Exosomes and Their Noncoding RNA Cargo Are Emerging as New Modulators for Diabetes Mellitus
Abstract
:1. Extracellular Vesicles and Exosomes
2. Exosome Uptake
3. Exosomes and Diabetes
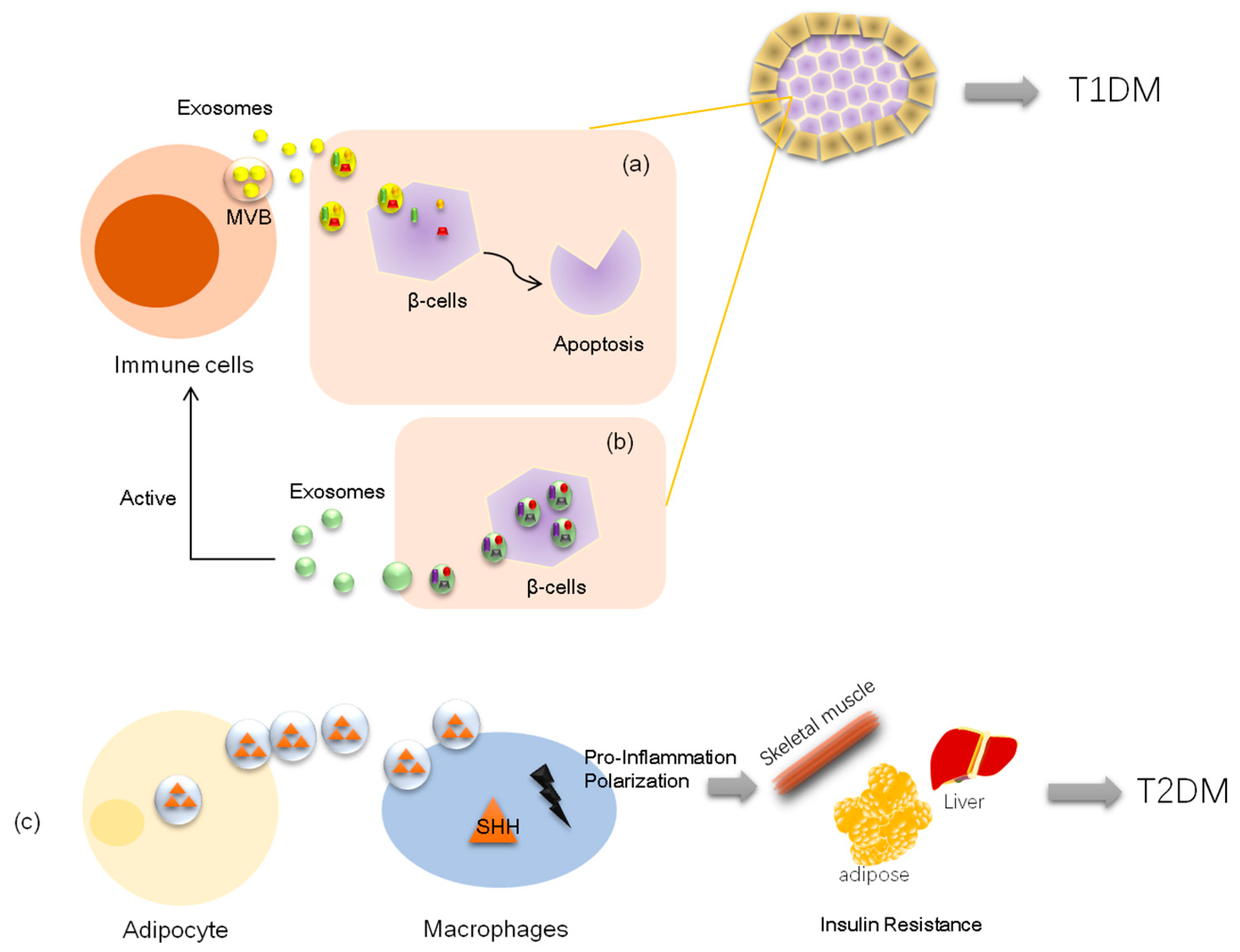
3.1. Exosomes and Type 1 Diabetes
3.2. Exosomes and Type 2 Diabetes
4. Exosome Noncoding RNA in Diabetes
4.1. MiRNA in Exosomes and Their Roles in Diabetes
4.2. Other Noncoding RNAs in Exosomes and Their Roles in Diabetes
5. Limitations and Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Regev-Rudzki, N.; Wilson, D.W.; Carvalho, T.G.; Sisquella, X.; Coleman, B.M.; Rug, M.; Bursac, D.; Angrisano, F.; Gee, M.; Hill, A.F.; et al. Cell-Cell Communication between Malaria-Infected Red Blood Cells via Exosome-like Vesicles. Cell 2013, 153, 1120–1133. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Kim, D.; Lim, R. Cell-cell communication in diabetic retinopathy. Vis. Res. 2017, 139, 115–122. [Google Scholar] [CrossRef] [PubMed]
- Ohyashiki, J.H.; Umezu, T.; Ohyashiki, K. Extracellular vesicle-mediated cell-cell communication in haematological neoplasms. Philos. Trans. R. Soc. B Biol. Sci. 2018, 373, 20160484. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, M.; Martin-Jaular, L.; Lavieu, G.; Théry, C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol. 2019, 21, 9–17. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Bermúdez, P.; Blesa, J.; Soriano, J.M.; Marcilla, A. Extracellular vesicles in food: Experimental evidence of their secretion in grape fruits. Eur. J. Pharm. Sci. 2017, 98, 40–50. [Google Scholar] [CrossRef] [PubMed]
- Todorova, D.; Simoncini, S.; Lacroix, R.; Sabatier, F.; Dignat-George, F. Extracellular Vesicles in Angiogenesis. Circ. Res. 2017, 120, 1658–1673. [Google Scholar] [CrossRef] [PubMed]
- Merchant, M.L.; Rood, I.M.; Deegens, J.K.J.; Klein, J.B. Isolation and characterization of urinary extracellular vesicles: Implications for biomarker discovery. Nat. Rev. Nephrol. 2017, 13, 731–749. [Google Scholar] [CrossRef] [PubMed]
- Choi, J.W.; Um, J.H.; Cho, J.H.; Lee, H.J. Tiny RNAs and their voyage via extracellular vesicles: Secretion of bacterial small RNA and eukaryotic microRNA. Exp. Biol. Med. 2017, 242, 1475–1481. [Google Scholar] [CrossRef]
- Parolini, I.; Federici, C.; Raggi, C.; Lugini, L.; Palleschi, S.; De Milito, A.; Coscia, C.; Iessi, E.; Logozzi, M.; Molinari, A.; et al. Microenvironmental pH Is a Key Factor for Exosome Traffic in Tumor Cells. J. Biol. Chem. 2009, 284, 34211–34222. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, M.; Song, W.; Tian, F.; Dai, Z.; Zhu, Q.; Ahmad, E.; Guo, S.; Zhu, C.; Zhong, H.; Yuan, Y.; et al. Temperature- and rigidity-mediated rapid transport of lipid nanovesicles in hydrogels. Proc. Natl. Acad. Sci. USA 2019, 116, 5362–5369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jelonek, K.; Widlak, P.; Pietrowska, M. The Influence of Ionizing Radiation on Exosome Composition, Secretion and Intercellular Communication. Protein Pept. Lett. 2016, 23, 656–663. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kucharzewska, P.; Belting, M. Emerging roles of extracellular vesicles in the adaptive response of tumour cells to microenvironmental stress. J. Extracell. Vesicles 2013, 2, 214074. [Google Scholar] [CrossRef] [PubMed]
- Cianciaruso, C.; Phelps, E.A.; Pasquier, M.; Hamelin, R.; Demurtas, D.; Ahmed, M.A.; Piemonti, L.; Hirosue, S.; Swartz, M.A.; De Palma, M.; et al. Primary Human and Rat beta-Cells Release the Intracellular Autoantigens GAD65, IA-2, and Proinsulin in Exosomes Together With Cytokine-Induced Enhancers of Immunity. Diabetes 2017, 66, 460–473. [Google Scholar] [CrossRef] [PubMed]
- Van der Pol, E.; Böing, A.N.; Harrison, P.; Sturk, A.; Nieuwland, R. Classification, functions, and clinical relevance of extracellular vesicles. Pharmacol. Rev. 2012, 64, 676–705. [Google Scholar] [CrossRef] [PubMed]
