Regulation and Function of C-Type Natriuretic Peptide (CNP) in Gonadotrope-Derived Cell Lines
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. Tissue Collection, RNA Extraction and Multiplex GeXP RT-qPCR Assay
2.4. Data Presentation & Statistical Analysis
3. Results
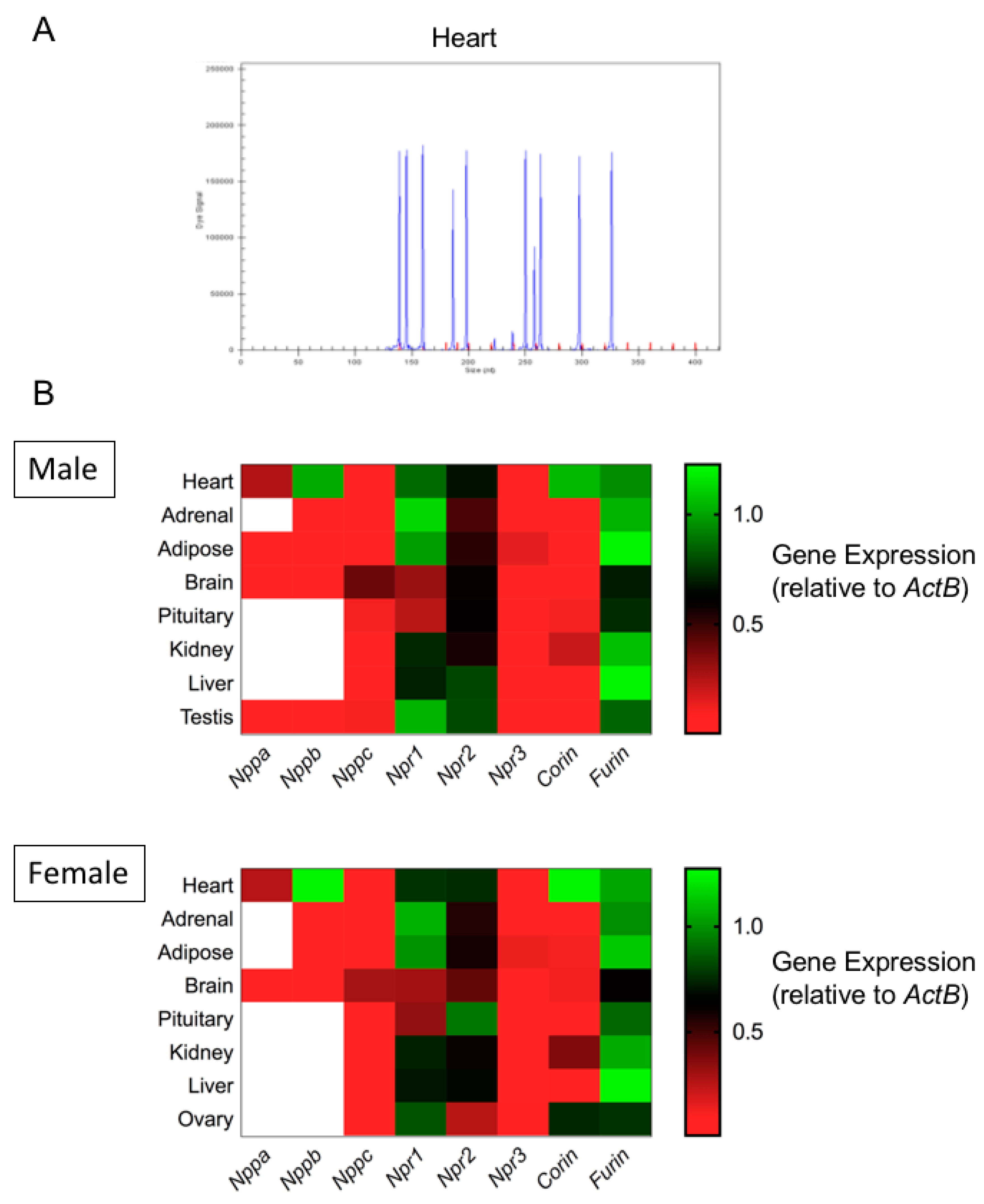
3.1. Expression Profiling of the Natriuretic Peptide System in Primary Mouse Endocrine Tissues by Multiplex RT-qPCR
3.2. Expression Profiling of the Natriuretic Peptide System in αT3-1 and LβT2 Gonadotrope Cell Lines
3.3. Effect of Continuous or Pulsatile Exposure to GnRH on Natriuretic Peptide Gene Expression
3.4. CNP Effects on Expression Levels of Gonadotrope Transcriptional Regulators and Signaling Genes
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Potter, L.R.; Abbey-Hosch, S.; Dickey, D.M. Natriuretic peptides, their receptors, and cyclic guanosine monophosphate-dependent signaling functions. Endocr. Rev. 2006, 27, 47–72. [Google Scholar] [CrossRef]
- Garbers, D.L.; Chrisman, T.D.; Wiegn, P.; Katafuchi, T.; Albanesi, J.P.; Bielinski, V.; Barylko, B.; Redfield, M.M.; Burnett, J.C., Jr. Membrane guanylyl cyclase receptors: An update. Trends Endocrinol. Metab. 2006, 17, 251–258. [Google Scholar] [CrossRef] [PubMed]
- Kuhn, M. Molecular Physiology of Membrane Guanylyl Cyclase Receptors. Physiol. Rev. 2016, 96, 751–804. [Google Scholar] [CrossRef] [PubMed]
- Fowkes, R.C.; McArdle, C.A. C-type natriuretic peptide: An important neuroendocrine regulator? Trends Endocrinol. Metab. 2000, 11, 333–338. [Google Scholar] [CrossRef]
- Komatsu, Y.; Nakao, K.; Suga, S.; Ogawa, Y.; Mukoyama, M.; Arai, H.; Shirakami, G.; Hosoda, K.; Nakagawa, O.; Hama, N.; et al. C-type natriuretic peptide (CNP) in rats and humans. Endocrinology 1991, 129, 1104–1106. [Google Scholar] [CrossRef] [PubMed]
- Grandclément, B.; Brisson, C.; Bayard, F.; Tremblay, J.; Gossard, F.; Morel, G. Localization of mRNA coding for the three subtypes of atrial natriuretic factor (ANF) receptors in rat anterior pituitary gland cells. J. Neuroendocrinol. 1995, 7, 939–948. [Google Scholar] [CrossRef] [PubMed]
- McArdle, C.A.; Poch, A.; Käppler, K. Cyclic guanosine monophosphate production in the pituitary: Stimulation by C-type natriuretic peptide and inhibition by gonadotropin-releasing hormone in αT3-1 cells. Endocrinology 1993, 132, 2065–2072. [Google Scholar] [CrossRef] [PubMed]
- Thompson, I.R.; Chand, A.N.; Jonas, K.C.; Burrin, J.M.; Steinhelper, M.E.; Wheeler-Jones, C.P.D.; McArdle, C.A.; Fowkes, R.C. Molecular characterization and functional interrogation of a local natriuretic peptide system in rodent pituitaries, αT3-1 and LβT2 gonadotrophs. J. Endocrinol. 2009, 203, 215–229. [Google Scholar] [CrossRef] [PubMed]
