Characterization of Cell Glycocalyx with Mass Spectrometry Methods
Abstract
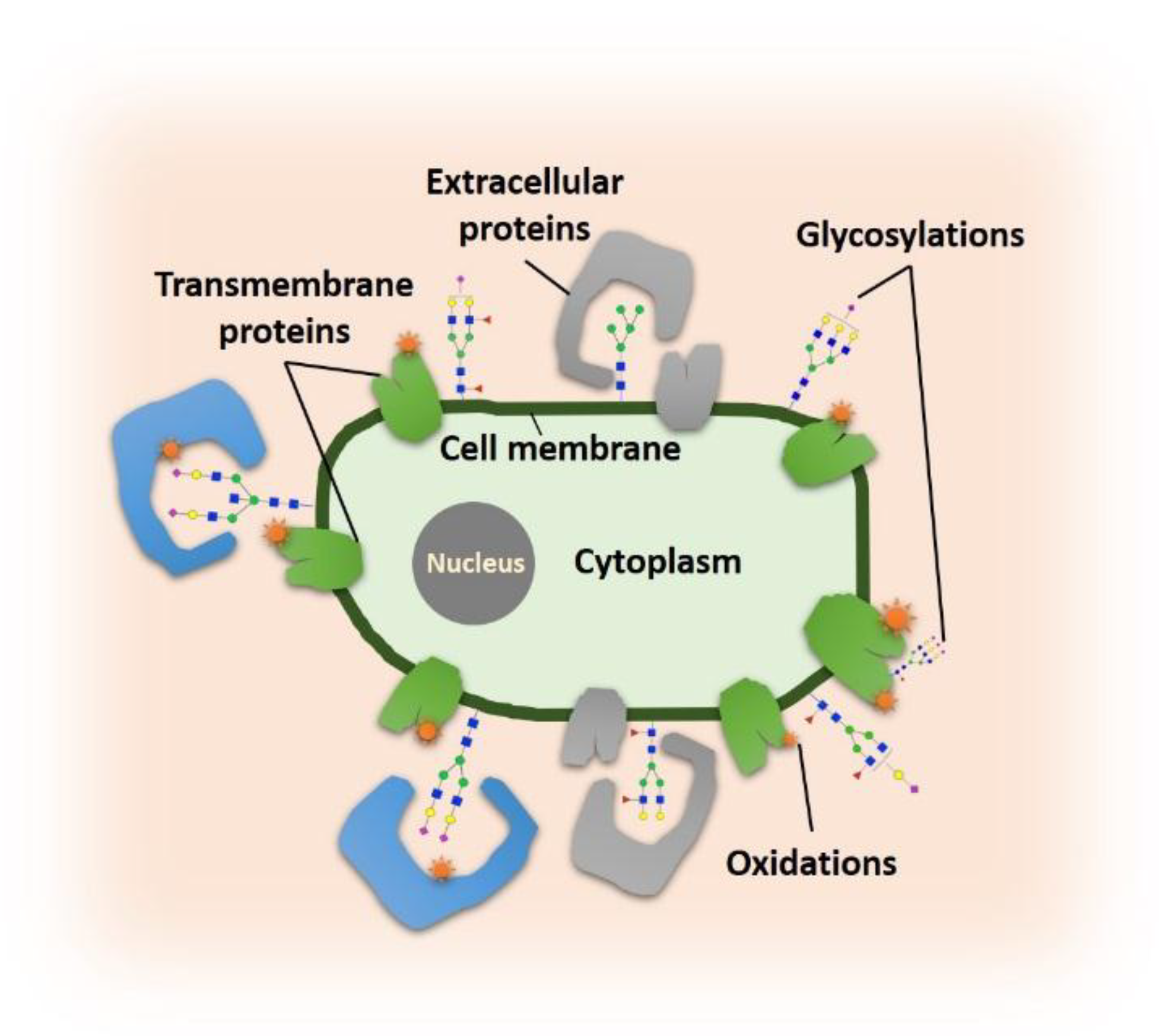
1. Introduction
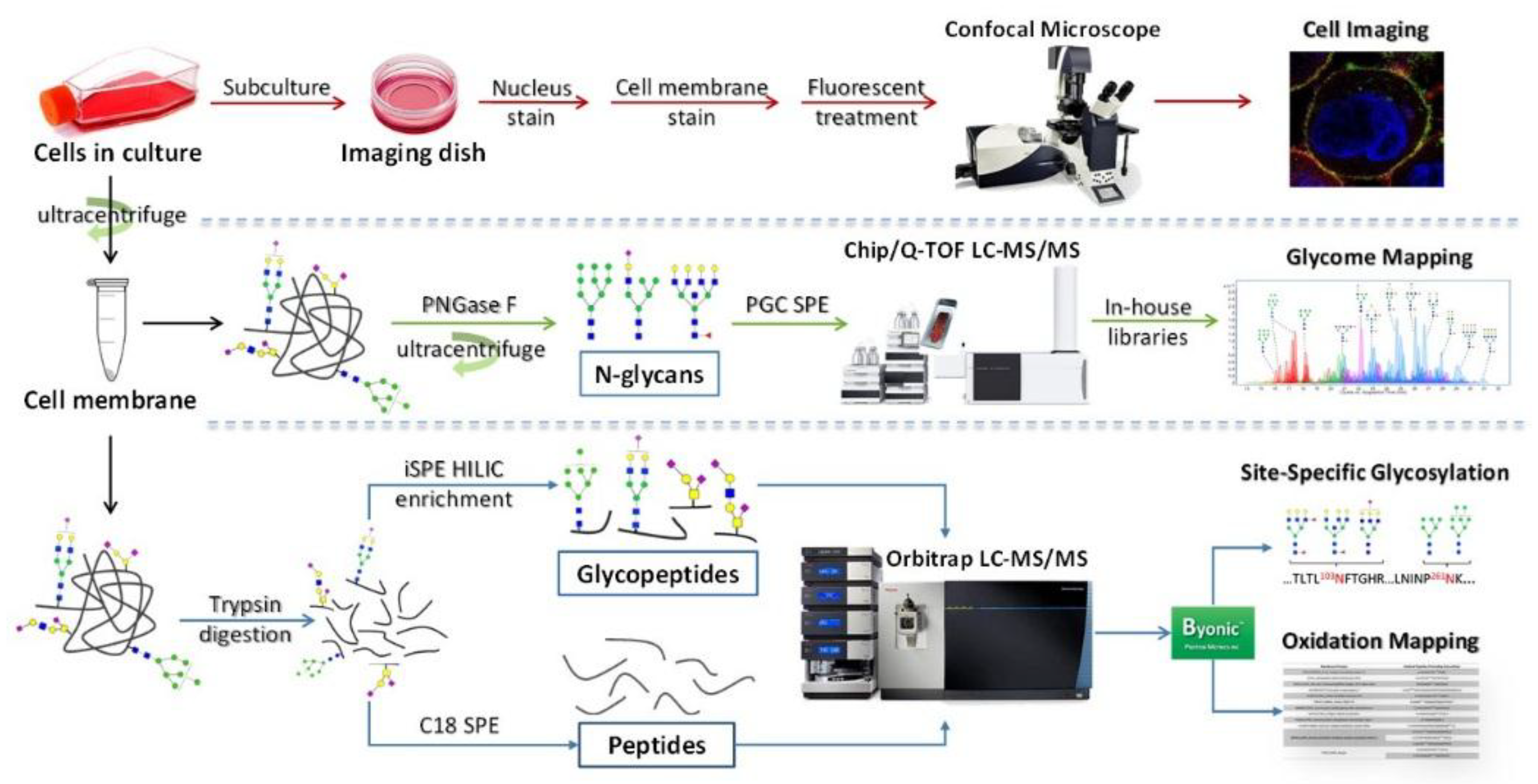
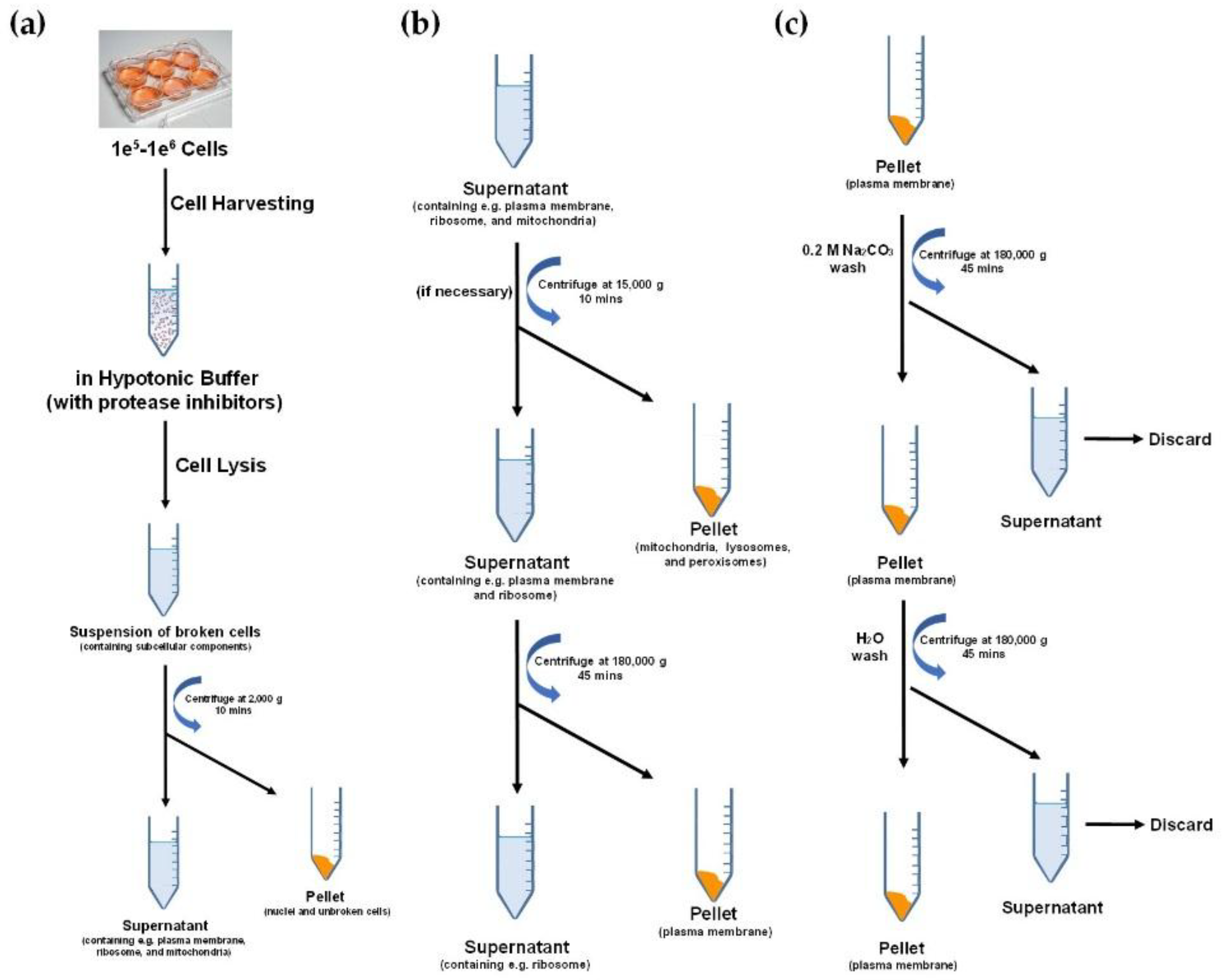
2. Cell Membrane Extraction Methods
3. Glycomic Analysis of Cell Membrane
3.1. Methods for Cell Membrane Glycan Analysis
3.2. Application of Glycomic Analysis with LC-MS/MS
4. Glycoproteomic Analysis of Cell Membrane
5. Glycosphingolipids Analysis of Cell Membrane
6. Glycosylation Studies with Isotopic Labeling
6.1. Metabolic Pathways of Glycans
6.2. Glycan Quantitation Using Isotope Labeling
6.3. Glycopeptide Quantitation Using Isotope-Labeling
7. Conclusions and Future Directions
Funding
Conflicts of Interest
References
- Hadley, B.; Maggioni, A.; Ashikov, A.; Day, C.J.; Haselhorst, T.; Tiralongo, J. Structure and function of nucleotide sugar transporters: Current progress. Comput. Struct. Biotechnol. J. 2014, 10, 23–32. [Google Scholar] [CrossRef] [PubMed]
- Lübke, T.; Marquardt, T.; von Figura, K.; Körner, C. A new type of carbohydrate-deficient glycoprotein syndrome due to a decreased import of GDP-fucose into the Golgi. J. Biol. Chem. 1999, 274, 25986–25989. [Google Scholar] [CrossRef] [PubMed]
- Lübke, T.; Marquardt, T.; Etzioni, A.; Hartmann, E.; von Figura, K.; Körner, C. Complementation cloning identifies CDG-IIc, a new type of congenital disorders of glycosylation, as a GDP-fucose transporter deficiency. Nat. Genet. 2001, 28, 73. [Google Scholar] [CrossRef] [PubMed]
- Lühn, K.; Wild, M.K.; Eckhardt, M.; Gerardy-Schahn, R.; Vestweber, D. The gene defective in leukocyte adhesion deficiency II encodes a putative GDP-fucose transporter. Nat. Genet. 2001, 28, 69. [Google Scholar] [CrossRef] [PubMed]
- Martinez-Duncker, I.; Dupré, T.; Piller, V.; Piller, F.; Candelier, J.-J.; Trichet, C.; Tchernia, G.; Oriol, R.; Mollicone, R. Genetic complementation reveals a novel human congenital disorder of glycosylation of type II, due to inactivation of the Golgi CMP–sialic acid transporter. Blood 2005, 105, 2671–2676. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.; Itoh, S.; Wang, X.; Isaji, T.; Miyoshi, E.; Kariya, Y.; Miyazaki, K.; Kawasaki, N.; Taniguchi, N.; Gu, J. Deletion of core fucosylation on α3β1 integrin down-regulates its functions. J. Biol. Chem. 2006, 281, 38343–38350. [Google Scholar] [CrossRef] [PubMed]
- Meesmann, H.M.; Fehr, E.-M.; Kierschke, S.; Herrmann, M.; Bilyy, R.; Heyder, P.; Blank, N.; Krienke, S.; Lorenz, H.-M.; Schiller, M. Decrease of sialic acid residues as an eat-me signal on the surface of apoptotic lymphocytes. J Cell Sci 2010, 123, 3347–3356. [Google Scholar] [CrossRef] [PubMed]
- Almaraz, R.T.; Tian, Y.; Bhattarcharya, R.; Tan, E.; Chen, S.-H.; Dallas, M.R.; Chen, L.; Zhang, Z.; Zhang, H.; Konstantopoulos, K.; et al. Metabolic Flux Increases Glycoprotein Sialylation: Implications for Cell Adhesion and Cancer Metastasis. Mol. Amp; Cell. Proteom. 2012, 11, M112.017558. [Google Scholar] [CrossRef] [PubMed]
- Hayashi, H.; Yamashita, Y. Role of N-glycosylation in cell surface expression and protection against proteolysis of the intestinal anion exchanger SLC26A3. Am. J. Physiol. Cell Physiol. 2011, 302, C781–C795. [Google Scholar] [CrossRef]
- Kundra, R.; Kornfeld, S. Asparagine-linked oligosaccharides protect Lamp-1 and Lamp-2 from intracellular proteolysis. J. Biol. Chem. 1999, 274, 31039–31046. [Google Scholar] [CrossRef]
- Pickard, J.M.; Maurice, C.F.; Kinnebrew, M.A.; Abt, M.C.; Schenten, D.; Golovkina, T.V.; Bogatyrev, S.R.; Ismagilov, R.F.; Pamer, E.G.; Turnbaugh, P.J. Rapid fucosylation of intestinal epithelium sustains host–commensal symbiosis in sickness. Nature 2014, 514, 638. [Google Scholar] [CrossRef] [PubMed]
- Shatnyeva, O.M.; Kubarenko, A.V.; Weber, C.E.; Pappa, A.; Schwartz-Albiez, R.; Weber, A.N.; Krammer, P.H.; Lavrik, I.N. Modulation of the CD95-induced apoptosis: The role of CD95 N-glycosylation. PLos ONE 2011, 6, e19927. [Google Scholar] [CrossRef] [PubMed]
- Swindall, A.F.; Bellis, S.L. Sialylation of the Fas death receptor by ST6Gal-I provides protection against Fas-mediated apoptosis in colon carcinoma cells. J. Biol. Chem. 2011, 286, 22982–22990. [Google Scholar] [CrossRef] [PubMed]
- Keppler, O.T.; Peter, M.E.; Hinderlich, S.; Moldenhauer, G.; Stehling, P.; Schmitz, I.; Schwartz-Albiez, R.; Reutter, W.; Pawlita, M. Differential sialylation of cell surface glycoconjugates in a human B lymphoma cell line regulates susceptibility for CD95 (APO-1/Fas)-mediated apoptosis and for infection by a lymphotropic virus. Glycobiology 1999, 9, 557–569. [Google Scholar] [CrossRef] [PubMed]
- Pretzlaff, R.K.; Xue, V.W.; Rowin, M.E. Sialidase Treatment Exposes the βT1-Integrin Active Ligand Binding Site on HL60 Cells and Increases Binding to Fibronectin. Cell Adhes. Commun. 2000, 7, 491–500. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Fukuda, T.; Hang, Q.; Hou, S.; Isaji, T.; Kameyama, A.; Gu, J. Inhibition of fucosylation by 2-fluorofucose suppresses human liver cancer HepG2 cell proliferation and migration as well as tumor formation. Sci. Rep. 2017, 7, 11563. [Google Scholar] [CrossRef] [PubMed]
- Mahdavi, J.; Sondén, B.; Hurtig, M.; Olfat, F.O.; Forsberg, L.; Roche, N.; Ångström, J.; Larsson, T.; Teneberg, S.; Karlsson, K.-A. Helicobacter pylori SabA adhesin in persistent infection and chronic inflammation. Science 2002, 297, 573–578. [Google Scholar] [CrossRef]
- Vagin, O.; Kraut, J.A.; Sachs, G. Role of N-glycosylation in trafficking of apical membrane proteins in epithelia. Am. J. Physiol. Ren. Physiol. 2009, 296, F459–F469. [Google Scholar] [CrossRef]
- Fan, H.; Meng, W.; Kilian, C.; Grams, S.; Reutter, W. Domain-specific N-glycosylation of the membrane glycoprotein dipeptidylpeptidase IV (CD26) influences its subcellular trafficking, biological stability, enzyme activity and protein folding. Eur. J. Biochem. 1997, 246, 243–251. [Google Scholar] [CrossRef]
- Mochizuki, K.; Kagawa, T.; Numari, A.; Harris, M.J.; Itoh, J.; Watanabe, N.; Mine, T.; Arias, I.M. Two N-linked glycans are required to maintain the transport activity of the bile salt export pump (ABCB11) in MDCK II cells. Am. J. Physiol. Gastrointest. Liver Physiol. 2007, 292, G818–G828. [Google Scholar] [CrossRef]
- Vagin, O.; Turdikulova, S.; Sachs, G. The H, K-ATPase β subunit as a model to study the role of N-glycosylation in membrane trafficking and apical sorting. J. Biol. Chem. 2004, 279, 39026–39034. [Google Scholar] [CrossRef]