- Pegtel, D.M.; Gould, S.J. Exosomes. Annu. Rev. Biochem. 2019, 88, 487–514. [Google Scholar] [CrossRef] [PubMed]
- Stahl, P.D.; Raposo, G. Extracellular Vesicles: Exosomes and Microvesicles, Integrators of Homeostasis. Physiology 2019, 34, 169–177. [Google Scholar] [CrossRef] [PubMed]
- Hauser, P.; Wang, S.; Didenko, V.V. Apoptotic Bodies: Selective Detection in Extracellular Vesicles. Methods Mol. Biol. 2017, 1554, 193–200. [Google Scholar]
- Reiter, K.; Aguilar, P.P.; Wetter, V.; Steppert, P.; Tover, A.; Jungbauer, A. Separation of virus-like particles and extracellular vesicles by flow-through and heparin affinity chromatography. J. Chromatogr. A 2019, 1588, 77–84. [Google Scholar] [CrossRef]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [Green Version]
- Colombo, M.; Raposo, G.; Théry, C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Biol. 2014, 30, 255–289. [Google Scholar] [CrossRef]
- Juan, T.; Furthauer, M. Biogenesis and function of ESCRT-dependent extracellular vesicles. Semin. Cell Dev. Biol. 2018, 74, 66–77. [Google Scholar] [CrossRef] [PubMed]
- Adell, M.A.Y.; Migliano, S.M.; Teis, D. ESCRT-III and Vps4: A dynamic multipurpose tool for membrane budding and scission. FEBS J. 2016, 283, 3288–3302. [Google Scholar] [CrossRef]
- Dreyer, F.; Baur, A.; Federico, M. Biogenesis and Functions of Exosomes and Extracellular Vesicles. Methods Mol. Biol. 2016, 1448, 201–216. [Google Scholar] [PubMed]
- Babst, M. MVB Vesicle Formation: ESCRT-Dependent, ESCRT-Independent and Everything in Between. Curr. Opin. Cell Biol. 2011, 23, 452–457. [Google Scholar] [CrossRef] [PubMed]
- Van Niel, G.; Charrin, S.; Simoes, S.; Romao, M.; Rochin, L.; Saftig, P.; Marks, M.S.; Rubinstein, E.; Raposo, G. The tetraspanin CD63 regulates ESCRT-independent and -dependent endosomal sorting during melanogenesis. Dev. Cell 2011, 21, 708–721. [Google Scholar] [CrossRef]
- Skotland, T.; Sandvig, K.; Llorente, A. Lipids in exosomes: Current knowledge and the way forward. Prog. Lipid Res. 2017, 66, 30–41. [Google Scholar] [CrossRef]
- Skotland, T.; Ekroos, K.; Kauhanen, D.; Simolin, H.; Seierstad, T.; Berge, V.; Sandvig, K.; Llorente, A. Molecular lipid species in urinary exosomes as potential prostate cancer biomarkers. Eur. J. Cancer 2017, 70, 122–132. [Google Scholar] [CrossRef]
- Li, W.; Li, C.; Zhou, T.; Liu, X.; Liu, X.; Li, X.; Chen, D. Role of exosomal proteins in cancer diagnosis. Mol. Cancer 2017, 16, 145. [Google Scholar] [CrossRef] [PubMed]
- Chen, T.; Xi, Q.Y.; Sun, J.J.; Ye, R.S.; Cheng, X.; Sun, R.P.; Wang, S.B.; Shu, G.; Wang, L.N.; Zhu, X.T.; et al. Revelation of mRNAs and proteins in porcine milk exosomes by transcriptomic and proteomic analysis. BMC Vet. Res. 2017, 13, 496. [Google Scholar] [CrossRef]
- Shurtleff, M.J.; Yao, J.; Qin, Y.; Nottingham, R.M.; Temoche-Diaz, M.M.; Schekman, R.; Lambowitz, A.M. Broad role for YBX1 in defining the small noncoding RNA composition of exosomes. Proc. Natl. Acad. Sci. USA 2017, 114, E8987–E8995. [Google Scholar] [CrossRef] [Green Version]
- Mulcahy, L.A.; Pink, R.C.; Carter, D.R.F. Routes and mechanisms of extracellular vesicle uptake. J. Extracell. Vesicles 2014, 3, 1093. [Google Scholar] [CrossRef] [PubMed]
- Kamerkar, S.; LeBleu, V.S.; Sugimoto, H.; Yang, S.; Ruivo, C.F.; Melo, S.A.; Lee, J.J.; Kalluri, R. Exosomes Facilitate Therapeutic Targeting of Oncogenic Kras in Pancreatic Cancer. Nature 2017, 546, 498–503. [Google Scholar] [CrossRef] [PubMed]
