How Cells Handle DNA Breaks during Mitosis: Detection, Signaling, Repair, and Fate Choice
Abstract
:1. Introduction
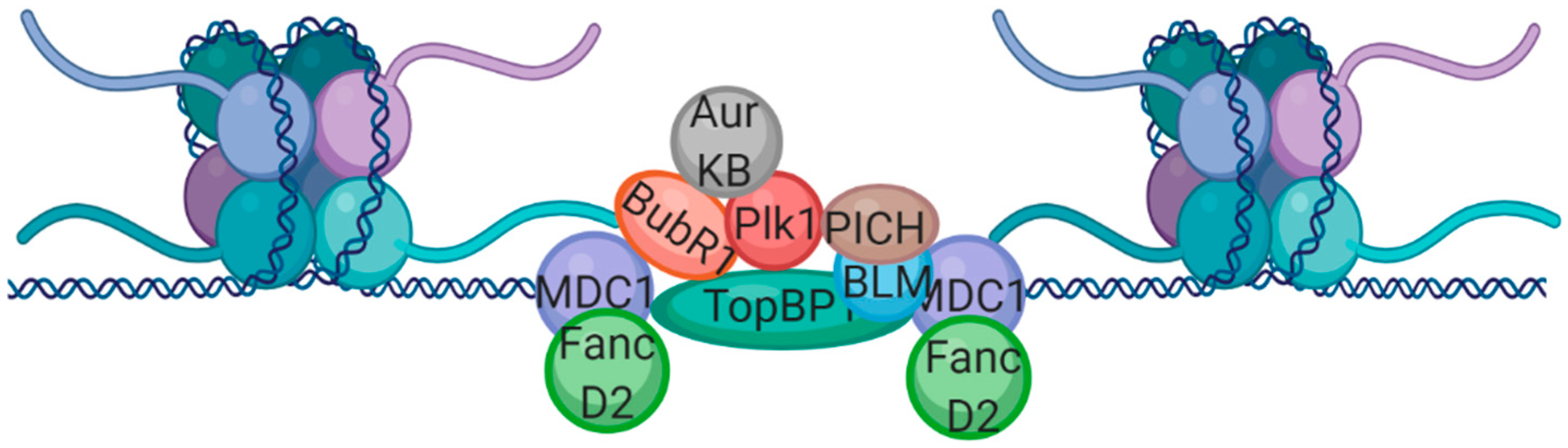
2. DNA Damage Signaling in Mitosis
3. Crosstalk between the SAC and DDR
4. Crosstalk between the SAC and Cell Death
5. The SAC and Senescence
DNA Damage, the SAC, and Inflammation
6. Conclusions
Funding
Conflicts of Interest
References
- Orthwein, A.; Fradet-Turcotte, A.; Noordermeer, S.M.; Canny, M.D.; Brun, C.M.; Strecker, J.; Escribano-Diaz, C.; Durocher, D. Mitosis inhibits DNA double-strand break repair to guard against telomere fusions. Science 2014, 344, 189–193. [Google Scholar] [CrossRef] [PubMed]
- Hustedt, N.; Durocher, D. The control of DNA repair by the cell cycle. Nat. Cell Biol. 2016, 19, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Kim, E.M.; Burke, D.J. DNA damage activates the SAC in an ATM/ATR-dependent manner, independently of the kinetochore. PLoS Genet. 2008, 4, e1000015. [Google Scholar] [CrossRef] [PubMed]
- Pines, J.; Rieder, C.L. Re-staging mitosis: A contemporary view of mitotic progression. Nat. Cell Biol. 2001, 3, E3–E6. [Google Scholar] [CrossRef] [PubMed]
- Mikhailov, A.; Cole, R.W.; Rieder, C.L. DNA damage during mitosis in human cells delays the metaphase/anaphase transition via the spindle-assembly checkpoint. Curr. Biol. 2002, 12, 1797–1806. [Google Scholar] [CrossRef]
- Mankouri, H.W.; Huttner, D.; Hickson, I.D. How unfinished business from S-phase affects mitosis and beyond. EMBO J. 2013, 32, 2661–2671. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Benada, J.; Burdova, K.; Lidak, T.; von Morgen, P.; Macurek, L. Polo-like kinase 1 inhibits DNA damage response during mitosis. Cell Cycle 2015, 14, 219–231. [Google Scholar] [CrossRef] [Green Version]
- Giunta, S.; Belotserkovskaya, R.; Jackson, S.P. DNA damage signaling in response to double-strand breaks during mitosis. J. Cell Biol. 2010, 190, 197–207. [Google Scholar] [CrossRef] [Green Version]
- Yang, S.S.; Yeh, E.; Salmon, E.D.; Bloom, K. Identification of a mid-anaphase checkpoint in budding yeast. J. Cell Biol. 1997, 136, 345–354. [Google Scholar] [CrossRef]
- Tinker-Kulberg, R.L.; Morgan, D.O. Pds1 and Esp1 control both anaphase and mitotic exit in normal cells and after DNA damage. Genes Dev. 1999, 13, 1936–1949. [Google Scholar] [CrossRef] [Green Version]
- Boettcher, B.; Barral, Y. The cell biology of open and closed mitosis. Nucleus 2013, 4, 160–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, N.; Costanzo, V. An ATM and ATR dependent pathway targeting centrosome dependent spindle assembly. Cell Cycle 2009, 8, 1997–2001. [Google Scholar] [CrossRef] [PubMed]
- Eliezer, Y.; Argaman, L.; Kornowski, M.; Roniger, M.; Goldberg, M. Interplay between the DNA damage proteins MDC1 and ATM in the regulation of the spindle assembly checkpoint. J. Biol. Chem. 2014, 289, 8182–8193. [Google Scholar] [CrossRef] [PubMed]
- Smits, V.A.; Klompmaker, R.; Arnaud, L.; Rijksen, G.; Nigg, E.A.; Medema, R.H. Polo-like kinase-1 is a target of the DNA damage checkpoint. Nat. Cell Biol. 2000, 2, 672–676. [Google Scholar] [CrossRef] [PubMed]
