MicroRNA-Based Combinatorial Cancer Therapy: Effects of MicroRNAs on the Efficacy of Anti-Cancer Therapies
Abstract
:1. Introduction
2. The Role of MiRNAs in Drug Efflux/Influx and Drug Sensitivity
2.1. Drug Transporters and Therapeutic Resistance
2.2. MiRNAs Directly Targeting Drug Transporter Genes
2.2.1. ABCB1
2.2.2. ABCC1
2.2.3. ABCC5
2.2.4. ABCF2 and Other Transporters
2.2.5. SLC19A1 and the Dual Role of a MiR-595
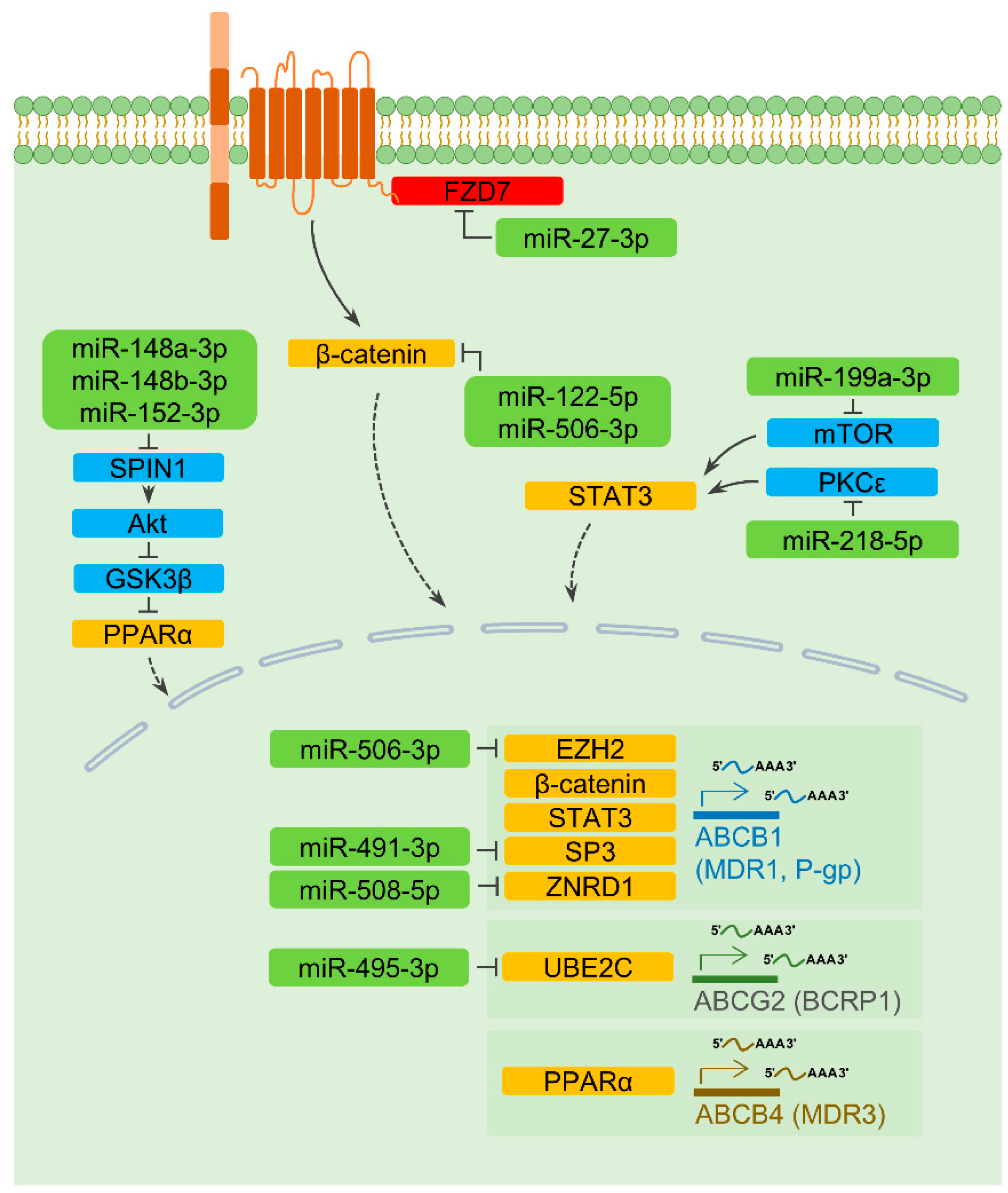
2.3. MiRNAs and the Transcription of Drug Transporter Genes
2.3.1. ABCB1
2.3.2. ABCB4 and ABCG2
2.4. A MiRNA Regulating Degradation of a Drug Transporter
3. MiRNAs in the Regulation of DNA Damage Repair and Therapeutic Resistance
3.1. DNA Damage Repair in Cancer
3.2. MiRNAs Negatively Regulating DNA Repair Mechanisms
3.2.1. MiR-7-5p
3.2.2. MiR-30-5p
3.2.3. MiR-138-5p
3.2.4. MiR-182-5p and MiR-4429
3.2.5. MiR-205-5p and MiR-211-5p
3.2.6. MiR-520g-3p and MiR-520h
3.3. MiRNAs Positively Regulating DNA Repair Mechanisms
3.3.1. MiR-488-3p
3.3.2. MiR-493-5p
4. Autophagy-Regulating MiRNAs and Therapeutic Resistance
4.1. General Mechanisms of Autophagy
4.2. Dual Roles of Autophagy in Cancer
4.3. MiRNAs Regulating mTOR and mTORC1
4.4. MiRNAs Regulating ULK1, BECLIN1, and ATG14
4.5. MiRNAs Regulating HMGBs
4.6. MiRNAs Regulating the Lipidation of LC3
4.7. MiRNAs and Other Autophagy-Regulating Genes
5. MiRNAs Involved in the Regulation of Therapeutic Resistance Associated with Cancer Stemness
5.1. Cancer Stem Cells
5.2. MiRNAs Regulating Wnt/β-Catenin Signaling
5.2.1. Wnt/β-Catenin Signaling and CSCs
5.2.2. Wnt/β-Catenin Signaling-Regulating MiRNAs
5.3. MiRNAs Regulating Notch Signaling
5.3.1. Notch Signaling and CSCs
5.3.2. MiRNAs Regulating Notch Receptor 1 and 3
5.3.3. A MiRNA-Regulating Notch Receptor 2 and its Downstream Signaling Factor
5.4. MiRNAs Regulating JAK/STAT3 Signaling
5.4.1. JAK/STAT3 Signaling and CSCs
5.4.2. MiRNAs Regulating Negative Regulators of JAK/STAT3 Signaling
5.5. A MiRNA and Hedgehog Signaling
5.6. MiRNAs Regulating NF-κB Signaling and PD-L1
5.7. Other MiRNAs Directly or Indirectly Regulating Stemness Factors
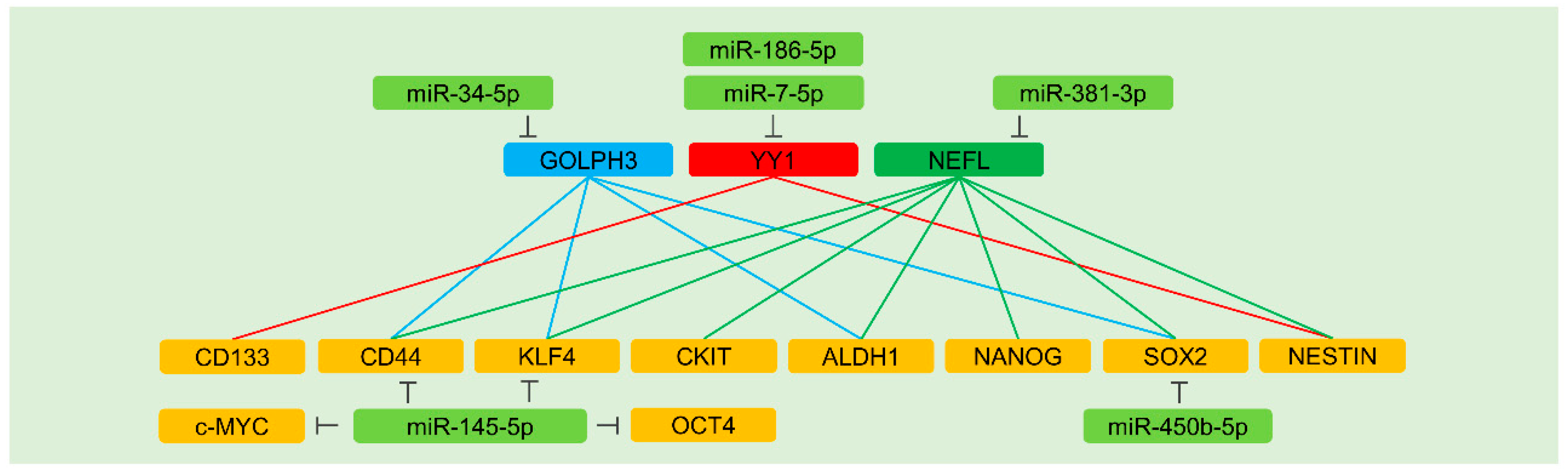
5.7.1. Direct Regulation of Stemness Factors
5.7.2. Indirect Regulation of Stemness Factors
GOLPH3
YY1
NEFL
6. MiRNAs Involved in the Regulation of Therapeutic Resistance Associated with Epithelial-Mesenchymal Transition (EMT)
6.1. EMT and Cancer Stemness
6.2. MiRNAs Regulating TGF-β Signaling
6.3. MiRNAs and HGF/c-MET
6.4. MiRNAs Directly Regulating EMT-Related Transcription Factors and Markers
6.5. MiRNAs Indirectly Regulating EMT-Related Transcription Factors
7. Extracellular Vesicle MiRNAs and Therapeutic Resistance
7.1. Extracellular Vesicles (EVs)
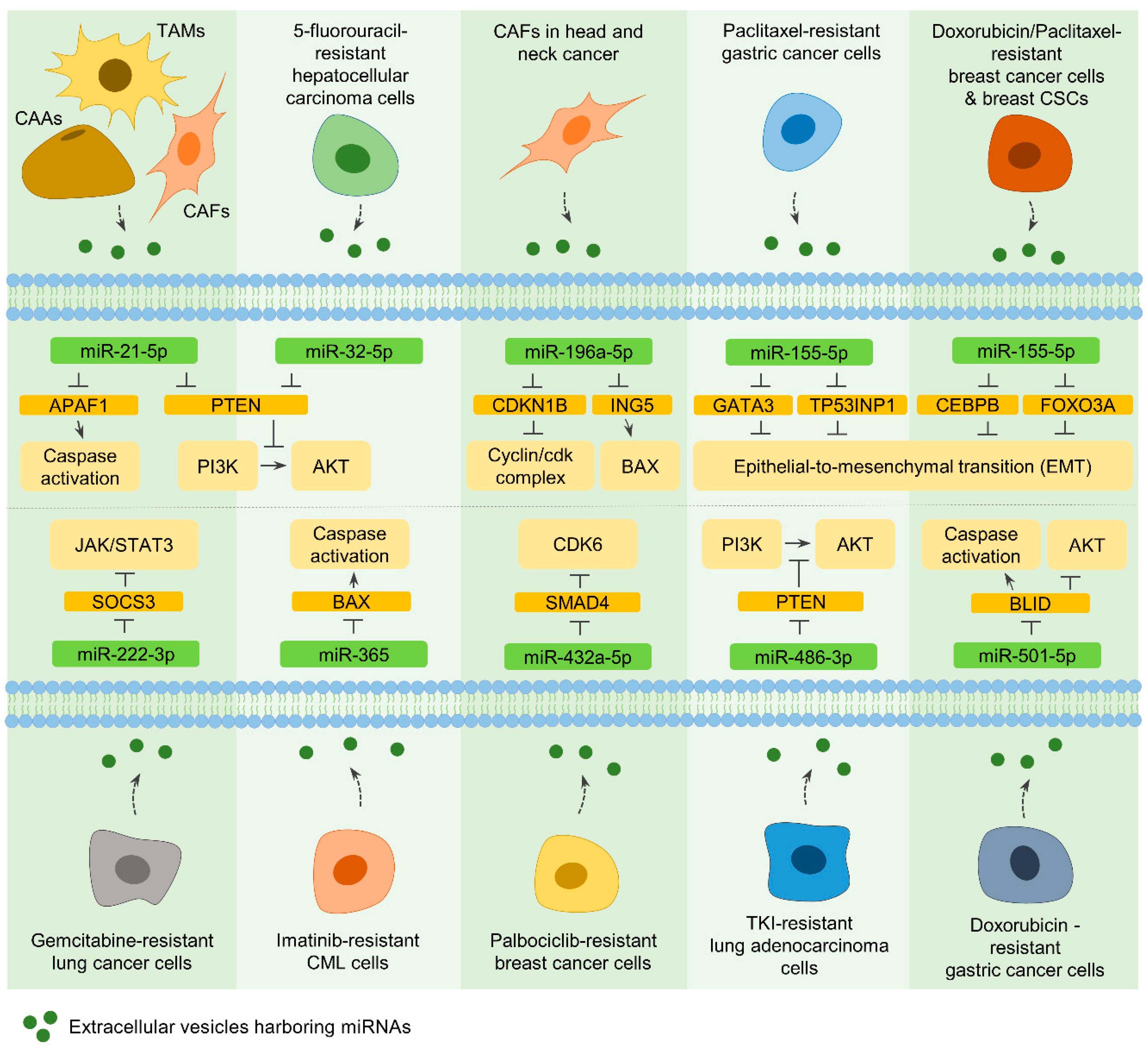
7.2. EVs from CAAs, TAMs, and CAFs
7.2.1. MiR-21-5p
7.2.2. MiR-196a-5p
7.3. EVs from Drug-Resistant Cancer Cells and CSCs
7.3.1. MiR-32-5p
7.3.2. MiR-155-5p
7.3.3. MiR-222-3p and MiR-365
7.3.4. MiR-432a-5p, MiR-486-3p, and MiR-501-5p
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Kathawala, R.J.; Gupta, P.; Ashby, C.R., Jr.; Chen, Z.S. The modulation of abc transporter-mediated multidrug resistance in cancer: A review of the past decade. Drug Resist. Updates 2015, 18, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Nagel, Z.D.; Kitange, G.J.; Gupta, S.K.; Joughin, B.A.; Chaim, I.A.; Mazzucato, P.; Lauffenburger, D.A.; Sarkaria, J.N.; Samson, L.D. DNA repair capacity in multiple pathways predicts chemoresistance in glioblastoma multiforme. Cancer Res. 2017, 77, 198–206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, S.F.; Wang, X.Y.; Fu, Z.Q.; Peng, Q.H.; Zhang, J.Y.; Ye, F.; Fu, Y.F.; Zhou, C.Y.; Lu, W.G.; Cheng, X.D.; et al. Txndc17 promotes paclitaxel resistance via inducing autophagy in ovarian cancer. Autophagy 2015, 11, 225–238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nunes, T.; Hamdan, D.; Leboeuf, C.; El Bouchtaoui, M.; Gapihan, G.; Nguyen, T.T.; Meles, S.; Angeli, E.; Ratajczak, P.; Lu, H.; et al. Targeting cancer stem cells to overcome chemoresistance. Int. J. Mol. Sci. 2018, 19, 4036. [Google Scholar] [CrossRef] [Green Version]