- Hoshino, A.; Costa-Silva, B.; Shen, T.L.; Rodrigues, G.; Hashimoto, A.; Mark, M.T.; Molina, H.; Kohsaka, S.; Di Giannatale, A.; Ceder, S.; et al. Tumour exosome integrins determine organotropic metastasis. Nature 2015, 527, 329–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Christianson, H.C.; Svensson, K.J.; Van Kuppevelt, T.H.; Li, J.P.; Belting, M. Cancer cell exosomes depend on cell-surface heparan sulfate proteoglycans for their internalization and functional activity. Proc. Natl. Acad. Sci. USA 2013, 110, 17380–17385. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, R.; Ding, Q.; Yaqoob, U.; De Assuncao, T.M.; Verma, V.K.; Hirsova, P.; Cao, S.; Mukhopadhyay, D.; Huebert, R.C.; Shah, V.H. Exosome Adherence and Internalization by Hepatic Stellate Cells Triggers Sphingosine 1-Phosphate-dependent Migration. J. Biol. Chem. 2015, 290, 30684–30696. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochieng, J.; Nangami, G.; Sakwe, A.; Rana, T.; Ingram, S.; Goodwin, J.S.; Moye, C.; Lammers, P.; Adunyah, S.E.; Goodwin, J.S. Extracellular histones are the ligands for the uptake of exosomes and hydroxyapatite-nanoparticles by tumor cells via syndecan-4. FEBS Lett. 2018, 592, 3274–3285. [Google Scholar] [CrossRef] [PubMed]
- Kulkarni, B.; Kirave, P.; Gondaliya, P.; Jash, K.; Jain, A.; Tekade, R.K.; Kalia, K. Exosomal miRNA in chemoresistance, immune evasion, metastasis and progression of cancer. Drug Discov. Today 2019. [Google Scholar] [CrossRef]
- Stefanius, K.; Servage, K.; Santos, M.D.S.; Gray, H.F.; Toombs, J.; Chimalapati, S.; Kim, M.S.; Malladi, V.S.; Brekken, R.; Orth, K. Human pancreatic cancer cell exosomes, but not human normal cell exosomes, act as an initiator in cell transformation. Elife 2019, 8, 40226. [Google Scholar] [CrossRef]
- Shi, Y.; Hu, F.B. The global implications of diabetes and cancer. Lancet 2014, 383, 1947–1948. [Google Scholar] [CrossRef]
- Cho, N.; Shaw, J.; Karuranga, S.; Huang, Y.; Fernandes, J.D.R.; Ohlrogge, A.; Malanda, B. IDF Diabetes Atlas: Global estimates of diabetes prevalence for 2017 and projections for 2045. Diabetes Res. Clin. Prat. 2018, 138, 271–281. [Google Scholar] [CrossRef]
- Naidoo, V.; Naidoo, M.; Ghai, M. Cell and tissue-specific epigenetic changes associated with chronic inflammation in insulin resistance and type 2 diabetes mellitus. Scand. J. Immunol. 2018, 88, e12723. [Google Scholar] [CrossRef] [PubMed]
- Yahagi, K.; Kolodgie, F.D.; Lutter, C.; Mori, H.; Romero, M.E.; Finn, A.V.; Virmani, R. Pathology of Human Coronary and Carotid Artery Atherosclerosis and Vascular Calcification in Diabetes Mellitus. Arterioscler. Thromb. Vasc. Biol. 2017, 37, 191–204. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ashcroft, F.M.; Rorsman, P. Diabetes mellitus and the beta cell: The last ten years. Cell 2012, 148, 1160–1171. [Google Scholar] [CrossRef] [PubMed]
- Buzzetti, R.; Zampetti, S.; Maddaloni, E. Adult-onset autoimmune diabetes: Current knowledge and implications for management. Nat. Rev. Endocrinol. 2017, 13, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Kakleas, K.; Soldatou, A.; Karachaliou, F.; Karavanaki, K.; Kostas, K.; Alexandra, S.; Feneli, K.; Kyriaki, K. Associated autoimmune diseases in children and adolescents with type 1 diabetes mellitus (T1DM). Autoimmun. Rev. 2015, 14, 781–797. [Google Scholar] [CrossRef]
- Bonifacio, E.; Mathieu, C.; Nepom, G.T.; Ziegler, A.G.; Anhalt, H.; Haller, M.J.; Harrison, L.C.; Hebrok, M.; Kushner, J.A.; Norris, J.M.; et al. Rebranding asymptomatic type 1 diabetes: The case for autoimmune beta cell disorder as a pathological and diagnostic entity. Diabetologia 2017, 60, 35–38. [Google Scholar] [CrossRef]
- Kahaly, G.J.; Hansen, M.P. Type 1 diabetes associated autoimmunity. Autoimmun. Rev. 2016, 15, 644–648. [Google Scholar] [CrossRef]
- Lukić, M.L.; Pejnovic, N.; Lukić, A. New Insight into Early Events in Type 1 Diabetes: Role for Islet Stem Cell Exosomes. Diabetes 2014, 63, 835–837. [Google Scholar] [CrossRef]
- Guay, C.; Kruit, J.K.; Rome, S.; Menoud, V.; Mulder, N.L.; Jurdzinski, A.; Mancarella, F.; Sebastiani, G.; Donda, A.; Gonzalez, B.J.; et al. Lymphocyte-Derived Exosomal MicroRNAs Promote Pancreatic beta Cell Death and May Contribute to Type 1 Diabetes Development. Cell Metab. 2019, 29, 348–361. [Google Scholar] [CrossRef]
