Abstract
In eukaryotic cells, autophagosomes and multivesicular bodies (MVBs) are two closely related partners in the lysosomal/vacuolar protein degradation system. Autophagosomes are double membrane-bound organelles that transport cytoplasmic components, including proteins and organelles for autophagic degradation in the lysosomes/vacuoles. MVBs are single-membrane organelles in the endocytic pathway that contain intraluminal vesicles whose content is either degraded in the lysosomes/vacuoles or recycled to the cell surface. In plants, both autophagosome and MVB pathways play important roles in plant responses to biotic and abiotic stresses. More recent studies have revealed that autophagosomes and MVBs also act together in plant stress responses in a variety of processes, including deployment of defense-related molecules, regulation of cell death, trafficking and degradation of membrane and soluble constituents, and modulation of plant hormone metabolism and signaling. In this review, we discuss these recent findings on the coordination and crosstalk between autophagosome and MVB pathways that contribute to the complex network of plant stress responses.
1. Introduction
The trafficking, degradation, and recycling of cellular components are essential for the growth and survival of all cellular organisms. Autophagy is an important pathway for the degradation of intracellular components, including organelles, proteins, and RNAs, in eukaryotic cells [1,2,3]. Under normal growth conditions, autophagy is suppressed and operates only at a low basal level. Upon exposure to stress conditions, such as nutrient starvation, autophagy is induced with the formation of a cup-shaped structure called a phagophore or isolation membrane, which can expand to engulf cytoplasmic components to form a closed double-membraned structure termed autophagosomes [4]. The autophagosomes fuse with the lysosomes/vacuoles to degrade the sequestered cargo. Multivesicular bodies (MVBs) are specialized endosomes in the endocytic pathway that function in the internalization, transport, sorting, and degradation of specific plasma membrane proteins, such as membrane receptors and ion transporters [5]. MVBs contain intraluminal vesicles generated from invagination and budding of the limiting membrane through the action of protein complexes named ESCRT-0, I, II, and III (endosomal sorting complex required for transport) [5]. The content of MVBs can be degraded through fusion with lysosomes/vacuoles or recycled into the cell surface after fusion with the plasma membrane. The endocytic and MVB pathway has a cellular housekeeping role and operates even under normal growth conditions but is subjected to tight regulation in response to developmental and environmental cues to ensure the correct delivery of membrane cargo [6,7].
Both autophagy and MVBs play a critical role in plant responses to a broad spectrum of biotic and abiotic stress conditions [1,8,9,10]. Both the formation of the autophagosomes and expression of autophagy-related genes (ATGs) are induced under nutrient starvation, biotic and abiotic stresses, such as oxidative, high salt, and osmotic stress conditions [1,11,12,13]. Arabidopsis autophagy mutants are hypersensitive to these abiotic stresses [8,12,14,15,16,17,18,19]. Autophagy also affects plant responses to microbial pathogens [13]. Depending on the age of plants and the stage of infection, autophagy can either promote or suppress defense-associated hypersensitive cell death against biotrophic pathogens [13,20,21,22,23]. Autophagy plays a critical role in plant resistance to necrotrophic fungal pathogens [11,20] and in plant–virus interactions [24,25]. Likewise, the endocytic and associated MVB pathways are essential for the regulation of the levels and activity of plasma membrane proteins in response to changing environmental conditions. The presence of excess levels of metal ions in the soil can trigger downregulation of their corresponding ion transporters in the plasma membrane through increased endocytosis [26,27]. Over the past decade or so, important progress has also been made in establishing the critical roles of MVBs and associated trafficking in pathogen recognition, defense signaling, and deployment of defense-related molecules during plant responses to microbial pathogens [6,9]. Both the endocytic activity and MVB biogenesis are induced in plant cells upon biotic and abiotic stresses [9,10]. In Arabidopsis, pathogen- and stress-induced endocytosis and MVB biogenesis are abolished in the mutants for LIP5 (lyst-interacting protein 5) [9,10]. LIP5 is an activator of the SKD1 (suppressor of K+ transport growth defect 1) ATPase, a core component in the ESCRT pathway central to MVB biogenesis [9,10]. Importantly, the lip5 mutants are compromised not only in plant disease resistance but also in plant tolerance to salt and heat stresses [9,10]. These results confirm a critical role of stress-induced, LIP5-dependent MVB biogenesis in plant responses to both biotic and abiotic stresses.
As two closely related partners in the lysosomal/vacuolar protein degradation system, there are extensive interactions in both the regulation and functions between the autophagosome and MVB pathways based on studies in yeast and animals [28,29,30,31]. For example, the maturation process of autophagosomes in some organisms often includes fusion events with early and late endosomal vesicles, including MVBs, to form a hybrid organelle called the amphisome prior to the fusion with lysosomes (forming an autolysosome) to degrade the incorporated materials [29]. The MVB pathway and autophagy are also coordinated in action to ensure cell survival during starvation in yeast cells [30]. In plants, extensive studies have uncovered not only conserved and unique components but also shared factors in the biogenesis and trafficking of autophagosomes and MVBs, which have been discussed in recent reviews [32,33]. With well-established common roles in plant stress responses, how the two related protein trafficking pathways act together to protect plants under environmental stresses is very important and has gained increasing interest in recent years. Progress has been made in the analysis of the interplay of signaling pathways that regulate stress-induced biogenesis of autophagosomes and MVBs [9,34]. More importantly, recent studies have provided strong evidence for the coordinated action of the autophagosome and MVB pathways during plant responses to biotic and abiotic stresses. In this review, we discuss these recent progresses in the analysis of the crosstalk and coordination between these two related trafficking pathways in plant stress responses.
2. Stress Regulation of Autophagosome and MVB Biogenesis
Autophagy occurs at a low basal level in plant cells under normal growth conditions but can be induced by biotic and abiotic stresses, such as nutrient starvation [35,36]. Upon nutrient, osmotic, or salt stress in plant cells, the Snf1 (sucrose non-fermenting 1)-related protein kinase 1 (SnRK1), a master regulator of metabolism, can activate autophagy by inhibiting the TOR (target of rapamycin) complex, a negative regulator of autophagy [4]. Inhibited TOR leads to activation of the ATG1 complex or the deactivation of the ribosomal p70 S6 kinase (S6K) and type 2A-phosphatase-associated protein 46 kD (Tap46), leading to autophagy activation [4]. Autophagy activation by oxidative and endoplasmic reticulum (ER) stress, however, is mediated by the TOR-independent pathways through direct activation of the ATG1 complex by SnRK1 [4]. Autophagy activation by ER stress can also be triggered by accumulated unfolded proteins through the activation of the IRE1b (inositol-requiring enzyme 1b) serine/threonine-protein kinase/endoribonuclease, a master regulator of ER stress responses [4,15]. After autophagy activation, autophagosome formation in plant cells follows a conserved process involving phagophore initiation, autophagosome expansion, maturation, and degradation. It has been shown that a number of ESCRT- and MVB-associated proteins play important roles in the process of autophagosome formation in plants [32,33]. For example, Arabidopsis SH3P2 (SH3 domain-containing protein 2), a ubiquitin-binding protein that acts with ESCRT-I and the deubiquitylating enzyme AMSH3 (associated molecule with the SH3 domain of STAM3) in the transfer of ubiquitinated proteins to the ESCRT machinery [37], also binds phosphatidylinositol 3-phosphate and ATG8 and regulates autophagosome formation in Arabidopsis [38,39]. Analysis using RNAi (RNA interference) and other approaches suggests that SH3P2 is a key regulator of plant autophagy most likely by facilitating membrane expansion or maturation during autophagosome formation [38,39]. FREE1 (FYVE domain protein required for endosomal sorting 1), another Arabidopsis protein that binds to phosphatidylinositol-3-phosphate and ubiquitin, is associated with MVBs through interaction with the ESCRT-I subunit VPS23 (vacuolar protein sorting 23) and ESCRT-III subunit Snf7 (sucrose non-fermenting protein 7) [40]. FREE1 also interacts with SH3P2 on autophagosomes [40]. The mutation of FREE1 leads to the formation of abnormal MVB-autophagosome hybrid structures, further supporting the interactions of the two related types of vesicles [40].
The endocytic and associated MVB pathway operates at high levels even under normal growth conditions and is essential for plant growth and development [41,42]. However, both the endocytic activity and MVB biogenesis are regulated in plant cells in responses to changes in environmental cues, particularly to stress conditions. Upon pathogen infection, plasma membrane-localized pattern recognition receptors, including the receptor-like kinases FLS2 (flagellin-sensitive 2) and CERK1 (chitin elicitor receptor kinase 1), are ubiquitinated and internalized through endocytosis for recycling or degradation upon binding to pathogen elicitors to modulate plant immune responses [43,44]. High levels of ions in soil can cause increased turnover through endocytosis of various plasma membrane-localized ion channels, including the boric transporter BOR1 (require high boron1) [45], the iron transporter IRT1 (iron-regulated transporter 1) [26,27], the phosphate transporters PHTs [46], and the nitrate transporter NRT1.7 [47]. Likewise, increased biogenesis of MVBs has been observed in plant cells upon infection by microbial pathogens or exposure to abiotic stress, such as high salt [9,10]. Therefore, there are constitutive MVB biogenesis and trafficking required for normal growth and development as well as induced MVB biogenesis and trafficking in response to altered environmental conditions. This notion of constitutive and induced MVB biogenesis is supported by the recent characterization of Arabidopsis LIP5 protein, a positive activator of the SKD1 AAAA ATPase, which catalyzes the disassembly of the ESCRT III complex during MVB biogenesis [9,10]. Unlike mutants for other ESCRT components, such as SKD ATPase, which are often lethal [48], knockout mutants for LIP5 grow and develop normally, indicating that the basal SKD ATPase activity is sufficient for the constitutive levels of MVB biogenesis required for plant growth and development [9,10]. However, the lip5 mutants are highly susceptible to microbial pathogens and severely compromised in tolerance to salt and heat stress [9,10]. The compromised disease resistance and stress tolerance of the lip5 mutants are associated with defects in pathogen- and stress-induced endocytosis and MVB biogenesis [9,10]. These results indicate that LIP5 is a key regulator of stress-induced MVB biogenesis in plant responses to both biotic and abiotic stresses.