- Thompson, I.R.; Chand, A.N.; King, P.J.; Ansorge, O.; Jones, C.A.; Kosinovic, V.; Karavitaki, N.; Wheeler-Jones, C.P.D.; McGonnell, I.M.; Korbonits, M.; et al. Expression and transcriptional regulation of guanylyl cyclase-B (GC-B) receptors in a range of human pituitary adenomas, normal human fetal pituitaries and anterior pituitary cell lines. Endocr. Relat. Cancer 2012, 19, 497–508. [Google Scholar] [CrossRef] [PubMed]
- Chusho, H.; Tamura, N.; Ogawa, Y.; Yasoda, A.; Suda, M.; Miyazawa, T.; Nakamura, K.; Nakao, K.; Kurihara, T.; Komatsu, Y.; et al. Dwarfism and early death in mice lacking C-type natriuretic peptide. Proc. Natl. Acad. Sci. USA 2001, 98, 4016–4021. [Google Scholar] [CrossRef] [Green Version]
- Tamura, N.; Doolittle, L.K.; Hammer, R.E.; Shelton, J.M.; Richardson, J.A.; Garbers, D.L. Critical roles of the guanylyl cyclase B receptor in endochondral ossification and development of female reproductive organs. Proc. Natl. Acad. Sci. USA 2004, 101, 17300–17305. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Denef, C. Paracrinicity: The story of 30 years of cellular pituitary crosstalk. J. Neuroendocrinol. 2008, 20, 1–70. [Google Scholar] [PubMed]
- Le Tissier, P.R.; Hodson, D.J.; Lafont, C.; Fontanaud, P.; Schaeffer, M.; Mollard, P. Anterior pituitary cell networks. Front. Neuroendocrinol. 2012, 33, 252–266. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bliss, S.P.; Navratil, A.M.; Xie, J.; Roberson, M.S. GnRH signaling, the gonadotrope and endocrine control of fertility. Front. Neuroendocrinol. 2010, 31, 322–340. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Acuff, C.G.; Huang, H.; Steinhelper, M.E. Estradiol induces C-type natriuretic peptide gene expression in mouse uterus. Am. J. Physiol. 1997, 273, H2672–H2677. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gutkowska, J.; Jankowski, M.; Sairam, M.R.; Fujio, N.; Reis, A.M.; Mukaddam-Daher, S.; Tremblay, J. Hormonal regulation of natriuretic peptide system during induced ovarian follicular development in the rat. Biol. Reprod. 1999, 61, 162–170. [Google Scholar] [CrossRef] [PubMed]
- McArdle, C.A.; Olcese, J.; Schmidt, C.; Poch, A.; Kratzmeier, M.; Middendorff, R. C-type natriuretic peptide (CNP) in the pituitary: Is CNP an autocrine regulator of gonadotropes? Endocrinology 1994, 135, 2794–2801. [Google Scholar] [CrossRef]
- Fowkes, R.C.; Forrest-Owen, W.; McArdle, C.A. C-type natriuretic peptide (CNP) effects in anterior pituitary cell lines: Evidence for homologous desensitisation of CNP-stimulated cGMP accumulation in αT3-1 gonadotroph-derived cells. J. Endocrinol. 2000, 166, 195–203. [Google Scholar] [CrossRef]
- Fowkes, R.C.; Forrest-Owen, W.; Williams, B.; McArdle, C.A. C-type natriuretic peptide (CNP) effects on intracellular calcium [Ca2+]i in mouse-derived αT3-1 cell line. Regul. Pept. 1999, 84, 43–49. [Google Scholar] [CrossRef]