- Zafra, F.; Gimenez, C. Molecular Determinants Involved in the Asymmetrical Distribution of Glycine Transporters in Polarized Cells; Portland Press Limited: London, UK, 2001. [Google Scholar]
- Hakomori, S.-i. Aberrant Glycosylation In Tumors And Tumor-Associated Carbohydrate Antigens. In Advances in Cancer Research; George, F.V.W., George, K., Eds.; Academic Press: Cambridge, MA, USA, 1989; Volume 52, pp. 257–331. [Google Scholar]
- Ju, T.; Lanneau, G.S.; Gautam, T.; Wang, Y.; Xia, B.; Stowell, S.R.; Willard, M.T.; Wang, W.; Xia, J.Y.; Zuna, R.E. Human tumor antigens Tn and sialyl Tn arise from mutations in Cosmc. Cancer Res. 2008, 68, 1636–1646. [Google Scholar] [CrossRef]
- Ju, T.; Wang, Y.; Aryal, R.P.; Lehoux, S.D.; Ding, X.; Kudelka, M.R.; Cutler, C.; Zeng, J.; Wang, J.; Sun, X. Tn and sialyl-Tn antigens, aberrant O-glycomics as human disease markers. Proteom. Clin. Appl. 2013, 7, 618–631. [Google Scholar]
- Achalli, S.; Madi, M.; Babu, S.G.; Shetty, S.R.; Kumari, S.; Bhat, S. Sialic acid as a biomarker of oral potentially malignant disorders and oral cancer. Indian J. Dent. Res. 2017, 28, 395. [Google Scholar] [CrossRef]
- Noda, K.; Miyoshi, E.; Gu, J.; Gao, C.-X.; Nakahara, S.; Kitada, T.; Honke, K.; Suzuki, K.; Yoshihara, H.; Yoshikawa, K. Relationship between elevated FX expression and increased production of GDP-L-fucose, a common donor substrate for fucosylation in human hepatocellular carcinoma and hepatoma cell lines. Cancer Res. 2003, 63, 6282–6289. [Google Scholar] [PubMed]
- Wang, X.; Inoue, S.; Gu, J.; Miyoshi, E.; Noda, K.; Li, W.; Mizuno-Horikawa, Y.; Nakano, M.; Asahi, M.; Takahashi, M. Dysregulation of TGF-β1 receptor activation leads to abnormal lung development and emphysema-like phenotype in core fucose-deficient mice. Proc. Natl. Acad. Sci. 2005, 102, 15791–15796. [Google Scholar] [CrossRef]
- Nie, H.; Liu, X.; Zhang, Y.; Li, T.; Zhan, C.; Huo, W.; He, A.; Yao, Y.; Jin, Y.; Qu, Y. Specific N-glycans of hepatocellular carcinoma cell surface and the abnormal increase of core-α-1, 6-fucosylated triantennary glycan via N-acetylglucosaminyltransferases-IVa regulation. Sci. Rep. 2015, 5, 16007. [Google Scholar] [CrossRef]
- Tian, Y.; Yao, Z.; Roden, R.B.; Zhang, H. Identification of glycoproteins associated with different histological subtypes of ovarian tumors using quantitative glycoproteomics. Proteomics 2011, 11, 4677–4687. [Google Scholar] [CrossRef]
- Tian, Y.; Bova, G.S.; Zhang, H. Quantitative glycoproteomic analysis of optimal cutting temperature-embedded frozen tissues identifying glycoproteins associated with aggressive prostate cancer. Anal. Chem. 2011, 83, 7013–7019. [Google Scholar] [CrossRef]
- Singh, R.; Bandyopadhyay, D. MUC1: A target molecule for cancer therapy. Cancer Biol. Ther. 2007, 6, 481–486. [Google Scholar] [CrossRef]
- Moore, A.; Medarova, Z.; Potthast, A.; Dai, G. In vivo targeting of underglycosylated MUC-1 tumor antigen using a multimodal imaging probe. Cancer Res. 2004, 64, 1821–1827. [Google Scholar] [CrossRef] [PubMed]
- Lo, S.-T.; Pantazopouos, P.; Medarova, Z.; Moore, A. Presentation of underglycosylated mucin 1 in pancreatic adenocarcinoma (PDAC) at early stages. Am. J. Cancer Res. 2016, 6, 1986. [Google Scholar] [PubMed]
- Chang, F.; Li, R.; Ladisch, S. Shedding of gangliosides by human medulloblastoma cells. Exp. Cell Res. 1997, 234, 341–346. [Google Scholar] [CrossRef] [PubMed]
- Valentino, L.; Moss, T.; Olson, E.; Wang, H.-J.; Elashoff, R.; Ladisch, S. Shed tumor gangliosides and progression of human neuroblastoma. Blood 1990, 75, 1564–1567. [Google Scholar] [PubMed]
- Li, G.; Li, L.; Joo, E.J.; Son, J.W.; Kim, Y.J.; Kang, J.K.; Lee, K.B.; Zhang, F.; Linhardt, R.J. Glycosaminoglycans and glycolipids as potential biomarkers in lung cancer. Glycoconj. J. 2017, 34, 661–669. [Google Scholar] [CrossRef] [PubMed]
- Holst, S.; Stavenhagen, K.; Balog, C.I.; Koeleman, C.A.; McDonnell, L.M.; Mayboroda, O.A.; Verhoeven, A.; Mesker, W.E.; Tollenaar, R.A.; Deelder, A.M. Investigations on aberrant glycosylation of glycosphingolipids in colorectal cancer tissues using liquid chromatography and matrix-assisted laser desorption time-of-flight mass spectrometry (MALDI-TOF-MS). Mol. Cell. Proteom. 2013, 12, 3081–3093. [Google Scholar] [CrossRef] [PubMed]
- Bethke, U.; Müthing, J.; Schauder, B.; Conradt, P.; Mühlradt, P.F. An improved semi-quantitative enzyme immunostaining procedure for glycosphingolipid antigens on high performance thin layer chromatograms. J. Immunol. Methods 1986, 89, 111–116. [Google Scholar] [CrossRef]
- Akama, T.O.; Fukuda, M.N. N-Glycan Structure Analysis Using Lectins and an α-Mannosidase Activity Assay. In Methods in Enzymology; Academic Press: Cambridge, MA, USA, 2006; Volum 416, pp. 304–314. [Google Scholar]
- Zhang, L.; Luo, S.; Zhang, B. The use of lectin microarray for assessing glycosylation of therapeutic proteins. mAbs 2016, 8, 524–535. [Google Scholar] [CrossRef]
- Cummings, R.D.; Etzler, M.E. Antibodies and lectins in glycan analysis. In Essentials of Glycobiology, 2nd ed.; Cold Spring Harbor Laboratory Press: Cold Sprin Harbor, NY, USA, 2009. [Google Scholar]
- Chandler, K.B.; Costello, C.E. Glycomics and glycoproteomics of membrane proteins and cell-surface receptors: Present trends and future opportunities. Electrophoresis 2016, 37, 1407–1419. [Google Scholar] [CrossRef]
- Simons, K.; Gerl, M.J. Revitalizing membrane rafts: New tools and insights. Nat. Rev. Mol. Cell Biol. 2010, 11, 688. [Google Scholar] [CrossRef]
- Clark, D.; Mao, L. Cancer Biomarker Discovery: Lectin-Based Strategies Targeting Glycoproteins. J. Dis. Markers 2012, 33. [Google Scholar] [CrossRef]
- Fitzgerald, J.; Leonard, P.; Darcy, E.; Sharma, S.; O’Kennedy, R. Immunoaffinity Chromatography: Concepts and Applications. In Protein Chromatography: Methods and Protocols; Walls, D., Loughran, S.T., Eds.; Springer: New York, NY, USA, 2017; pp. 27–51. [Google Scholar]
- Grammel, M.; Hang, H.C. Chemical reporters for biological discovery. Nat. Chem. Biol. 2013, 9, 475. [Google Scholar] [CrossRef] [PubMed]
- Hubbard, S.C.; Boyce, M.; McVaugh, C.T.; Peehl, D.M.; Bertozzi, C.R. Cell surface glycoproteomic analysis of prostate cancer-derived PC-3 cells. Bioorganic Med. Chem. Lett. 2011, 21, 4945–4950. [Google Scholar] [CrossRef] [PubMed]
- Besanceney-Webler, C.; Jiang, H.; Wang, W.; Baughn, A.D.; Wu, P. Metabolic labeling of fucosylated glycoproteins in Bacteroidales species. Bioorganic Med. Chem. Lett. 2011, 21, 4989–4992. [Google Scholar] [CrossRef] [PubMed]
