Endoglin Protein Interactome Profiling Identifies TRIM21 and Galectin-3 as New Binding Partners
Abstract
:1. Introduction
2. Materials and Methods
2.1. Cell-free Protein Microarrays
2.2. Cell Culture
2.3. Plasmids and Cell Transfections
2.4. Immunoprecipitation Assays
2.5. Mass Spectrometry and Data Analysis
2.6. Western Blot Analysis
2.7. Immunofluorescence Microscopy
2.8. Bio-Layer Interferometry Measurements
2.9. Statistical Analysis
3. Results
3.1. Identification of Human Proteins Interacting with Endoglin Using Cell-Free Protein Microarray Technology
3.2. Galectin-3 Interacts with Endoglin in Cells
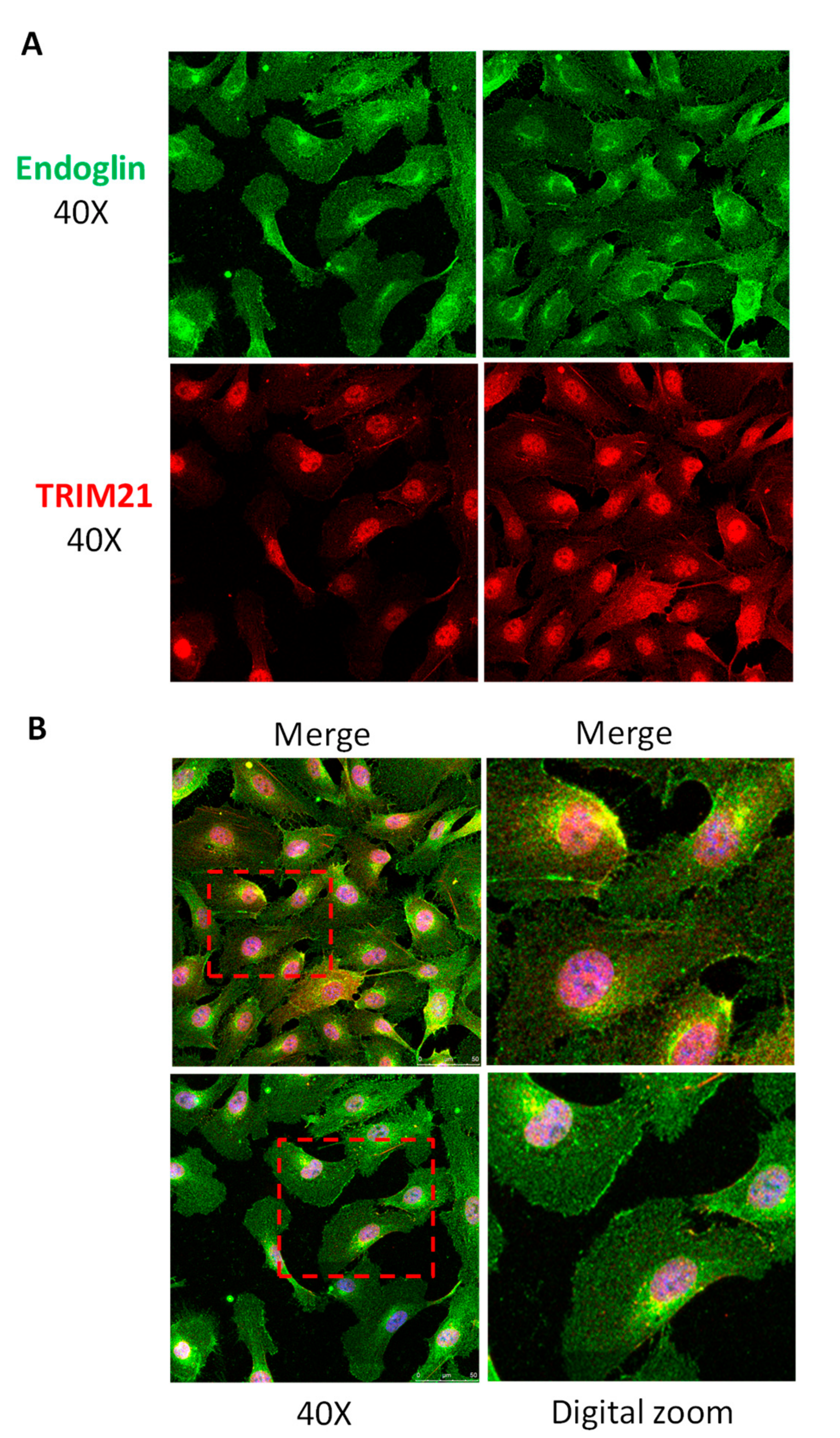
3.3. TRIM21 Interacts with Endoglin in Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Lebrin, F.; Mummery, C.L. Endoglin-mediated vascular remodeling: Mechanisms underlying hereditary hemorrhagic telangiectasia. Trends Cardiovasc. Med. 2008, 18, 25–32. [Google Scholar] [CrossRef] [PubMed]
- López-Novoa, J.M.; Bernabeu, C. The physiological role of endoglin in the cardiovascular system. Am. J. Physiol. Heart Circ. Physiol. 2010. [Google Scholar] [CrossRef]
- Núñez-Gómez, E.; Pericacho, M.; Ollauri-Ibáñez, C.; Bernabéu, C.; López-Novoa, J.M. The role of endoglin in post-ischemic revascularization. Angiogenesis 2017, 20, 1–24. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Llorente, L.; Gallardo-Vara, E.; Rossi, E.; Smadja, D.M.; Botella, L.M.; Bernabeu, C. Endoglin and alk1 as therapeutic targets for hereditary hemorrhagic telangiectasia. Expert Opin. Ther. Targets 2017, 21, 933–947. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- McAllister, K.A.; Grogg, K.M.; Johnson, D.W.; Gallione, C.J.; Baldwin, M.A.; Jackson, C.E.; Helmbold, E.A.; Markel, D.S.; McKinnon, W.C.; Murrell, J.; et al. Endoglin, a TGF-beta binding protein of endothelial cells, is the gene for hereditary haemorrhagic telangiectasia type 1. Nat. Genet. 1994, 8, 345–351. [Google Scholar] [CrossRef] [PubMed]
- Shovlin, C.L. Hereditary haemorrhagic telangiectasia: Pathophysiology, diagnosis and treatment. Blood Rev. 2010, 24, 203–219. [Google Scholar] [CrossRef] [Green Version]
- Tual-Chalot, S.; Oh, S.P.; Arthur, H.M. Mouse models of hereditary hemorrhagic telangiectasia: Recent advances and future challenges. Front. Genet. 2015. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Guo, B.; Bernabeu, C.; Kumar, S. Angiogenesis in breast cancer: The role of transforming growth factor beta and CD105. Microsc. Res. Tech. 2001, 52, 437–449. [Google Scholar] [CrossRef]
- Bernabeu, C.; Lopez-Novoa, J.M.; Quintanilla, M. The emerging role of TGF-beta superfamily coreceptors in cancer. Biochim. Biophys. Acta 2009, 1792, 954–973. [Google Scholar] [CrossRef]
- Fonsatti, E.; Nicolay, H.J.; Altomonte, M.; Covre, A.; Maio, M. Targeting cancer vasculature via endoglin/CD105: A novel antibody-based diagnostic and therapeutic strategy in solid tumours. Cardiovasc. Res. 2010, 86, 12–19. [Google Scholar] [CrossRef]
- Paauwe, M.; Ten Dijke, P.; Hawinkels, L.J. Endoglin for tumor imaging and targeted cancer therapy. Expert Opin. Ther. Targets 2013, 17, 421–435. [Google Scholar] [CrossRef] [PubMed]
- Rosen, L.S.; Gordon, M.S.; Robert, F.; Matei, D.E. Endoglin for targeted cancer treatment. Curr. Oncol. Rep. 2014. [Google Scholar] [CrossRef] [PubMed]
- Wong, V.C.; Chan, P.L.; Bernabeu, C.; Law, S.; Wang, L.D.; Li, J.L.; Tsao, S.W.; Srivastava, G.; Lung, M.L. Identification of an invasion and tumor-suppressing gene, Endoglin (ENG), silenced by both epigenetic inactivation and allelic loss in esophageal squamous cell carcinoma. Int. J. Cancer. 2008, 123, 2816–2823. [Google Scholar] [CrossRef]
- Henry, L.A.; Johnson, D.A.; Sarrió, D.; Lee, S.; Quinlan, P.R.; Crook, T.; Thompson, A.M.; Reis-Filho, J.S.; Isacke, C.M. Endoglin expression in breast tumor cells suppresses invasion and metastasis and correlates with improved clinical outcome. Oncogene 2011, 30, 1046–1058. [Google Scholar] [CrossRef] [PubMed]