- Nitta, M.; Kobayashi, O.; Honda, S.; Hirota, T.; Kuninaka, S.; Marumoto, T.; Ushio, Y.; Saya, H. Spindle checkpoint function is required for mitotic catastrophe induced by DNA-damaging agents. Oncogene 2004, 23, 6548–6558. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, R.; Shah, R.B.; Liu, P.H.; Gupta, Y.K.; Ando, K.; Aggarwal, A.K.; Sidi, S. An Inhibitor of PIDDosome Formation. Mol. Cell 2015, 58, 767–779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nelson, G.; Buhmann, M.; von Zglinicki, T. DNA damage foci in mitosis are devoid of 53BP1. Cell Cycle 2009, 8, 3379–3383. [Google Scholar] [CrossRef] [Green Version]
- van Vugt, M.A.; Gardino, A.K.; Linding, R.; Ostheimer, G.J.; Reinhardt, H.C.; Ong, S.E.; Tan, C.S.; Miao, H.; Keezer, S.M.; Li, J.; et al. A mitotic phosphorylation feedback network connects Cdk1, Plk1, 53BP1, and Chk2 to inactivate the G(2)/M DNA damage checkpoint. PLoS Biol. 2010, 8, e1000287. [Google Scholar] [CrossRef]
- Lok, G.T.; Sy, S.M.; Dong, S.S.; Ching, Y.P.; Tsao, S.W.; Thomson, T.M.; Huen, M.S. Differential regulation of RNF8-mediated Lys48- and Lys63-based poly-ubiquitylation. Nucleic Acids Res. 2012, 40, 196–205. [Google Scholar] [CrossRef]
- Terasawa, M.; Shinohara, A.; Shinohara, M. Canonical non-homologous end joining in mitosis induces genome instability and is suppressed by M-phase-specific phosphorylation of XRCC4. PLoS Genet. 2014, 10, e1004563. [Google Scholar] [CrossRef]
- Mamely, I.; van Vugt, M.A.; Smits, V.A.; Semple, J.I.; Lemmens, B.; Perrakis, A.; Medema, R.H.; Freire, R. Polo-like kinase-1 controls proteasome-dependent degradation of Claspin during checkpoint recovery. Curr. Biol. 2006, 16, 1950–1955. [Google Scholar] [CrossRef] [PubMed]
- Mailand, N.; Bekker-Jensen, S.; Bartek, J.; Lukas, J. Destruction of Claspin by SCFbetaTrCP restrains Chk1 activation and facilitates recovery from genotoxic stress. Mol. Cell 2006, 23, 307–318. [Google Scholar] [CrossRef] [PubMed]
- Peschiaroli, A.; Dorrello, N.V.; Guardavaccaro, D.; Venere, M.; Halazonetis, T.; Sherman, N.E.; Pagano, M. SCFbetaTrCP-mediated degradation of Claspin regulates recovery from the DNA replication checkpoint response. Mol. Cell 2006, 23, 319–329. [Google Scholar] [CrossRef] [PubMed]
- McClintock, B. The Stability of Broken Ends of Chromosomes in Zea Mays. Genetics 1941, 26, 234–282. [Google Scholar] [PubMed]
- Hoffelder, D.R.; Luo, L.; Burke, N.A.; Watkins, S.C.; Gollin, S.M.; Saunders, W.S. Resolution of anaphase bridges in cancer cells. Chromosoma 2004, 112, 389–397. [Google Scholar] [CrossRef] [PubMed]
- Acilan, C.; Potter, D.M.; Saunders, W.S. DNA repair pathways involved in anaphase bridge formation. Genes Chromosomes Cancer 2007, 46, 522–531. [Google Scholar] [CrossRef] [PubMed]
- Coelho, P.A.; Queiroz-Machado, J.; Sunkel, C.E. Condensin-dependent localisation of topoisomerase II to an axial chromosomal structure is required for sister chromatid resolution during mitosis. J. Cell Sci. 2003, 116, 4763–4776. [Google Scholar] [CrossRef] [Green Version]
