinger, M.; Wick, N.; Hantusch, B.; Novatchkova, M.; Taubenschmid, J.; Hämmerle, M.; Esk, C.; Bagley, J.A.; et al. Human blood vessel organoids as a model of diabetic vasculopathy. Nature 2019, 565, 505–510. [Google Scholar] [CrossRef]
- Van Hoof, D.; Krijgsveld, J.; Mummery, C. Proteomic analysis of stem cell differentiation and early development. Cold Spring Harb. Perspect. Biol. 2012, 4, a008177. [Google Scholar] [CrossRef] [PubMed]
- Lam, A.Q.; Freedman, B.S.; Bonventre, J.V. Directed differentiation of pluripotent stem cells to kidney cells. Semin. Nephrol. 2014, 34, 445–461. [Google Scholar] [CrossRef] [PubMed]
- Fujita, J.; Tohyama, S.; Kishino, Y.; Okada, M.; Morita, Y. Concise review: Genetic and epigenetic regulation of cardiac differentiation from human pluripotent stem cells. Stem Cells 2019. [Google Scholar] [CrossRef] [PubMed]
- Pacitti, D.; Privolizzi, R.; Bax, B.E. Organs to Cells and Cells to Organoids: The Evolution of Central Nervous System Modelling. Front. Cell. Neurosci. 2019, 13, 129. [Google Scholar] [CrossRef] [PubMed]
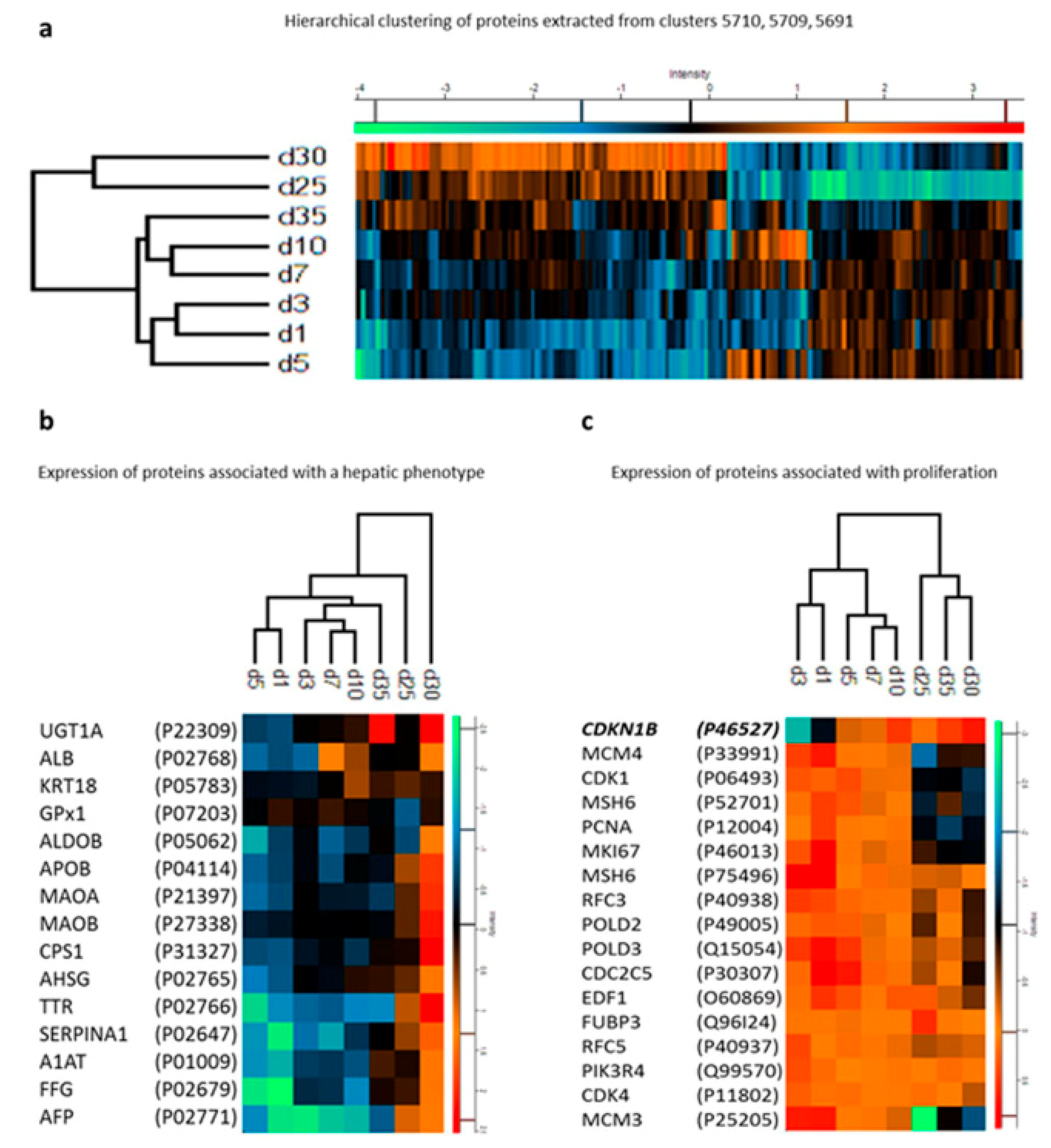
- Hurrell, T.; Segeritz, C.-P.; Vallier, L.; Lilley, K.S.; Cromarty, A.D. A proteomic time course through the differentiation of human induced pluripotent stem cells into hepatocyte-like cells. Sci. Rep. 2019, 9, 3270. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, S.; Holton, K.L.; Lanza, R. Efficient differentiation of functional hepatocytes from human embryonic stem cells. Stem Cells 2008, 26, 1117–1127. [Google Scholar] [CrossRef] [PubMed]
- Baxter, M.; Withey, S.; Harrison, S.; Segeritz, C.-P.; Zhang, F.; Atkinson-Dell, R.; Rowe, C.; Gerrard, D.T.; Sison-Young, R.; Jenkins, R.; et al. Phenotypic and functional analyses show stem cell-derived hepatocyte-like cells better mimic fetal rather than adult hepatocytes. J. Hepatol. 2015, 62, 581–589. [Google Scholar] [CrossRef] [PubMed]
- Bronte, F.; Bronte, G.; Fanale, D.; Caruso, S.; Bronte, E.; Bavetta, M.G.; Fiorentino, E.; Rolfo, C.; Bazan, V.; Di Marco, V.; et al. HepatomiRNoma: The proposal of a new network of targets for diagnosis, prognosis and therapy in hepatocellular carcinoma. Crit. Rev. Oncol. Hematol. 2016, 97, 312–321. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Loya, K.; Rani, B.; Möbus, S.; Balakrishnan, A.; Lamle, J.; Cathomen, T.; Vogel, A.; Manns, M.P.; Ott, M.; et al. MicroRNA-221 overexpression accelerates hepatocyte proliferation during liver regeneration. Hepatology 2013, 57, 299–310. [Google Scholar] [CrossRef]