- Lakshman, M.; Huang, X.; Ananthanarayanan, V.; Jovanovic, B.; Liu, Y.; Craft, C.S.; Romero, D.; Vary, C.P.; Bergan, R.C. Endoglin suppresses human prostate cancer metastasis. Clin. Exp. Metastasis 2011, 28, 39–53. [Google Scholar] [CrossRef] [PubMed]
- Pérez-Gómez, E.; Villa-Morales, M.; Santos, J.; Fernández-Piqueras, J.; Gamallo, C.; Dotor, J.; Bernabéu, C.; Quintanilla, M. A role for endoglin as a suppressor of malignancy during mouse skin carcinogenesis. Cancer Res. 2007, 67, 10268–10277. [Google Scholar] [CrossRef] [PubMed]
- Pardali, E.; van der Schaft, D.W.; Wiercinska, E.; Gorter, A.; Hogendoorn, P.C.; Griffioen, A.W.; ten Dijke, P. Critical role of endoglin in tumor cell plasticity of Ewing sarcoma and melanoma. Oncogene 2011, 30, 334–345. [Google Scholar] [CrossRef]
- Venkatesha, S.; Toporsian, M.; Lam, C.; Hanai, J.; Mammoto, T.; Kim, Y.M.; Bdolah, Y.; Lim, K.H.; Yuan, H.T.; Libermann, T.A.; et al. Soluble endoglin contributes to the pathogenesis of preeclampsia. Nat. Med. 2006, 12, 642–649. [Google Scholar] [CrossRef]
- Gregory, A.L.; Xu, G.; Sotov, V.; Letarte, M. Review: The enigmatic role of endoglin in the placenta. Placenta 2014. [Google Scholar] [CrossRef]
- Aristorena, M.; Gallardo-Vara , E.; Vicen, M.; de Las Casas-Engel , M.; Ojeda-Fernandez , L.; Nieto, C.; Blanco, F.J.; Valbuena-Diez , A.C.; Botella, L.M.; Nachtigal, P.; et al. MMP-12, secreted by pro-inflammatory macrophages, targets endoglin in human macrophages and endothelial cells. Int. J. Mol. Sci. 2019. [Google Scholar] [CrossRef]
- Oujo, B.; Perez-Barriocanal, F.; Bernabeu, C.; Lopez-Novoa, J.M. Membrane and soluble forms of endoglin in preeclampsia. Curr. Mol. Med. 2013, 13, 1345–1357. [Google Scholar] [CrossRef] [PubMed]
- Hawinkels, L.J.; Kuiper, P.; Wiercinska, E.; Verspaget, H.W.; Liu, Z.; Pardali, E.; Sier, C.F.; ten Dijke, P. Matrix metalloproteinase-14 (MT1-MMP)-mediated endoglin shedding inhibits tumor angiogenesis. Cancer Res. 2010, 70, 4141–4150. [Google Scholar] [CrossRef]
- Valbuena-Diez, A.C.; Blanco, F.J.; Oujo, B.; Langa, C.; Gonzalez-Nuñez, M.; Llano, E.; Pendas, A.M.; Díaz, M.; Castrillo, A.; Lopez-Novoa, J.M.; et al. Oxysterol-induced soluble endoglin release and its involvement in hypertension. Circulation 2012, 126, 2612–2624. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Sanz-Rodriguez, F.; Eleno, N.; Düwell, A.; Blanco, F.J.; Langa, C.; Botella, L.M.; Cabañas, C.; Lopez-Novoa, J.M.; Bernabeu, C. Endothelial endoglin is involved in inflammation: Role in leukocyte adhesion and transmigration. Blood 2013, 121, 403–415. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Smadja, D.M.; Boscolo, E.; Langa, C.; Arevalo, M.A.; Pericacho, M.; Gamella-Pozuelo, L.; Kauskot, A.; Botella, L.M.; Gaussem, P.; et al. Endoglin regulates mural cell adhesion in the circulatory system. Cell. Mol. Life Sci. 2016, 73, 1715–1739. [Google Scholar] [CrossRef] [PubMed]
- Vitverova, B.; Blazickova, K.; Najmanova, I.; Vicen, M.; Hyšpler, R.; Dolezelova, E.; Nemeckova, I.; Tebbens, J.D.; Bernabeu, C.; Pericacho, M.; et al. Soluble endoglin and hypercholesterolemia aggravate endothelial and vessel wall dysfunction in mouse aorta. Atherosclerosis 2018, 271, 15–25. [Google Scholar] [CrossRef] [PubMed]
- Vicen, M.; Vitverova, B.; Havelek, R.; Blazickova, K.; Machacek, M.; Rathouska, J.; Najmanová, I.; Dolezelova, E.; Prasnicka, A.; Sternak, M.; et al. Regulation and role of endoglin in cholesterol-induced endothelial and vascular dysfunction in vivo and in vitro. FASEB J. 2019, 33, 6099–6114. [Google Scholar] [CrossRef]
- Gougos, A.; Letarte, M. Primary structure of endoglin, an RGD-containing glycoprotein of human endothelial cells. J. Biol. Chem. 1990, 265, 8361–8364. [Google Scholar]