- Hande, M.P.; Samper, E.; Lansdorp, P.; Blasco, M.A. Telomere length dynamics and chromosomal instability in cells derived from telomerase null mice. J. Cell Biol. 1999, 144, 589–601. [Google Scholar] [CrossRef]
- Leimbacher, P.A.; Jones, S.E.; Shorrocks, A.K.; de Marco Zompit, M.; Day, M.; Blaauwendraad, J.; Bundschuh, D.; Bonham, S.; Fischer, R.; Fink, D.; et al. MDC1 Interacts with TOPBP1 to Maintain Chromosomal Stability during Mitosis. Mol. Cell 2019, 74, 571–583. [Google Scholar] [CrossRef]
- Royou, A.; Gagou, M.E.; Karess, R.; Sullivan, W. BubR1- and Polo-coated DNA tethers facilitate poleward segregation of acentric chromatids. Cell 2010, 140, 235–245. [Google Scholar] [CrossRef]
- Lukas, C.; Savic, V.; Bekker-Jensen, S.; Doil, C.; Neumann, B.; Pedersen, R.S.; Grofte, M.; Chan, K.L.; Hickson, I.D.; Bartek, J.; et al. 53BP1 nuclear bodies form around DNA lesions generated by mitotic transmission of chromosomes under replication stress. Nat. Cell Biol. 2011, 13, 243–253. [Google Scholar] [CrossRef] [PubMed]
- Durkin, S.G.; Glover, T.W. Chromosome fragile sites. Annu. Rev. Genet. 2007, 41, 169–192. [Google Scholar] [CrossRef] [PubMed]
- Baumann, C.; Korner, R.; Hofmann, K.; Nigg, E.A. PICH, a centromere-associated SNF2 family ATPase, is regulated by Plk1 and required for the spindle checkpoint. Cell 2007, 128, 101–114. [Google Scholar] [CrossRef] [PubMed]
- Chan, K.L.; North, P.S.; Hickson, I.D. BLM is required for faithful chromosome segregation and its localization defines a class of ultrafine anaphase bridges. EMBO J. 2007, 26, 3397–3409. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Minocherhomji, S.; Ying, S.; Bjerregaard, V.A.; Bursomanno, S.; Aleliunaite, A.; Wu, W.; Mankouri, H.W.; Shen, H.; Liu, Y.; Hickson, I.D. Replication stress activates DNA repair synthesis in mitosis. Nature 2015, 528, 286–290. [Google Scholar] [CrossRef] [PubMed]
- Pedersen, R.T.; Kruse, T.; Nilsson, J.; Oestergaard, V.H.; Lisby, M. TopBP1 is required at mitosis to reduce transmission of DNA damage to G1 daughter cells. J. Cell Biol. 2015, 210, 565–582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Izumi, H.; Matsumoto, Y.; Ikeuchi, T.; Saya, H.; Kajii, T.; Matsuura, S. BubR1 localizes to centrosomes and suppresses centrosome amplification via regulating Plk1 activity in interphase cells. Oncogene 2009, 28, 2806–2820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttlin, E.L.; Ting, L.; Bruckner, R.J.; Gebreab, F.; Gygi, M.P.; Szpyt, J.; Tam, S.; Zarraga, G.; Colby, G.; Baltier, K.; et al. The BioPlex Network: A Systematic Exploration of the Human Interactome. Cell 2015, 162, 425–440. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Chen, J.; Gong, Z. TopBP1 controls BLM protein level to maintain genome stability. Mol. Cell 2013, 52, 667–678. [Google Scholar] [CrossRef]
- Kumar, R.; Cheok, C.F. RIF1: A novel regulatory factor for DNA replication and DNA damage response signaling. DNA Repair 2014, 15, 54–59. [Google Scholar] [CrossRef] [Green Version]