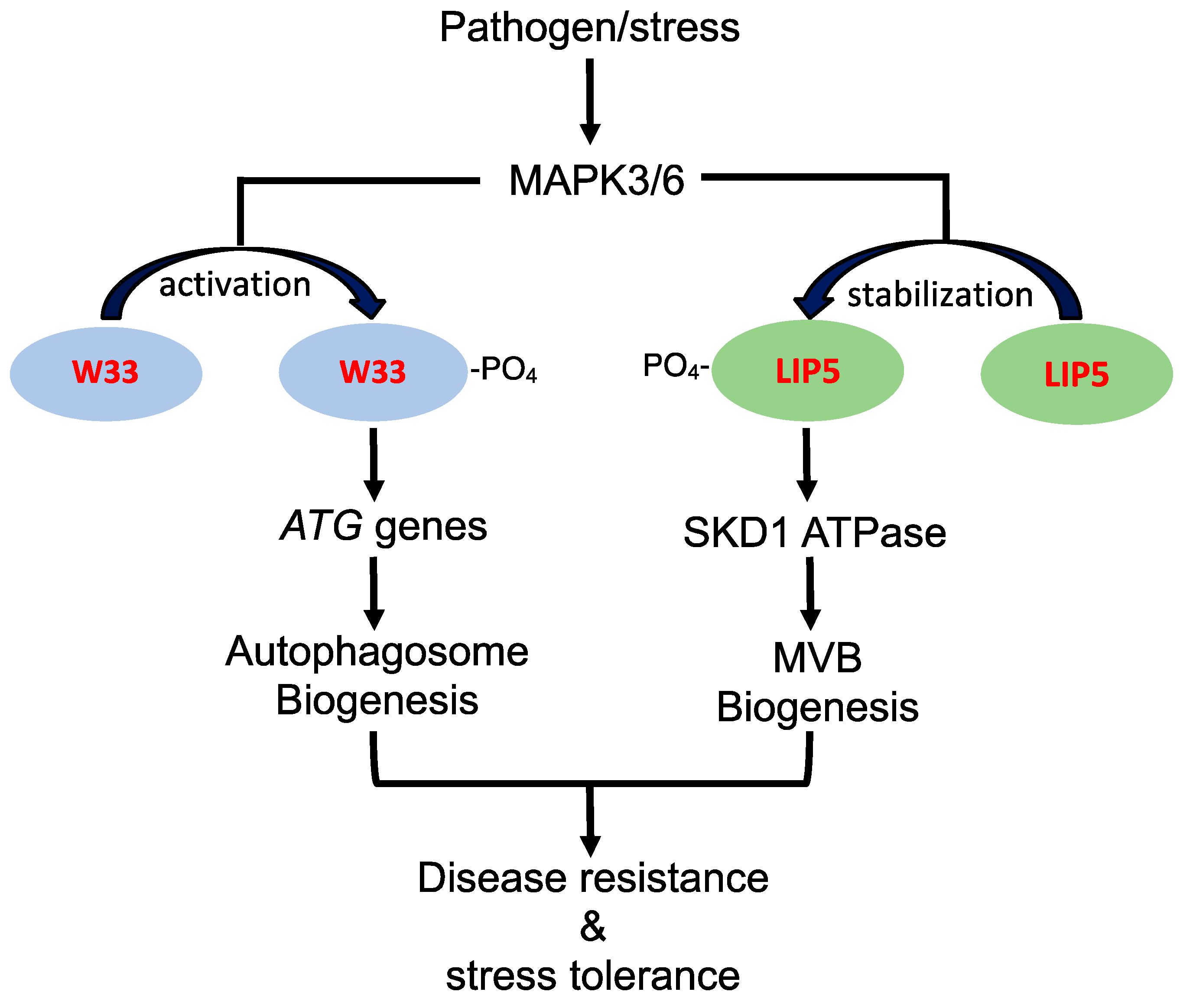
There are also studies supporting that the regulatory pathways for stress-induced autophagy and MVB pathways intersects in the complex network of plant stress responses. In Arabidopsis, both autophagy and WRKY33 transcription factor, which belongs to a family of DNA-binding proteins containing a conserved WRKYGQK motif, play an important role in plant resistance to necrotrophic pathogens [49,50]. WRKY33 interacts with autophagy-related protein ATG18a and is required for sustained induction of ATG18a gene expression by pathogen infection [11]. Furthermore, sustained induction autophagosome formation by pathogen infection is compromised in the wrky33 mutants [11]. These results indicate that the critical role of WRKY33 in plant immune response is in part mediated through its positive regulation of pathogen-induced autophagy. WRKY33 is a target of stress- and pathogen-responsive MAPK3 and 6 (mitogen-activated protein kinase 3 and 6), which phosphorylate and activate the transcription factor [34] (Figure 1). Interestingly, the LIP5 positive regulator of stress-induced MVB biogenesis is also an interacting protein and target of MAPK3 and 6 (Figure 1). Under normal growth conditions, unphosphorylated LIP5 is unstable and subjected to degradation [9,10]. Upon pathogen infection or salt treatment, activated MAPK3 and 6 phosphorylate LIP5 and increase its stability [9,10]. As a result, LIP5 protein levels increase in plant cells upon exposure to stress conditions, leading to stimulation of SKD1 ATPase and induced MVB biogenesis (Figure 1). Therefore, the stress-responsive MAPK3/6 signaling cascade, which plays a central role in plant growth, development, and stress responses [51], appears to regulate pathogen- and stress-induced formation of both autophagosomes and MVBs, supporting coordinated regulation of the two related trafficking pathways during plant stress responses (Figure 1).
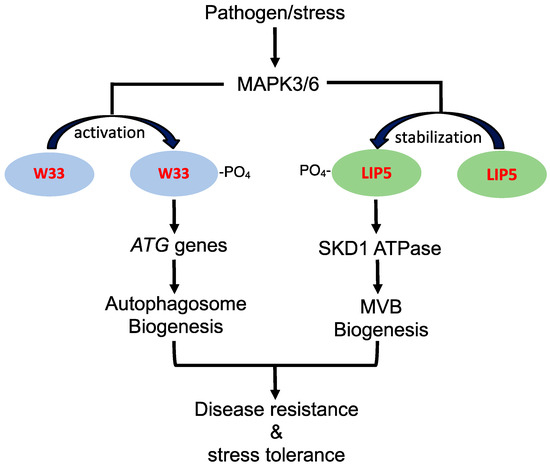
Figure 1.
Role of pathogen/stress-responsive MAPK3/6 signaling cascade in stress-induced autophagosome and MVB biogenesis. Pathogen/stress-activated MAPK3 and 6 phosphorylate and activate WRKY33 (W33) transcription factor to activates ATG gene transcription for sustained autophagy induction. Pathogen/stress-activated MAPK3 and 6 also phosphorylate LIP5 to improve its stability for stimulation of SKD1 required for stressed-induced MVB biogenesis. SDK1 is an AAA ATPase that catalyzes the release of the ESCRT-III complex from the membrane during the biogenesis of MVBs. Induced autophagosome and MVB biogenesis promote plant disease resistance and stress tolerance.
3. Autophagy and MVB Coordination in Plant Biotic Stress Response
Both autophagy and MVB pathways play important and complex roles in plant immune responses to microbial pathogens [6,13]. One of the critical roles of vesicle trafficking in the plant immune system is the mobilization and trafficking of defense-related molecules to the plant cell surface against invading pathogens. MVBs plays a critical role in the cell surface defense in plant cells. When plants are infected by filamentous pathogens, the germinating spores of the pathogens develop infection pegs on the leaf surface to invade the epidermal cells, which can induce plant defense responses, including the formation of local cell wall appositions (papilla) at the attack sites [52]. MVBs and cell wall-associated paramural bodies (PMBs) accumulate in the vicinity of pathogen-induced papillae [53,54,55]. PMBs are situated between the cell wall and the plasma membrane, likely resulting from the fusion of MVBs with the plasma membrane [55]. The accumulation of MVBs and PMBs occurs near papillae in plant cells infected not only by pathogenic fungal pathogens, but also by bacteria, and nematodes for the delivery of defense-related molecules, including phytoalexins, callose, and reactive oxygen species (ROS), to papillae [56,57,58]. The fusion of MVBs with the plasma membrane can also lead to the generation of plant extracellular vesicles (EVs). EVs from Arabidopsis leaves contain proteins involved in the metabolism and transport of defense-related molecules, including small RNAs, and their secretion is enhanced in Arabidopsis after Pseudomonas syringae infection [59,60,61]. EV protein profiles are most similar to those from the TGN(trans Golgi network)/MVB compartments, supporting that EVs are derived from the intralumenal vesicles of MVBs [60,61]. We have also shown that in Arabidopsis, both the endocytosis and MVB biogenesis were induced in an LIP5-dependent manner after infection by the bacterial pathogen P. syringae [9]. Pathogen infection resulted in increased numbers of MVBs and PMBs in wild-type plants but not in the lip5 mutants [9]. These results provide genetic evidence for a critical role of pathogen-induced, LIP5-dependent MVB biogenesis in the formation of defense-related vesicles at the infection sites (Figure 2).
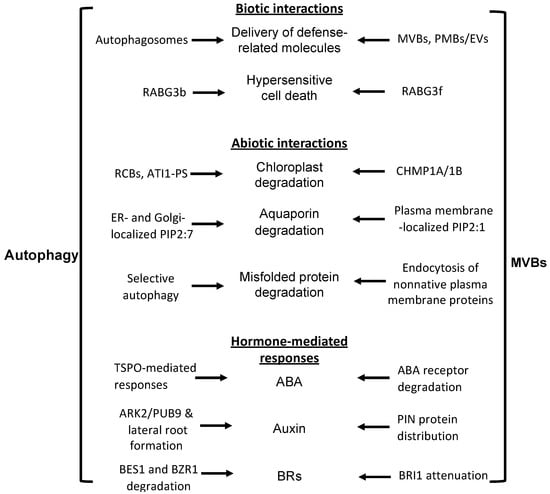
Figure 2.