- Vuckovic, D.; Dagley, L.F.; Purcell, A.W.; Emili, A. Membrane proteomics by high performance liquid chromatography–tandem mass spectrometry: Analytical approaches and challenges. Proteomics 2013, 13, 404–423. [Google Scholar] [CrossRef] [PubMed]
- Målen, H.; Pathak, S.; Søfteland, T.; de Souza, G.A.; Wiker, H.G. Definition of novel cell envelope associated proteins in Triton X-114 extracts of Mycobacterium tuberculosis H37Rv. BMC Microbiol. 2010, 10, 132. [Google Scholar] [CrossRef] [PubMed]
- Cao, R.; Li, X.; Liu, Z.; Peng, X.; Hu, W.; Wang, X.; Chen, P.; Xie, J.; Liang, S. Integration of a Two-Phase Partition Method into Proteomics Research on Rat Liver Plasma Membrane Proteins. J. Proteome Res. 2006, 5, 634–642. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.; Elschenbroich, S.; Sharma, P.; Sepiashvili, L.; Gramolini, A.O.; Kislinger, T. Use of Colloidal Silica-Beads for the Isolation of Cell-Surface Proteins for Mass Spectrometry-Based Proteomics. In Immune Receptors: Methods and Protocols; Rast, J.P., Booth, J.W.D., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 227–241. [Google Scholar]
- Yavuz, H.; Kattan, I.; Hernandez, J.M.; Hofnagel, O.; Witkowska, A.; Raunser, S.; Walla, P.J.; Jahn, R. Arrest of trans-SNARE zippering uncovers loosely and tightly docked intermediates in membrane fusion. J. Biol. Chem. 2018, 293, 8645–8655. [Google Scholar] [CrossRef] [PubMed]
- Elia, G. Cell Surface Protein Biotinylation for SDS-PAGE Analysis. In Protein Electrophoresis: Methods and Protocols; Kurien, B.T., Scofield, R.H., Eds.; Humana Press: Totowa, NJ, USA, 2012; pp. 361–372. [Google Scholar]
- Pan, P.-W.; Zhang, Q.; Hou, J.; Liu, Z.; Bai, F.; Cao, M.-r.; Sun, T.; Bai, G. Cell surface glycoprotein profiling of cancer cells based on bioorthogonal chemistry. Anal. Bioanal. Chem. 2012, 403, 1661–1670. [Google Scholar] [CrossRef] [PubMed]
- Cowell, J.; Buck, M.; Essa, A.H.; Clarke, R.; Vollmer, W.; Vollmer, D.; Hilkens, C.M.; Isaacs, J.D.; Hall, M.J.; Gray, J. Traceless Cleavage of Protein–Biotin Conjugates under Biologically Compatible Conditions. ChemBioChem 2017, 18, 1688–1691. [Google Scholar] [CrossRef] [PubMed]
- Hacker, S.M.; Backus, K.M.; Lazear, M.R.; Forli, S.; Correia, B.E.; Cravatt, B.F. Global profiling of lysine reactivity and ligandability in the human proteome. Nat. Chem. 2017, 9, 1181. [Google Scholar] [CrossRef] [PubMed]
- Mi, W.; Jia, W.; Zheng, Z.; Wang, J.; Cai, Y.; Ying, W.; Qian, X. Surface glycoproteomic analysis of hepatocellular carcinoma cells by affinity enrichment and mass spectrometric identification. Glycoconj. J. 2012, 29, 411–424. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.-Y.; Hua, X.-W.; Jia, H.-R.; Liu, P.; Gu, N.; Chen, Z.; Wu, F.-G. Enhanced cell membrane enrichment and subsequent cellular internalization of quantum dots via cell surface engineering: Illuminating plasma membranes with quantum dots. J. Mater. Chem. B 2016, 4, 834–843. [Google Scholar] [CrossRef]
- Zhang, L.; Xie, J.; Wang, X.e.; Liu, X.; Tang, X.; Cao, R.; Hu, W.; Nie, S.; Fan, C.; Liang, S. Proteomic analysis of mouse liver plasma membrane: Use of differential extraction to enrich hydrophobic membrane proteins. Proteomics 2005, 5, 4510–4524. [Google Scholar] [CrossRef] [PubMed]
- Suski, J.M.; Lebiedzinska, M.; Wojtala, A.; Duszynski, J.; Giorgi, C.; Pinton, P.; Wieckowski, M.R. Isolation of plasma membrane–associated membranes from rat liver. Nat. Protoc. 2014, 9, 312. [Google Scholar] [CrossRef] [PubMed]
- Lund, R.; Leth-Larsen, R.; Jensen, O.N.; Ditzel, H.J. Efficient Isolation and Quantitative Proteomic Analysis of Cancer Cell Plasma Membrane Proteins for Identification of Metastasis-Associated Cell Surface Markers. J. Proteome Res. 2009, 8, 3078–3090. [Google Scholar] [CrossRef]
- Park, D.D.; Xu, G.; Wong, M.; Phoomak, C.; Liu, M.; Haigh, N.E.; Wongkham, S.; Yang, P.; Maverakis, E.; Lebrilla, C.B. Membrane glycomics reveal heterogeneity and quantitative distribution of cell surface sialylation. Chem. Sci. 2018, 9, 6271–6285. [Google Scholar] [CrossRef]
- Mattow, J.; Siejak, F.; Hagens, K.; Schmidt, F.; Koehler, C.; Treumann, A.; Schaible, U.E.; Kaufmann, S.H.E. An improved strategy for selective and efficient enrichment of integral plasma membrane proteins of mycobacteria. Proteomics 2007, 7, 1687–1701. [Google Scholar] [CrossRef]
- Ishibashi, Y.; Kobayashi, U.; Hijikata, A.; Sakaguchi, K.; Goda, H.M.; Tamura, T.; Okino, N.; Ito, M. Preparation and characterization of EGCase I, applicable to the comprehensive analysis of GSLs, using a rhodococcal expression system. J. Lipid Res. 2012, 53, 2242–2251. [Google Scholar] [CrossRef]
- Wang, C.; Wen, Y.; Yang, M.; Huang, L.; Wang, Z.; Fan, J. High-sensitivity quantification of glycosphingolipid glycans by ESI-MS utilizing ozonolysis-based release and isotopic Girard’s reagent labeling. Anal. Biochem. 2019, 582, 113355. [Google Scholar] [CrossRef]
- Guan, F.; Tan, Z.; Li, X.; Pang, X.; Zhu, Y.; Li, D.; Yang, G. A lectin-based isolation/enrichment strategy for improved coverage of N-glycan analysis. Carbohydr. Res. 2015, 416, 7–13. [Google Scholar] [CrossRef]
- Zhu, F.; Clemmer, D.E.; Trinidad, J.C. Characterization of lectin binding affinities via direct LC-MS profiling: Implications for glycopeptide enrichment and separation strategies. Analyst 2017, 142, 65–74. [Google Scholar] [CrossRef] [PubMed]
- de Boer, A.R.; Hokke, C.H.; Deelder, A.M.; Wuhrer, M. Serum antibody screening by surface plasmon resonance using a natural glycan microarray. Glycoconj. J. 2008, 25, 75–84. [Google Scholar] [CrossRef]
- Yang, S.; Zhang, H. Solid-phase glycan isolation for glycomics analysis. Proteom. Clin. Appl. 2012, 6, 596–608. [Google Scholar] [CrossRef] [PubMed]
- Abrahams, J.L.; Campbell, M.P.; Packer, N.H. Building a PGC-LC-MS N-glycan retention library and elution mapping resource. Glycoconj. J. 2018, 35, 15–29. [Google Scholar] [CrossRef] [PubMed]
- Domann, P.J.; Pardos-Pardos, A.C.; Fernandes, D.L.; Spencer, D.I.R.; Radcliffe, C.M.; Royle, L.; Dwek, R.A.; Rudd, P.M. Separation-based Glycoprofiling Approaches using Fluorescent Labels. Proteomics 2007, 7, 70–76. [Google Scholar] [CrossRef] [PubMed]
- Ruhaak, L.; Zauner, G.; Huhn, C.; Bruggink, C.; Deelder, A.; Wuhrer, M. Glycan labeling strategies and their use in identification and quantification. Anal. Bioanal. Chem. 2010, 397, 3457–3481. [Google Scholar] [CrossRef] [PubMed]