- Maycotte, P.; Jones, K.L.; Goodall, M.L.; Thorburn, J.; Thorburn, A. Autophagy supports breast cancer stem cell maintenance by regulating il6 secretion. Mol. Cancer Res. 2015, 13, 651–658. [Google Scholar] [CrossRef] [Green Version]
- Sharif, T.; Martell, E.; Dai, C.; Kennedy, B.E.; Murphy, P.; Clements, D.R.; Kim, Y.; Lee, P.W.; Gujar, S.A. Autophagic homeostasis is required for the pluripotency of cancer stem cells. Autophagy 2017, 13, 264–284. [Google Scholar] [CrossRef] [Green Version]
- Katsuno, Y.; Meyer, D.S.; Zhang, Z.; Shokat, K.M.; Akhurst, R.J.; Miyazono, K.; Derynck, R. Chronic tgf-beta exposure drives stabilized emt, tumor stemness, and cancer drug resistance with vulnerability to bitopic mtor inhibition. Sci. Signal. 2019, 12, eaau8544. [Google Scholar] [CrossRef]
- Au Yeung, C.L.; Co, N.N.; Tsuruga, T.; Yeung, T.L.; Kwan, S.Y.; Leung, C.S.; Li, Y.; Lu, E.S.; Kwan, K.; Wong, K.K.; et al. Exosomal transfer of stroma-derived mir21 confers paclitaxel resistance in ovarian cancer cells through targeting apaf1. Nat. Commun. 2016, 7, 11150. [Google Scholar] [CrossRef] [Green Version]
- Amodio, N.; Gallo Cantafio, M.E.; Botta, C.; Agosti, V.; Federico, C.; Caracciolo, D.; Ronchetti, D.; Rossi, M.; Driessen, C.; Neri, A.; et al. Replacement of mir-155 elicits tumor suppressive activity and antagonizes bortezomib resistance in multiple myeloma. Cancers 2019, 11, 236. [Google Scholar] [CrossRef] [Green Version]
- Clarke, P.A.; Roe, T.; Swabey, K.; Hobbs, S.M.; McAndrew, C.; Tomlin, K.; Westwood, I.; Burke, R.; van Montfort, R.; Workman, P. Dissecting mechanisms of resistance to targeted drug combination therapy in human colorectal cancer. Oncogene 2019, 38, 5076–5090. [Google Scholar] [CrossRef] [Green Version]
- Su, Y.; Li, X.; Ma, J.; Zhao, J.; Liu, S.; Wang, G.; Edwards, H.; Taub, J.W.; Lin, H.; Ge, Y. Targeting pi3k, mtor, erk, and bcl-2 signaling network shows superior antileukemic activity against aml ex vivo. Biochem. Pharmacol. 2018, 148, 13–26. [Google Scholar] [CrossRef] [PubMed]
- Park, J.K.; Doseff, A.I.; Schmittgen, T.D. Micrornas targeting caspase-3 and -7 in panc-1 cells. Int. J. Mol. Sci. 2018, 19, 1206. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, J.K.; Kogure, T.; Nuovo, G.J.; Jiang, J.; He, L.; Kim, J.H.; Phelps, M.A.; Papenfuss, T.L.; Croce, C.M.; Patel, T.; et al. Mir-221 silencing blocks hepatocellular carcinoma and promotes survival. Cancer Res. 2011, 71, 7608–7616. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Elgamal, O.A.; Park, J.K.; Gusev, Y.; Azevedo-Pouly, A.C.; Jiang, J.; Roopra, A.; Schmittgen, T.D. Tumor suppressive function of mir-205 in breast cancer is linked to hmgb3 regulation. PLoS ONE 2013, 8, e76402. [Google Scholar] [CrossRef] [PubMed]
- Barbato, L.; Bocchetti, M.; Di Biase, A.; Regad, T. Cancer stem cells and targeting strategies. Cells 2019, 8, 926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Q.; Shu, Y. Role of solute carriers in response to anticancer drugs. Mol. Cell Ther. 2014, 2, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, C.; Shan, Z.; Li, C.; Yang, L. Mir-129 regulates cisplatin-resistance in human gastric cancer cells by targeting p-gp. Biomed. Pharmacother. 2017, 86, 450–456. [Google Scholar] [CrossRef]
- Yang, T.; Zheng, Z.M.; Li, X.N.; Li, Z.F.; Wang, Y.; Geng, Y.F.; Bai, L.; Zhang, X.B. Mir-223 modulates multidrug resistance via downregulation of abcb1 in hepatocellular carcinoma cells. Exp. Biol. Med. 2013, 238, 1024–1032. [Google Scholar] [CrossRef]