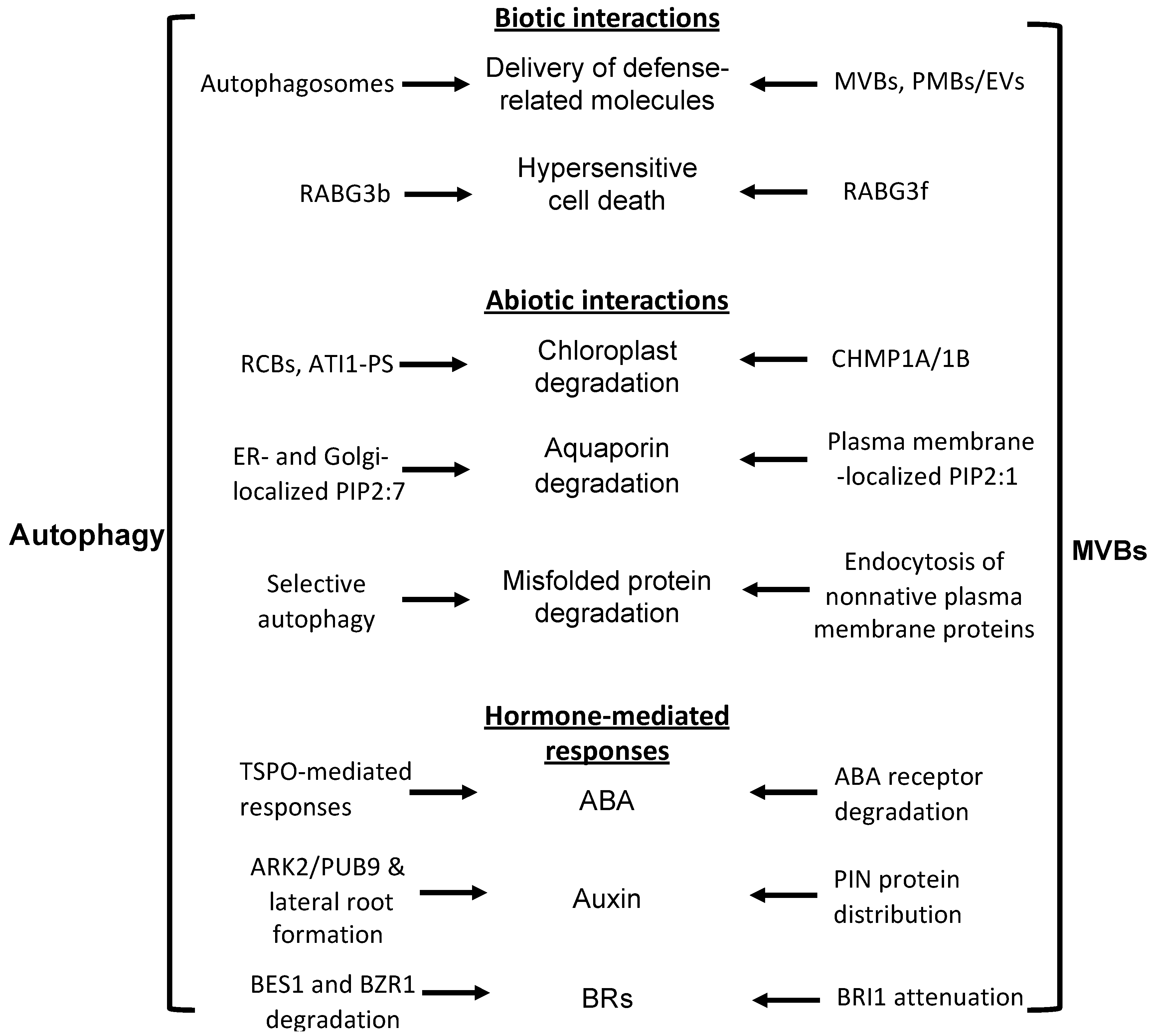
Coordination and crosstalk between the autophagosome and MVB pathways in plant biotic and abiotic interactions and in plant hormone-regulated stress responses.
Upon successful penetration, filamentous pathogens can develop haustoria, a special feeding structure, into plant cells. Each haustorium is surrounded by the plasma membrane of the plant cell termed extrahaustorial membrane (EHM), which is likely synthesized de novo [62]. In Arabidopsis, Rab5 GTPase is an MVB marker that accumulates in the EHM after infection with a powdery mildew fungus [63]. Cell surface immune receptors, such as FLS2 and RPW8 (resistance to powdery mildew 8) resistance proteins, are also recruited to the EHM upon pathogen infection, likely as a host border control mechanism at the plant–pathogen interface [62]. In tobacco (Nicotiana benthamiana) cells invaded by the oomycete pathogen Phytophthora infestans, RabG3c, an Rab7 GTPase and an MVB marker, but not a tonoplast-localized sucrose transporter, is also recruited to the EHM [64]. Specific rerouting of MVBs from the vacuole to the host–pathogen interface may participate in the formation of the EHM and surface defense. Interestingly, recent studies have revealed that perihaustorial compartments are also the hotspots for autophagosome biogenesis. Defense-related autophagosomes labeled by the ATG8CL and NBR1(neighbor of BRCA1 gene 1) autophagy markers are diverted to the EHM in tobacco cells upon infection of P. infestans [65]. Overexpression of both ATG9 and NBR1 led to increased resistance while silencing of NBR1 caused increased disease lesions [65,66]. These results indicate that increased autophagy activity at the plant–pathogen interface contributes to plant defense. The roles of pathogen-induced NBR1-mediated selective autophagy in the plant interaction with the pathogen are further supported by the finding that the PexRD54 effector of P. infestans binds host autophagy protein ATG8CL to deplete the autophagy cargo receptor NBR1 out of ATG8CL complexes, thereby antagonizing NBR1′s positive effect on pathogen defense [66]. Therefore, both defense-related autophagosomes and MVBs are mobilized to the plant–pathogen interface to restrict pathogens during plant interactions with filamentous pathogens. These observations support the coordinated roles of autophagy and MVB pathways in plant surface defense against invading pathogens (Figure 2).
Pathogen-induced host hypersensitive cell death is a critical process that can often determine the outcomes of plant interactions with microbial pathogens [67]. Both the autophagosome and MVB pathways participate in the regulation of plant hypersensitive cell death upon pathogen infection. Studies on plants Rab GTPases, regulators of vesicle trafficking, also indicate crosstalk between autophagosome and MVB pathways in immunity-associated hypersensitive cell death. In endocytosis in yeast and animals, proteins endocytosed from the plasma membrane pass through the early endosomes to the MVB late endosomes before the fusion with the lysosomes/vacuoles. The maturation of the early endosomes to MVBs involves Rab5 to Rab7 GTPase conversion. This conversion requires Mon1 (monensin sensitivity 1) protein, which acts together with CCZ1 (calcium caffeine zinc sensitivity 1) as a guanine-nucleotide exchange factor for Rab7 activation and Rab5 inactivation [68]. In Arabidopsis, there are eight genes encoding putative Rab7 proteins [68]. Genetic analysis with different combinations of rab7 mutants indicate that their functions are highly redundant [68]. Analysis of RABG3f, a highly expressed member of the Arabidopsis Rab7 family, showed that it is localized to MVBs and the vacuoles [68]. Expression of a dominant-negative form of the Rab7 protein in Arabidopsis led to the formation of enlarged MVBs, altered vacuole morphology, inhibited vacuolar trafficking, blocked degradation of storage proteins in the protein storage vacuole, and caused seedling death [68]. These results indicate that Rab7 proteins are required for both MVB-to-vacuole trafficking and vacuole biogenesis. On the other hand, analysis of another Rab7 protein family member, RabG3b, indicates a positive role in autophagy and hypersensitive cell death in response to pathogen infection [69,70]. Expression of a constitutively active RabG3b (RabG3bCA) in transgenic Arabidopsis plants led to accelerated, unrestricted programmed cell death within one day of infection by avirulent strains of the bacterial pathogen P. syringae [69,70]. By contrast, the autophagy-defective atg5-1 mutant gradually developed chlorotic cell death through uninfected sites over several days [69,70]. Microscopic analyses showed the accumulation of autophagic structures during hypersensitive cell death in RabG3bCA cells [69,70]. These results suggest that RabG3b contributes to hypersensitive cell death through the activation of autophagy, which plays a positive role in plant immunity-triggered programmed cell death during the hypersensitive response. Thus, members of the Rab7 protein family regulate both MVB and autophagosome pathways during plant responses to pathogen infection (Figure 2).
4. Autophagy and MVB Coordination in Plant Abiotic Stress Response
Autophagy plays a critical role in nutrient recycling during leaf senescence or under nutrient starvation [1,71]. Due to the abundance in proteins and other molecules, chloroplasts are dismantled during leaf senescence or under N or C starvation and their constituents are transported by autophagosomes to vacuoles for degradation through several pathways [72]. Chloroplast stromal proteins, including Rubisco, are transported into the small double membrane structures called Rubisco-containing bodies (RCBs). The autophagic nature of RCBs is supported by the colocalization of RCBs labeled by a chloroplast-targeted red fluorescent protein with the GFP-ATG8 autophagosome marker [73]. Delivery of small starch granules (SSGs) from chloroplasts to vacuoles is carried out by plastid-derived small spherical structures called SSG-like bodies in an autophagy-dependent manner [74]. The selective autophagy receptor ATI1 (ATG8-interacting Protein 1) also mediates the delivery of chloroplast components to vacuoles for degradation [75]. ATI1-plastid associated bodies (ATI-PS), which contain thylakoid membrane proteins and chlorophylls, are detected in the periphery and inside of plastids [75]. ATI1-PS bodies are released from chloroplasts into the cytosol independent of the autophagic machinery [75]. However, their fusion with the central vacuole requires functional autophagy [75]. In addition, when cells are subjected to UV-induced damage, entire chloroplasts can be engulfed by autophagosomal structures [76].