- Anumula, K.R.; Dhume, S.T. High resolution and high sensitivity methods for oligosaccharide mapping and characterization by normal phase high performance liquid chromatography following derivatization with highly fluorescent anthranilic acid. Glycobiology 1998, 8, 685–694. [Google Scholar] [CrossRef]
- Yu, Y.Q.; Gilar, M.; Kaska, J.; Gebler, J.C. A rapid sample preparation method for mass spectrometric characterization of N-linked glycans. Rapid Commun. Mass Spectrom. 2005, 19, 2331–2336. [Google Scholar] [CrossRef]
- Ruhaak, L.R.; Xu, G.; Li, Q.; Goonatilleke, E.; Lebrilla, C.B. Mass Spectrometry Approaches to Glycomic and Glycoproteomic Analyses. Chem. Rev. 2018, 118, 7886–7930. [Google Scholar] [CrossRef]
- Honda, S.; Akao, E.; Suzuki, S.; Okuda, M.; Kakehi, K.; Nakamura, J. High-performance liquid chromatography of reducing carbohydrates as strongly ultraviolet-absorbing and electrochemically sensitive 1-phenyl-3-methyl5-pyrazolone derivatives. Anal. Biochem. 1989, 180, 351–357. [Google Scholar] [CrossRef]
- Chai, W.; Piskarev, V.; Lawson, A.M. Negative-ion electrospray mass spectrometry of neutral underivatized oligosaccharides. Anal. Chem. 2001, 73, 651–657. [Google Scholar] [CrossRef] [PubMed]
- De Leoz, M.L.A.; Simón-Manso, Y.; Woods, R.J.; Stein, S.E. Cross-Ring Fragmentation Patterns in the Tandem Mass Spectra of Underivatized Sialylated Oligosaccharides and Their Special Suitability for Spectrum Library Searching. J. Am. Soc. Mass Spectrom. 2019, 30, 426–438. [Google Scholar] [CrossRef] [PubMed]
- Balaguer, E.; Neusüss, C. Glycoprotein Characterization Combining Intact Protein and Glycan Analysis by Capillary Electrophoresis-Electrospray Ionization-Mass Spectrometry. Anal. Chem. 2006, 78, 5384–5393. [Google Scholar] [CrossRef] [PubMed]
- de Leoz, M.L.A.; An, H.J.; Kronewitter, S.; Kim, J.; Beecroft, S.; Vinall, R.; Miyamoto, S.; de Vere White, R.; Lam, K.S.; Lebrilla, C. Glycomic approach for potential biomarkers on prostate cancer: Profiling of N-linked glycans in human sera and pRNS cell lines. Dis. Markers 2008, 25, 243–258. [Google Scholar] [CrossRef] [PubMed]
- Hung, W.-T.; Wang, S.-H.; Chen, Y.-T.; Yu, H.-M.; Chen, C.-H.; Yang, W.-B. MALDI-TOF MS analysis of native and permethylated or benzimidazole-derivatized polysaccharides. Molecules 2012, 17, 4950–4961. [Google Scholar] [CrossRef] [PubMed]
- Reiding, K.R.; Blank, D.; Kuijper, D.M.; Deelder, A.M.; Wuhrer, M. High-throughput profiling of protein N-glycosylation by MALDI-TOF-MS employing linkage-specific sialic acid esterification. Anal. Chem. 2014, 86, 5784–5793. [Google Scholar] [CrossRef]
- You, X.; Qin, H.; Ye, M. Recent advances in methods for the analysis of protein O-glycosylation at proteome level. J. Sep. Sci. 2018, 41, 248–261. [Google Scholar] [CrossRef]
- Iwase, H. Release of O-glycans by Enzymatic Methods. In Experimental Glycoscience: Glycochemistry; Taniguchi, N., Suzuki, A., Ito, Y., Narimatsu, H., Kawasaki, T., Hase, S., Eds.; Springer Japan: Tokyo, Japan, 2008; pp. 14–17. [Google Scholar]
- Jensen, P.H.; Karlsson, N.G.; Kolarich, D.; Packer, N.H. Structural analysis of N-and O-glycans released from glycoproteins. Nat. Protoc. 2012, 7, 1299. [Google Scholar] [CrossRef]
- Miura, Y.; Kato, K.; Takegawa, Y.; Kurogochi, M.; Furukawa, J.-i.; Shinohara, Y.; Nagahori, N.; Amano, M.; Hinou, H.; Nishimura, S.-I. Glycoblotting-Assisted O-Glycomics: Ammonium Carbamate Allows for Highly Efficient O-Glycan Release from Glycoproteins. Anal. Chem. 2010, 82, 10021–10029. [Google Scholar] [CrossRef]
- Song, X.; Ju, H.; Lasanajak, Y.; Kudelka, M.R.; Smith, D.F.; Cummings, R.D. Oxidative release of natural glycans for functional glycomics. Nat. Methods 2016, 13, 528. [Google Scholar] [CrossRef]
- Banerjee, P.S.; Hart, G.W.; Cho, J.W. Chemical approaches to study O-GlcNAcylation. Chem. Soc. Rev. 2013, 42, 4345–4357. [Google Scholar] [CrossRef]
- Ma, J.; Hart, G.W. O-GlcNAc profiling: From proteins to proteomes. Clin. Proteom. 2014, 11, 8. [Google Scholar] [CrossRef]
- An, H.J.; Lebrilla, C.B. A glycomics approach to the discovery of potential cancer biomarkers. In Functional Glycomics; Springer: Berlin, Germany, 2010; pp. 199–213. [Google Scholar]
- Bensing, B.A.; Li, Q.; Park, D.; Lebrilla, C.B.; Sullam, P.M. Streptococcal Siglec-like adhesins recognize different subsets of human plasma glycoproteins: Implications for infective endocarditis. Glycobiology 2018, 1, 11. [Google Scholar] [CrossRef]
- Lee, S.H.; Yu, S.-Y.; Nakayama, J.; Khoo, K.-H.; Stone, E.L.; Fukuda, M.N.; Marth, J.D.; Fukuda, M. Core2 O-glycan structure is essential for the cell surface expression of sucrase isomaltase and dipeptidyl peptidase-IV during intestinal cell differentiation. J. Biol. Chem. 2010, 285, 37683–37692. [Google Scholar] [CrossRef]
- Amano, M.; Eriksson, H.; Manning, J.C.; Detjen, K.M.; André, S.; Nishimura, S.I.; Lehtiö, J.; Gabius, H.J. Tumour suppressor p16INK4a–anoikis-favouring decrease in N/O-glycan/cell surface sialylation by down-regulation of enzymes in sialic acid biosynthesis in tandem in a pancreatic carcinoma model. Febs J. 2012, 279, 4062–4080. [Google Scholar] [CrossRef]
- An, H.J.; Gip, P.; Kim, J.; Wu, S.; Park, K.W.; McVaugh, C.T.; Schaffer, D.V.; Bertozzi, C.R.; Lebrilla, C.B. Extensive determination of glycan heterogeneity reveals an unusual abundance of high mannose glycans in enriched plasma membranes of human embryonic stem cells. Mol. Cell. Proteom. 2012, 11, M111. [Google Scholar] [CrossRef]
- Park, D.; Brune, K.A.; Mitra, A.; Marusina, A.I.; Maverakis, E.; Lebrilla, C.B. Characteristic Changes in Cell Surface Glycosylation Accompany Intestinal Epithelial Cell (IEC) Differentiation: High Mannose Structures Dominate the Cell Surface Glycome of Undifferentiated Enterocytes. Mol. Cell. Proteom. Mcp 2015, 14, 2910–2921. [Google Scholar] [CrossRef]
- Hidalgo, I.J.; Raub, T.J.; Borchardt, R.T. Characterization of the human colon carcinoma cell line (Caco-2) as a model system for intestinal epithelial permeability. Gastroenterology 1989, 96, 736–749. [Google Scholar] [CrossRef]
- Park, D.; Xu, G.; Wong, M.; Lebrilla, C.B.; Barboza, M.; Raybould, H.; Mills, D.A.; Shah, I.M. Enterocyte glycosylation is responsive to changes in extracellular conditions: Implications for membrane functions. Glycobiology 2017, 27, 847–860. [Google Scholar] [CrossRef]