- Zhao, Y.; Qi, X.; Chen, J.; Wei, W.; Yu, C.; Yan, H.; Pu, M.; Li, Y.; Miao, L.; Li, C.; et al. The mir-491-3p/sp3/abcb1 axis attenuates multidrug resistance of hepatocellular carcinoma. Cancer Lett. 2017, 408, 102–111. [Google Scholar] [CrossRef]
- Zou, Z.; Zou, R.; Zong, D.; Shi, Y.; Chen, J.; Huang, J.; Zhu, J.; Chen, L.; Bao, X.; Liu, Y.; et al. Mir-495 sensitizes mdr cancer cells to the combination of doxorubicin and taxol by inhibiting mdr1 expression. J. Cell. Mol. Med. 2017, 21, 1929–1943. [Google Scholar] [CrossRef]
- Shang, Y.; Zhang, Z.; Liu, Z.; Feng, B.; Ren, G.; Li, K.; Zhou, L.; Sun, Y.; Li, M.; Zhou, J.; et al. Mir-508-5p regulates multidrug resistance of gastric cancer by targeting abcb1 and znrd1. Oncogene 2014, 33, 3267–3276. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shang, Y.; Feng, B.; Zhou, L.; Ren, G.; Zhang, Z.; Fan, X.; Sun, Y.; Luo, G.; Liang, J.; Wu, K.; et al. The mir27b-ccng1-p53-mir-508-5p axis regulates multidrug resistance of gastric cancer. Oncotarget 2016, 7, 538–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, J.; Wang, T.; Guo, R.; Yang, X.; Yin, J.; Yu, J.; Xiang, Q.; Pan, X.; Tang, H.; Lei, X. Involvement of mir-133a and mir-326 in adm resistance of hepg2 through modulating expression of abcc1. J. Drug Target. 2015, 23, 519–524. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Liu, Y.; Ren, J.; Deng, S.; Yi, G.; Guo, M.; Shu, S.; Zhao, L.; Peng, Y.; Qi, S. Mir-1268a regulates abcc1 expression to mediate temozolomide resistance in glioblastoma. J. Neurooncol. 2018, 138, 499–508. [Google Scholar] [CrossRef] [PubMed]
- Oguri, T.; Achiwa, H.; Sato, S.; Bessho, Y.; Takano, Y.; Miyazaki, M.; Muramatsu, H.; Maeda, H.; Niimi, T.; Ueda, R. The determinants of sensitivity and acquired resistance to gemcitabine differ in non-small cell lung cancer: A role of abcc5 in gemcitabine sensitivity. Mol. Cancer Ther. 2006, 5, 1800–1806. [Google Scholar] [CrossRef] [Green Version]
- Amponsah, P.S.; Fan, P.; Bauer, N.; Zhao, Z.; Gladkich, J.; Fellenberg, J.; Herr, I. Microrna-210 overexpression inhibits tumor growth and potentially reverses gemcitabine resistance in pancreatic cancer. Cancer Lett. 2017, 388, 107–117. [Google Scholar] [CrossRef]
- Bao, L.; Wu, J.; Dodson, M.; Rojo de la Vega, E.M.; Ning, Y.; Zhang, Z.; Yao, M.; Zhang, D.D.; Xu, C.; Yi, X. Abcf2, an nrf2 target gene, contributes to cisplatin resistance in ovarian cancer cells. Mol. Carcinog. 2017, 56, 1543–1553. [Google Scholar] [CrossRef]
- Xiao, S.; Zhang, M.; Liu, C.; Wang, D. Mir-514 attenuates proliferation and increases chemoresistance by targeting atp binding cassette subfamily in ovarian cancer. Mol. Genet. Genom. 2018, 293, 1159–1167. [Google Scholar] [CrossRef]
- Li, X.; Tian, Y.; Tu, M.J.; Ho, P.Y.; Batra, N.; Yu, A.M. Bioengineered mir-27b-3p and mir-328-3p modulate drug metabolism and disposition via the regulation of target adme gene expression. Acta Pharm. Sin. B 2019, 9, 639–647. [Google Scholar] [CrossRef]
- Wang, S.M.; Sun, L.L.; Wu, W.S.; Yan, D. Mir-595 suppresses the cellular uptake and cytotoxic effects of methotrexate by targeting slc19a1 in cem/c1 cells. Basic Clin. Pharmacol. Toxicol. 2018, 123, 8–13. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Zhang, M.; Chen, X.; Liu, Y.; Lou, G. Microrna-595 sensitizes ovarian cancer cells to cisplatin by targeting abcb1. Oncotarget 2016, 7, 87091–87099. [Google Scholar] [CrossRef] [PubMed]
- Shen, D.Y.; Zhang, W.; Zeng, X.; Liu, C.Q. Inhibition of wnt/beta-catenin signaling downregulates p-glycoprotein and reverses multi-drug resistance of cholangiocarcinoma. Cancer Sci. 2013, 104, 1303–1308. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.; Ma, T.; Huang, C.; Zhang, L.; Lv, X.; Xu, T.; Hu, T.; Li, J. Mir-27a modulates the mdr1/p-glycoprotein expression by inhibiting fzd7/beta-catenin pathway in hepatocellular carcinoma cells. Cell. Signal. 2013, 25, 2693–2701. [Google Scholar] [CrossRef] [PubMed]