The ESCRT-III subunit paralogs CHMP1A (charged MVB protein1) and CHMP1B play a direct role in the autophagic degradation of plastid proteins in Arabidopsis [77]. Specifically, the two homologs of the ESCRT-III component are required for the transport of RCB cargo into the vacuoles [77]. Like autophagy mutants, chmp1 mutant plants hyperaccumulated plastid clusters with plastid proteins, including proteins involved in plastid division [77]. Autophagy was increased in chmp1 based on an increase in vacuolar GFP cleavage from the autophagic reporter GFP-ATG8. However, autophagic degradation of the stromal cargo RECA-GFP was greatly reduced in the chmp1 plants upon starvation [77]. Thus, it appears that the autophagy machinery is responsible for the release of RCB bodies containing plastid material into the cytoplasm, whereas CHMP1 proteins are required for the delivery of RCB bodies to the vacuole. Therefore, ESCRT components and the autophagy machinery act in coordination in the delivery of chloroplast proteins to the vacuoles for degradation and nutrient recycling under starvation (Figure 2).
Coordination and crosstalk between endocytic MVB and autophagosome pathways are also involved in plant responses to water-related abiotic stresses caused by salt, osmotic, and drought conditions. Water flow across plant cell membranes is modulated by aquaporins in the plasma membrane and the tonoplast to regulate growth and transpiration [78]. The protein levels and activities of aquaporins are constantly regulated at the levels of transcription, protein stability, subcellular trafficking, and gating [78]. Generally speaking, the expression of most aquaporin genes is downregulated upon exposure to water-related stress, such as drought or salt stress conditions, probably to decrease the water permeability of the root, thereby limiting water loss and potentially creating a hydraulic signal for the induction of stomatal closure [78]. Recent studies have shown that both MVB and autophagosome pathways are involved in the degradation of plasma membrane-localized aquaporin proteins in response to abiotic stress responses. In Arabidopsis, salt stress reduces the fluorescence of GFP(green fluorescence protein)-labeled aquaporin PIP2;1 (plasma membrane intrinsic protein 2;1) at the plasma membrane and increased it in the vacuolar lumen [79]. The internalization of PIP2;1 is inhibited by inhibitors of clathrin-mediated endocytosis and phosphatidylinositol 3-kinase (PI3K) and phosphatidylinositol 4-kinase (PI4K) associated with vesicle trafficking [79]. Inhibiting PI4K and PI3K suppresses salt-induced endocytosis at the plasma membrane and trafficking after internalization of GFP-PIP2;1 [79]. These results suggest that salt stress induces the internalization of PIP2;1 from the plasma membrane through endocytosis to the vacuolar lumen though MVBs in a manner that is dependent on clathrin, PI3K, and PI4K.
Interestingly, a recent study has shown that another plasma membrane aquaporin, PIP2;7, is downregulated by the multi-stress regulator TSPO (outer membrane tryptophan-rich sensory protein) through a selective autophagic pathway during plant stress responses [80]. In Arabidopsis, the polytopic and stress-responsive membrane protein TSPO is localized in the Golgi apparatus, where its levels are tightly regulated by various mechanisms, including selective autophagy [80]. TSPO interacts with the plasma membrane aquaporin PIP2;7 at the ER and Golgi membranes in planta [80]. Overexpression of TSPO reduced the accumulation of overexpressed PIP2;7 in the plasma membrane and abolished the membrane water permeability mediated by transgenic PIP2;7 [80]. Inhibition of autophagy increased the stability of both TSPO and PIP2;7, suggesting that the autophagic pathway is responsible for the degradation of the complex containing both TSPO and PIP2;7 [80]. These results support a critical role for TSPO through a selective autophagy pathway in regulating the protein levels of PIP2;7 at the plasma membrane during abiotic stress conditions. Thus, the autophagosome and MVB pathways act coordinately in downregulating the levels of aquaporin proteins at the plasma membrane to reduce water transport under abiotic stresses. Selective autophagy increases the degradation of newly synthesized aquaporin proteins at the ER and Golgi apparatus to reduce their transport to the plasma membrane. On the other hand, the MVBs pathway promotes degradation of those aquaporin proteins already localized in the plasma membrane through increased endocytosis. Through coordinated degradation of both newly synthesized aquaporin proteins at the ER and Golgi apparatus by autophagy and plasma membrane-localized aquaporin proteins by the endocytosis/MVB pathway, plant root cells can rapidly downregulate the water channel proteins at the cell surface to reduce root cell water permeability upon exposure to abiotic stresses (Figure 2).
Arabidopsis autophagy-deficient mutants are hypersensitive to heat stress, indicating an important role of autophagy in plant heat stress responses [18,81]. High temperature increases misfolded proteins and causes proteotoxic stress. Cellular responses to proteotoxic stress include chaperone-dependent refolding of misfolded proteins, degradation through the 26S proteasome system, and sequestration through aggregation [82]. Protein aggregates are potentially cytotoxic and are removed primarily by a specific type of selective autophagy termed aggrephagy [83]. In plants, misfolded protein aggregates are ubiquitinated by unknown E3 ligases and recognized by the selective autophagy receptor NBR1 through its ubiquitin-associated (UBA) domain [18,19,84]. NBR1 also interacts with ATG8, thereby tethering the protein aggregates to ATG8 for autophagic clearance in the vacuole. Arabidopsis nbr1 mutants are hypersensitive to abiotic stresses, including high temperature, and this phenotype is associated with increased accumulation of ubiquitinated insoluble protein aggregates under heat stress [18,19,84]. These results indicate that NBR1-mediated aggrephagy plays a critical role in plant heat stress responses. There are additional pathways of selective autophagy that target misfolded proteins in different subcellular compartments under heat stress. In the ER, for example, an increase in misfolded proteins results in ER stress and triggers autophagy [15]. Arabidopsis ATG8-interacting ATI3 proteins also play a role in plant heat stress responses [85]. ATI3s interact with two closely related ER proteins UBAC2A (ubiquitin-associated protein 2a) and UBAC2B implicated in ER-associated protein degradation [85]. The ati3 and ubac2 mutants are compromised in both the heat tolerance and sensitivity to an ER stress-inducing agent [85]. These results support that ATI3 and UBAC2 participate in plant heat stress responses by targeting specific unknown ER components for autophagic degradation.
Heat stress increases misfolded proteins not only in intracellular compartments but also in the plasma membrane. Extensive studies in different eukaryotic systems have established that protein conformational surveillance and quality control of plasma-membrane proteins are necessary for cellular and organismal survival [86]. Like their intracellular counterparts, nonnative plasma membrane proteins are subjected to degradative quality control through four steps: (1) Recognition and ubiquitination, (2) endocytosis, (3) sorting into the intralumenal vesicles of MVBs, and (4) fusion of MVBs with the lysosomal/vacuolar compartment for degradation [86]. In plants, ubiquitination-mediated internalization and vacuolar clearance of plasma membrane proteins have also been demonstrated [87]. Furthermore, Arabidopsis lip5 mutants are compromised in tolerance to heat stress and the heat sensitivity of the lip5 mutant is associated with increased accumulation of ubiquitinated insoluble protein aggregates under heat stress [10]. Therefore, while autophagy plays a critical role in the clearance of nonnative protein aggregrates in the intracellular compartments, the endocytic and MVB pathway is critical in removal of nonnative plasma membrane proteins under heat stress (Figure 2). Ubiquitination of nonnative plasma membrane proteins in other eukaryotic organisms involves the conserved CHIP (C-terminus of Hsc70 interacting protein) E3 ligase [86]. Genetic analysis has shown that Arabidopsis mutants for the conserved CHIP E3 ligase are also compromised in heat tolerance and this phenotype is enhanced in the autophagy-deficient mutants [19]. The additive roles of CHIP- and autophagy-dependent protein degradation pathways further support the collective action of multiple protein degradation pathways under heat stress.
5. Autophagy and MVB Coordination in Plant Hormone-Mediated Regulation of Stress Responses
Plant hormones play important roles not only in plant growth and development but also in plant responses to biotic and abiotic stresses [19]. Among well-studied plant hormones, ABA (abscisic acid), ET (ethylene), JA (jasmonic acid), and SA (salicylic acid) are closely associated with plant stress responses, while others including auxin, CK (cytokinins), GA (gibberellic acid), and BRs (brassinosteroids) also participate in stress-triggered signaling [35]. Plant hormones can regulate autophagy and endocytic pathways in response to developmental and environmental cues. Autophagy genes, for example, can be regulated transcriptionally and posttranscriptionally by plant hormones, including ET, auxin, ABA, and SA [35]. Recent studies have also revealed that the critical roles of the endocytosis, MVB, and autophagy pathways in plant stress responses are in part mediated through their coordinated action in the metabolism, distribution, and signaling of important plant hormones, such as ABA, auxin, and BRs [35,88,89] (Figure 2).
ABA is an important plant hormone in seed maturation and germination and in plant response to both biotic and abiotic stresses. As discussed earlier, the multi-stress regulator TSPO is a selective autophagy receptor that mediates autophagic degradation of specific plasma membrane proteins, such as PIP2;7, aquaporin during plant stress responses [80]. TSPO is induced by ABA [90] and, thus, links the action of selective autophagy with ABA-regulated plant stress responses. Likewise, selective autophagy receptors ATI1 and 2 play a role not only in salt tolerance but also in seed maturation in response to ABA [91]. Therefore, the turnover of specific proteins through selective autophagy is a critical mechanism of ABA-regulated stress responses. The MVB pathway also plays a critical role in ABA-regulated stress responses through modulation of ABA signaling. ABA signaling involves a molecular module consisting of the PYR/PYL (pyrabactin resistance/pyrabactin resistance-like) ABA receptors, type 2C protein phosphatases (PP2Cs), and members of the sucrose non-fermenting-related kinase group 2 (SnRK2) family [89]. ABA binding of PYR/PYL ABA receptors promotes inhibition of PP2Cs [89]. When PP2Cs are inhibited, SnRK2s will remain phosphorylated and can activate ABA-responsive transcription factors [89]. PYR/PYL ABA receptors are soluble but are present at the membrane, most likely through interaction with a family of small proteins called CAR (C2-domain ABA-related) proteins that contain a lipid-binding C2 domain [92]. The E3 ligase RSL1 (ring finger of seed longevity 1), which contains a transmembrane domain and is localized to the plasma membrane and TGN, can ubiquitinate the PYR1 and PYL ABA receptors [93,94]. Furthermore, MVB ESCRT-I components FREE1 and VPS23A interact with PYR1/PYL4 and the levels of the ABA receptors increase in the free1 and vps23a mutants [93,95]. These results indicate that ESCRT-I components FREE1 and VPS23a participate in ABA signaling by recognizing those PYR1/PYL4 ABA receptors ubiquitinated by the RSL1 E3 ligase and target their turnover by the MVB pathway. Very recently, it has been reported that FREE1 can also be phosphorylated by SnRK2 and relocate to the nucleus, where it interacts with ABA-responsive transcription factors and inhibits their DNA-binding activity [96]. Thus, both components from both autophagy and MVB pathways modulate the turnover and activity of ABA receptors and downstream regulators, including ABA-responsive transcription factors (Figure 2).