- Bernabeu, C.; Conley, B.A.; Vary, C.P. Novel biochemical pathways of endoglin in vascular cell physiology. J. Cell. Biochem. 2007, 102, 1375–1388. [Google Scholar] [CrossRef] [Green Version]
- Llorca, O.; Trujillo, A.; Blanco, F.J.; Bernabeu, C. Structural model of human endoglin, a transmembrane receptor responsible for hereditary hemorrhagic telangiectasia. J. Mol. Biol. 2007, 365, 694–705. [Google Scholar] [CrossRef]
- Saito, T.; Bokhove, M.; Croci, R.; Zamora-Caballero, S.; Han, L.; Letarte, M.; de Sanctis, D.; Jovine, L. Structural Basis of the Human Endoglin-BMP9 Interaction: Insights into BMP Signaling and HHT1. Cell Rep. 2017, 19, 1917–1928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bokhove, M.; Jovine, L. Structure of Zona Pellucida Module Proteins. Curr. Top. Dev. Biol. 2018, 130, 413–442. [Google Scholar] [CrossRef] [PubMed]
- Rossi, E.; Pericacho, M.; Bachelot-Loza, C.; Pidard, D.; Gaussem, P.; Poirault-Chassac, S.; Blanco, F.J.; Langa, C.; González-Manchón, C.; Lopez-Novoa, J.M.; et al. Human endoglin as a potential new partner involved in platelet-endothelium interactions. Cell. Mol. Life Sci. 2018, 75, 1269–1284. [Google Scholar] [CrossRef] [PubMed]
- Castonguay, R.; Werner, E.D.; Matthews, R.G.; Presman, E.; Mulivor, A.W.; Solban, N.; Sako, D.; Pearsall, R.S.; Underwood, K.W.; Seehra, J.; et al. Soluble endoglin specifically binds bone morphogenetic proteins 9 and 10 via its orphan domain, inhibits blood vessel formation, and suppresses tumor growth. J. Biol. Chem. 2011, 286, 30034–30046. [Google Scholar] [CrossRef] [PubMed]
- Alt, A.; Miguel-Romero, L.; Donderis, J.; Aristorena, M.; Blanco, F.J.; Round, A.; Rubio, V.; Bernabeu, C.; Marina, A. Structural and functional insights into endoglin ligand recognition and binding. PLoS ONE 2012. [Google Scholar] [CrossRef] [PubMed]
- Cheifetz, S.; Bellón, T.; Calés, C.; Vera, S.; Bernabeu, C.; Massagué, J.; Letarte, M. Endoglin is a component of the transforming growth factor-beta receptor system in human endothelial cells. J. Biol. Chem. 1992, 267, 19027–19030. [Google Scholar] [PubMed]
- Bellón, T.; Corbí, A.; Lastres, P.; Calés, C.; Cebrián, M.; Vera, S.; Cheifetz, S.; Massague, S.; Letarte, J.; Bernabéu, C. Identification and expression of two forms of the human transforming growth factor-beta-binding protein endoglin with distinct cytoplasmic regions. Eur. J. Immunol. 1993, 23, 2340–2345. [Google Scholar] [CrossRef] [PubMed]
- Lastres, P.; Letamendía, A.; Zhang, H.; Rius, C.; Almendro, N.; Raab, U.; López, L.A.; Langa, C.; Fabra, A.; Letarte, M.; et al. Endoglin modulates cellular responses to TGF-beta 1. J. Cell Biol. 1996, 133, 1109–1121. [Google Scholar] [CrossRef] [PubMed]
- Mahmoud, M.; Upton, P.D.; Arthur, H.M. Angiogenesis regulation by TGFβ signalling: Clues from an inherited vascular disease. Biochem. Soc. Trans. 2011, 39, 1659–1666. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.J.; Santibanez, J.F.; Guerrero-Esteo, M.; Langa, C.; Vary, C.P.; Bernabeu, C. Interaction and functional interplay between endoglin and ALK-1, two components of the endothelial transforming growth factor-beta receptor complex. J. Cell. Physiol. 2005, 204, 574–584. [Google Scholar] [CrossRef] [PubMed]
- Guerrero-Esteo, M.; Sanchez-Elsner, T.; Letamendia, A.; Bernabeu, C. Extracellular and cytoplasmic domains of endoglin interact with the transforming growth factor-beta receptors I and II. J. Biol. Chem. 2002, 277, 29197–29209. [Google Scholar] [CrossRef] [PubMed]