- Cao, F.; Yin, L.X. Mir-122 enhances sensitivity of hepatocellular carcinoma to oxaliplatin via inhibiting mdr1 by targeting wnt/beta-catenin pathway. Exp. Mol. Pathol. 2019, 106, 34–43. [Google Scholar] [CrossRef] [PubMed]
- Zhou, H.; Lin, C.; Zhang, Y.; Zhang, X.; Zhang, C.; Zhang, P.; Xie, X.; Ren, Z. Mir-506 enhances the sensitivity of human colorectal cancer cells to oxaliplatin by suppressing mdr1/p-gp expression. Cell Prolif. 2017, 50, e12341. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Zhang, Y.; Liang, R.; Gao, Z.; Sun, D.; Wang, L. Rnai-mediated ezh2 depletion decreases mdr1 expression and sensitizes multidrug-resistant hepatocellular carcinoma cells to chemotherapy. Oncol. Rep. 2013, 29, 1037–1042. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Lin, C.; Liao, G.; Liu, S.; Ding, J.; Tang, F.; Wang, Z.; Liang, X.; Li, B.; Wei, Y.; et al. Microrna-506 suppresses tumor proliferation and metastasis in colon cancer by directly targeting the oncogene ezh2. Oncotarget 2015, 6, 32586–32601. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Zhan, M.; Xu, S.W.; Chen, W.; Long, M.M.; Shi, Y.H.; Liu, Q.; Mohan, M.; Wang, J. Mir-218-5p restores sensitivity to gemcitabine through prkce/mdr1 axis in gallbladder cancer. Cell Death Dis. 2017, 8, e2770. [Google Scholar] [CrossRef] [Green Version]
- Li, Q.; Xia, X.; Ji, J.; Ma, J.; Tao, L.; Mo, L.; Chen, W. Mir-199a-3p enhances cisplatin sensitivity of cholangiocarcinoma cells by inhibiting mtor signaling pathway and expression of mdr1. Oncotarget 2017, 8, 33621–33630. [Google Scholar] [CrossRef] [Green Version]
- Zhou, J.; Wulfkuhle, J.; Zhang, H.; Gu, P.; Yang, Y.; Deng, J.; Margolick, J.B.; Liotta, L.A.; Petricoin, E., 3rd; Zhang, Y. Activation of the pten/mtor/stat3 pathway in breast cancer stem-like cells is required for viability and maintenance. Proc. Natl. Acad. Sci. USA 2007, 104, 16158–16163. [Google Scholar] [CrossRef] [Green Version]
- Jain, K.; Basu, A. The multifunctional protein kinase c-epsilon in cancer development and progression. Cancers 2014, 6, 860–878. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, X.; Xiao, W.; Wang, L.; Tian, Z.; Zhang, J. Deactivation of signal transducer and activator of transcription 3 reverses chemotherapeutics resistance of leukemia cells via down-regulating p-gp. PLoS ONE 2011, 6, e20965. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hong, L.; Piao, Y.; Han, Y.; Wang, J.; Zhang, X.; Du, Y.; Cao, S.; Qiao, T.; Chen, Z.; Fan, D. Zinc ribbon domain-containing 1 (znrd1) mediates multidrug resistance of leukemia cells through regulation of p-glycoprotein and bcl-2. Mol. Cancer Ther. 2005, 4, 1936–1942. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Si, W.; Shen, J.; Du, C.; Chen, D.; Gu, X.; Li, C.; Yao, M.; Pan, J.; Cheng, J.; Jiang, D.; et al. A mir-20a/mapk1/c-myc regulatory feedback loop regulates breast carcinogenesis and chemoresistance. Cell Death Differ. 2018, 25, 406–420. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Wang, Y.W.; Gao, P. Spin1, negatively regulated by mir-148/152, enhances adriamycin resistance via upregulating drug metabolizing enzymes and transporter in breast cancer. J. Exp. Clin. Cancer Res. 2018, 37, 100. [Google Scholar] [CrossRef]
- Guo, J.; Jin, D.; Wu, Y.; Yang, L.; Du, J.; Gong, K.; Chen, W.; Dai, J.; Miao, S.; Xi, S. The mir 495-ube2c-abcg2/ercc1 axis reverses cisplatin resistance by downregulating drug resistance genes in cisplatin-resistant non-small cell lung cancer cells. EBioMedicine 2018, 35, 204–221. [Google Scholar] [CrossRef]
- Ghonem, N.S.; Ananthanarayanan, M.; Soroka, C.J.; Boyer, J.L. Peroxisome proliferator-activated receptor alpha activates human multidrug resistance transporter 3/atp-binding cassette protein subfamily b4 transcription and increases rat biliary phosphatidylcholine secretion. Hepatology 2014, 59, 1030–1042. [Google Scholar] [CrossRef] [Green Version]