Auxin is a plant hormone with a cardinal role in the coordination of plant growth in response to developmental and environmental signals. One such auxin-regulated plastic plant body development in response to environmental stress is the formation of lateral roots under nutrient deficiency to increase nutrient acquisition from soil [97]. Lateral root development can also be inhibited as an avoidance strategy when plants are confronted with nutrient stress, high salinity, or heavy metals. Auxin is an important positive regulator of lateral root development and its directional transport through auxin efflux carriers, such as PIN (pin-formed), is critical [97]. Directional auxin transport promotes the establishment of auxin concentration gradients, and polar root cell growth during lateral root development is linked to the dynamic redistribution of PINs at the plasma membrane, which is tightly controlled by several protein trafficking pathways [98]. The plasma membrane-localized PINs can be internalized through clathrin-mediated endocytosis and can then be either recycled back to the plasma membrane or delivered to the vacuole for degradation through MVBs [98]. Using chemical biology, Perez-Henriquez and colleagues reported that the bioactive compound Sortin2 increases endosomal trafficking of plasma membrane recycling proteins, including PIN2, to the vacuole through MVBs and this action is important for Sortin2-induced lateral root initiation that is independent of the auxin receptor SCFTIR [97]. These results support a pivotal role of endocytic trafficking of late endosome/MVB towards the vacuole in the induction of lateral roots. There are also reports on a role of autophagy in stress-induced lateral root formation. In Arabidopsis, lateral root formation under phosphate starvation requires the S-domain receptor kinase ARK2 and U box/Armadillo Repeat-containing E3 ligase PUB9 module [99,100]. The ark2/pub9 mutant plants are defective in both lateral root formation and auxin accumulation in the root tips under phosphate starvation [99,100]. Interestingly, PUB9 is localized to autophagosomes upon phosphorylation by ARK2 or under phosphate starvation [99,100]. Inhibition of autophagic responses in Arabidopsis also leads to inhibition of both lateral root formation and auxin accumulation in the root tips [99,100]. These results indicate that autophagy is involved in the action of ARK2/PUB9 module in the regulation of lateral root development under phosphate starvation, likely through selective degradation of repressors of auxin accumulation and signaling (Figure 2).
Like auxin, BRs play important roles not only in plant growth and development but also in plant responses to environmental stresses, such as extreme temperatures and drought [88]. BRs are perceived at the cell surface by plasma membrane-localized receptors BR-INSENSITIVE-1 (BRI1), and its homologs, BRI1-LIKE-1 (BRL1) and BRL3. The binding triggers interaction with BRI1-ASSOCIATED-KINASE-1 (BAK1) (or members from the SOMATICEMBRYOGENESIS-RECEPTOR-KINASE (SERK) family) and the transphosphorylation of their kinase domains [88]. Activation of receptor complexes leads to a signaling cascade that relays BR signals to the accumulation of BRASSINAZOLERESISTANT-1 (BZR1) and BR-INSENSITIVE-EMS-SUPPRESSOR-1 (BES1) transcription factors in the nucleus, which control expression of BR-regulated genes [88]. Genetic analysis with BR-deficient or insensitive mutants have revealed BRs have a negative role in plant stress tolerance [101,102,103]. Both the endocytic and autophagy pathways can modulate components in BR signaling for downregulation of BR-regulated growth and upregulation of stress responses. First, the activity of BRI1 is under tight positive and negative regulation through phosphorylation, ubiquitination, endocytosis, and vacuolar degradation [104]. Upon BR perception, activated BRI1 phosphorylates PUB13 E3 ligase and stimulates their association and PUB13-dependent ubiquitination of BRI1 [104]. Ubiquitinated BRI1 undergoes clathrin-mediated endocytosis that is delivered through MVB trafficking to the vacuole for degradation [104]. Impaired internalization of BRI1 results in increased accumulation of BRI1 proteins in the plasma membrane and BR hypersensitivity [88]. Therefore, the MVB pathway plays a role in the attenuation of BR signaling (Figure 2). Second, autophagy also modulates BR responses through degradation of the BR-responsive transcription factors BES1 and BZR1 (Figure 2). Autophagy-dependent turnover of BEST1 is mediated by the autophagy receptor DSK2 (dominant suppressor of KAR2), which also interacts with ATG8 [102]. The interaction of DSK2 with ATG8 is activated upon phosphorylation of DSK2 by the GSK3-like kinase BIN2, a negative regulator in the BR pathway [102]. This mode of DSK2 regulation and turnover of BES1 integrates BR and autophagy pathways to achieve balances between growth and stress responses [88,102]. Similarly, carbon starvation leads to TOR inactivation, autophagy induction, and degradation of BZR1 transcription factor in the BR pathway [105]. By promoting the degradation of both BES1 and BZR1 transcription factors under stress conditions, autophagy helps modulate BR-promoted growth to promote stress responses. Intriguingly, studies using the application of exogenous BRs have indicated a positive role of BRs in plant tolerance to several stresses [106,107,108]. A recent study has reported that exogenous application of BR promotes autophagosome formation, decreases accumulation of ubiquitinated proteins, and enhances chlorophyll content under N starvation in tomato [109]. The protective role of exogenous BR in the plant response to N starvation is in part mediated by the action of the BZR1 transcription factor through upregulation of autophagy gene expression [109]. Therefore, the roles of BRs in plant stress responses are complex, which may reflect the dynamic nature of the activity of particular components in the BR and other associated signal transduction pathways that are subject to tight regulation by cellular processes, including autophagy and MVB trafficking.
6. Conclusions and Prospects
Studies in non-plant eukaryotic organisms, such as yeast, have established that autophagy and MVB pathways interact and coordinate at multiple levels in the regulation of cell growth and survival [28,29,30,31]. Recent studies have also made significant progress in revealing crosstalk and coordination of the two important trafficking pathways in plant cells [32,33]. In particular, a number of reported studies have identified components commonly associated with MVB and related trafficking pathways to be important in the biogenesis, maturation, and trafficking of autophagosomes in plant cells [32,33]. Both the autophagy and MVB pathways play critical roles in plant responses to a broad spectrum of biotic and abiotic stresses and, given the closely related nature of the two protein degradation pathways, it is highly expected that these two close protein degradation pathways are coordinated in the regulation and action in plant stress responses. In this review, we discussed recent progress in the analysis of a substantial number of components that are involved in the crosstalk and coordination between autophagy and MVB pathways in both their regulation and functions during plant responses to biotic and abiotic stresses (Figure 2, Table 1). Despite this progress, our understanding of the complex nature of the interactions between the autophagy and MVB pathways during the plant stress response is still very limited. In contrast to the large number of studies on stress-responsive autophagy, the role of the MVB pathway in the plant stress response has been examined only to a very limited extent, most likely due to la ack of appropriate mutants because of the essential role of MVBs in plant growth and development. With the identification of key regulators, such as LIP5, required for stress-induced MVB biogenesis, it is now possible to examine genetically the functional and mechanistic interactions between the autophagy and MVB pathways in the plant stress response through the generation and analysis of genetic mutants that are deficient in both stress-induced autophagosome and MVB biogenesis. Such studies will further our understanding of the important and dynamic roles of the cellular vesicle trafficking system in the complex network of plant stress responses.

Table 1.
Important Arabidopsis genes in autophagy and MVB (multivesicular body) coordination in plant stress responses.
Author Contributions
C.Z. and Z.C. conceived the idea. M.W., X.L., S.L., B.F., C.Z. and Z.C. wrote and evaluated the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the Natural Science Foundation of Zhejiang Province grant LY19C020001 and National Research and Development Plan of the Ministry of Science of Technology of China grant SQ2018YFE010025 to C.Z. and the U.S. National Science Foundation grant IOS1456300 to Z.C.