- Gallardo-Vara, E.; Blanco, F.J.; Roqué, M.; Friedman, S.L.; Suzuki, T.; Botella, L.M.; Bernabeu, C. Transcription factor KLF6 upregulates expression of metalloprotease MMP14 and subsequent release of soluble endoglin during vascular injury. Angiogenesis 2016, 19, 155–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ermini, L.; Ausman, J.; Melland-Smith, M.; Yeganeh, B.; Rolfo, A.; Litvack, M.L.; Todros, T.; Letarte, M.; Post, M.; Caniggia, I. A Single Sphingomyelin Species Promotes Exosomal Release of Endoglin into the Maternal Circulation in Preeclampsia. Sci. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- del Castillo, G.; Sánchez-Blanco, E.; Martín-Villar, E.; Valbuena-Diez, A.C.; Langa, C.; Pérez-Gómez, E.; Renart, J.; Bernabéu, C.; Quintanilla, M. Soluble endoglin antagonizes Met signaling in spindle carcinoma cells. Carcinogenesis 2015, 36, 212–222. [Google Scholar] [CrossRef] [PubMed]
- Díez, P.; González-González, M.; Lourido, L.; Dégano, R.M.; Ibarrola, N.; Casado-Vela, J.; LaBaer, J.; Fuentes, M. NAPPA as a Real New Method for Protein Microarray Generation. Microarrays 2015, 4, 214–227. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Díez, P.; Dasilva, N.; González-González, M.; Matarraz, S.; Casado-Vela, J.; Orfao, A.; Fuentes, M. Data Analysis Strategies for Protein Microarrays. Microarrays 2012, 1, 64–83. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Das, A.; Dinh, P.X.; Pattnaik, A.K. Trim21 regulates Nmi-IFI35 complex-mediated inhibition of innate antiviral response. Virology 2015, 485, 383–392. [Google Scholar] [CrossRef] [Green Version]
- Calvier, L.; Miana, M.; Reboul, P.; Cachofeiro, V.; Martinez-Martinez, E.; de Boer, R.A.; Poirier, F.; Lacolley, P.; Zannad, F.; Rossignol, P.; et al. Galectin-3 mediates aldosterone-induced vascular fibrosis. Arterioscler. Thromb. Vasc. Biol. 2013, 33, 67–75. [Google Scholar] [CrossRef]
- Casado-Vela, J.; Ruiz, E.J.; Nebreda, A.R.; Casal, J.I. A combination of neutral loss and targeted product ion scanning with two enzymatic digestions facilitates the comprehensive mapping of phosphorylation sites. Proteomics 2007, 7, 2522–2529. [Google Scholar] [CrossRef]
- Cristobo, I.; Larriba, M.J.; de los Ríos, V.; García, F.; Muñoz, A.; Casal, J.I. Proteomic analysis of 1α, 25-Dihydroxyvitamin D3 action on human colon cancer cells reveals a link to splicing regulation. J. Proteomics 2011, 75, 384–397. [Google Scholar] [CrossRef]
- Käll, L.; Canterbury, J.D.; Weston, J.; Noble, W.S.; MacCoss, M.J. Semi-supervised learning for peptide identification from shotgun proteomics datasets. Nat. Methods 2007, 4, 923–925. [Google Scholar] [CrossRef] [PubMed]
- Sciacchitano, S.; Lavra, L.; Morgante, A.; Ulivieri, A.; Magi, F.; de Francesco, G.P.; Bellotti, C.; Salehi, L.B.; Ricci, A. Galectin-3: One Molecule for an Alphabet of Diseases, from A to Z. Int. J. Mol. Sci. 2018. [Google Scholar] [CrossRef] [PubMed]
- Abdiche, Y.; Malashock, D.; Pinkerton, A.; Pons, J. Determining kinetics and affinities of protein interactions using a parallel real-time label-free biosensor, the Octet. Anal. Biochem. 2008, 377, 209–217. [Google Scholar] [CrossRef] [PubMed]
- Do, T.; Ho, F.; Heidecker, B.; Witte, K.; Chang, L.; Lerner, L. A rapid method for determining dynamic binding capacity of resins for the purification of proteins. Protein Expr. Purif. 2008, 60, 147–150. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J. The Role of Tripartite Motif Family Proteins in TGF-β Signaling Pathway and Cancer. J. Cancer Prev. 2018, 23, 162–169. [Google Scholar] [CrossRef] [PubMed]
- Fletcher, A.J.; Mallery, D.L.; Watkinson, R.E.; Dickson, C.F.; James, L.C. Sequential ubiquitination and deubiquitination enzymes synchronize the dual sensor and effector functions of TRIM21. Proc. Natl. Acad. Sci. USA 2015, 112, 10014–10019. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anandapadamanaban, M.; Kyriakidis, N.C.; Csizmók, V.; Wallenhammar, A.; Espinosa, A.C.; Ahlner, A.; Round, A.R.; Trewhella, J.; Moche, M.; Wahren-Herlenius, M.; et al. E3 ubiquitin-protein ligase TRIM21-mediated lysine capture by UBE2E1 reveals substrate-targeting mode of a ubiquitin-conjugating E2. J. Biol. Chem. 2019, 294, 11404–11419. [Google Scholar] [CrossRef] [PubMed]