- Hengeveld, R.C.; de Boer, H.R.; Schoonen, P.M.; de Vries, E.G.; Lens, S.M.; van Vugt, M.A. Rif1 Is Required for Resolution of Ultrafine DNA Bridges in Anaphase to Ensure Genomic Stability. Dev. Cell 2015, 34, 466–474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stewart, G.S.; Wang, B.; Bignell, C.R.; Taylor, A.M.; Elledge, S.J. MDC1 is a mediator of the mammalian DNA damage checkpoint. Nature 2003, 421, 961–966. [Google Scholar] [CrossRef] [PubMed]
- Musacchio, A. The Molecular Biology of Spindle Assembly Checkpoint Signaling Dynamics. Curr. Biol. 2015, 25, R1002–R1018. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tipton, A.R.; Wang, K.; Link, L.; Bellizzi, J.J.; Huang, H.; Yen, T.; Liu, S.T. BUBR1 and closed MAD2 (C-MAD2) interact directly to assemble a functional mitotic checkpoint complex. J. Biol. Chem. 2011, 286, 21173–21179. [Google Scholar] [CrossRef] [PubMed]
- Thornton, B.R.; Toczyski, D.P. Securin and B-cyclin/CDK are the only essential targets of the APC. Nat. Cell Biol. 2003, 5, 1090–1094. [Google Scholar] [CrossRef] [PubMed]
- Smith, E.; Dejsuphong, D.; Balestrini, A.; Hampel, M.; Lenz, C.; Takeda, S.; Vindigni, A.; Costanzo, V. An ATM- and ATR-dependent checkpoint inactivates spindle assembly by targeting CEP63. Nat. Cell Biol. 2009, 11, 278–285. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, C.; Hao, J.; Kong, D.; Cui, X.; Zhang, W.; Wang, H.; Guo, X.; Ma, S.; Liu, X.; Pu, P.; et al. ATM-mediated Mad1 Serine 214 phosphorylation regulates Mad1 dimerization and the spindle assembly checkpoint. Carcinogenesis 2014, 35, 2007–2013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, Y.; Wang, H.T.; Zhai, Y.; Russell, P.; Du, L.L. Mdb1, a fission yeast homolog of human MDC1, modulates DNA damage response and mitotic spindle function. PLoS ONE 2014, 9, e97028. [Google Scholar] [CrossRef] [PubMed]
- Townsend, K.; Mason, H.; Blackford, A.N.; Miller, E.S.; Chapman, J.R.; Sedgwick, G.G.; Barone, G.; Turnell, A.S.; Stewart, G.S. Mediator of DNA damage checkpoint 1 (MDC1) regulates mitotic progression. J. Biol. Chem. 2009, 284, 33939–33948. [Google Scholar] [CrossRef]
- Coster, G.; Hayouka, Z.; Argaman, L.; Strauss, C.; Friedler, A.; Brandeis, M.; Goldberg, M. The DNA damage response mediator MDC1 directly interacts with the anaphase-promoting complex/cyclosome. J. Biol. Chem. 2007, 282, 32053–32064. [Google Scholar] [CrossRef]
- Zachos, G.; Black, E.J.; Walker, M.; Scott, M.T.; Vagnarelli, P.; Earnshaw, W.C.; Gillespie, D.A. Chk1 is required for spindle checkpoint function. Dev. Cell 2007, 12, 247–260. [Google Scholar] [CrossRef] [PubMed]
- Royou, A.; Macias, H.; Sullivan, W. The Drosophila Grp/Chk1 DNA damage checkpoint controls entry into anaphase. Curr. Biol. 2005, 15, 334–339. [Google Scholar] [CrossRef] [PubMed]
- Di Paolo, A.; Racca, C.; Calsou, P.; Larminat, F. Loss of BRCA1 impairs centromeric cohesion and triggers chromosomal instability. FASEB J. 2014, 28, 5250–5261. [Google Scholar] [CrossRef] [PubMed]
- Xu, R.; Xu, Y.; Huo, W.; Lv, Z.; Yuan, J.; Ning, S.; Wang, Q.; Hou, M.; Gao, G.; Ji, J.; et al. Mitosis-specific MRN complex promotes a mitotic signaling cascade to regulate spindle dynamics and chromosome segregation. Proc. Natl. Acad. Sci. USA 2018, 115, E10079–E10088. [Google Scholar] [CrossRef] [PubMed]