- Rong, M.; Chen, G.; Dang, Y. Increased miR-221 expression in hepatocellular carcinoma tissues and its role in enhancing cell growth and inhibiting apoptosis in vitro. BMC Cancer 2013, 13, 21. [Google Scholar] [CrossRef] [PubMed]
- Franco, C.; Soares, R.; Pires, E.; Koci, K.; Almeida, A.M.; Santos, R.; Coelho, A.V. Understanding regeneration through proteomics. Proteomics 2013, 13, 686–709. [Google Scholar] [CrossRef] [PubMed]
- Dambach, D.M.; Andrews, B.A.; Moulin, F. New technologies and screening strategies for hepatotoxicity: Use of in vitro models. Toxicol. Pathol. 2005, 33, 17–26. [Google Scholar] [CrossRef] [PubMed]
- Cyganek, L.; Tiburcy, M.; Sekeres, K.; Gerstenberg, K.; Bohnenberger, H.; Lenz, C.; Henze, S.; Stauske, M.; Salinas, G.; Zimmermann, W.-H.; et al. Deep phenotyping of human induced pluripotent stem cell-derived atrial and ventricular cardiomyocytes. JCI Insight 2018, 3, 99941. [Google Scholar] [CrossRef]
- Doll, S.; Dreßen, M.; Geyer, P.E.; Itzhak, D.N.; Braun, C.; Doppler, S.A.; Meier, F.; Deutsch, M.-A.; Lahm, H.; Lange, R.; et al. Region and cell-type resolved quantitative proteomic map of the human heart. Nat. Commun. 2017, 8, 1469. [Google Scholar] [CrossRef] [PubMed]
- Lancaster, M.A.; Knoblich, J.A. Organogenesis in a dish: Modeling development and disease using organoid technologies. Science 2014, 345, 1247125. [Google Scholar] [CrossRef]
- Takahashi, T. Organoids for drug discovery and personalized medicine. Annu. Rev. Pharmacol. Toxicol. 2019, 59, 447–462. [Google Scholar] [CrossRef]
- Doudna, J.A.; Charpentier, E. The new frontier of genome engineering with CRISPR-Cas9. Science 2014, 346, 1258096. [Google Scholar] [CrossRef]
- Rinschen, M.M.; Gödel, M.; Grahammer, F.; Zschiedrich, S.; Helmstädter, M.; Kretz, O.; Zarei, M.; Braun, D.A.; Dittrich, S.; Pahmeyer, C.; et al. A multi-layered quantitative in vivo Expression Atlas of the Podocyte unravels kidney disease candidate genes. Cell Rep. 2018, 23, 2495–2508. [Google Scholar] [CrossRef]
- Hale, L.J.; Howden, S.E.; Phipson, B.; Lonsdale, A.; Er, P.X.; Ghobrial, I.; Hosawi, S.; Wilson, S.; Lawlor, K.T.; Khan, S.; et al. 3D organoid-derived human glomeruli for personalised podocyte disease modelling and drug screening. Nat. Commun. 2018, 9, 5167. [Google Scholar] [CrossRef]
- Sison, K.; Eremina, V.; Baelde, H.; Min, W.; Hirashima, M.; George Fantus, I.; Quaggin, S.E. Glomerular structure and function require paracrine, not autocrine, VEGF–VEGFR-2 signaling. J. Am. Soc. Nephrol. 2010, 21, 1691–1701. [Google Scholar] [CrossRef]