- Jin, Y.; Zhang, Y.; Li, B.; Zhang, J.; Dong, Z.; Hu, X.; Wan, Y. TRIM21 mediates ubiquitination of Snail and modulates epithelial to mesenchymal transition in breast cancer cells. Int. J. Biol. Macromol. 2019, 124, 846–853. [Google Scholar] [CrossRef]
- Nguyen, J.Q.; Irby, R.B. TRIM21 is a novel regulator of Par-4 in colon and pancreatic cancer cells. Cancer Biol. Ther. 2017, 18, 16–25. [Google Scholar] [CrossRef]
- Zhou, W.; Zhang, Y.; Zhong, C.; Hu, J.; Hu, H.; Zhou, D.; Cao, M. Decreased expression of TRIM21 indicates unfavorable outcome and promotes cell growth in breast cancer. Cancer Manag. Res. 2018, 10, 3687–3696. [Google Scholar] [CrossRef]
- Ahn, Y.; Hwang, J.H.; Zheng, Z.; Bang, D.; Kim, D.Y. Enhancement of Th1/Th17 inflammation by TRIM21 in Behçet’s disease. Sci. Rep. 2017. [Google Scholar] [CrossRef] [PubMed]
- Vinter, H.; Langkilde, A.; Ottosson, V.; Espinosa, A.; Wahren-Herlenius, M.; Raaby, L.; Johansen, C.; Iversen, L. TRIM21 is important in the early phase of inflammation in the imiquimod-induced psoriasis-like skin inflammation mouse model. Exp. Dermatol. 2017, 26, 713–720. [Google Scholar] [CrossRef] [PubMed]
- Gómez-Martín, D.; Galindo-Feria, A.S.; Barrera-Vargas, A.; Merayo-Chalico, J.; Juárez-Vega, G.; Torres-Ruiz, J.; Alcocer-Varela, J. Ro52/TRIM21-deficient expression and function in different subsets of peripheral blood mononuclear cells is associated with a proinflammatory cytokine response in patients with idiopathic inflammatory myopathies. Clin. Exp. Immunol. 2017, 188, 154–162. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhou, G.; Wu, W.; Yu, L.; Yu, T.; Yang, W.; Wang, P.; Zhang, X.; Cong, Y.; Liu, Z. Tripartite motif-containing (TRIM) 21 negatively regulates intestinal mucosal inflammation through inhibiting TH1/TH17 cell differentiation in patients with inflammatory bowel diseases. J. Allergy Clin. Immunol. 2018, 142, 1218–1228. [Google Scholar] [CrossRef]
- Foss, S.; Watkinson, R.; Sandlie, I.; James, L.C.; Andersen, J.T. TRIM21: A cytosolic Fc receptor with broad antibody isotype specificity. Immunol. Rev. 2015, 268, 328–339. [Google Scholar] [CrossRef] [PubMed]
- Rhodes, D.A.; Isenberg, D.A. TRIM21 and the Function of Antibodies inside Cells. Trends Immunol. 2017, 38, 916–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Oke, V.; Wahren-Herlenius, M. The immunobiology of Ro52 (TRIM21) in autoimmunity: A critical review. J. Autoimmun. 2012, 39, 77–82. [Google Scholar] [CrossRef]
- Sanz-Rodriguez, F.; Guerrero-Esteo, M.; Botella, L.M.; Banville, D.; Vary, C.P.; Bernabéu, C. Endoglin regulates cytoskeletal organization through binding to ZRP-1, a member of the Lim family of proteins. J. Biol. Chem. 2004, 279, 32858–32868. [Google Scholar] [CrossRef]
- Conley, B.A.; Koleva, R.; Smith, J.D.; Kacer, D.; Zhang, D.; Bernabéu, C.; Vary, C.P. Endoglin controls cell migration and composition of focal adhesions: Function of the cytosolic domain. J. Biol. Chem. 2004, 279, 27440–27449. [Google Scholar] [CrossRef]
- Meng, Q.; Lux, A.; Holloschi, A.; Li, J.; Hughes, J.M.; Foerg, T.; McCarthy, J.E.; Heagerty, A.M.; Kioschis, P.; Hafner, M.; et al. Identification of Tctex2beta, a novel dynein light chain family member that interacts with different transforming growth factor-beta receptors. J. Biol. Chem. 2006, 281, 37069–37080. [Google Scholar] [CrossRef]