- Park, D.; Arabyan, N.; Williams, C.C.; Song, T.; Mitra, A.; Weimer, B.C.; Maverakis, E.; Lebrilla, C.B. Salmonella Typhimurium Enzymatically Landscapes the Host Intestinal Epithelial Cell (IEC) Surface Glycome to Increase Invasion. Mol. Amp; Cell. Proteom. 2016, 15, 3653–3664. [Google Scholar] [CrossRef]
- Awan, B.; Turkov, D.; Schumacher, C.; Jacobo, A.; McEnerney, A.; Ramsey, A.; Xu, G.; Park, D.; Kalomoiris, S.; Yao, W.; et al. FGF2 Induces Migration of Human Bone Marrow Stromal Cells by Increasing Core Fucosylations on N-Glycans of Integrins. Stem Cell Rep. 2018, 11, 325–333. [Google Scholar] [CrossRef]
- Ruiz-May, E.; Catalá, C.; Rose, J.K. N-glycoprotein enrichment by lectin affinity chromatography. In Plant Proteomics; Springer: Berlin, Germany, 2014; pp. 633–643. [Google Scholar]
- Xu, Y.; Wu, Z.; Zhang, L.; Lu, H.; Yang, P.; Webley, P.A.; Zhao, D. Highly specific enrichment of glycopeptides using boronic acid-functionalized mesoporous silica. Anal. Chem. 2008, 81, 503–508. [Google Scholar] [CrossRef]
- Haun, R.S.; Quick, C.M.; Siegel, E.R.; Raju, I.; Mackintosh, S.G.; Tackett, A.J. Bioorthogonal labeling cell-surface proteins expressed in pancreatic cancer cells to identify potential diagnostic/therapeutic biomarkers. Cancer Biol. Ther. 2015, 16, 1557–1565. [Google Scholar] [CrossRef]
- Xie, R.; Dong, L.; Du, Y.; Zhu, Y.; Hua, R.; Zhang, C.; Chen, X. In vivo metabolic labeling of sialoglycans in the mouse brain by using a liposome-assisted bioorthogonal reporter strategy. Proc. Natl. Acad. Sci. 2016, 113, 5173–5178. [Google Scholar] [CrossRef]
- Hsu, T.-L.; Hanson, S.R.; Kishikawa, K.; Wang, S.-K.; Sawa, M.; Wong, C.-H. Alkynyl sugar analogs for the labeling and visualization of glycoconjugates in cells. Proc. Natl. Acad. Sci. 2007, 104, 2614–2619. [Google Scholar] [CrossRef]
- Rabuka, D.; Hubbard, S.C.; Laughlin, S.T.; Argade, S.P.; Bertozzi, C.R. A chemical reporter strategy to probe glycoprotein fucosylation. J. Am. Chem. Soc. 2006, 128, 12078–12079. [Google Scholar] [CrossRef]
- Zhang, H.; Li, X.-j.; Martin, D.B.; Aebersold, R. Identification and quantification of N-linked glycoproteins using hydrazide chemistry, stable isotope labeling and mass spectrometry. Nat. Biotechnol. 2003, 21, 660. [Google Scholar] [CrossRef]
- Huang, J.; Wan, H.; Yao, Y.; Li, J.; Cheng, K.; Mao, J.; Chen, J.; Wang, Y.; Qin, H.; Zhang, W.; et al. Highly Efficient Release of Glycopeptides from Hydrazide Beads by Hydroxylamine Assisted PNGase F Deglycosylation for N-Glycoproteome Analysis. Anal. Chem. 2015, 87, 10199–10204. [Google Scholar] [CrossRef]
- Totten, S.M.; Feasley, C.L.; Bermudez, A.; Pitteri, S.J. Parallel comparison of N-linked glycopeptide enrichment techniques reveals extensive glycoproteomic analysis of plasma enabled by SAX-ERLIC. J. Proteome Res. 2017, 16, 1249–1260. [Google Scholar] [CrossRef]
- Li, B.; An, H.J.; Kirmiz, C.; Lebrilla, C.B.; Lam, K.S.; Miyamoto, S. Glycoproteomic Analyses of Ovarian Cancer Cell Lines and Sera from Ovarian Cancer Patients Show Distinct Glycosylation Changes in Individual Proteins. J. Proteome Res. 2008, 7, 3776–3788. [Google Scholar] [CrossRef]
- Cao, L.; Qu, Y.; Zhang, Z.; Wang, Z.; Prytkova, I.; Wu, S. Intact glycopeptide characterization using mass spectrometry. Expert Rev. Proteom. 2016, 13, 513–522. [Google Scholar] [CrossRef]
- Yang, H.; Yang, C.; Sun, T. Characterization of glycopeptides using a stepped higher-energy C-trap dissociation approach on a hybrid quadrupole orbitrap. Rapid Commun. Mass Spectrom. 2018, 32, 1353–1362. [Google Scholar] [CrossRef]
- Hogan, J.M.; Pitteri, S.J.; Chrisman, P.A.; McLuckey, S.A. Complementary structural information from a tryptic N-linked glycopeptide via electron transfer ion/ion reactions and collision-induced dissociation. J. Proteome Res. 2005, 4, 628–632. [Google Scholar] [CrossRef]
- Yu, Q.; Wang, B.; Chen, Z.; Urabe, G.; Glover, M.S.; Shi, X.; Guo, L.-W.; Kent, K.C.; Li, L. Electron-transfer/higher-energy collision dissociation (EThcD)-enabled intact glycopeptide/glycoproteome characterization. J. Am. Soc. Mass Spectrom. 2017, 28, 1751–1764. [Google Scholar] [CrossRef]
- Chen, Z.; Yu, Q.; Hao, L.; Liu, F.; Johnson, J.; Tian, Z.; Kao, W.J.; Xu, W.; Li, L. Site-specific characterization and quantitation of N-glycopeptides in PKM2 knockout breast cancer cells using DiLeu isobaric tags enabled by electron-transfer/higher-energy collision dissociation (EThcD). Analyst 2018, 143, 2508–2519. [Google Scholar] [CrossRef]
- Singh, C.; Zampronio, C.G.; Creese, A.J.; Cooper, H.J. Higher energy collision dissociation (HCD) product ion-triggered electron transfer dissociation (ETD) mass spectrometry for the analysis of N-linked glycoproteins. J. Proteome Res. 2012, 11, 4517–4525. [Google Scholar] [CrossRef]
- Bern, M.; Kil, Y.J.; Becker, C. Byonic: Advanced Peptide and Protein Identification Software. In Current Protocols in Bioinformatics; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2002. [Google Scholar] [CrossRef]
- Zeng, W.-F.; Liu, M.-Q.; Zhang, Y.; Wu, J.-Q.; Fang, P.; Peng, C.; Nie, A.; Yan, G.; Cao, W.; Liu, C. pGlyco: A pipeline for the identification of intact N-glycopeptides by using HCD-and CID-MS/MS and MS3. Sci. Rep. 2016, 6, 25102. [Google Scholar] [CrossRef]
- Liu, M.-Q.; Zeng, W.-F.; Fang, P.; Cao, W.-Q.; Liu, C.; Yan, G.-Q.; Zhang, Y.; Peng, C.; Wu, J.-Q.; Zhang, X.-J. pGlyco 2.0 enables precision N-glycoproteomics with comprehensive quality control and one-step mass spectrometry for intact glycopeptide identification. Nat. Commun. 2017, 8, 538–542. [Google Scholar] [CrossRef]
- Park, G.W.; Kim, J.Y.; Hwang, H.; Lee, J.Y.; Ahn, Y.H.; Lee, H.K.; Ji, E.S.; Kim, K.H.; Jeong, H.K.; Yun, K.N.; et al. Integrated GlycoProteome Analyzer (I-GPA) for Automated Identification and Quantitation of Site-Specific N-Glycosylation. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Nasir, W.; Toledo, A.G.; Noborn, F.; Nilsson, J.; Wang, M.X.; Bandeira, N.; Larson, G. SweetNET: A Bioinformatics Workflow for Glycopeptide MS/MS Spectral Analysis. J. Proteome Res. 2016, 15, 2826–2840. [Google Scholar] [CrossRef]
- Kim, J.W.; Hwang, H.; Lim, J.S.; Lee, H.J.; Jeong, S.K.; Yoo, J.S.; Paik, Y.K. gFinder: A Web-Based Bioinformatics Tool for the Analysis of N-Glycopeptides. J. Proteome Res. 2016, 15, 4116–4125. [Google Scholar] [CrossRef]
- Li, Q.; Xie, Y.; Xu, G.; Lebrilla, C.B. Identification of potential sialic acid binding proteins on cell membrane by proximity chemical labeling. Chem. Sci. 2019. [Google Scholar] [CrossRef]