- Dai, Y.D.; Sheng, H.; Dias, P.; Rahman, M.J.; Bashratyan, R.; Regn, D.; Marquardt, K. Autoimmune Responses to Exosomes and Candidate Antigens Contribute to Type 1 Diabetes in Non-Obese Diabetic Mice. Curr. Diabetes Rep. 2017, 17, 130. [Google Scholar] [CrossRef]
- Sheng, H.; Hassanali, S.; Nugent, C.; Wen, L.; Hamilton-Williams, E.; Dias, P.; Dai, Y.D. Insulinoma-released exosomes or microparticles are immunostimulatory and can activate autoreactive T cells spontaneously developed in non-obese diabetes mice1. J. Immunol. 2011, 187, 1591–1600. [Google Scholar] [CrossRef] [PubMed]
- Klein, L.; Hinterberger, M.; Von Rohrscheidt, J.; Aichinger, M. Autonomous versus dendritic cell-dependent contributions of medullary thymic epithelial cells to central tolerance. Trends Immunol. 2011, 32, 188–193. [Google Scholar] [CrossRef]
- Malhotra, D.; Fletcher, A.L.; Astarita, J.; Lukacs-Kornek, V.; Tayalia, P.; Gonzalez, S.F.; Elpek, K.G.; Chang, S.K.; Knoblich, K.; Hemler, M.E.; et al. Transcriptional profiling of stroma from inflamed and resting lymph nodes defines immunological hallmarks. Nat. Immunol. 2012, 13, 499–510. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nojehdehi, S.; Soudi, S.; Hesampour, A.; Rasouli, S.; Soleimani, M.; Hashemi, S.M. Immunomodulatory effects of mesenchymal stem cell-derived exosomes on experimental type-1 autoimmune diabetes. J. Cell. Biochem. 2018, 119, 9433–9443. [Google Scholar] [CrossRef]
- Jin, J.; Shi, Y.; Gong, J.; Zhao, L.; Li, Y.; He, Q.; Huang, H. Exosome secreted from adipose-derived stem cells attenuates diabetic nephropathy by promoting autophagy flux and inhibiting apoptosis in podocyte. Stem Cell Res. Ther. 2019, 10, 95. [Google Scholar] [CrossRef]
- Zhu, L.L.; Huang, X.; Yu, W.; Chen, H.; Chen, Y.; Dai, Y.T. Transplantation of adipose tissue-derived stem cell-derived exosomes ameliorates erectile function in diabetic rats. Andrologia 2018, 50, e12871. [Google Scholar] [CrossRef] [PubMed]
- Wen, D.; Peng, Y.; Liu, D.; Weizmann, Y.; Mahato, R.I. Mesenchymal stem cell and derived exosome as small RNA carrier and Immunomodulator to improve islet transplantation. J. Control. Release 2016, 238, 166–175. [Google Scholar] [CrossRef] [PubMed]
- Korutla, L.; Rickels, M.R.; Hu, R.W.; Freas, A.; Reddy, S.; Habertheuer, A.; Harmon, J.; Korutla, V.; Ram, C.; Naji, A.; et al. Noninvasive diagnosis of recurrent autoimmune type 1 diabetes after islet cell transplantation. Am. J. Transplant. 2019, 19, 1852–1858. [Google Scholar] [CrossRef] [PubMed]
- Shah, M.; Vella, A. What is type 2 diabetes? Medicine 2014, 42, 687–691. [Google Scholar] [CrossRef]
- Czech, M.P. Insulin action and resistance in obesity and type 2 diabetes. Nat. Med. 2017, 23, 804–814. [Google Scholar] [CrossRef]
- Tsai, S.; Clemente-Casares, X.; Revelo, X.S.; Winer, S.; Winer, D.A. Are Obesity-Related Insulin Resistance and Type 2 Diabetes Autoimmune Diseases? Diabetes 2015, 64, 1886–1897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar] [CrossRef] [PubMed]
- Song, M.; Han, L.; Chen, F.F.; Wang, D.; Wang, F.; Zhang, L.; Wang, Z.H.; Zhong, M.; Tang, M.X.; Zhang, W. Adipocyte-Derived Exosomes Carrying Sonic Hedgehog Mediate M1 Macrophage Polarization-Induced Insulin Resistance via Ptch and PI3K Pathways. Cell. Physiol. Biochem. 2018, 48, 1416–1432. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Chen, F.F.; Shang, Y.Y.; Li, Y.; Wang, Z.H.; Han, L.; Li, Y.H.; Zhang, L.; Ti, Y.; Zhang, W.; et al. Insulin resistance adipocyte-derived exosomes aggravate atherosclerosis by increasing vasa vasorum angiogenesis in diabetic ApoE mice. Int. J. Cardiol. 2018, 265, 181–187. [Google Scholar] [CrossRef] [PubMed]