- Xu, G.; Barrios-Rodiles, M.; Jerkic, M.; Turinsky, A.L.; Nadon, R.; Vera, S.; Voulgaraki, D.; Wrana, J.L.; Toporsian, M.; Letarte, M. Novel protein interactions with endoglin and activin receptor-like kinase 1: Potential role in vascular networks. Mol. Cell. Proteom. 2014, 13, 489–502. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, -L.A.; Garrido-Martin, E.M.; Sanz-Rodriguez, F.; Pericacho, M.; Rodriguez-Barbero, A.; Eleno, N.; Lopez-Novoa, J.M.; Düwell, A.; Vega, M.A.; Bernabeu, C.; et al. Gene expression fingerprinting for human hereditary hemorrhagic telangiectasia. Hum. Mol. Genet. 2007, 16, 1515–1533. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, B.; Eyries, M.; Montagne, K.; Martin, S.; Agrapart, M.; Simerman-François, R.; Letarte, M.; Soubrier, F. Altered endothelial gene expression associated with hereditary haemorrhagic telangiectasia. Eur. J. Clin. Investig. 2007, 37, 580–588. [Google Scholar] [CrossRef] [PubMed]
- Blanco, F.J.; Ojeda-Fernandez, L.; Aristorena, M.; Gallardo-Vara, E.; Benguria, A.; Dopazo, A.; Langa, C.; Botella, L.M.; Bernabeu, C. Genome-wide transcriptional and functional analysis of endoglin isoforms in the human promonocytic cell line U937. J. Cell. Physiol. 2015, 230, 947–958. [Google Scholar] [CrossRef] [PubMed]
- Fernandez-García, C.E.; Tarin, C.; Roldan-Montero, R.; Martinez-Lopez, D.; Torres-Fonseca, M.; Lindhot, J.S.; Vega de Ceniga, M.; Egido, J.; Lopez-Andres, N.; Blanco-Colio, L.M.; et al. Increased galectin-3 levels are associated with abdominal aortic aneurysm progression and inhibition of galectin-3 decreases elastase-induced AAA development. Clin. Sci. 2017, 131, 2707–2719. [Google Scholar] [CrossRef] [PubMed]
- Arrieta, V.; Martinez-Martinez, E.; Ibarrola, J.; Alvarez, V.; Sádaba, R.; Garcia-Peña, A.; Fernández-Celis, A.; Cachofeiro, V.; Rossignol, P.; López-Andrés, N. A role for galectin-3 in the development of early molecular alterations in short-term aortic stenosis. Clin. Sci. 2017, 131, 935–949. [Google Scholar] [CrossRef] [PubMed]
- Dos Santos, S.N.; Sheldon, H.; Pereira, J.X.; Paluch, C.; Bridges, E.M.; El-Cheikh, M.C.; Harris, A.L.; Bernardes, E.S. Galectin-3 acts as an angiogenic switch to induce tumor angiogenesis via Jagged-1/Notch activation. Oncotarget 2017, 8, 49484–49501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Barman, S.A.; Chen, F.; Li, X.; Haigh, S.; Stepp, D.W.; Kondrikov, D.; Mahboubi, K.; Bordan, Z.; Traber, P.; Su, Y.; et al. Galectin-3 Promotes Vascular Remodeling and Contributes to Pulmonary Hypertension. Am. J. Respir. Crit. Care Med. 2018, 197, 1488–1492. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Lin, J.; Hu, T.; Ren, Z.; Li, L.; Hameed, I.; Zhang, X.; Men, C.; Guo, Y.; Xu, D.; et al. Galectin-3 exacerbates ox-LDL-mediated endothelial injury by inducing inflammation via integrin β1-RhoA-JNK signaling activation. J. Cell. Physiol. 2019, 234, 10990–11000. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Martínez, E.; Brugnolaro, C.; Ibarrola, J.; Ravassa, S.; Buonafine, M.; López, B.; Fernández-Celis, A.; Querejeta, R.; Santamaria, E.; Fernández-Irigoyen, J.; et al. CT-1 (Cardiotrophin-1)-Gal-3 (Galectin-3) Axis in Cardiac Fibrosis and Inflammation. Hypertension 2019, 73, 602–611. [Google Scholar] [CrossRef]
- Pérez, L.; Vallejos, A.; Echeverria, C.; Varela, D.; Cabello-Verrugio, C.; Simon, F. OxHDL controls LOX-1 expression and plasma membrane localization through a mechanism dependent on NOX/ROS/NF-κB pathway on endothelial cells. Lab. Investig. 2019, 99, 421–437. [Google Scholar] [CrossRef] [PubMed]
- Jeschke, U.; Mayr, D.; Schiessl, B.; Mylonas, I.; Schulze, S.; Kuhn, C.; Friese, K.; Walzel, H. Expression of galectin-1, -3 (gal-1, gal-3) and the Thomsen-Friedenreich (TF) antigen in normal, IUGR, preeclamptic and HELLP placentas. Placenta 2007, 28, 1165–1173. [Google Scholar] [CrossRef] [PubMed]
- Nangia-Makker, P.; Honjo, Y.; Sarvis, R.; Akahani, S.; Hogan, V.; Pienta, K.J.; Raz, A. Galectin-3 induces endothelial cell morphogenesis and angiogenesis. Am. J. Pathol. 2000, 156, 899–909. [Google Scholar] [CrossRef]