- Kailemia, M.J.; Xu, G.; Wong, M.; Li, Q.; Goonatilleke, E.; Leon, F.; Lebrilla, C.B. Recent advances in the mass spectrometry methods for glycomics and cancer. Anal. Chem. 2017, 90, 208–224. [Google Scholar] [CrossRef]
- Merrill, A.H. Sphingolipid and Glycosphingolipid Metabolic Pathways in the Era of Sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef]
- Scandroglio, F.; Loberto, N.; Valsecchi, M.; Chigorno, V.; Prinetti, A.; Sonnino, S. Thin layer chromatography of gangliosides. Glycoconj. J. 2008, 26, 961. [Google Scholar] [CrossRef]
- Kundu, S.K.; Dunn Scott, D. Rapid separation of gangliosides by high-performance liquid chromatography. J. Chromatogr. B: Biomed. Sci. Appl. 1982, 232, 19–27. [Google Scholar] [CrossRef]
- Rinaldi, S.; Brennan, K.M.; Goodyear, C.S.; O’Leary, C.; Schiavo, G.; Crocker, P.R.; Willison, H.J. Analysis of lectin binding to glycolipid complexes using combinatorial glycoarrays. Glycobiology 2009, 19, 789–796. [Google Scholar] [CrossRef]
- Distler, U.; Souady, J.; Hülsewig, M.; Drmić-Hofman, I.; Haier, J.; Denz, A.; Grützmann, R.; Pilarsky, C.; Senninger, N.; Dreisewerd, K. Tumor-associated CD75s-and iso-CD75s-gangliosides are potential targets for adjuvant therapy in pancreatic cancer. Mol. Cancer Ther. 2008, 7, 2464–2475. [Google Scholar] [CrossRef]
- Albrecht, S.; Vainauskas, S.; Stöckmann, H.; McManus, C.; Taron, C.H.; Rudd, P.M. Comprehensive Profiling of Glycosphingolipid Glycans Using a Novel Broad Specificity Endoglycoceramidase in a High-Throughput Workflow. Anal. Chem. 2016, 88, 4795–4802. [Google Scholar] [CrossRef][Green Version]
- Fujitani, N.; Takegawa, Y.; Ishibashi, Y.; Araki, K.; Furukawa, J.-i.; Mitsutake, S.; Igarashi, Y.; Ito, M.; Shinohara, Y. Qualitative and quantitative cellular glycomics of glycosphingolipids based on rhodococcal endoglycosylceramidase-assisted glycan cleavage, glycoblotting-assisted sample preparation, and matrix-assisted laser desorption ionization tandem time-of-flight mass spectrometry analysis. J. Biol. Chem. 2011, 286, 41669–41679. [Google Scholar]
- Kirsch, S.; Souady, J.; Mormann, M.; Bindila, L.; Peter-Katalinić, J. Ceramide Profiles of Human Serum Gangliosides GM2 and GD1a exhibit Cancer-associated Alterations. J. Glycom. Lipidom. 2012, S2, 005. [Google Scholar] [CrossRef]
- Vesper, H.; Schmelz, E.-M.; Nikolova-Karakashian, M.N.; Dillehay, D.L.; Lynch, D.V.; Merrill Jr, A.H. Sphingolipids in food and the emerging importance of sphingolipids to nutrition. J. Nutr. 1999, 129, 1239–1250. [Google Scholar] [CrossRef]
- Zancada, L.; Sánchez-Juanes, F.; Alonso, J.; Hueso, P. Neutral glycosphingolipid content of ovine milk. J. Dairy Sci. 2010, 93, 19–26. [Google Scholar] [CrossRef]
- Sanchez-Juanes, F.; Alonso, J.M.; Zancada, L.; Hueso, P. Glycosphingolipids from bovine milk and milk fat globule membranes: A comparative study. Adhesion to enterotoxigenic Escherichia coli strains. Biol. Chem. 2009, 390, 31–40. [Google Scholar] [CrossRef]
- Nagafuku, M.; Kabayama, K.; Oka, D.; Kato, A.; Tani-Ichi, S.; Shimada, Y.; Ohno-Iwashita, Y.; Yamasaki, S.; Saito, T.; Iwabuchi, K. Reduction of glycosphingolipid levels in lipid rafts affects the expression state and function of glycosylphosphatidylinositol-anchored proteins but does not impair signal transduction via the T cell receptor. J. Biol. Chem. 2003, 278, 51920–51927. [Google Scholar] [CrossRef]
- Yu, R.K.; Nakatani, Y.; Yanagisawa, M. The role of glycosphingolipid metabolism in the developing brain. J. Lipid Res. 2009, 50, S440–S445. [Google Scholar] [CrossRef]
- Zarei, M.; Bindila, L.; Souady, J.; Dreisewerd, K.; Berkenkamp, S.; Müthing, J.; Peter-Katalinić, J. A sialylation study of mouse brain gangliosides by MALDI a-TOF and o-TOF mass spectrometry. J. Mass Spectrom. 2008, 43, 716–725. [Google Scholar] [CrossRef]
- Sarbu, M.; Vukelić, Ž.; Clemmer, D.E.; Zamfir, A.D. Electrospray ionization ion mobility mass spectrometry provides novel insights into the pattern and activity of fetal hippocampus gangliosides. Biochimie 2017, 139, 81–94. [Google Scholar] [CrossRef]
- Sarbu, M.; Vukelić, Ž.; Clemmer, D.E.; Zamfir, A.D. Ion mobility mass spectrometry provides novel insights into the expression and structure of gangliosides in the normal adult human hippocampus. Analyst 2018, 143, 5234–5246. [Google Scholar] [CrossRef]
- Jones, E.E.; Zhang, W.; Zhao, X.; Quiason, C.; Dale, S.; Shahidi-Latham, S.; Grabowski, G.A.; Setchell, K.D.; Drake, R.R.; Sun, Y. Tissue localization of glycosphingolipid accumulation in a Gaucher disease mouse brain by LC-ESI-MS/MS and high-resolution MALDI imaging mass spectrometry. Slas Discov. Adv. Life Sci. RD 2017, 22, 1218–1228. [Google Scholar] [CrossRef]
- Wong, M.; Xu, G.; Park, D.; Barboza, M.; Lebrilla, C.B. Intact glycosphingolipidomic analysis of the cell membrane during differentiation yields extensive glycan and lipid changes. Sci. Rep. 2018, 8, 10993. [Google Scholar] [CrossRef]
- Jang, C.; Chen, L.; Rabinowitz, J.D. Metabolomics and Isotope Tracing. Cell 2018, 173, 822–837. [Google Scholar] [CrossRef]
- Antoniewicz, M.R. A guide to 13C metabolic flux analysis for the cancer biologist. Exp. Mol. Med. 2018, 50, 19. [Google Scholar] [CrossRef]
- Dong, W.; Keibler, M.A.; Stephanopoulos, G. Review of metabolic pathways activated in cancer cells as determined through isotopic labeling and network analysis. Metab. Eng. 2017, 43, 113–124. [Google Scholar] [CrossRef]
- Gonzalez, P.S.; O’Prey, J.; Cardaci, S.; Barthet, V.J.A.; Sakamaki, J.-i.; Beaumatin, F.; Roseweir, A.; Gay, D.M.; Mackay, G.; Malviya, G.; et al. Mannose impairs tumour growth and enhances chemotherapy. Nature 2018, 563, 719–723. [Google Scholar] [CrossRef]
- Shi, Y.-B.; Yin, D. A good sugar, d-mannose, suppresses autoimmune diabetes. Cell Biosci. 2017, 7, 48. [Google Scholar] [CrossRef]
- Ichikawa, M.; Scott, D.A.; Losfeld, M.-E.; Freeze, H.H. The Metabolic Origins of Mannose in Glycoproteins. J. Biol. Chem. 2014, 289, 6751–6761. [Google Scholar] [CrossRef]
- Rillahan, C.D.; Antonopoulos, A.; Lefort, C.T.; Sonon, R.; Azadi, P.; Ley, K.; Dell, A.; Haslam, S.M.; Paulson, J.C. Global metabolic inhibitors of sialyl- and fucosyltransferases remodel the glycome. Nat. Chem. Biol. 2012, 8, 661. [Google Scholar] [CrossRef]
- Yurchenco, P.D.; Atkinson, P.H. Fucosyl-glycoprotein and precursor polls in HeLa cells. Biochemistry 1975, 14, 3107–3114. [Google Scholar] [CrossRef]