Acknowledgments
The authors wish to thank members of the Zhu and Chen labs for their discussion and feedback on the review.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Bassham, D.C.; Laporte, M.; Marty, F.; Moriyasu, Y.; Ohsumi, Y.; Olsen, L.J.; Yoshimoto, K. Autophagy in development and stress responses of plants. Autophagy 2006, 2, 2–11. [Google Scholar] [CrossRef] [PubMed]
- Levine, B.; Mizushima, N.; Virgin, H.W. Autophagy in immunity and inflammation. Nature 2011, 469, 323–335. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Xu, M.; Wang, G.; Galili, G. Autophagy: An Important Biological Process That Protects Plants from Stressful Environments. Front. Plant Sci. 2016, 7, 2030. [Google Scholar] [CrossRef] [PubMed]
- Soto-Burgos, J.; Zhuang, X.; Jiang, L.; Bassham, D.C. Dynamics of Autophagosome Formation. Plant Physiol. 2018, 176, 219–229. [Google Scholar] [CrossRef]
- Hanson, P.I.; Cashikar, A. Multivesicular body morphogenesis. Annu. Rev. Cell Dev. Biol. 2012, 28, 337–362. [Google Scholar] [CrossRef]
- Li, X.; Bao, H.; Wang, Z.; Wang, M.; Fan, B.; Zhu, C.; Chen, Z. Biogenesis and Function of Multivesicular Bodies in Plant Immunity. Front. Plant Sci. 2018, 9, 979. [Google Scholar] [CrossRef]
- Paez Valencia, J.; Goodman, K.; Otegui, M.S. Endocytosis and Endosomal Trafficking in Plants. Annu. Rev. Plant Biol. 2016, 67, 309–335. [Google Scholar] [CrossRef]
- Bassham, D.C. Plant autophagy—More than a starvation response. Curr. Opin. Plant Biol. 2007, 10, 587–593. [Google Scholar] [CrossRef]
- Wang, F.; Shang, Y.; Fan, B.; Yu, J.Q.; Chen, Z. Arabidopsis LIP5, a positive regulator of multivesicular body biogenesis, is a critical target of pathogen-responsive MAPK cascade in plant basal defense. PLoS Pathog. 2014, 10, e1004243. [Google Scholar] [CrossRef]
- Wang, F.; Yang, Y.; Wang, Z.; Zhou, J.; Fan, B.; Chen, Z. A Critical Role of Lyst-Interacting Protein5, a Positive Regulator of Multivesicular Body Biogenesis, in Plant Responses to Heat and Salt Stresses. Plant Physiol. 2015, 169, 497–511. [Google Scholar] [CrossRef]
- Lai, Z.; Wang, F.; Zheng, Z.; Fan, B.; Chen, Z. A critical role of autophagy in plant resistance to necrotrophic fungal pathogens. Plant J. Cell Mol. Biol. 2011, 66, 953–968. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.; Xiong, Y.; Bassham, D.C. Autophagy is required for tolerance of drought and salt stress in plants. Autophagy 2009, 5, 954–963. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Yu, J.Q.; Chen, Z. The Perplexing Role of Autophagy in Plant Innate Immune Responses. Mol. Plant Pathol. 2014. [Google Scholar] [CrossRef] [PubMed]
- Michaeli, S.; Galili, G.; Genschik, P.; Fernie, A.R.; Avin-Wittenberg, T. Autophagy in Plants—What’s New on the Menu? Trends Plant Sci. 2016, 21, 134–144. [Google Scholar] [CrossRef]
- Pu, Y.; Bassham, D.C. Links between ER stress and autophagy in plants. Plant Signal. Behav. 2013, 8. [Google Scholar] [CrossRef]
- Slavikova, S.; Shy, G.; Yao, Y.; Glozman, R.; Levanony, H.; Pietrokovski, S.; Elazar, Z.; Galili, G. The autophagy-associated Atg8 gene family operates both under favourable growth conditions and under starvation stresses in Arabidopsis plants. J. Exp. Bot. 2005, 56, 2839–2849. [Google Scholar] [CrossRef]
- Wang, Y.; Wu, Y.; Tang, D. The autophagy gene, ATG18a, plays a negative role in powdery mildew resistance and mildew-induced cell death in Arabidopsis. Plant Signal. Behav. 2011, 6, 1408–1410. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Cheng, Y.; Chi, Y.J.; Fan, B.; Yu, J.Q.; Chen, Z. NBR1-Mediated Selective Autophagy Targets Insoluble Ubiquitinated Protein Aggregates in Plant Stress Responses. PLoS Genet. 2013, 9, e1003196. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Y.; Qi, J.; Chi, Y.; Fan, B.; Yu, J.Q.; Chen, Z. E3 Ubiquitin Ligase CHIP and NBR1-Mediated Selective Autophagy Protect Additively against Proteotoxicity in Plant Stress Responses. PLoS Genet. 2014, 10, e1004116. [Google Scholar] [CrossRef]
- Lenz, H.D.; Haller, E.; Melzer, E.; Kober, K.; Wurster, K.; Stahl, M.; Bassham, D.C.; Vierstra, R.D.; Parker, J.E.; Bautor, J.; et al. Autophagy differentially controls plant basal immunity to biotrophic and necrotrophic pathogens. Plant J. Cell Mol. Biol. 2011, 66, 818–830. [Google Scholar] [CrossRef]
- Liu, Y.; Schiff, M.; Czymmek, K.; Talloczy, Z.; Levine, B.; Dinesh-Kumar, S.P. Autophagy regulates programmed cell death during the plant innate immune response. Cell 2005, 121, 567–577. [Google Scholar] [CrossRef] [PubMed]
- Seay, M.D.; Dinesh-Kumar, S.P. Life after death: Are autophagy genes involved in cell death and survival during plant innate immune responses? Autophagy 2005, 1, 185–186. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Ustun, S.; Hafren, A.; Liu, Q.; Marshall, R.S.; Minina, E.A.; Bozhkov, P.V.; Vierstra, R.D.; Hofius, D. Bacteria Exploit Autophagy for Proteasome Degradation and Enhanced Virulence in Plants. Plant Cell 2018, 30, 668–685. [Google Scholar] [CrossRef] [PubMed]
- Haxim, Y.; Ismayil, A.; Jia, Q.; Wang, Y.; Zheng, X.; Chen, T.; Qian, L.; Liu, N.; Wang, Y.; Han, S.; et al. Autophagy functions as an antiviral mechanism against geminiviruses in plants. Elife 2017, 6. [Google Scholar] [CrossRef]
- Ismayil, A.; Yang, M.; Liu, Y. Role of autophagy during plant-virus interactions. Semin. Cell Dev. Biol. 2019. [Google Scholar] [CrossRef]
- Dubeaux, G.; Neveu, J.; Zelazny, E.; Vert, G. Metal Sensing by the IRT1 Transporter-Receptor Orchestrates Its Own Degradation and Plant Metal Nutrition. Mol. Cell 2018, 69, 953–964. e5. [Google Scholar] [CrossRef]
- Shin, L.J.; Lo, J.C.; Chen, G.H.; Callis, J.; Fu, H.; Yeh, K.C. IRT1 degradation factor1, a ring E3 ubiquitin ligase, regulates the degradation of iron-regulated transporter1 in Arabidopsis. Plant Cell 2013, 25, 3039–3051. [Google Scholar] [CrossRef]
- Fader, C.M.; Colombo, M.I. Multivesicular bodies and autophagy in erythrocyte maturation. Autophagy 2006, 2, 122–125. [Google Scholar] [CrossRef]
- Fader, C.M.; Colombo, M.I. Autophagy and multivesicular bodies: Two closely related partners. Cell Death Differ. 2009, 16, 70–78. [Google Scholar] [CrossRef]
- Muller, M.; Schmidt, O.; Angelova, M.; Faserl, K.; Weys, S.; Kremser, L.; Pfaffenwimmer, T.; Dalik, T.; Kraft, C.; Trajanoski, Z.; et al. The coordinated action of the MVB pathway and autophagy ensures cell survival during starvation. Elife 2015, 4, e07736. [Google Scholar] [CrossRef]
- Shin, H.; Bang, S.; Kim, J.; Jun, J.H.; Song, H.; Lim, H.J. The formation of multivesicular bodies in activated blastocysts is influenced by autophagy and FGF signaling in mice. Sci. Rep. 2017, 7, 41986. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; He, Y.; Cao, W.; Gao, J.; Jiang, L. The Multivesicular Body and Autophagosome Pathways in Plants. Front. Plant Sci. 2018, 9, 1837. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Cui, Y.; Gao, C.; Jiang, L. Endocytic and autophagic pathways crosstalk in plants. Curr Opin Plant Biol. 2015, 28, 39–47. [Google Scholar] [CrossRef] [PubMed]
- Mao, G.; Meng, X.; Liu, Y.; Zheng, Z.; Chen, Z.; Zhang, S. Phosphorylation of a WRKY transcription factor by two pathogen-responsive MAPKs drives phytoalexin biosynthesis in Arabidopsis. Plant Cell 2011, 23, 1639–1653. [Google Scholar] [CrossRef] [PubMed]
- Liao, C.Y.; Bassham, D.C. Combating stress: The interplay between hormone signaling and autophagy in plants. J. Exp. Bot. 2019. [Google Scholar] [CrossRef] [PubMed]
- Signorelli, S.; Tarkowski, L.P.; Van den Ende, W.; Bassham, D.C. Linking Autophagy to Abiotic and Biotic Stress Responses. Trends Plant Sci. 2019, 24, 413–430. [Google Scholar] [CrossRef]
- Nagel, M.K.; Kalinowska, K.; Vogel, K.; Reynolds, G.D.; Wu, Z.; Anzenberger, F.; Ichikawa, M.; Tsutsumi, C.; Sato, M.H.; Kuster, B.; et al. Arabidopsis SH3P2 is an ubiquitin-binding protein that functions together with ESCRT-I and the deubiquitylating enzyme AMSH3. Proc. Natl. Acad. Sci. USA 2017, 114, E7197–E7204. [Google Scholar] [CrossRef]
- Zhuang, X.; Jiang, L. Autophagosome biogenesis in plants: Roles of SH3P2. Autophagy 2014, 10, 704–705. [Google Scholar] [CrossRef]
- Zhuang, X.; Wang, H.; Lam, S.K.; Gao, C.; Wang, X.; Cai, Y.; Jiang, L. A BAR-domain protein SH3P2, which binds to phosphatidylinositol 3-phosphate and ATG8, regulates autophagosome formation in Arabidopsis. Plant Cell 2013, 25, 4596–4615. [Google Scholar] [CrossRef]