- Hinds, T.D., Jr.; Burns, K.A.; Hosick, P.A.; McBeth, L.; Nestor-Kalinoski, A.; Drummond, H.A.; AlAmodi, A.A.; Hankins, M.W.; Vanden Heuvel, J.P.; Stec, D.E. Biliverdin reductase a attenuates hepatic steatosis by inhibition of glycogen synthase kinase (gsk) 3beta phosphorylation of serine 73 of peroxisome proliferator-activated receptor (ppar) alpha. J. Biol. Chem. 2016, 291, 25179–25191. [Google Scholar] [CrossRef] [Green Version]
- Chen, X.; Wang, Y.W.; Xing, A.Y.; Xiang, S.; Shi, D.B.; Liu, L.; Li, Y.X.; Gao, P. Suppression of spin1-mediated pi3k-akt pathway by mir-489 increases chemosensitivity in breast cancer. J. Pathol. 2016, 239, 459–472. [Google Scholar] [CrossRef]
- Katayama, K.; Fujiwara, C.; Noguchi, K.; Sugimoto, Y. Rsk1 protects p-glycoprotein/abcb1 against ubiquitin-proteasomal degradation by downregulating the ubiquitin-conjugating enzyme e2 r1. Sci. Rep. 2016, 6, 36134. [Google Scholar] [CrossRef] [Green Version]
- Katayama, K.; Yoshioka, S.; Tsukahara, S.; Mitsuhashi, J.; Sugimoto, Y. Inhibition of the mitogen-activated protein kinase pathway results in the down-regulation of p-glycoprotein. Mol. Cancer Ther. 2007, 6, 2092–2102. [Google Scholar] [CrossRef] [Green Version]
- Nikolaev, A.; Yang, E.S. The impact of DNA repair pathways in cancer biology and therapy. Cancers 2017, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Gil Del Alcazar, C.R.; Todorova, P.K.; Habib, A.A.; Mukherjee, B.; Burma, S. Augmented hr repair mediates acquired temozolomide resistance in glioblastoma. Mol. Cancer Res. 2016, 14, 928–940. [Google Scholar] [CrossRef] [Green Version]
- Chen, G.; Chen, J.; Qiao, Y.; Shi, Y.; Liu, W.; Zeng, Q.; Xie, H.; Shi, X.; Sun, Y.; Liu, X.; et al. Znf830 mediates cancer chemoresistance through promoting homologous-recombination repair. Nucleic Acids Res. 2018, 46, 1266–1279. [Google Scholar] [CrossRef]
- Lai, J.; Yang, H.; Zhu, Y.; Ruan, M.; Huang, Y.; Zhang, Q. Mir-7-5p-mediated downregulation of parp1 impacts DNA homologous recombination repair and resistance to doxorubicin in small cell lung cancer. BMC Cancer 2019, 19, 602. [Google Scholar] [CrossRef] [Green Version]
- Lin, S.; Yu, L.; Song, X.; Bi, J.; Jiang, L.; Wang, Y.; He, M.; Xiao, Q.; Sun, M.; Olopade, O.I.; et al. Intrinsic adriamycin resistance in p53-mutated breast cancer is related to the mir-30c/fancf/rev1-mediated DNA damage response. Cell Death Dis. 2019, 10, 666. [Google Scholar] [CrossRef]
- Liu, J.; Li, H.; Sun, L.; Feng, X.; Wang, Z.; Yuan, Y.; Xing, C. The differential expression of core genes in nucleotide excision repair pathway indicates colorectal carcinogenesis and prognosis. Biomed. Res. Int. 2018, 2018, 9651320. [Google Scholar] [CrossRef] [Green Version]
- Ning, J.; Jiao, Y.; Xie, X.; Deng, X.; Zhang, Y.; Yang, Y.; Zhao, C.; Wang, H.; Gu, K. Mir1385p modulates the expression of excision repair crosscomplementing proteins ercc1 and ercc4, and regulates the sensitivity of gastric cancer cells to cisplatin. Oncol. Rep. 2019, 41, 1131–1139. [Google Scholar]
- Ma, F.; Zhang, M.; Gong, W.; Weng, M.; Quan, Z. Mir-138 suppresses cell proliferation by targeting bag-1 in gallbladder carcinoma. PLoS ONE 2015, 10, e0126499. [Google Scholar] [CrossRef]
- Sun, D.K.; Wang, J.M.; Zhang, P.; Wang, Y.Q. Microrna-138 regulates metastatic potential of bladder cancer through zeb2. Cell. Physiol. Biochem. 2015, 37, 2366–2374. [Google Scholar] [CrossRef]
- Chan, X.H.; Nama, S.; Gopal, F.; Rizk, P.; Ramasamy, S.; Sundaram, G.; Ow, G.S.; Ivshina, A.V.; Tanavde, V.; Haybaeck, J.; et al. Targeting glioma stem cells by functional inhibition of a prosurvival oncomir-138 in malignant gliomas. Cell Rep. 2012, 2, 591–602. [Google Scholar] [CrossRef] [Green Version]