- Ng, B.G.; Xia, Z.-J.; Scott, D.; Freeze, H.H. Fucose Metabolism: Rethinking Old Concepts and Identifying New Mechanisms; Oxford University Press Inc.: London, UK, 2018; Volume 28, p. 1026. [Google Scholar]
- Xu, G.; Wong, M.; Li, Q.; Park, D.D.; Cheng, Z.; Lebrilla, C.B. Unveiling the metabolic fate of monosaccharides in cell membranes with glycomic and glycoproteomic analyses. Chem. Sci. 2019. [Google Scholar] [CrossRef]
- Banazadeh, A.; Veillon, L.; Wooding, K.M.; Zabet-moghaddam, M.; Mechref, Y. Recent advances in mass spectrometric analysis of glycoproteins. Electrophoresis 2017, 38, 162–189. [Google Scholar] [CrossRef]
- Alvarez-Manilla, G.; Warren, N.L.; Abney, T.; Atwood, J., III; Azadi, P.; York, W.S.; Pierce, M.; Orlando, R. Tools for glycomics: Relative quantitation of glycans by isotopic permethylation using 13CH3I. Glycobiology 2007, 17, 677–687. [Google Scholar] [CrossRef]
- Michael, C.; Rizzi, A.M. Tandem mass spectrometry of isomeric aniline-labeled N-glycans separated on porous graphitic carbon: Revealing the attachment position of terminal sialic acids and structures of neutral glycans. Rapid Commun. Mass Spectrom. 2015, 29, 1268–1278. [Google Scholar] [CrossRef]
- Prien, J.M.; Prater, B.D.; Qin, Q.; Cockrill, S.L. Mass Spectrometric-Based Stable Isotopic 2-Aminobenzoic Acid Glycan Mapping for Rapid Glycan Screening of Biotherapeutics. Anal. Chem. 2010, 82, 1498–1508. [Google Scholar] [CrossRef]
- Walker, S.H.; Taylor, A.D.; Muddiman, D.C. Individuality Normalization when Labeling with Isotopic Glycan Hydrazide Tags (INLIGHT): A Novel Glycan-Relative Quantification Strategy. J. Am. Soc. Mass Spectrom. 2013, 24, 1376–1384. [Google Scholar] [CrossRef]
- Wang, C.; Wu, Z.; Yuan, J.; Wang, B.; Zhang, P.; Zhang, Y.; Wang, Z.; Huang, L. Simplified Quantitative Glycomics Using the Stable Isotope Label Girard’s Reagent P by Electrospray Ionization Mass Spectrometry. J. Proteome Res. 2014, 13, 372–384. [Google Scholar] [CrossRef]
- Shah, P.; Yang, S.; Sun, S.; Aiyetan, P.; Yarema, K.J.; Zhang, H. Mass Spectrometric Analysis of Sialylated Glycans with Use of Solid-Phase Labeling of Sialic Acids. Anal. Chem. 2013, 85, 3606–3613. [Google Scholar] [CrossRef]
- Wei, L.; Cai, Y.; Yang, L.; Zhang, Y.; Lu, H. Duplex Stable Isotope Labeling (DuSIL) for Simultaneous Quantitation and Distinction of Sialylated and Neutral N-Glycans by MALDI-MS. Anal. Chem. 2018, 90, 10442–10449. [Google Scholar] [CrossRef]
- McAlister, G.C.; Huttlin, E.L.; Haas, W.; Ting, L.; Jedrychowski, M.P.; Rogers, J.C.; Kuhn, K.; Pike, I.; Grothe, R.A.; Blethrow, J.D.; et al. Increasing the Multiplexing Capacity of TMTs Using Reporter Ion Isotopologues with Isobaric Masses. Anal. Chem. 2012, 84, 7469–7478. [Google Scholar] [CrossRef]
- Yang, S.; Yuan, W.; Yang, W.; Zhou, J.; Harlan, R.; Edwards, J.; Li, S.; Zhang, H. Glycan Analysis by Isobaric Aldehyde Reactive Tags and Mass Spectrometry. Anal. Chem. 2013, 85, 8188–8195. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.; Wang, M.; Chen, L.; Yin, B.; Song, G.; Turko, I.V.; Phinney, K.W.; Betenbaugh, M.J.; Zhang, H.; Li, S. QUANTITY: An Isobaric Tag for Quantitative Glycomics. Sci. Rep. 2015, 5, 17585. [Google Scholar] [CrossRef] [PubMed]
- Zhong, X.; Chen, Z.; Snovida, S.; Liu, Y.; Rogers, J.C.; Li, L. Capillary Electrophoresis-Electrospray Ionization-Mass Spectrometry for Quantitative Analysis of Glycans Labeled with Multiplex Carbonyl-Reactive Tandem Mass Tags. Anal. Chem. 2015, 87, 6527–6534. [Google Scholar] [CrossRef] [PubMed]
- Cai, Y.; Jiao, J.; Bin, Z.; Zhang, Y.; Yang, P.; Lu, H. Glycan reductive isotope-coded amino acid labeling (GRIAL) for mass spectrometry-based quantitative N-glycomics. Chem. Commun. 2015, 51, 772–775. [Google Scholar] [CrossRef] [PubMed]
- Cao, W.; Zhang, W.; Huang, J.; Jiang, B.; Zhang, L.; Yang, P. Glycan reducing end dual isotopic labeling (GREDIL) for mass spectrometry-based quantitative N-glycomics. Chem. Commun. 2015, 51, 13603–13606. [Google Scholar] [CrossRef] [PubMed]
- Chen, B.; Zhong, X.; Feng, Y.; Snovida, S.; Xu, M.; Rogers, J.; Li, L. Targeted MultiNotch MS3 Approach for Relative Quantification of N-Glycans Using Multiplexed Carbonyl-Reactive Isobaric Tags. Anal. Chem. 2018, 90, 1129–1135. [Google Scholar] [CrossRef]
- Feng, Y.; Chen, B.; Yu, Q.; Zhong, X.; Frost, D.C.; Ikonomidou, C.; Li, L. Isobaric Multiplex Labeling Reagents for Carbonyl-Containing Compound (SUGAR) Tags: A Probe for Quantitative Glycomic Analysis. Anal. Chem. 2019, 91, 3141–3146. [Google Scholar] [CrossRef]
- Barrientos, R.C.; Zhang, Q. Isobaric Labeling of Intact Gangliosides toward Multiplexed LC–MS/MS-Based Quantitative Analysis. Anal. Chem. 2018, 90, 2578–2586. [Google Scholar] [CrossRef]
- Zhang, W.; Cao, W.; Huang, J.; Wang, H.; Wang, J.; Xie, C.; Yang, P. PNGase F-mediated incorporation of 18O into glycans for relative glycan quantitation. Analyst 2015, 140, 1082–1089. [Google Scholar] [CrossRef]
- Shi, Q.; Hashimoto, R.; Otsubo, T.; Ikeda, K.; Todoroki, K.; Mizuno, H.; Jin, D.; Toyo’oka, T.; Jiang, Z.; Min, J.Z. A novel, simplified strategy of relative quantification N-glycan: Quantitative glycomics using electrospray ionization mass spectrometry through the stable isotopic labeling by transglycosylation reaction of mutant enzyme Endo-M-N175Q. J. Pharm. Biomed. Anal. 2018, 149, 365–373. [Google Scholar] [CrossRef]
- Echeverria, B.; Etxebarria, J.; Ruiz, N.; Hernandez, Á.; Calvo, J.; Haberger, M.; Reusch, D.; Reichardt, N.-C. Chemo-Enzymatic Synthesis of 13C Labeled Complex N-Glycans As Internal Standards for the Absolute Glycan Quantification by Mass Spectrometry. Anal. Chem. 2015, 87, 11460–11467. [Google Scholar] [CrossRef]
- Calderon, A.D.; Liu, Y.; Li, X.; Wang, X.; Chen, X.; Li, L.; Wang, P.G. Substrate specificity of FUT8 and chemoenzymatic synthesis of core-fucosylated asymmetric N-glycans. Org. Biomol. Chem. 2016, 14, 4027–4031. [Google Scholar] [CrossRef]
- Yang, W.; Ramadan, S.; Orwenyo, J.; Kakeshpour, T.; Diaz, T.; Eken, Y.; Sanda, M.; Jackson, J.E.; Wilson, A.K.; Huang, X. Chemoenzymatic synthesis of glycopeptides bearing rare N-glycan sequences with or without bisecting GlcNAc. Chem. Sci. 2018, 9, 8194–8206. [Google Scholar] [CrossRef]
- Gagarinov, I.A.; Li, T.; Toraño, J.S.; Caval, T.; Srivastava, A.D.; Kruijtzer, J.A.W.; Heck, A.J.R.; Boons, G.-J. Chemoenzymatic Approach for the Preparation of Asymmetric Bi-, Tri-, and Tetra-Antennary N-Glycans from a Common Precursor. J. Am. Chem. Soc. 2017, 139, 1011–1018. [Google Scholar] [CrossRef]