- Haisenleder, D.J.; Dalkin, A.C.; Ortolano, G.A.; Marshall, J.C.; Shupnik, M.A. A pulsatile gonadotropin-releasing hormone stimulus is required to increase transcription of the gonadotropin subunit genes: Evidence for differential regulation of transcription by pulse frequency in vivo. Endocrinology 1991, 128, 509–517. [Google Scholar] [CrossRef]
- Thompson, I.R.; Ciccone, N.A.; Zhou, Q.; Xu, S.; Khogeer, A.; Carroll, R.S.; Kaiser, U.B. GnRH Pulse Frequency Control of Fshb Gene Expression Is Mediated via ERK1/2 Regulation of ICER. Mol. Endocrinol. 2016, 30, 348–360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thompson, I.R.; Kaiser, U.B. GnRH pulse frequency-dependent differential regulation of LH and FSH gene expression. Mol. Cell. Endocrinol. 2014, 385, 28–35. [Google Scholar] [CrossRef] [PubMed]
- Thompson, I.R.; Ciccone, N.A.; Xu, S.; Zaytseva, S.; Carroll, R.S.; Kaiser, U.B. GnRH pulse frequency-dependent stimulation of FSHβ transcription is mediated via activation of PKA and CREB. Mol. Endocrinol. 2013, 27, 606–618. [Google Scholar] [CrossRef] [PubMed]
- Kanasaki, H.; Purwana, I.; Oride, A.; Mijiddorj, T.; Miyazaki, K. Extracellular Signal-Regulated Kinase (ERK) Activation and Mitogen-Activated Protein Kinase Phosphatase 1 Induction by Pulsatile Gonadotropin-Releasing Hormone in Pituitary Gonadotrophs. J. Signal Transduct. 2012, 2012, 198527. [Google Scholar] [CrossRef] [PubMed]
- Purwana, I.N.; Kanasaki, H.; Mijiddorj, T.; Oride, A.; Miyazaki, K. Induction of dual-specificity phosphatase 1 (DUSP1) by pulsatile gonadotropin-releasing hormone stimulation: Role for gonadotropin subunit expression in mouse pituitary LbetaT2 cells. Biol. Reprod. 2011, 84, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Perrett, R.M.; Voliotis, M.; Armstrong, S.P.; Fowkes, R.C.; Pope, G.R.; Tsaneva-Atanasova, K.; McArdle, C.A. Pulsatile hormonal signaling to extracellular signal-regulated kinase: Exploring system sensitivity to gonadotropin-releasing hormone pulse frequency and width. J. Biol. Chem. 2014, 289, 7873–7883. [Google Scholar] [CrossRef] [PubMed]
- Armstrong, S.P.; Caunt, C.J.; Fowkes, R.C.; Tsaneva-Atanasova, K.; McArdle, C.A. Pulsatile and sustained gonadotropin-releasing hormone (GnRH) receptor signaling: Does the Ca2+/NFAT signaling pathway decode GnRH pulse frequency? J. Biol. Chem. 2009, 284, 35746–35757. [Google Scholar] [CrossRef]
- Armstrong, S.P.; Caunt, C.J.; Fowkes, R.C.; Tsaneva-Atanasova, K.; McArdle, C.A. Pulsatile and sustained gonadotropin-releasing hormone (GnRH) receptor signaling: Does the ERK signaling pathway decode GnRH pulse frequency? J. Biol. Chem. 2010, 285, 24360–24371. [Google Scholar] [CrossRef] [PubMed]