- Su, T.; Xiao, Y.; Xiao, Y.; Guo, Q.; Li, C.; Huang, Y.; Deng, Q.; Wen, J.; Zhou, F.; Luo, X.H. Bone Marrow Mesenchymal Stem Cells-Derived Exosomal MiR-29b-3p Regulates Aging-Associated Insulin Resistance. ACS Nano 2019, 13, 2450–2462. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Jayabalan, N.; Guanzon, D.; Palma, C.; Scholz-Romero, K.; Elfeky, O.; Zuñiga, F.; Ormazabal, V.; Diaz, E.; Rice, G.E.; et al. Human placental exosomes in gestational diabetes mellitus carry a specific set of miRNAs associated with skeletal muscle insulin sensitivity. Clin. Sci. 2018, 132, 2451–2467. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Gu, H.; Huang, W.; Peng, J.; Li, Y.; Yang, L.; Qin, D.; Essandoh, K.; Wang, Y.; Peng, T.; et al. Hsp20-Mediated Activation of Exosome Biogenesis in Cardiomyocytes Improves Cardiac Function and Angiogenesis in Diabetic Mice. Diabetes 2016, 65, 3111–3128. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, S.; Luo, F.; Feng, Y.M.; Wei, X.; Miao, H.; Lu, Y.B.; Tang, Y.; Ding, D.F.; Jin, J.F.; Zhu, Q. Neutral Ceramidase Secreted Via Exosome Protects Against Palmitate-Induced Apoptosis in INS-1 Cells. Exp. Clin. Endocrinol. Diabetes 2017, 125, 130–135. [Google Scholar] [CrossRef]
- Sun, Y.; Shi, H.; Yin, S.; Ji, C.; Zhang, X.; Zhang, B.; Wu, P.; Shi, Y.; Mao, F.; Yan, Y.; et al. Human Mesenchymal Stem Cell Derived Exosomes Alleviate Type 2 Diabetes Mellitus by Reversing Peripheral Insulin Resistance and Relieving beta-Cell Destruction. ACS Nano 2018, 12, 7613–7628. [Google Scholar] [CrossRef]
- Nagaishi, K.; Mizue, Y.; Chikenji, T.; Otani, M.; Nakano, M.; Saijo, Y.; Tsuchida, H.; Ishioka, S.; Nishikawa, A.; Saito, T.; et al. Umbilical cord extracts improve diabetic abnormalities in bone marrow-derived mesenchymal stem cells and increase their therapeutic effects on diabetic nephropathy. Sci. Rep. 2017, 7, 8484. [Google Scholar] [CrossRef]
- Li, G.; Liu, H.; Ma, C.; Chen, Y.; Wang, J.; Yang, Y. Exosomes are the novel players involved in the beneficial effects of exercise on type 2 diabetes. J. Cell. Physiol. 2019, 234, 14896–14905. [Google Scholar] [CrossRef] [PubMed]
- Safdar, A.; Saleem, A.; Tarnopolsky, M.A. The potential of endurance exercise-derived exosomes to treat metabolic diseases. Nat. Rev. Endocrinol. 2016, 12, 504–517. [Google Scholar] [CrossRef] [PubMed]
- Witczak, J.K.; Min, T.; Prior, S.L.; Stephens, J.W.; James, P.; Rees, A. Bariatric Surgery Is Accompanied by Changes in Extracellular Vesicle-Associated and Plasma Fatty Acid Binding Protein 4. Obes. Surg. 2018, 28, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Chang, W. Non-coding RNAs and Berberine: A new mechanism of its anti-diabetic activities. Eur. J. Pharmacol. 2017, 795, 8–12. [Google Scholar] [CrossRef] [PubMed]
- Wong, W.K.; Sørensen, A.E.; Joglekar, M.V.; Hardikar, A.A.; Dalgaard, L.T. Non-Coding RNA in Pancreas and beta-Cell Development. Noncoding RNA 2018, 4, E41. [Google Scholar] [PubMed]
- Vishnoi, A.; Rani, S. MiRNA Biogenesis and Regulation of Diseases: An Overview. Methods Mol. Biol. 2017, 1509, 1–10. [Google Scholar]
- Brennan, E.; Wang, B.; McClelland, A.; Mohan, M.; Marai, M.; Beuscart, O.; Derouiche, S.; Gray, S.; Pickering, R.; Tikellis, C.; et al. Protective Effect of let-7 miRNA Family in Regulating Inflammation in Diabetes-Associated Atherosclerosis. Diabetes 2017, 66, 2266–2277. [Google Scholar] [CrossRef] [Green Version]
- Jiang, L.Q.; Franck, N.; Egan, B.; Sjögren, R.J.O.; Katayama, M.; Duque-Guimaraes, D.; Arner, P.; Zierath, J.R.; Krook, A. Autocrine role of interleukin-13 on skeletal muscle glucose metabolism in type 2 diabetic patients involves microRNA let-7. Am. J. Physiol. Metab. 2013, 305, E1359–E1366. [Google Scholar] [CrossRef] [Green Version]
- Vickers, K.C.; Landstreet, S.R.; Levin, M.G.; Shoucri, B.M.; Toth, C.L.; Taylor, R.C.; Palmisano, B.T.; Tabet, F.; Cui, H.L.; Rye, K.A.; et al. MicroRNA-223 coordinates cholesterol homeostasis. Proc. Natl. Acad. Sci. USA 2014, 111, 14518–14523. [Google Scholar] [CrossRef] [Green Version]
- Massart, J.; Sjögren, R.J.; Lundell, L.S.; Mudry, J.M.; Franck, N.; O’Gorman, D.J.; Egan, B.; Zierath, J.R.; Krook, A. Altered miR-29 Expression in Type 2 Diabetes Influences Glucose and Lipid Metabolism in Skeletal Muscle. Diabetes 2017, 66, 1807–1818. [Google Scholar] [CrossRef] [Green Version]