- Lazar, I.M.; Deng, J.; Ikenishi, F.; Lazar, A.C. Exploring the glycoproteomics landscape with advanced MS technologies. ELECTROPHORESIS 2015, 36, 225–237. [Google Scholar] [CrossRef]
- Zhou, Y.; Shan, Y.; Zhang, L.; Zhang, Y. Recent advances in stable isotope labeling based techniques for proteome relative quantification. J. Chromatogr. A 2014, 1365, 1–11. [Google Scholar] [CrossRef]
- Pichler, P.; Köcher, T.; Holzmann, J.; Möhring, T.; Ammerer, G.; Mechtler, K. Improved Precision of iTRAQ and TMT Quantification by an Axial Extraction Field in an Orbitrap HCD Cell. Anal. Chem. 2011, 83, 1469–1474. [Google Scholar] [CrossRef]
- Zhang, S.; Liu, X.; Kang, X.; Sun, C.; Lu, H.; Yang, P.; Liu, Y. iTRAQ plus 18O: A new technique for target glycoprotein analysis. Talanta 2012, 91, 122–127. [Google Scholar] [CrossRef]
- Shah, P.; Wang, X.; Yang, W.; Toghi Eshghi, S.; Sun, S.; Hoti, N.; Chen, L.; Yang, S.; Pasay, J.; Rubin, A.; et al. Integrated Proteomic and Glycoproteomic Analyses of Prostate Cancer Cells Reveal Glycoprotein Alteration in Protein Abundance and Glycosylation. Mol. Cell. Proteom. 2015, 14, 2753. [Google Scholar] [CrossRef]
- Kalxdorf, M.; Gade, S.; Eberl, H.C.; Bantscheff, M. Monitoring Cell-surface N-Glycoproteome Dynamics by Quantitative Proteomics Reveals Mechanistic Insights into Macrophage Differentiation. Mol. Amp Amp Cell. Proteom. 2017, 16, 770. [Google Scholar] [CrossRef]
- Boersema, P.J.; Raijmakers, R.; Lemeer, S.; Mohammed, S.; Heck, A.J.R. Multiplex peptide stable isotope dimethyl labeling for quantitative proteomics. Nat. Protoc. 2009, 4, 484. [Google Scholar] [CrossRef]
- Kovanich, D.; Cappadona, S.; Raijmakers, R.; Mohammed, S.; Scholten, A.; Heck, A.J.R. Applications of stable isotope dimethyl labeling in quantitative proteomics. Anal. Bioanal. Chem. 2012, 404, 991–1009. [Google Scholar] [CrossRef]
- Weng, Y.; Qu, Y.; Jiang, H.; Wu, Q.; Zhang, L.; Yuan, H.; Zhou, Y.; Zhang, X.; Zhang, Y. An integrated sample pretreatment platform for quantitative N-glycoproteome analysis with combination of on-line glycopeptide enrichment, deglycosylation and dimethyl labeling. Anal. Chim. Acta 2014, 833, 1–8. [Google Scholar] [CrossRef]
- Lin, C.-Y.; Ma, Y.-C.; Pai, P.-J.; Her, G.-R. A comparative study of glycoprotein concentration, glycoform profile and glycosylation site occupancy using isotope labeling and electrospray linear ion trap mass spectrometry. Anal. Chim. Acta 2012, 728, 49–56. [Google Scholar] [CrossRef]
- Chen, R.; Wang, F.; Tan, Y.; Sun, Z.; Song, C.; Ye, M.; Wang, H.; Zou, H. Development of a combined chemical and enzymatic approach for the mass spectrometric identification and quantification of aberrant N-glycosylation. J. Proteom. 2012, 75, 1666–1674. [Google Scholar] [CrossRef]
- Sun, Z.; Qin, H.; Wang, F.; Cheng, K.; Dong, M.; Ye, M.; Zou, H. Capture and Dimethyl Labeling of Glycopeptides on Hydrazide Beads for Quantitative Glycoproteomics Analysis. Anal. Chem. 2012, 84, 8452–8456. [Google Scholar] [CrossRef]
- Huang, J.; Qin, H.; Sun, Z.; Huang, G.; Mao, J.; Cheng, K.; Zhang, Z.; Wan, H.; Yao, Y.; Dong, J.; et al. A peptide N-terminal protection strategy for comprehensive glycoproteome analysis using hydrazide chemistry based method. Sci. Rep. 2015, 5, 10164. [Google Scholar] [CrossRef]
- Woo, C.M.; Iavarone, A.T.; Spiciarich, D.R.; Palaniappan, K.K.; Bertozzi, C.R. Isotope-targeted glycoproteomics (IsoTaG): A mass-independent platform for intact N- and O-glycopeptide discovery and analysis. Nat. Methods 2015, 12, 561. [Google Scholar] [CrossRef]
- Spiciarich, D.R.; Nolley, R.; Maund, S.L.; Purcell, S.C.; Herschel, J.; Iavarone, A.T.; Peehl, D.M.; Bertozzi, C.R. Bioorthogonal Labeling of Human Prostate Cancer Tissue Slice Cultures for Glycoproteomics. Angew. Chem. Int. Ed. 2017, 56, 8992–8997. [Google Scholar] [CrossRef]
- Woo, C.M.; Felix, A.; Zhang, L.; Elias, J.E.; Bertozzi, C.R. Isotope-targeted glycoproteomics (IsoTaG) analysis of sialylated N- and O-glycopeptides on an Orbitrap Fusion Tribrid using azido and alkynyl sugars. Anal. Bioanal. Chem. 2017, 409, 579–588. [Google Scholar] [CrossRef]
- Woo, C.M.; Lund, P.J.; Huang, A.C.; Davis, M.M.; Bertozzi, C.R.; Pitteri, S.J. Mapping and Quantification of Over 2000 O-linked Glycopeptides in Activated Human T Cells with Isotope-Targeted Glycoproteomics (Isotag). Mol. Cell. Proteom. 2018, 17, 764. [Google Scholar] [CrossRef]
- Ong, S.-E.; Blagoev, B.; Kratchmarova, I.; Kristensen, D.B.; Steen, H.; Pandey, A.; Mann, M. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol. Cell. Proteom. 2002, 1, 376–386. [Google Scholar] [CrossRef]
- Boonen, K.; De Haes, W.; Van Houtven, J.; Verdonck, R.; Baggerman, G.; Valkenborg, D.; Schoofs, L. Quantitative peptidomics with isotopic and isobaric tags. In Peptidomics; Springer: Berlin, Germany, 2018; pp. 141–159. [Google Scholar]
- Patel, V.J.; Thalassinos, K.; Slade, S.E.; Connolly, J.B.; Crombie, A.; Murrell, J.C.; Scrivens, J.H. A comparison of labeling and label-free mass spectrometry-based proteomics approaches. J. Proteome Res. 2009, 8, 3752–3759. [Google Scholar] [CrossRef]
- Li, Q.; Kailemia, M.J.; Merleev, A.A.; Xu, G.; Serie, D.; Danan, L.M.; Haj, F.G.; Maverakis, E.M.; Lebrilla, C.B. Site-specific glycosylation quantitation of 50 serum glycoproteins enhanced by predictive glycopeptidomics for improved disease biomarker discovery. Anal. Chem. 2019, 91, 5433–5445. [Google Scholar] [CrossRef]
- Hong, Q.; Ruhaak, L.R.; Stroble, C.; Parker, E.; Huang, J.; Maverakis, E.; Lebrilla, C.B. A method for comprehensive glycosite-mapping and direct quantitation of plasma glycoproteins. J. Proteome Res. 2015, 14, 5179–5192. [Google Scholar] [CrossRef]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Q.; Xie, Y.; Wong, M.; Lebrilla, C.B. Characterization of Cell Glycocalyx with Mass Spectrometry Methods. Cells 2019, 8, 882. https://doi.org/10.3390/cells8080882
Li Q, Xie Y, Wong M, Lebrilla CB. Characterization of Cell Glycocalyx with Mass Spectrometry Methods. Cells. 2019; 8(8):882. https://doi.org/10.3390/cells8080882
Chicago/Turabian StyleLi, Qiongyu, Yixuan Xie, Maurice Wong, and Carlito B. Lebrilla. 2019. "Characterization of Cell Glycocalyx with Mass Spectrometry Methods" Cells 8, no. 8: 882. https://doi.org/10.3390/cells8080882
APA StyleLi, Q., Xie, Y., Wong, M., & Lebrilla, C. B. (2019). Characterization of Cell Glycocalyx with Mass Spectrometry Methods. Cells, 8(8), 882. https://doi.org/10.3390/cells8080882