- Garner, K.L.; Voliotis, M.; Alobaid, H.; Perrett, R.M.; Pham, T.; Tsaneva-Atanasova, K.; McArdle, C.A. Information Transfer via Gonadotropin-Releasing Hormone Receptors to ERK and NFAT: Sensing GnRH and Sensing Dynamics. J. Endocr. Soc. 2017, 1, 260–277. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casteel, D.E.; Zhang, T.; Zhuang, S.; Pilz, R.B. cGMP-dependent protein kinase anchoring by IRAG regulates its nuclear translocation and transcriptional activity. Cell. Signal. 2008, 20, 1392–1399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broderick, K.E.; Zhang, T.; Rangaswami, H.; Zeng, Y.; Zhao, X.; Boss, G.R.; Pilz, R.B. Guanosine 3′,5′-cyclic monophosphate (cGMP)/cGMP-dependent protein kinase induce interleukin-6 transcription in osteoblasts. Mol. Endocrinol. 2007, 21, 1148–1162. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhuang, S.; Chen, Y.; Boss, G.R.; Pilz, R.B. Cyclic GMP-dependent protein kinase regulates CCAAT enhancer-binding protein beta functions through inhibition of glycogen synthase kinase-3. J. Biol. Chem. 2005, 280, 32683–32692. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Zhuang, S.; Cassenaer, S.; Casteel, D.E.; Gudi, T.; Boss, G.R.; Pilz, R.B. Synergism between calcium and cyclic GMP in cyclic AMP response element-dependent transcriptional regulation requires cooperation between CREB and C/EBP-beta. Mol. Cell. Biol. 2003, 23, 4066–4082. [Google Scholar] [CrossRef] [PubMed]
- Gudi, T.; Hong, G.K.; Vaandrager, A.B.; Lohmann, S.M.; Pilz, R.B. Nitric oxide and cGMP regulate gene expression in neuronal and glial cells by activating type II cGMP-dependent protein kinase. FASEB J. 1999, 13, 2143–2152. [Google Scholar] [CrossRef] [PubMed]
- McArdle, C.A.; Bunting, R.; Mason, W.T. Dynamic video imaging of cystolic Ca(2+) in the alphaT3-1, gonadotrope-derived cell line. Mol. Cell. Neurosci. 1992, 3, 124–132. [Google Scholar] [CrossRef]
- Staines, K.A.; Madi, K.; Mirczuk, S.M.; Parker, S.; Burleigh, A.; Poulet, B.; Hopkinson, M.; Bodey, A.J.; Fowkes, R.C.; Farquharson, C.; et al. Endochondral Growth Defect and Deployment of Transient Chondrocyte Behaviors Underlie Osteoarthritis Onset in a Natural Murine Model. Arthritis Rheumatol. 2016, 68, 880–891. [Google Scholar] [CrossRef] [PubMed]
- Walther, T.; Stepan, H. C-type natriuretic peptide in reproduction, pregnancy and fetal development. J. Endocrinol. 2004, 180, 17–22. [Google Scholar] [CrossRef] [Green Version]
- Zhang, M.; Su, Y.Q.; Sugiura, K.; Xia, G.; Eppig, J.J. Granulosa cell ligand NPPC and its receptor NPR2 maintain meiotic arrest in mouse oocytes. Science 2010, 330, 366–369. [Google Scholar] [CrossRef]
- Huang, D.H.; Zhang, S.W.; Zhao, H.; Zhang, L. The role of C-type natriuretic peptide in rat testes during spermatogenesis. Asian J. Androl. 2011, 13, 275–280. [Google Scholar] [CrossRef]