- Markowska, A.I.; Liu, F.T.; Panjwani, N. Galectin-3 is an important mediator of VEGF- and bFGF-mediated angiogenic response. J. Exp. Med. 2010, 207, 1981–1993. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Griffioen, A.W.; Thijssen, V.L. Galectins in tumor angiogenesis. Ann. Transl. Med. 2014, 2, 90. [Google Scholar] [CrossRef]
- Gallardo-Vara, E.; Tual-Chalot, S.; Botella, L.M.; Arthur, H.M.; Bernabeu, C. Soluble endoglin regulates expression of angiogenesis-related proteins and induction of arteriovenous malformations in a mouse model of hereditary hemorrhagic telangiectasia. Dis. Model. Mech. 2018. [Google Scholar] [CrossRef]
- Colomb, F.; Wang, W.; Simpson, D.; Zafar, M.; Beynon, R.; Rhodes, J.M.; Yu, L.G. Galectin-3 interacts with the cell-surface glycoprotein CD146 (MCAM, MUC18) and induces secretion of metastasis-promoting cytokines from vascular endothelial cells. J. Biol. Chem. 2017, 292, 8381–8389. [Google Scholar] [CrossRef] [Green Version]
- Ruvolo, P.P. Galectin 3 as a guardian of the tumor microenvironment. Biochim. Biophys. Acta 2016, 1863, 427–437. [Google Scholar] [CrossRef]
- Pérez-Gómez, E.; Del Castillo, G.; Santibanez, J.F.; López-Novoa, J.M.; Bernabéu, C.; Quintanilla, M. The role of the TGF-β coreceptor endoglin in cancer. Sci. World J. 2010, 10, 2367–2384. [Google Scholar] [CrossRef]
- Clift, D.; So, C.; McEwan, W.A.; James, L.C.; Schuh, M. Acute and rapid degradation of endogenous proteins by Trim-Away. Nat. Protoc. 2018, 13, 2149–2175. [Google Scholar] [CrossRef]
- Iyengar, P.V. Regulation of Ubiquitin Enzymes in the TGF-β Pathway. Int. J. Mol. Sci. 2017. [Google Scholar] [CrossRef] [PubMed]
- Paquet, M.E.; Pece-Barbara, N.; Vera, S.; Cymerman, U.; Karabegovic, A.; Shovlin, C.; Letarte, M. Analysis of several endoglin mutants reveals no endogenous mature or secreted protein capable of interfering with normal endoglin function. Hum. Mol. Genet. 2001, 10, 1347–1357. [Google Scholar] [CrossRef]
- Wang, H.; Zhou, Y.; Oyang, L.; Han, Y.; Xia, L.; Lin, J.; Tang, Y.; Su, M.; Tan, S.; Tian, Y.; et al. LPLUNC1 stabilises PHB1 by counteracting TRIM21-mediated ubiquitination to inhibit NF-κB activity in nasopharyngeal carcinoma. Oncogene 2019, 38, 5062–5075. [Google Scholar] [CrossRef]
- Hu, J.; Guan, W.; Yan, L.; Ye, Z.; Wu, L.; Xu, H. Cancer Stem Cell Marker Endoglin (CD105) Induces Epithelial Mesenchymal Transition (EMT) but Not Metastasis in Clear Cell Renal Cell Carcinoma. Stem Cells Int. 2019. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Li, Y.M.; Zeng, X.X.; Wang, X.Y.; Chen, S.K.; Gui, L.X.; Lin, M.J. Galectin-3- Mediated Transdifferentiation of Pulmonary Artery Endothelial Cells Contributes to Hypoxic Pulmonary Vascular Remodeling. Cell. Physiol. Biochem. 2018, 51, 763–777. [Google Scholar] [CrossRef] [PubMed]
- Li, L.; Wei, J.; Mallampalli, R.K.; Zhao, Y.; Zhao, J. TRIM21 mitigates human lung microvascular endothelial cells inflammatory responses to lipopolysaccharide. Am. J. Respir. Cell Mol. Biol. 2019. [Epub ahead of print]. [Google Scholar] [CrossRef]
- Natarajan, V. Mind the Gap between the Endothelium and E3 Ubiquitin Ligase: TRIM21 is a Viable Therapeutic Target in Sepsis-Induced Endothelial Dysfunction. Am. J. Respir. Cell Mol. Biol. 2019. [Epub ahead of print]. [Google Scholar] [CrossRef]
- Torsney, E.; Charlton, R.; Parums, D.; Collis, M.; Arthur, H.M. Inducible expression of human endoglin during inflammation and wound healing in vivo. Inflamm. Res. 2002, 51, 464–470. [Google Scholar] [CrossRef]
- Van Laake, L.W.; van den Driesche, S.; Post, S.; Feijen, A.; Jansen, M.A.; Driessens, M.H.; Mager, J.J.; Snijder, R.J.; Westermann, C.J.; Doevendans, P.A.; et al. Endoglin has a crucial role in blood cell-mediated vascular repair. Circulation 2006, 114, 2288–2297. [Google Scholar] [CrossRef]