- Gao, C.; Zhuang, X.; Cui, Y.; Fu, X.; He, Y.; Zhao, Q.; Zeng, Y.; Shen, J.; Luo, M.; Jiang, L. Dual roles of an Arabidopsis ESCRT component FREE1 in regulating vacuolar protein transport and autophagic degradation. Proc. Natl. Acad. Sci. USA 2015, 112, 1886–1891. [Google Scholar] [CrossRef]
- Cui, Y.; Shen, J.; Gao, C.; Zhuang, X.; Wang, J.; Jiang, L. Biogenesis of Plant Prevacuolar Multivesicular Bodies. Mol. Plant 2016, 9, 774–786. [Google Scholar] [CrossRef] [PubMed]
- Spitzer, C.; Reyes, F.C.; Buono, R.; Sliwinski, M.K.; Haas, T.J.; Otegui, M.S. The ESCRT-related CHMP1A and B proteins mediate multivesicular body sorting of auxin carriers in Arabidopsis and are required for plant development. Plant Cell 2009, 21, 749–766. [Google Scholar] [CrossRef] [PubMed]
- Erwig, J.; Ghareeb, H.; Kopischke, M.; Hacke, R.; Matei, A.; Petutschnig, E.; Lipka, V. Chitin-induced and CHITIN ELICITOR RECEPTOR KINASE1 (CERK1) phosphorylation-dependent endocytosis of Arabidopsis thaliana LYSIN MOTIF-CONTAINING RECEPTOR-LIKE KINASE5 (LYK5). New Phytol. 2017, 215, 382–396. [Google Scholar] [CrossRef] [PubMed]
- Spallek, T.; Beck, M.; Ben Khaled, S.; Salomon, S.; Bourdais, G.; Schellmann, S.; Robatzek, S. ESCRT-I mediates FLS2 endosomal sorting and plant immunity. PLoS Genet. 2013, 9, e1004035. [Google Scholar] [CrossRef] [PubMed]
- Kasai, K.; Takano, J.; Fujiwara, T. Analysis of endocytosis and ubiquitination of the BOR1 transporter. Methods Mol. Biol. 2014, 1209, 203–217. [Google Scholar] [CrossRef] [PubMed]
- Cardona-Lopez, X.; Cuyas, L.; Marin, E.; Rajulu, C.; Irigoyen, M.L.; Gil, E.; Puga, M.I.; Bligny, R.; Nussaume, L.; Geldner, N.; et al. ESCRT-III-Associated Protein ALIX Mediates High-Affinity Phosphate Transporter Trafficking to Maintain Phosphate Homeostasis in Arabidopsis. Plant Cell 2015, 27, 2560–2581. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Sun, Q.; Wang, K.; Du, Q.; Li, W.X. Nitrogen Limitation Adaptation (NLA) is involved in source-to-sink remobilization of nitrate by mediating the degradation of NRT1.7 in Arabidopsis. New Phytol. 2017, 214, 734–744. [Google Scholar] [CrossRef]
- Haas, T.J.; Sliwinski, M.K.; Martinez, D.E.; Preuss, M.; Ebine, K.; Ueda, T.; Nielsen, E.; Odorizzi, G.; Otegui, M.S. The Arabidopsis AAA ATPase SKD1 is involved in multivesicular endosome function and interacts with its positive regulator LYST-INTERACTING PROTEIN5. Plant Cell 2007, 19, 1295–1312. [Google Scholar] [CrossRef]
- Lai, Z.; Li, Y.; Wang, F.; Cheng, Y.; Fan, B.; Yu, J.Q.; Chen, Z. Arabidopsis sigma factor binding proteins are activators of the WRKY33 transcription factor in plant defense. Plant Cell 2011, 23, 3824–3841. [Google Scholar] [CrossRef]
- Zheng, Z.; Qamar, S.A.; Chen, Z.; Mengiste, T. Arabidopsis WRKY33 transcription factor is required for resistance to necrotrophic fungal pathogens. Plant J. 2006, 48, 592–605. [Google Scholar] [CrossRef]
- Xu, J.; Zhang, S. Mitogen-activated protein kinase cascades in signaling plant growth and development. Trends Plant Sci. 2015, 20, 56–64. [Google Scholar] [CrossRef] [PubMed]
- Schulze-Lefert, P. Knocking on the heaven’s wall: Pathogenesis of and resistance to biotrophic fungi at the cell wall. Curr. Opin. Plant Biol. 2004, 7, 377–383. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Ehlers, K.; Kogel, K.H.; van Bel, A.J.; Huckelhoven, R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 2006, 172, 563–576. [Google Scholar] [CrossRef] [PubMed]
- An, Q.; Huckelhoven, R.; Kogel, K.H.; van Bel, A.J. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell MicroBiol. 2006, 8, 1009–1019. [Google Scholar] [CrossRef]
- An, Q.; van Bel, A.J.; Huckelhoven, R. Do plant cells secrete exosomes derived from multivesicular bodies? Plant Signal. Behav. 2007, 2, 4–7. [Google Scholar] [CrossRef]
- Bohlenius, H.; Morch, S.M.; Godfrey, D.; Nielsen, M.E.; Thordal-Christensen, H. The multivesicular body-localized GTPase ARFA1b/1c is important for callose deposition and ROR2 syntaxin-dependent preinvasive basal defense in barley. Plant Cell 2010, 22, 3831–3844. [Google Scholar] [CrossRef]
- Meyer, D.; Pajonk, S.; Micali, C.; O’Connell, R.; Schulze-Lefert, P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 2009, 57, 986–999. [Google Scholar] [CrossRef]
- Nielsen, M.E.; Feechan, A.; Bohlenius, H.; Ueda, T.; Thordal-Christensen, H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc. Natl. Acad. Sci. USA 2012, 109, 11443–11448. [Google Scholar] [CrossRef]
- Baldrich, P.; Rutter, B.D.; Karimi, H.Z.; Podicheti, R.; Meyers, B.C.; Innes, R.W. Plant Extracellular Vesicles Contain Diverse Small RNA Species and Are Enriched in 10- to 17-Nucleotide “Tiny” RNAs. Plant Cell 2019, 31, 315–324. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Extracellular Vesicles Isolated from the Leaf Apoplast Carry Stress-Response Proteins. Plant Physiol. 2017, 173, 728–741. [Google Scholar] [CrossRef]
- Rutter, B.D.; Innes, R.W. Extracellular vesicles as key mediators of plant-microbe interactions. Curr Opin Plant Biol. 2018, 44, 16–22. [Google Scholar] [CrossRef]
- Berkey, R.; Zhang, Y.; Ma, X.; King, H.; Zhang, Q.; Wang, W.; Xiao, S. Homologues of the RPW8 Resistance Protein Are Localized to the Extrahaustorial Membrane that Is Likely Synthesized De Novo. Plant Physiol. 2017, 173, 600–613. [Google Scholar] [CrossRef] [PubMed]
- Nielsen, M.E.; Jurgens, G.; Thordal-Christensen, H. VPS9a Activates the Rab5 GTPase ARA7 to Confer Distinct Pre- and Postinvasive Plant Innate Immunity. Plant Cell 2017, 29, 1927–1937. [Google Scholar] [CrossRef] [PubMed]
- Bozkurt, T.O.; Belhaj, K.; Dagdas, Y.F.; Chaparro-Garcia, A.; Wu, C.H.; Cano, L.M.; Kamoun, S. Rerouting of plant late endocytic trafficking toward a pathogen interface. Traffic 2015, 16, 204–226. [Google Scholar] [CrossRef] [PubMed]
- Dagdas, Y.F.; Pandey, P.; Tumtas, Y.; Sanguankiattichai, N.; Belhaj, K.; Duggan, C.; Leary, A.Y.; Segretin, M.E.; Contreras, M.P.; Savage, Z.; et al. Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. Elife 2018, 7. [Google Scholar] [CrossRef]
- Dagdas, Y.F.; Belhaj, K.; Maqbool, A.; Chaparro-Garcia, A.; Pandey, P.; Petre, B.; Tabassum, N.; Cruz-Mireles, N.; Hughes, R.K.; Sklenar, J.; et al. An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. Elife 2016, 5. [Google Scholar] [CrossRef]
- Jones, J.D.; Dangl, J.L. The plant immune system. Nature 2006, 444, 323–329. [Google Scholar] [CrossRef]
- Cui, Y.; Zhao, Q.; Gao, C.; Ding, Y.; Zeng, Y.; Ueda, T.; Nakano, A.; Jiang, L. Activation of the Rab7 GTPase by the MON1-CCZ1 Complex Is Essential for PVC-to-Vacuole Trafficking and Plant Growth in Arabidopsis. Plant Cell 2014, 26, 2080–2097. [Google Scholar] [CrossRef]
- Kwon, S.I.; Cho, H.J.; Kim, S.R.; Park, O.K. The Rab GTPase RabG3b positively regulates autophagy and immunity-associated hypersensitive cell death in Arabidopsis. Plant Physiol. 2013, 161, 1722–1736. [Google Scholar] [CrossRef]
- Kwon, S.I.; Cho, H.J.; Lee, J.S.; Jin, H.; Shin, S.J.; Kwon, M.; Noh, E.W.; Park, O.K. Overexpression of constitutively active Arabidopsis RabG3b promotes xylem development in transgenic poplars. Plant Cell Env. 2011, 34, 2212–2224. [Google Scholar] [CrossRef]
- Li, F.Q.; Vierstra, R.D. Autophagy: A multifaceted intracellular system for bulk and selective recycling. Trends Plant Sci. 2012, 17, 526–537. [Google Scholar] [CrossRef] [PubMed]
- Zhuang, X.; Jiang, L. Chloroplast Degradation: Multiple Routes Into the Vacuole. Front. Plant Sci. 2019, 10, 359. [Google Scholar] [CrossRef] [PubMed]
- Ishida, H.; Yoshimoto, K. Chloroplasts are partially mobilized to the vacuole by autophagy. Autophagy 2008, 4, 961–962. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Yu, B.; Zhao, J.; Guo, J.; Li, Y.; Han, S.; Huang, L.; Du, Y.; Hong, Y.; Tang, D.; et al. Autophagy contributes to leaf starch degradation. Plant Cell 2013, 25, 1383–1399. [Google Scholar] [CrossRef]
- Michaeli, S.; Honig, A.; Levanony, H.; Peled-Zehavi, H.; Galili, G. Arabidopsis ATG8-INTERACTING PROTEIN1 is involved in autophagy-dependent vesicular trafficking of plastid proteins to the vacuole. Plant Cell 2014, 26, 4084–4101. [Google Scholar] [CrossRef]