- Robinson, J.W.; Zhang, M.; Shuhaibar, L.C.; Norris, R.P.; Geerts, A.; Wunder, F.; Eppig, J.J.; Potter, L.R.; Jaffe, L.A. Luteinizing hormone reduces the activity of the NPR2 guanylyl cyclase in mouse ovarian follicles, contributing to the cyclic GMP decrease that promotes resumption of meiosis in oocytes. Dev. Biol. 2012, 366, 308–316. [Google Scholar] [CrossRef] [Green Version]
- Rai, A.J.; Kamath, R.M.; Gerald, W.; Fleisher, M. Analytical validation of the GeXP analyzer and design of a workflow for cancer-biomarker discovery using multiplexed gene-expression profiling. Anal. Bioanal. Chem. 2009, 393, 1505–1511. [Google Scholar] [CrossRef]
- Kutyrev, I.; Cleveland, B.; Leeds, T.; Wiens, G.D. Dataset of proinflammatory cytokine and cytokine receptor gene expression in rainbow trout (Oncorhynchus mykiss) measured using a novel GeXP multiplex, RT-PCR assay. Data Brief 2017, 11, 192–196. [Google Scholar] [CrossRef]
- DiCicco-Bloom, E.; Lelièvre, V.; Zhou, X.; Rodriguez, W.; Tam, J.; Waschek, J.A. Embryonic expression and multifunctional actions of the natriuretic peptides and receptors in the developing nervous system. Dev. Biol. 2004, 271, 161–175. [Google Scholar] [CrossRef] [Green Version]
- Surendran, K.; Simon, T.C. CNP gene expression is activated by Wnt signaling and correlates with Wnt4 expression during renal injury. Am. J. Physiol. Renal. Physiol. 2003, 284, F653–F662. [Google Scholar] [CrossRef]
- Sellitti, D.F.; Koles, N.; Mendonça, M.C. Regulation of C-type natriuretic peptide expression. Peptides. 2011, 32, 1964–1971. [Google Scholar] [CrossRef] [Green Version]
- Jankowski, M.; Reis, A.M.; Mukaddam-Daher, S.; Dam, T.V.; Farookhi, R.; Gutkowska, J. C-type natriuretic peptide and the guanylyl cyclase receptors in the rat ovary are modulated by the estrous cycle. Biol. Reprod. 1997, 56, 59–66. [Google Scholar] [CrossRef]
- Wang, X.; Wang, H.; Liu, W.; Zhang, Z.; Zhang, Y.; Zhang, W.; Chen, Z.; Xia, G.; Wang, C. High level of C-type natriuretic peptide induced by hyperandrogen-mediated anovulation in polycystic ovary syndrome mice. Clin. Sci. 2018, 132, 759–776. [Google Scholar] [CrossRef] [Green Version]
- Turgeon, J.L.; Kimura, Y.; Waring, D.W.; Mellon, P.L. Steroid and pulsatile gonadotropin-releasing hormone (GnRH) regulation of luteinizing hormone and GnRH receptor in a novel gonadotrope cell line. Mol. Endocrinol. 1996, 10, 439–450. [Google Scholar]
- Ruf-Zamojski, F.; Fribourg, M.; Ge, Y.; Nair, V.; Pincas, H.; Zaslavsky, E.; Nudelman, G.; Tuminello, S.J.; Watanabe, H.; Turgeon, J.L.; et al. Regulatory Architecture of the LβT2 Gonadotrope Cell Underlying the Response to Gonadotropin-Releasing Hormone. Front. Endocrinol. 2018, 9, 34. [Google Scholar] [CrossRef]