- Zhao, W.; Steinfeld, J.B.; Liang, F.; Chen, X.; Maranon, D.G.; Jian Ma, C.; Kwon, Y.; Rao, T.; Wang, W.; Sheng, C.; et al. Brca1-bard1 promotes rad51-mediated homologous DNA pairing. Nature 2017, 550, 360–365. [Google Scholar] [CrossRef]
- Lai, T.H.; Ewald, B.; Zecevic, A.; Liu, C.; Sulda, M.; Papaioannou, D.; Garzon, R.; Blachly, J.S.; Plunkett, W.; Sampath, D. Hdac inhibition induces microrna-182, which targets rad51 and impairs hr repair to sensitize cells to sapacitabine in acute myelogenous leukemia. Clin. Cancer Res. 2016, 22, 3537–3549. [Google Scholar] [CrossRef] [Green Version]
- Moskwa, P.; Buffa, F.M.; Pan, Y.; Panchakshari, R.; Gottipati, P.; Muschel, R.J.; Beech, J.; Kulshrestha, R.; Abdelmohsen, K.; Weinstock, D.M.; et al. Mir-182-mediated downregulation of brca1 impacts DNA repair and sensitivity to parp inhibitors. Mol. Cell. 2011, 41, 210–220. [Google Scholar] [CrossRef]
- Sun, H.; Fan, G.; Deng, C.; Wu, L. Mir-4429 sensitized cervical cancer cells to irradiation by targeting rad51. J. Cell. Physiol. 2019, 235, 185–193. [Google Scholar] [CrossRef]
- El Bezawy, R.; Cominetti, D.; Fenderico, N.; Zuco, V.; Beretta, G.L.; Dugo, M.; Arrighetti, N.; Stucchi, C.; Rancati, T.; Valdagni, R.; et al. Mir-875-5p counteracts epithelial-to-mesenchymal transition and enhances radiation response in prostate cancer through repression of the egfr-zeb1 axis. Cancer Lett. 2017, 395, 53–62. [Google Scholar] [CrossRef]
- Wanner, G.; Mayer, C.; Kehlbach, R.; Rodemann, H.P.; Dittmann, K. Activation of protein kinase cepsilon stimulates DNA-repair via epidermal growth factor receptor nuclear accumulation. Radiother. Oncol. 2008, 86, 383–390. [Google Scholar] [CrossRef]
- El Bezawy, R.; Tinelli, S.; Tortoreto, M.; Doldi, V.; Zuco, V.; Folini, M.; Stucchi, C.; Rancati, T.; Valdagni, R.; Gandellini, P.; et al. Mir-205 enhances radiation sensitivity of prostate cancer cells by impairing DNA damage repair through pkcepsilon and zeb1 inhibition. J. Exp. Clin. Cancer Res. 2019, 38, 51. [Google Scholar] [CrossRef]
- Yuan, X.; Ma, R.; Yang, S.; Jiang, L.; Wang, Z.; Zhu, Z.; Li, H. Mir-520g and mir-520h overcome bortezomib resistance in multiple myeloma via suppressing ape1. Cell Cycle 2019, 18, 1660–1669. [Google Scholar] [CrossRef]
- Roychoudhury, S.; Nath, S.; Song, H.; Hegde, M.L.; Bellot, L.J.; Mantha, A.K.; Sengupta, S.; Ray, S.; Natarajan, A.; Bhakat, K.K. Human apurinic/apyrimidinic endonuclease (ape1) is acetylated at DNA damage sites in chromatin, and acetylation modulates its DNA repair activity. Mol. Cell. Biol. 2017, 37, e00401–e00416. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.Y.; Dong, Z.; Liu, J.; Yin, J.Y.; Zhou, L.; Wu, X.; Yang, Y.; Mo, W.; Huang, W.; Khoo, S.K.; et al. Role of eif3a in regulating cisplatin sensitivity and in translational control of nucleotide excision repair of nasopharyngeal carcinoma. Oncogene 2011, 30, 4814–4823. [Google Scholar] [CrossRef]
- Yin, J.Y.; Shen, J.; Dong, Z.Z.; Huang, Q.; Zhong, M.Z.; Feng, D.Y.; Zhou, H.H.; Zhang, J.T.; Liu, Z.Q. Effect of eif3a on response of lung cancer patients to platinum-based chemotherapy by regulating DNA repair. Clin. Cancer Res. 2011, 17, 4600–4609. [Google Scholar] [CrossRef] [Green Version]
- Fang, C.; Chen, Y.X.; Wu, N.Y.; Yin, J.Y.; Li, X.P.; Huang, H.S.; Zhang, W.; Zhou, H.H.; Liu, Z.Q. Mir-488 inhibits proliferation and cisplatin sensibility in non-small-cell lung cancer (nsclc) cells by activating the eif3a-mediated ner signaling pathway. Sci. Rep. 2017, 7, 40384. [Google Scholar] [CrossRef] [Green Version]
- Guillemette, S.; Serra, R.W.; Peng, M.; Hayes, J.A.; Konstantinopoulos, P.A.; Green, M.R.; Cantor, S.B. Resistance to therapy in brca2 mutant cells due to loss of the nucleosome remodeling factor chd4. Genes Dev. 2015, 29, 489–494. [Google Scholar] [CrossRef]