- Foley, N.H.; O’Neill, L.A. miR-107: A Toll-like receptor-regulated miRNA dysregulated in obesity and type II diabetes. J. Leukoc. Biol. 2012, 92, 521–527. [Google Scholar] [CrossRef] [PubMed]
- Nicholson, K.M.; Anderson, N.G. The protein kinase B/Akt signalling pathway in human malignancy. Cell. Signal. 2002, 14, 381–395. [Google Scholar] [CrossRef]
- Katayama, M.; Wiklander, O.P.; Fritz, T.; Caidahl, K.; Andaloussi, S.E.; Zierath, J.R.; Krook, A. Circulating Exosomal miR-20b-5p is Elevated in Type 2 Diabetes and Could Impair Insulin Action in Human Skeletal Muscle. Diabetes 2018, 68, 515–526. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Zhang, B.; Zheng, W.; Kang, M.; Chen, Q.; Qin, W.; Li, C.; Zhang, Y.; Shao, Y.; Wu, Y. Exosomes derived from pancreatic cancer cells induce insulin resistance in C2C12 myotube cells through the PI3K/Akt/FoxO1 pathway. Sci. Rep. 2017, 7, 5384. [Google Scholar] [CrossRef] [PubMed]
- Ying, W.; Riopel, M.; Bandyopadhyay, G.; Dong, Y.; Birmingham, A.; Seo, J.B.; Ofrecio, J.M.; Wollam, J.; Hernandez-Carretero, A.; Fu, W.; et al. Adipose Tissue Macrophage-Derived Exosomal miRNAs Can Modulate In Vivo and In Vitro Insulin Sensitivity. Cell 2017, 171, 372–384. [Google Scholar] [CrossRef] [PubMed]
- Castaño, C.; Kalko, S.; Novials, A.; Párrizas, M. Obesity-associated exosomal miRNAs modulate glucose and lipid metabolism in mice. Proc. Natl. Acad. Sci. USA 2018, 115, 12158–12163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Safwat, A.; Sabry, D.; Ragiae, A.; Amer, E.; Mahmoud, R.; Shamardan, R. Adipose mesenchymal stem cells–derived exosomes attenuate retina degeneration of streptozotocin-induced diabetes in rabbits. J. Circ. Biomark. 2018, 7, 1849454418807827. [Google Scholar] [CrossRef] [PubMed]
- Tsukita, S.; Yamada, T.; Takahashi, K.; Munakata, Y.; Hosaka, S.; Takahashi, H.; Gao, J.; Shirai, Y.; Kodama, S.; Asai, Y.; et al. MicroRNAs 106b and 222 Improve Hyperglycemia in a Mouse Model of Insulin-Deficient Diabetes via Pancreatic beta-Cell Proliferation. EBioMedicine 2017, 15, 163–172. [Google Scholar] [CrossRef]
- Kubota, K.; Nakano, M.; Kobayashi, E.; Mizue, Y.; Chikenji, T.; Otani, M.; Nagaishi, K.; Fujimiya, M. An enriched environment prevents diabetes-induced cognitive impairment in rats by enhancing exosomal miR-146a secretion from endogenous bone marrow-derived mesenchymal stem cells. PLoS ONE 2018, 13, e0204252. [Google Scholar] [CrossRef]
- Eissa, S.; Matboli, M.; Bekhet, M.M. Clinical verification of a novel urinary microRNA panal: 133b, 342 and 30 as biomarkers for diabetic nephropathy identified by bioinformatics analysis. Biomed. Pharmacother. 2016, 83, 92–99. [Google Scholar] [CrossRef]
- Fu, Q.; Jiang, H.; Yang, T. Injury Factors Alter miRNAs Profiles of Exosomes Derived from Islets and Circulation. Diabetes 2018, 67, 3986–3999. [Google Scholar] [CrossRef]
- Lakhter, A.J.; Pratt, R.E.; Moore, R.E.; Doucette, K.K.; Maier, B.F.; DiMeglio, L.A.; Sims, E.K. Beta cell extracellular vesicle miR-21-5p cargo is increased in response to inflammatory cytokines and serves as a biomarker of type 1 diabetes. Diabetologia 2018, 61, 1124–1134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, W.; Yang, S.; Qiao, R.; Zhang, J. Potential Value of Urinary Exosome-Derived let-7c-5p in the Diagnosis and Progression of Type II Diabetic Nephropathy. Clin. Lab. 2018, 64, 709–718. [Google Scholar] [CrossRef] [PubMed]
- Mohan, A.; Singh, R.S.; Kumari, M.; Garg, D.; Upadhyay, A.; Ecelbarger, C.M.; Tripathy, S.; Tiwari, S. Urinary Exosomal microRNA-451-5p Is a Potential Early Biomarker of Diabetic Nephropathy in Rats. PLoS ONE 2016, 11, e0154055. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.; Jia, Y.; Cuihua, X.; Hu, F.; Xue, M.; Xue, Y. Urinary Exosomal MicroRNA Profiling in Incipient Type 2 Diabetic Kidney Disease. J. Diabetes Res. 2017, 2017, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Fang, Y.; Wang, X.; Li, W.; Han, J.; Jin, J.; Su, F.; Zhang, J.; Huang, W.; Xiao, F.; Pan, Q.; et al. Screening of circular RNAs and validation of circANKRD36 associated with inflammation in patients with type 2 diabetes mellitus. Int. J. Mol. Med. 2018, 42, 1865–1874. [Google Scholar] [CrossRef] [PubMed]