- Roberson, M.S.; Misra-Press, A.; Laurance, M.E.; Stork, P.J.; Maurer, R.A. A role for mitogen-activated protein kinase in mediating activation of the glycoprotein hormone alpha-subunit promoter by gonadotropin-releasing hormone. Mol. Cell. Biol. 1995, 15, 3531–3539. [Google Scholar] [CrossRef] [Green Version]
- Sundaresan, S.; Colin, I.M.; Pestell, R.G.; Jameson, J.L. Stimulation of mitogen-activated protein kinase by gonadotropin-releasing hormone: Evidence for the involvement of protein kinase C. Endocrinology 1996, 137, 304–311. [Google Scholar] [CrossRef]
- Mulvaney, J.M.; Roberson, M.S. Divergent signaling pathways requiring discrete calcium signals mediate concurrent activation of two mitogen-activated protein kinases by gonadotropin-releasing hormone. J. Biol. Chem. 2000, 275, 14182–14189. [Google Scholar] [CrossRef]
- Caunt, C.J.; Finch, A.R.; Sedgley, K.R.; McArdle, C.A. GnRH receptor signalling to ERK: Kinetics and compartmentalization. Trends Endocrinol. Metab. 2006, 17, 308–313. [Google Scholar] [CrossRef]
- Fowkes, R.C.; King, P.; Burrin, J.M. Regulation of human glycoprotein hormone alpha-subunit gene transcription in LbetaT2 gonadotropes by protein kinase C and extracellular signal-regulated kinase 1/2. Biol. Reprod. 2002, 67, 725–734. [Google Scholar] [CrossRef]
- Kanasaki, H.; Bedecarrats, G.Y.; Kam, K.Y.; Xu, S.; Kaiser, U.B. Gonadotropin-releasing hormone pulse frequency-dependent activation of extracellular signal-regulated kinase pathways in perifused LbetaT2 cells. Endocrinology 2005, 146, 5503–5513. [Google Scholar] [CrossRef]
- Alarid, E.T.; Windle, J.J.; Whyte, D.B.; Mellon, P.L. Immortalization of pituitary cells at discrete stages of development by directed oncogenesis in transgenic mice. Development 1996, 122, 3319–3329. [Google Scholar]
- Thomas, P.; Mellon, P.L.; Turgeon, J.; Waring, D.W. The L beta T2 clonal gonadotrope: A model for single cell studies of endocrine cell secretion. Endocrinology 1996, 137, 2979–2989. [Google Scholar] [CrossRef]
- Müller, D.; Hida, B.; Guidone, G.; Speth, R.C.; Michurina, T.V.; Enikolopov, G.; Middendorff, R. Expression of guanylyl cyclase (GC)-A and GC-B during brain development: Evidence for a role of GC-B in perinatal neurogenesis. Endocrinology 2009, 150, 5520–5529. [Google Scholar] [CrossRef]
- Gudi, T.; Huvar, I.; Meinecke, M.; Lohmann, S.M.; Boss, G.R.; Pilz, R.B. Regulation of gene expression by cGMP-dependent protein kinase. Transactivation of the c-fos promoter. J. Biol. Chem. 1996, 271, 4597–4600. [Google Scholar]
- Pilz, R.B.; Casteel, D.E. Regulation of gene expression by cyclic GMP. Circ. Res. 2003, 93, 1034–1046. [Google Scholar] [CrossRef]