- Mc Gee, M.M. Targeting the Mitotic Catastrophe Signaling Pathway in Cancer. Mediat. Inflamm. 2015, 2015, 146282. [Google Scholar] [CrossRef] [PubMed]
- Tinel, A.; Tschopp, J. The PIDDosome, a protein complex implicated in activation of caspase-2 in response to genotoxic stress. Science 2004, 304, 843–846. [Google Scholar] [CrossRef] [PubMed]
- Bouchier-Hayes, L.; Green, D.R. Caspase-2: The orphan caspase. Cell Death Differ. 2012, 19, 51–57. [Google Scholar] [CrossRef] [PubMed]
- Krumschnabel, G.; Manzl, C.; Villunger, A. Caspase-2: Killer, savior and safeguard--emerging versatile roles for an ill-defined caspase. Oncogene 2009, 28, 3093–3096. [Google Scholar] [CrossRef]
- Sidi, S.; Sanda, T.; Kennedy, R.D.; Hagen, A.T.; Jette, C.A.; Hoffmans, R.; Pascual, J.; Imamura, S.; Kishi, S.; Amatruda, J.F.; et al. Chk1 suppresses a caspase-2 apoptotic response to DNA damage that bypasses p53, Bcl-2, and caspase-3. Cell 2008, 133, 864–877. [Google Scholar] [CrossRef]
- Ando, K.; Kernan, J.L.; Liu, P.H.; Sanda, T.; Logette, E.; Tschopp, J.; Look, A.T.; Wang, J.; Bouchier-Hayes, L.; Sidi, S. PIDD death-domain phosphorylation by ATM controls prodeath versus prosurvival PIDDosome signaling. Mol. Cell 2012, 47, 681–693. [Google Scholar] [CrossRef]
- Ando, K.; Parsons, M.J.; Shah, R.B.; Charendoff, C.I.; Paris, S.L.; Liu, P.H.; Fassio, S.R.; Rohrman, B.A.; Thompson, R.; Oberst, A.; et al. NPM1 directs PIDDosome-dependent caspase-2 activation in the nucleolus. J. Cell Biol. 2017, 216, 1795–1810. [Google Scholar] [CrossRef] [PubMed]
- Shah, R.B.; Thompson, R.; Sidi, S. A mitosis-sensing caspase activation platform? New insights into the PIDDosome. Mol. Cell Oncol. 2016, 3, e1059921. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meitinger, F.; Anzola, J.V.; Kaulich, M.; Richardson, A.; Stender, J.D.; Benner, C.; Glass, C.K.; Dowdy, S.F.; Desai, A.; Shiau, A.K.; et al. 53BP1 and USP28 mediate p53 activation and G1 arrest after centrosome loss or extended mitotic duration. J. Cell Biol. 2016, 214, 155–166. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Ruter, D.L.; Kushner, E.J.; Bautch, V.L. Excess centrosomes induce p53-dependent senescence without DNA damage in endothelial cells. FASEB J. 2017, 31, 4295–4304. [Google Scholar] [CrossRef] [PubMed]
- Fava, L.L.; Schuler, F.; Sladky, V.; Haschka, M.D.; Soratroi, C.; Eiterer, L.; Demetz, E.; Weiss, G.; Geley, S.; Nigg, E.A.; et al. The PIDDosome activates p53 in response to supernumerary centrosomes. Genes Dev. 2017, 31, 34–45. [Google Scholar] [CrossRef] [PubMed]
- Vicker, M.G.; Bultmann, H.; Glade, U.; Hafker, T. Ionizing radiation at low doses induces inflammatory reactions in human blood. Radiat. Res. 1991, 128, 251–257. [Google Scholar] [CrossRef] [PubMed]