- Johnstone, M.; Vasistha, N.A.; Barbu, M.C.; Dando, O.; Burr, K.; Christopher, E.; Glen, S.; Robert, C.; Fetit, R.; Macleod, K.G.; et al. Reversal of proliferation deficits caused by chromosome 16p13.11 microduplication through targeting NFκB signaling: An integrated study of patient-derived neuronal precursor cells, cerebral organoids and in vivo brain imaging. Mol. Psychiatry 2019, 24, 294–311. [Google Scholar] [CrossRef] [PubMed]
- Bradshaw, N.J.; Hayashi., M.A. NDE1 and NDEL1 from genes to (mal)functions: parallel but distinct roles impacting on neurodevelopmental disorders and psychiatric illness. Cell. Mol. Life Sci. 2017, 74, 1191–1210. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, R.M. Cell and environment interactions in tumor microregions: The multicell spheroid model. Science 1988, 240, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Kunz-Schughart, L.A.; Freyer, J.P.; Hofstaedter, F.; Ebner, R. The use of 3-D cultures for high-throghput screening: The multicellular spheroid model. J. Biomol. Screen. 2004, 273, 273–285. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Lee, H.-K.; Moo, L.; Hanlon, E.; Stein, T.; Xia, W. Common proteomic profiles of induced pluripotent stem cell-derived three-dimensional neurons and brain tissue from Alzheimer patients. J. Proteomics 2018, 182, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Abe, Y.; Tada, A.; Isoyama, J.; Nagayama, S.; Yao, R.; Adachi, J.; Tomonaga, T.; Nagayama, S.; Yao, R.; Adachi, J.; et al. Improved phosphoproteomic analysis for phosphosignaling and active-kinome profiling in Matrigel-embedded spheroids and patient-derived organoids. Sci. Rep. 2018, 8, 11401. [Google Scholar] [CrossRef] [PubMed]
- Schick, R.; Mekies, L.N.; Shemer, Y.; Eisen, B.; Hallas, T.; Ben Jehuda, R.; Ben-Ari, M.; Szantai, A.; Willi, L.; Shulman, R.; et al. Functional abnormalities in induced pluripotent stem cell-derived cardiomyocytes generated from titin-mutated patients with dilated cardiomyopathy. PLoS ONE 2018, 13, e0205719. [Google Scholar] [CrossRef]
- Arvanitis, D.A.; Vafiadaki, E.; Sanoudou, D.; Kranias, E.G. Histidine-rich calcium binding protein: The new regulator of sarcoplasmic reticulum calcium cycling. J. Mol. Cell. Cardiol. 2011, 50, 43–49. [Google Scholar] [CrossRef]
- Burns, A.; Iliffe, S. Alzheimer’s disease. BMJ 2009, 338, b158. [Google Scholar] [CrossRef]
- Van der Zee, J.; Sleegers, K.; Van Broeckhoven, C. Invited article: The Alzheimer disease-frontotemporal lobar degeneration spectrum. Neurology 2008, 71, 1191–1197. [Google Scholar] [CrossRef]
- Lindoso, R.S.; Sandim, V.; Collino, F.; Carvalho, A.B.; Dias, J.; da Costa, M.R.; Zingali, R.B.; Vieyra, A. Proteomics of cell-cell interactions in health and disease. Proteomics 2016, 16, 328–344. [Google Scholar] [CrossRef] [PubMed]