- Faiotto, V.B.; Franci, D.; Enz Hubert, R.M.; de Souza, G.R.; Fiusa, M.M.L.; Hounkpe, B.W.; Santos, T.M.; Carvalho-Filho, M.A.; De Paula, E.V. Circulating levels of the angiogenesis mediators endoglin, HB-EGF, BMP-9 and FGF-2 in patients with severe sepsis and septic shock. J. Crit. Care. 2017, 42, 162–167. [Google Scholar] [CrossRef]
- Perez-Riverol, Y.; Csordas, A.; Bai, J.; Bernal-Llinares, M.; Hewapathirana, S.; Kundu, D.J.; Inuganti, A.; Griss, J.; Mayer, G.; Eisenacher, M.; et al. The PRIDE database and related tools and resources in 2019: improving support for quantification data. Nucleic Acids Res. 2019, 47, D442–D450. [Google Scholar] [CrossRef] [PubMed]



| Accession # | Protein Name | Cellular Compartment |
|---|---|---|
| NM_172160.1 | Potassium voltage-gated channel, shaker-related subfamily, beta member 1 (KCNAB1), transcript variant 1 | Plasma membrane |
| Q14722 | ||
| NM_138565.1 | Cortactin (CTTN), transcript variant 2 | Plasma membrane |
| Q14247 | ||
| BC036123.1 | Stromal membrane-associated protein 1 (SMAP1) | Plasma membrane |
| Q8IYB5 | ||
| NM_173822.1 | Family with sequence similarity 126, member B (FAM126B) | Plasma membrane, cytosol |
| Q8IXS8 | ||
| BC047536.1 | Sciellin (SCEL) | Plasma membrane, extracellular or secreted |
| O95171 | ||
| BC068068.1 | Galectin-3 | Plasma membrane, mitochondrion, nucleus, extracellular or secreted |
| P17931 | ||
| BC001247.1 | Actin-binding LIM protein 1 (ABLIM1) | Cytoskeleton |
| O14639 | ||
| NM_198943.1 | Family with sequence similarity 39, member B (FAM39B) | Endosome, cytoskeleton |
| Q6VEQ5 | ||
| NM_005898.4 | Cell cycle associated protein 1 (CAPRIN1), transcript variant 1 | Cytosol |
| Q14444 | ||
| BC002559.1 | YTH domain family, member 2 (YTHDF2) | Nucleus, cytosol |
| Q9Y5A9 | ||
| NM_003141.2 | Tripartite motif-containing 21 (TRIM21) | Nucleus, cytosol |
| P19474 | ||
| BC025279.1 | Scaffold attachment factor B2 (SAFB2) | Nucleus |
| Q14151 | ||
| BC031650.1 | Putative E3 ubiquitin-protein ligase SH3RF2 | Nucleus |
| Q8TEC5 | ||
| BC034488.2 | ATP-binding cassette, sub-family F (GCN20), member 1 (ABCF1) | Nucleus |
| Q8NE71 | ||
| BC040946.1 | Spliceosome-associated protein CWC15 homolog (HSPC148) | Nucleus |
| Q9P013 | ||
| NM_003609.2 | HIRA interacting protein 3 (HIRIP3) | Nucleus |
| Q9BW71 | ||
| NM_005572.1 | Lamin A/C (LMNA), transcript variant 2 | Nucleus |
| P02545 | ||
| NM_006479.2 | RAD51 associated protein 1 (RAD51AP1) | Nucleus |
| Q96B01 | ||
| NM_014321.2 | Origin recognition complex, subunit 6 like (yeast) (ORC6L) | Nucleus |
| Q9Y5N6 | ||
| NM_015138.2 | RNA polymerase-associated protein RTF1 homolog (RTF1) | Nucleus |
| Q92541 | ||
| NM_032141.1 | Coiled-coil domain containing 55 (CCDC55), transcript variant 1 | Nucleus |
| Q9H0G5 | ||
| BC012289.1 | Protein PRRC2B, KIAA0515 | Data not available |
| Q5JSZ5 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gallardo-Vara, E.; Ruiz-Llorente, L.; Casado-Vela, J.; Ruiz-Rodríguez, M.J.; López-Andrés, N.; Pattnaik, A.K.; Quintanilla, M.; Bernabeu, C. Endoglin Protein Interactome Profiling Identifies TRIM21 and Galectin-3 as New Binding Partners. Cells 2019, 8, 1082. https://doi.org/10.3390/cells8091082
Gallardo-Vara E, Ruiz-Llorente L, Casado-Vela J, Ruiz-Rodríguez MJ, López-Andrés N, Pattnaik AK, Quintanilla M, Bernabeu C. Endoglin Protein Interactome Profiling Identifies TRIM21 and Galectin-3 as New Binding Partners. Cells. 2019; 8(9):1082. https://doi.org/10.3390/cells8091082
Chicago/Turabian StyleGallardo-Vara, Eunate, Lidia Ruiz-Llorente, Juan Casado-Vela, María J. Ruiz-Rodríguez, Natalia López-Andrés, Asit K. Pattnaik, Miguel Quintanilla, and Carmelo Bernabeu. 2019. "Endoglin Protein Interactome Profiling Identifies TRIM21 and Galectin-3 as New Binding Partners" Cells 8, no. 9: 1082. https://doi.org/10.3390/cells8091082