- Izumi, M.; Ishida, H.; Nakamura, S.; Hidema, J. Entire Photodamaged Chloroplasts Are Transported to the Central Vacuole by Autophagy. Plant Cell 2017, 29, 377–394. [Google Scholar] [CrossRef]
- Spitzer, C.; Li, F.; Buono, R.; Roschzttardtz, H.; Chung, T.; Zhang, M.; Osteryoung, K.W.; Vierstra, R.D.; Otegui, M.S. The endosomal protein CHARGED MULTIVESICULAR BODY PROTEIN1 regulates the autophagic turnover of plastids in Arabidopsis. Plant Cell 2015, 27, 391–402. [Google Scholar] [CrossRef]
- Chaumont, F.; Tyerman, S.D. Aquaporins: Highly regulated channels controlling plant water relations. Plant Physiol. 2014, 164, 1600–1618. [Google Scholar] [CrossRef]
- Ueda, M.; Tsutsumi, N.; Fujimoto, M. Salt stress induces internalization of plasma membrane aquaporin into the vacuole in Arabidopsis thaliana. Biochem. Biophys. Res. Commun. 2016, 474, 742–746. [Google Scholar] [CrossRef]
- Hachez, C.; Veljanovski, V.; Reinhardt, H.; Guillaumot, D.; Vanhee, C.; Chaumont, F.; Batoko, H. The Arabidopsis abiotic stress-induced TSPO-related protein reduces cell-surface expression of the aquaporin PIP2;7 through protein-protein interactions and autophagic degradation. Plant Cell 2014, 26, 4974–4990. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, J.; Yu, J.Q.; Chen, Z. Role and regulation of autophagy in heat stress responses of tomato plants. Front. Plant Sci. 2014, 5, 174. [Google Scholar] [CrossRef]
- Harper, J.W.; Bennett, E.J. Proteome complexity and the forces that drive proteome imbalance. Nature 2016, 537, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Wurzer, B.; Zaffagnini, G.; Fracchiolla, D.; Turco, E.; Abert, C.; Romanov, J.; Martens, S. Oligomerization of p62 allows for selection of ubiquitinated cargo and isolation membrane during selective autophagy. Elife 2015, 4, e08941. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.; Lee, H.N.; Marshall, R.S.; Lomax, A.W.; Yoon, M.J.; Kim, J.; Kim, J.H.; Vierstra, R.D.; Chung, T. NBR1 Mediates Selective Autophagy of Defective Proteins in Arabidopsis. J. Exp. Bot. 2019. [Google Scholar] [CrossRef]
- Zhou, J.; Wang, Z.; Wang, X.; Li, X.; Zhang, Z.; Fan, B.; Zhu, C.; Chen, Z. Dicot-specific ATG8-interacting ATI3 proteins interact with conserved UBAC2 proteins and play critical roles in plant stress responses. Autophagy 2018. [Google Scholar] [CrossRef]
- Apaja, P.M.; Lukacs, G.L. Protein homeostasis at the plasma membrane. Physiology 2014, 29, 265–277. [Google Scholar] [CrossRef]
- Scheuring, D.; Kunzl, F.; Viotti, C.; Yan, M.S.; Jiang, L.; Schellmann, S.; Robinson, D.G.; Pimpl, P. Ubiquitin initiates sorting of Golgi and plasma membrane proteins into the vacuolar degradation pathway. BMC Plant Biol. 2012, 12, 164. [Google Scholar] [CrossRef]
- Nolan, T.; Vukasinovic, N.; Liu, D.; Russinova, E.; Yin, Y. Brassinosteroids: Multi-Dimensional Regulators of Plant Growth, Development, and Stress Responses. Plant Cell 2019. [Google Scholar] [CrossRef]
- Yu, F.; Xie, Q. Non-26S Proteasome Endomembrane Trafficking Pathways in ABA Signaling. Trends Plant Sci. 2017, 22, 976–985. [Google Scholar] [CrossRef]
- Guillaumot, D.; Guillon, S.; Morsomme, P.; Batoko, H. ABA, porphyrins and plant TSPO-related protein. Plant Signal. Behav. 2009, 4, 1087–1090. [Google Scholar] [CrossRef]
- Honig, A.; Avin-Wittenberg, T.; Ufaz, S.; Galili, G. A New Type of Compartment, Defined by Plant-Specific Atg8-Interacting Proteins, Is Induced upon Exposure of Arabidopsis Plants to Carbon Starvation. Plant Cell 2012, 24, 288–303. [Google Scholar] [CrossRef] [PubMed]
- Diaz, M.; Sanchez-Barrena, M.J.; Gonzalez-Rubio, J.M.; Rodriguez, L.; Fernandez, D.; Antoni, R.; Yunta, C.; Belda-Palazon, B.; Gonzalez-Guzman, M.; Peirats-Llobet, M.; et al. Calcium-dependent oligomerization of CAR proteins at cell membrane modulates ABA signaling. Proc. Natl. Acad. Sci. USA 2016, 113, E396–E405. [Google Scholar] [CrossRef] [PubMed]
- Belda-Palazon, B.; Rodriguez, L.; Fernandez, M.A.; Castillo, M.C.; Anderson, E.M.; Gao, C.; Gonzalez-Guzman, M.; Peirats-Llobet, M.; Zhao, Q.; De Winne, N.; et al. FYVE1/FREE1 Interacts with the PYL4 ABA Receptor and Mediates Its Delivery to the Vacuolar Degradation Pathway. Plant Cell 2016, 28, 2291–2311. [Google Scholar] [CrossRef] [PubMed]
- Fernandez, M.A.; Belda-Palazon, B.; Julian, J.; Coego, A.; Lozano-Juste, J.; Inigo, S.; Rodriguez, L.; Bueso, E.; Goossens, A.; Rodriguez, P.L. RBR-type E3 ligases and the Ub-conjugating enzyme UBC26 regulate ABA receptor levels and signaling. Plant Physiol. 2019. [Google Scholar] [CrossRef]
- Yu, F.; Lou, L.; Tian, M.; Li, Q.; Ding, Y.; Cao, X.; Wu, Y.; Belda-Palazon, B.; Rodriguez, P.L.; Yang, S.; et al. ESCRT-I Component VPS23A Affects ABA Signaling by Recognizing ABA Receptors for Endosomal Degradation. Mol. Plant 2016, 9, 1570–1582. [Google Scholar] [CrossRef]
- Li, H.; Li, Y.; Zhao, Q.; Li, T.; Wei, J.; Li, B.; Shen, W.; Yang, C.; Zeng, Y.; Rodriguez, P.L.; et al. The plant ESCRT component FREE1 shuttles to the nucleus to attenuate abscisic acid signalling. Nat. Plants 2019, 5, 512–524. [Google Scholar] [CrossRef]
- Perez-Henriquez, P.; Raikhel, N.V.; Norambuena, L. Endocytic trafficking towards the vacuole plays a key role in the auxin receptor SCF(TIR)-independent mechanism of lateral root formation in A. thaliana. Mol. Plant 2012, 5, 1195–1209. [Google Scholar] [CrossRef]
- Luschnig, C.; Vert, G. The dynamics of plant plasma membrane proteins: PINs and beyond. Development 2014, 141, 2924–2938. [Google Scholar] [CrossRef]
- Deb, S.; Sankaranarayanan, S.; Wewala, G.; Widdup, E.; Samuel, M.A. The S-Domain Receptor Kinase Arabidopsis Receptor Kinase2 and the U Box/Armadillo Repeat-Containing E3 Ubiquitin Ligase9 Module Mediates Lateral Root Development under Phosphate Starvation in Arabidopsis. Plant Physiol. 2014, 165, 1647–1656. [Google Scholar] [CrossRef]
- Sankaranarayanan, S.; Samuel, M.A. A proposed role for selective autophagy in regulating auxin-dependent lateral root development under phosphate starvation in Arabidopsis. Plant Signal. Behav. 2015, 10, e989749. [Google Scholar] [CrossRef]
- Feng, Y.; Yin, Y.; Fei, S. Down-regulation of BdBRI1, a putative brassinosteroid receptor gene produces a dwarf phenotype with enhanced drought tolerance in Brachypodium distachyon. Plant Sci. 2015, 234, 163–173. [Google Scholar] [CrossRef] [PubMed]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective Autophagy of BES1 Mediated by DSK2 Balances Plant Growth and Survival. Dev. Cell 2017, 41, 33–46.e7. [Google Scholar] [CrossRef] [PubMed]
- Northey, J.G.; Liang, S.; Jamshed, M.; Deb, S.; Foo, E.; Reid, J.B.; McCourt, P.; Samuel, M.A. Farnesylation mediates brassinosteroid biosynthesis to regulate abscisic acid responses. Nat. Plants 2016, 2, 16114. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Liu, D.; Wang, P.; Ma, X.; Lin, W.; Chen, S.; Mishev, K.; Lu, D.; Kumar, R.; Vanhoutte, I.; et al. Regulation of Arabidopsis brassinosteroid receptor BRI1 endocytosis and degradation by plant U-box PUB12/PUB13-mediated ubiquitination. Proc. Natl. Acad. Sci. USA 2018, 115, E1906–E1915. [Google Scholar] [CrossRef]
- Zhang, Z.; Zhu, J.Y.; Roh, J.; Marchive, C.; Kim, S.K.; Meyer, C.; Sun, Y.; Wang, W.; Wang, Z.Y. TOR Signaling Promotes Accumulation of BZR1 to Balance Growth with Carbon Availability in Arabidopsis. Curr. Biol. 2016, 26, 1854–1860. [Google Scholar] [CrossRef]
- Nie, W.F.; Wang, M.M.; Xia, X.J.; Zhou, Y.H.; Shi, K.; Chen, Z.; Yu, J.Q. Silencing of tomato RBOH1 and MPK2 abolishes brassinosteroid-induced H(2)O(2) generation and stress tolerance. Plant Cell Env. 2013, 36, 789–803. [Google Scholar] [CrossRef]
- Xia, X.J.; Wang, Y.J.; Zhou, Y.H.; Tao, Y.; Mao, W.H.; Shi, K.; Asami, T.; Chen, Z.; Yu, J.Q. Reactive oxygen species are involved in brassinosteroid-induced stress tolerance in cucumber. Plant Physiol. 2009, 150, 801–814. [Google Scholar] [CrossRef]
- Xia, X.J.; Zhou, Y.H.; Ding, J.; Shi, K.; Asami, T.; Chen, Z.; Yu, J.Q. Induction of systemic stress tolerance by brassinosteroid in Cucumis sativus. New Phytol. 2011, 191, 706–720. [Google Scholar] [CrossRef]
- Wang, Y.; Cao, J.J.; Wang, K.X.; Xia, X.J.; Shi, K.; Zhou, Y.H.; Yu, J.Q.; Zhou, J. BZR1 Mediates Brassinosteroid-Induced Autophagy and Nitrogen Starvation in Tomato. Plant Physiol. 2019, 179, 671–685. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).