- Hosokawa, K.; Dantes, A.; Schere-Levy, C.; Barash, A.; Yoshida, Y.; Kotsuji, F.; Vlodavsky, I.; Amsterdam, A. Induction of Ad4BP/SF-1, steroidogenic acute regulatory protein, and cytochrome P450scc enzyme system expression in newly established human granulosa cell lines. Endocrinology 1998, 139, 4679–4687. [Google Scholar] [CrossRef] [PubMed]
- Lai, W.A.; Yeh, Y.T.; Fang, W.L.; Wu, L.S.; Harada, N.; Wang, P.H.; Ke, F.C.; Lee, W.L.; Hwang, J.J. Calcineurin and CRTC2 mediate FSH and TGFβ1 upregulation of Cyp19a1 and Nr5a in ovary granulosa cells. J. Mol. Endocrinol. 2014, 53, 259–270. [Google Scholar] [CrossRef] [PubMed]
- Vaccarino, F.M.; Hayward, M.D.; Le, H.N.; Hartigan, D.J.; Duman, R.S.; Nestler, E.J. Induction of immediate early genes by cyclic AMP in primary cultures of neurons from rat cerebral cortex. Mol. Brain Res. 1993, 19, 76–82. [Google Scholar] [CrossRef]
- Duan, W.R.; Ito, M.; Park, Y.; Maizels, E.T.; Hunzicker-Dunn, M.; Jameson, J.L. GnRH regulates early growth response protein 1 transcription through multiple promoter elements. Mol. Endocrinol. 2002, 16, 221–233. [Google Scholar] [CrossRef] [PubMed]
- Jonas, K.C.; Melrose, T.; Thompson, I.R.; Baxter, G.F.; Lipscomb, V.J.; Niessen, S.J.; Lawson, C.; McArdle, C.A.; Roberson, M.S.; McGonnell, I.M.; et al. Natriuretic peptide activation of extracellular regulated kinase 1/2 (ERK1/2) pathway by particulate guanylyl cyclases in GH3 somatolactotropes. Cell Tissue Res. 2017, 369, 567–578. [Google Scholar] [CrossRef] [PubMed]
- Dhayade, S.; Kaesler, S.; Sinnberg, T.; Dobrowinski, H.; Peters, S.; Naumann, U.; Liu, H.; Hunger, R.E.; Thunemann, M.; Biedermann, T.; et al. Sildenafil Potentiates a cGMP-Dependent Pathway to Promote Melanoma Growth. Cell Rep. 2016, 14, 2599–2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]




© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mirczuk, S.M.; Lessey, A.J.; Catterick, A.R.; Perrett, R.M.; Scudder, C.J.; Read, J.E.; Lipscomb, V.J.; Niessen, S.J.; Childs, A.J.; McArdle, C.A.; et al. Regulation and Function of C-Type Natriuretic Peptide (CNP) in Gonadotrope-Derived Cell Lines. Cells 2019, 8, 1086. https://doi.org/10.3390/cells8091086
Mirczuk SM, Lessey AJ, Catterick AR, Perrett RM, Scudder CJ, Read JE, Lipscomb VJ, Niessen SJ, Childs AJ, McArdle CA, et al. Regulation and Function of C-Type Natriuretic Peptide (CNP) in Gonadotrope-Derived Cell Lines. Cells. 2019; 8(9):1086. https://doi.org/10.3390/cells8091086
Chicago/Turabian StyleMirczuk, Samantha M, Andrew J Lessey, Alice R Catterick, Rebecca M Perrett, Christopher J Scudder, Jordan E Read, Victoria J Lipscomb, Stijn J Niessen, Andrew J Childs, Craig A McArdle, and et al. 2019. "Regulation and Function of C-Type Natriuretic Peptide (CNP) in Gonadotrope-Derived Cell Lines" Cells 8, no. 9: 1086. https://doi.org/10.3390/cells8091086