- Meghani, K.; Fuchs, W.; Detappe, A.; Drane, P.; Gogola, E.; Rottenberg, S.; Jonkers, J.; Matulonis, U.; Swisher, E.M.; Konstantinopoulos, P.A.; et al. Multifaceted impact of microrna 493-5p on genome-stabilizing pathways induces platinum and parp inhibitor resistance in brca2-mutated carcinomas. Cell Rep. 2018, 23, 100–111. [Google Scholar] [CrossRef] [Green Version]
- Levine, B.; Kroemer, G. Biological functions of autophagy genes: A disease perspective. Cell 2019, 176, 11–42. [Google Scholar] [CrossRef] [Green Version]
- Gallagher, L.E.; Williamson, L.E.; Chan, E.Y. Advances in autophagy regulatory mechanisms. Cells 2016, 5, 24. [Google Scholar] [CrossRef]
- Yu, L.; Chen, Y.; Tooze, S.A. Autophagy pathway: Cellular and molecular mechanisms. Autophagy 2018, 14, 207–215. [Google Scholar] [CrossRef] [Green Version]
- Walczak, M.; Martens, S. Dissecting the role of the atg12-atg5-atg16 complex during autophagosome formation. Autophagy 2013, 9, 424–425. [Google Scholar] [CrossRef] [Green Version]
- Peng, Q.; Qin, J.; Zhang, Y.; Cheng, X.; Wang, X.; Lu, W.; Xie, X.; Zhang, S. Autophagy maintains the stemness of ovarian cancer stem cells by foxa2. J. Exp. Clin. Cancer Res. 2017, 36, 171. [Google Scholar] [CrossRef] [Green Version]
- Wang, K.; Chen, B.; Yin, T.; Zhan, Y.; Lu, Y.; Zhang, Y.; Chen, J.; Wu, W.; Zhou, S.; Mao, W.; et al. N-methylparoxetine blocked autophagic flux and induced apoptosis by activating ros-mapk pathway in non-small cell lung cancer cells. Int. J. Mol. Sci. 2019, 20, 3415. [Google Scholar] [CrossRef] [Green Version]
- Jang, J.E.; Eom, J.I.; Jeung, H.K.; Cheong, J.W.; Lee, J.Y.; Kim, J.S.; Min, Y.H. Targeting ampk-ulk1-mediated autophagy for combating bet inhibitor resistance in acute myeloid leukemia stem cells. Autophagy 2017, 13, 761–762. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pietrocola, F.; Pol, J.; Vacchelli, E.; Rao, S.; Enot, D.P.; Baracco, E.E.; Levesque, S.; Castoldi, F.; Jacquelot, N.; Yamazaki, T.; et al. Caloric restriction mimetics enhance anticancer immunosurveillance. Cancer Cell 2016, 30, 147–160. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Galluzzi, L.; Green, D.R. Autophagy-independent functions of the autophagy machinery. Cell 2019, 177, 1682–1699. [Google Scholar] [CrossRef] [PubMed]
- Levy, J.M.M.; Towers, C.G.; Thorburn, A. Targeting autophagy in cancer. Nat. Rev. Cancer 2017, 17, 528–542. [Google Scholar] [CrossRef]
- Rosenfeldt, M.T.; O’Prey, J.; Morton, J.P.; Nixon, C.; MacKay, G.; Mrowinska, A.; Au, A.; Rai, T.S.; Zheng, L.; Ridgway, R.; et al. P53 status determines the role of autophagy in pancreatic tumour development. Nature 2013, 504, 296–300. [Google Scholar] [CrossRef]
- Sun, W.; Yi, Y.; Xia, G.; Zhao, Y.; Yu, Y.; Li, L.; Hua, C.; He, B.; Yang, B.; Yu, C.; et al. Nrf2-mir-129-3p-mtor axis controls an mirna regulatory network involved in hdaci-induced autophagy. Mol. Ther. 2019, 27, 1039–1050. [Google Scholar] [CrossRef]
- Mitra, R.; Lin, C.C.; Eischen, C.M.; Bandyopadhyay, S.; Zhao, Z. Concordant dysregulation of mir-5p and mir-3p arms of the same precursor microrna may be a mechanism in inducing cell proliferation and tumorigenesis: A lung cancer study. RNA 2015, 21, 1055–1065. [Google Scholar] [CrossRef] [Green Version]
- Li, B.; Wang, W.; Li, Z.; Chen, Z.; Zhi, X.; Xu, J.; Li, Q.; Wang, L.; Huang, X.; Wang, L.; et al. Microrna-148a-3p enhances cisplatin cytotoxicity in gastric cancer through mitochondrial fission induction and cyto-protective autophagy suppression. Cancer Lett. 2017, 410, 212–227. [Google Scholar] [CrossRef]
- An, Y.; Zhang, Z.; Shang, Y.; Jiang, X.; Dong, J.; Yu, P.; Nie, Y.; Zhao, Q. Mir-23b-3p regulates the chemoresistance of gastric cancer cells by targeting atg12 and hmgb2. Cell Death Dis. 2015, 6, e1766. [Google Scholar] [CrossRef] [Green Version]