- Stoll, L.; Sobel, J.; Rodriguez-Trejo, A.; Guay, C.; Lee, K.; Venø, M.T.; Kjems, J.; Laybutt, D.R.; Regazzi, R. Circular RNAs as novel regulators of beta-cell functions in normal and disease conditions. Mol. Metab. 2018, 9, 69–83. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Guo, S.; Li, W.; Yu, P. The circular RNA Cdr1as, via miR-7 and its targets, regulates insulin transcription and secretion in islet cells. Sci. Rep. 2015, 5, 12453. [Google Scholar] [CrossRef] [PubMed]
- Henaoui, I.S.; Jacovetti, C.; Mollet, I.G.; Guay, C.; Sobel, J.; Eliasson, L.; Regazzi, R. PIWI-interacting RNAs as novel regulators of pancreatic beta cell function. Diabetologia 2017, 60, 1977–1986. [Google Scholar] [CrossRef] [PubMed]
- Kong, Y.; Hsieh, C.H.; Alonso, L.C. ANRIL: A lncRNA at the CDKN2A/B Locus with Roles in Cancer and Metabolic Disease. Front. Endocrinol. 2018, 9, 405. [Google Scholar] [CrossRef]
- Gao, Y.; Wu, F.; Zhou, J.; Yan, L.; Jurczak, M.J.; Lee, H.Y.; Yang, L.; Mueller, M.; Zhou, X.B.; Dandolo, L.; et al. The H19/let-7 double-negative feedback loop contributes to glucose metabolism in muscle cells. Nucleic Acids Res. 2014, 42, 13799–13811. [Google Scholar] [CrossRef] [PubMed]
- Hu, M.; Wang, R.; Li, X.; Fan, M.; Lin, J.; Zhen, J.; Chen, L.; Lv, Z. LncRNA MALAT1 is dysregulated in diabetic nephropathy and involved in high glucose-induced podocyte injury via its interplay with beta-catenin. J. Cell. Mol. Med. 2017, 21, 2732–2747. [Google Scholar] [CrossRef] [PubMed]
- Li, C.P.; Wang, S.H.; Wang, W.Q.; Song, S.G.; Liu, X.M. Long Noncoding RNA-Sox2OT Knockdown Alleviates Diabetes Mellitus-Induced Retinal Ganglion Cell (RGC) injury. Cell. Mol. Neurobiol. 2017, 37, 361–369. [Google Scholar] [CrossRef] [PubMed]
- Kameswaran, V.; Bramswig, N.C.; McKenna, L.B.; Penn, M.; Schug, J.; Hand, N.J.; Chen, Y.; Choi, I.; Vourekas, A.; Won, K.J.; et al. Epigenetic regulation of the DLK1-MEG3 microRNA cluster in human type 2 diabetic islets. Cell Metab. 2014, 19, 135–145. [Google Scholar] [CrossRef] [PubMed]
- Ruan, Y.; Lin, N.; Ma, Q.; Chen, R.; Zhang, Z.; Wen, W.; Chen, H.; Sun, J. Circulating LncRNAs Analysis in Patients with Type 2 Diabetes Reveals Novel Genes Influencing Glucose Metabolism and Islet beta-Cell Function. Cell. Physiol. Biochem. 2018, 46, 335–350. [Google Scholar] [CrossRef] [PubMed]
- Robbins, P.D.; Morelli, A.E. Regulation of immune responses by extracellular vesicles. Nat. Rev. Immunol. 2014, 14, 195–208. [Google Scholar] [CrossRef] [Green Version]
- Tao, S.C.; Rui, B.Y.; Wang, Q.Y.; Zhou, D.; Zhang, Y.; Guo, S.C. Extracellular vesicle-mimetic nanovesicles transport LncRNA-H19 as competing endogenous RNA for the treatment of diabetic wounds. Drug Deliv. 2018, 25, 241–255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yamashita, T.; Takahashi, Y.; Nishikawa, M.; Takakura, Y. Effect of exosome isolation methods on physicochemical properties of exosomes and clearance of exosomes from the blood circulation. Eur. J. Pharm. Biopharm. 2016, 98, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, P.; Kaslan, M.; Lee, S.H.; Yao, J.; Gao, Z. Progress in Exosome Isolation Techniques. Theranostics 2017, 7, 789–804. [Google Scholar] [CrossRef]
- Fitts, C.A.; Ji, N.; Li, Y.; Tan, C. Exploiting Exosomes in Cancer Liquid Biopsies and Drug Delivery. Adv. Healthc. Mater. 2019, 8, e1801268. [Google Scholar] [CrossRef]
- Kim, M.S.; Haney, M.J.; Zhao, Y.; Mahajan, V.; Deygen, I.; Klyachko, N.L.; Inskoe, E.; Piroyan, A.; Sokolsky, M.; Okolie, O.; et al. Development of exosome-encapsulated paclitaxel to overcome MDR in cancer cells. Nanomedicine 2016, 12, 655–664. [Google Scholar] [CrossRef] [PubMed]