- Harding, S.M.; Benci, J.L.; Irianto, J.; Discher, D.E.; Minn, A.J.; Greenberg, R.A. Mitotic progression following DNA damage enables pattern recognition within micronuclei. Nature 2017, 548, 466–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackenzie, K.J.; Carroll, P.; Martin, C.A.; Murina, O.; Fluteau, A.; Simpson, D.J.; Olova, N.; Sutcliffe, H.; Rainger, J.K.; Leitch, A.; et al. cGAS surveillance of micronuclei links genome instability to innate immunity. Nature 2017, 548, 461–465. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bakhoum, S.F.; Ngo, B.; Laughney, A.M.; Cavallo, J.A.; Murphy, C.J.; Ly, P.; Shah, P.; Sriram, R.K.; Watkins, T.B.K.; Taunk, N.K.; et al. Chromosomal instability drives metastasis through a cytosolic DNA response. Nature 2018, 553, 467–472. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dou, Z.; Ghosh, K.; Vizioli, M.G.; Zhu, J.; Sen, P.; Wangensteen, K.J.; Simithy, J.; Lan, Y.; Lin, Y.; Zhou, Z.; et al. Cytoplasmic chromatin triggers inflammation in senescence and cancer. Nature 2017, 550, 402–406. [Google Scholar] [CrossRef] [Green Version]
- Gluck, S.; Guey, B.; Gulen, M.F.; Wolter, K.; Kang, T.W.; Schmacke, N.A.; Bridgeman, A.; Rehwinkel, J.; Zender, L.; Ablasser, A. Innate immune sensing of cytosolic chromatin fragments through cGAS promotes senescence. Nat. Cell Biol. 2017, 19, 1061–1070. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Wang, H.; Ren, J.; Chen, Q.; Chen, Z.J. cGAS is essential for cellular senescence. Proc. Natl. Acad. Sci. USA 2017, 114, E4612–E4620. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- White, M.J.; McArthur, K.; Metcalf, D.; Lane, R.M.; Cambier, J.C.; Herold, M.J.; van Delft, M.F.; Bedoui, S.; Lessene, G.; Ritchie, M.E.; et al. Apoptotic caspases suppress mtDNA-induced STING-mediated type I IFN production. Cell 2014, 159, 1549–1562. [Google Scholar] [CrossRef] [PubMed]
- Rongvaux, A.; Jackson, R.; Harman, C.C.; Li, T.; West, A.P.; de Zoete, M.R.; Wu, Y.; Yordy, B.; Lakhani, S.A.; Kuan, C.Y.; et al. Apoptotic caspases prevent the induction of type I interferons by mitochondrial DNA. Cell 2014, 159, 1563–1577. [Google Scholar] [CrossRef] [PubMed]
- Sarhan, J.; Liu, B.C.; Muendlein, H.I.; Weindel, C.G.; Smirnova, I.; Tang, A.Y.; Ilyukha, V.; Sorokin, M.; Buzdin, A.; Fitzgerald, K.A.; et al. Constitutive interferon signaling maintains critical threshold of MLKL expression to license necroptosis. Cell Death Differ. 2019, 26, 332–347. [Google Scholar] [CrossRef]


© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Thompson, R.; Gatenby, R.; Sidi, S. How Cells Handle DNA Breaks during Mitosis: Detection, Signaling, Repair, and Fate Choice. Cells 2019, 8, 1049. https://doi.org/10.3390/cells8091049
Thompson R, Gatenby R, Sidi S. How Cells Handle DNA Breaks during Mitosis: Detection, Signaling, Repair, and Fate Choice. Cells. 2019; 8(9):1049. https://doi.org/10.3390/cells8091049
Chicago/Turabian StyleThompson, Ruth, Rachel Gatenby, and Samuel Sidi. 2019. "How Cells Handle DNA Breaks during Mitosis: Detection, Signaling, Repair, and Fate Choice" Cells 8, no. 9: 1049. https://doi.org/10.3390/cells8091049
APA StyleThompson, R., Gatenby, R., & Sidi, S. (2019). How Cells Handle DNA Breaks during Mitosis: Detection, Signaling, Repair, and Fate Choice. Cells, 8(9), 1049. https://doi.org/10.3390/cells8091049