- Krasny, L.; Bland, P.; Kogata, N.; Wai, P.; Howard, B.A.; Natrajan, R.C.; Huang, P.H. SWATH mass spectrometry as a tool for quantitative profiling of the matrisome. J. Proteomics 2018, 189, 11–22. [Google Scholar] [CrossRef]
- Abecasis, B.; Gomes-Alves, P.; Rosa, S.; Gouveia, P.J.; Ferreira, L.; Serra, M.; Alves, P.M. Unveiling the molecular crosstalk in a human induced pluripotent stem cell-derived cardiac model. Biotechnol. Bioeng. 2019, 116, 1245–1252. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Deng, Y.; Liu, Y.; Gong, W.; Deng, W. Stem cell models for drug discovery and toxicology studies. J. Biochem. Mol. Toxicol. 2013, 27, 17–27. [Google Scholar] [CrossRef] [PubMed]
- Csöbönyeiová, M.; Polák, Š.; Danišovič, L. Toxicity testing and drug screening using iPSC-derived hepatocytes, cardiomyocytes, and neural cells. Can. J. Physiol. Pharm. 2016, 94, 687–694. [Google Scholar] [CrossRef]
- Zhao, W.-N.; Cheng, C.; Theriault, K.M.; Sheridan, S.D.; Tsai, L.-H.; Haggarty, S.J. A high-throughput screen for Wnt/β-catenin signaling pathway modulators in human iPSC-derived neural progenitors. J. Biomol. Screen. 2012, 17, 1252–1263. [Google Scholar] [CrossRef] [PubMed]
- Wetmore, B.A.; Merrick, B.A. Toxicoproteomics: Proteomics applied to toxicology and pathology. Toxicol. Pathol. 2004, 32, 619–642. [Google Scholar] [CrossRef] [PubMed]
- Schirle, M.; Bantscheff, M.; Kuster, B. Mass spectrometry-based proteomics in preclinical drug discovery. Chem. Biol. 2012, 19, 72–84. [Google Scholar] [CrossRef] [PubMed]
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lindoso, R.S.; Kasai-Brunswick, T.H.; Monnerat Cahli, G.; Collino, F.; Bastos Carvalho, A.; Campos de Carvalho, A.C.; Vieyra, A. Proteomics in the World of Induced Pluripotent Stem Cells. Cells 2019, 8, 703. https://doi.org/10.3390/cells8070703
Lindoso RS, Kasai-Brunswick TH, Monnerat Cahli G, Collino F, Bastos Carvalho A, Campos de Carvalho AC, Vieyra A. Proteomics in the World of Induced Pluripotent Stem Cells. Cells. 2019; 8(7):703. https://doi.org/10.3390/cells8070703
Chicago/Turabian StyleLindoso, Rafael Soares, Tais H. Kasai-Brunswick, Gustavo Monnerat Cahli, Federica Collino, Adriana Bastos Carvalho, Antonio Carlos Campos de Carvalho, and Adalberto Vieyra. 2019. "Proteomics in the World of Induced Pluripotent Stem Cells" Cells 8, no. 7: 703. https://doi.org/10.3390/cells8070703
APA StyleLindoso, R. S., Kasai-Brunswick, T. H., Monnerat Cahli, G., Collino, F., Bastos Carvalho, A., Campos de Carvalho, A. C., & Vieyra, A. (2019). Proteomics in the World of Induced Pluripotent Stem Cells. Cells, 8(7), 703. https://doi.org/10.3390/cells8070703