- Wiklander, O.P.B.; Nordin, J.Z.; O’Loughlin, A.; Gustafsson, Y.; Corso, G.; Mäger, I.; Vader, P.; Lee, Y.; Sork, H.; Seow, Y.; et al. Extracellular vesicle in vivo biodistribution is determined by cell source, route of administration and targeting. J. Extracell. Vesicles 2015, 4, 26316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tamura, R.; Uemoto, S.; Tabata, Y. Augmented liver targeting of exosomes by surface modification with cationized pullulan. Acta Biomater. 2017, 57, 274–284. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fang, X.; Duan, Y.; Adkins, G.B.; Pan, S.; Wang, H.; Liu, Y.; Zhong, W. Highly Efficient Exosome Isolation and Protein Analysis by an Integrated Nanomaterial-Based Platform. Anal. Chem. 2018, 90, 2787–2795. [Google Scholar] [CrossRef] [PubMed]


| Exosome MiRNA Name | Main Indication for Diabetes | Expression in Diabetic Statue | Exosome Extracted From | Effected Cells | Experimental Model | Reference |
|---|---|---|---|---|---|---|
| MiR-20b-5p | Pathological factor | upregulate | Serum | Primary human skeletal muscle cells | T2D patients | Katayama, M et al. 2018. [83] |
| MiR-122,miR-192,miR-27a-3p, miR-27b-3p | Pathological factor | upregulate | Serum | White adipose tissue | Diet-induced obesity mice | Castano, C et al. 2018 [86] |
| MiR-142-3p, miR-142-5p, miR-155 | Pathological factor | upregulate | Lymphocyte | Pancreatic β cells | Non-obese diabetic mice | Guay, C et al. 2018 [49] |
| MiR-155 | Pathological factor | upregulate | Adipose tissue macrophage | Liver, adipose tissue, and muscle | Obese mice | Ying, W et al. 2017 [85] |
| MiR-29b-3p | Pathological factor | upregulate | Bone marrow mesenchymal stem cells | Adipocytes, C2C12 myocytes, and primary cultured hepatocytes | Aging-related insulin resistance mice | Sun, T et al. 2019 [65] |
| MiR-486 | Therapy agent | upregulate | Adipose derived stem cells | Podocyte cells | Spontaneous diabetic mice | Jin, J et al. 2019 [55] |
| MiR-222 | Therapy agent | downregulate | Adipose tissue mesenchymal stem cells | Retinal tissue | T1D rabbit | Safwat, A et al. 2018 [87] |
| MiR-146a | Therapy agent | downregulate | Bone narrow stem /stromal cell | Astrocytes | T1D mice | Kubota, K et al. 2018 [89] |
| MiR-106b, miR-222 | Therapy agent | downregulate | Bone narrow stem cell | Pancreatic β cells | T1D mice | Tsukita, S et al. 2017 [88] |
| MiR-21-5p | Biomarker | upregulate | Serum | NR | T1D patients | Lakhter, A et al. 2018 [92] |
| MiR-375-3p | Biomarker | upregulate | Serum | NR | New-onset T1D and T2D patients | Fu, Q et al. 2018. [91] |
| MiR-133b, miR-342, miR-30 | Biomarker | upregulate | Serum | NR | T2D patients | Eissa, S et al. 2017 [90] |
| MiR-451-5p | Biomarker | upregulate | Urinary | NR | T1D rats | Mohan, A et al. 2016 [94] |
| Let-7c-5p | Biomarker | upregulate | Urinary | NR | T2D patients with kidney disease | Li, W et al. 2018 [93] |
| MiR-362-3p, miR-877-3p, miR-150-5p, miR-15a-5p | Biomarker | upregulate | Urinary | NR | T2D patients with kidney disease | Xie, Y et al. 2017 [95] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chang, W.; Wang, J. Exosomes and Their Noncoding RNA Cargo Are Emerging as New Modulators for Diabetes Mellitus. Cells 2019, 8, 853. https://doi.org/10.3390/cells8080853
Chang W, Wang J. Exosomes and Their Noncoding RNA Cargo Are Emerging as New Modulators for Diabetes Mellitus. Cells. 2019; 8(8):853. https://doi.org/10.3390/cells8080853
Chicago/Turabian StyleChang, Wenguang, and Jianxun Wang. 2019. "Exosomes and Their Noncoding RNA Cargo Are Emerging as New Modulators for Diabetes Mellitus" Cells 8, no. 8: 853. https://doi.org/10.3390/cells8080853
APA StyleChang, W., & Wang, J. (2019). Exosomes and Their Noncoding RNA Cargo Are Emerging as New Modulators for Diabetes Mellitus. Cells, 8(8), 853. https://doi.org/10.3390/cells8080853