- Jin, F.; Wang, Y.; Li, M.; Zhu, Y.; Liang, H.; Wang, C.; Wang, F.; Zhang, C.Y.; Zen, K.; Li, L. Mir-26 enhances chemosensitivity and promotes apoptosis of hepatocellular carcinoma cells through inhibiting autophagy. Cell Death Dis. 2017, 8, e2540. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; Huang, J.; Xie, M.; Yu, Y.; Zhu, S.; Kang, R.; Cao, L.; Tang, D.; Duan, X. Mir34a regulates autophagy and apoptosis by targeting hmgb1 in the retinoblastoma cell. Autophagy 2014, 10, 442–452. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, Y.; An, Y.; Wang, Y.; Zhang, C.; Zhang, H.; Huang, C.; Jiang, H.; Wang, X.; Li, X. Mir-101 inhibits autophagy and enhances cisplatin-induced apoptosis in hepatocellular carcinoma cells. Oncol. Rep. 2013, 29, 2019–2024. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankel, L.B.; Wen, J.; Lees, M.; Hoyer-Hansen, M.; Farkas, T.; Krogh, A.; Jaattela, M.; Lund, A.H. Microrna-101 is a potent inhibitor of autophagy. EMBO J. 2011, 30, 4628–4641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Z.C.; Huang, F.Z.; Xu, H.B.; Sun, J.C.; Wang, C.F. Microrna-137 inhibits autophagy and chemosensitizes pancreatic cancer cells by targeting atg5. Int. J. Biochem. Cell Biol. 2019, 111, 63–71. [Google Scholar] [CrossRef] [PubMed]
- Meng, Y.; Gao, R.; Ma, J.; Zhao, J.; Xu, E.; Wang, C.; Zhou, X. Microrna-140-5p regulates osteosarcoma chemoresistance by targeting hmgn5 and autophagy. Sci. Rep. 2017, 7, 416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, K.; Chen, J.; Zhou, H.; Chen, Y.; Zhi, Y.; Zhang, B.; Chen, L.; Chu, X.; Wang, R.; Zhang, C. Pu.1/microrna-142-3p targets atg5/atg16l1 to inactivate autophagy and sensitize hepatocellular carcinoma cells to sorafenib. Cell Death Dis. 2018, 9, 312. [Google Scholar] [CrossRef]
- He, J.; Yu, J.J.; Xu, Q.; Wang, L.; Zheng, J.Z.; Liu, L.Z.; Jiang, B.H. Downregulation of atg14 by egr1-mir152 sensitizes ovarian cancer cells to cisplatin-induced apoptosis by inhibiting cyto-protective autophagy. Autophagy 2015, 11, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Hu, J.L.; He, G.Y.; Lan, X.L.; Zeng, Z.C.; Guan, J.; Ding, Y.; Qian, X.L.; Liao, W.T.; Ding, Y.Q.; Liang, L. Inhibition of atg12-mediated autophagy by mir-214 enhances radiosensitivity in colorectal cancer. Oncogenesis 2018, 7, 16. [Google Scholar] [CrossRef]
- Huang, S.; Qi, P.; Zhang, T.; Li, F.; He, X. The hif1alpha/mir2243p/atg5 axis affects cell mobility and chemosensitivity by regulating hypoxiainduced protective autophagy in glioblastoma and astrocytoma. Oncol. Rep. 2019, 41, 1759–1768. [Google Scholar]
- Tan, S.; Shi, H.; Ba, M.; Lin, S.; Tang, H.; Zeng, X.; Zhang, X. Mir-409-3p sensitizes colon cancer cells to oxaliplatin by inhibiting beclin-1-mediated autophagy. Int. J. Mol. Med. 2016, 37, 1030–1038. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiong, J.; Wang, D.; Wei, A.; Ke, N.; Wang, Y.; Tang, J.; He, S.; Hu, W.; Liu, X. Microrna-410-3p attenuates gemcitabine resistance in pancreatic ductal adenocarcinoma by inhibiting hmgb1-mediated autophagy. Oncotarget 2017, 8, 107500–107512. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gao, A.M.; Zhang, X.Y.; Hu, J.N.; Ke, Z.P. Apigenin sensitizes hepatocellular carcinoma cells to doxorubic through regulating mir-520b/atg7 axis. Chem. Biol. Interact. 2018, 280, 45–50. [Google Scholar] [CrossRef] [PubMed]
- Huang, H.; Tang, J.; Zhang, L.; Bu, Y.; Zhang, X. Mir-874 regulates multiple-drug resistance in gastric cancer by targeting atg16l1. Int. J. Oncol. 2018, 53, 2769–2779. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, D.; Kang, R.; Livesey, K.M.; Cheh, C.W.; Farkas, A.; Loughran, P.; Hoppe, G.; Bianchi, M.E.; Tracey, K.J.; Zeh, H.J., 3rd; et al. Endogenous hmgb1 regulates autophagy. J. Cell Biol. 2010, 190, 881–892. [Google Scholar] [CrossRef] [PubMed]
- Tang, D.; Kang, R.; Cheh, C.W.; Livesey, K.M.; Liang, X.; Schapiro, N.E.; Benschop, R.; Sparvero, L.J.; Amoscato, A.A.; Tracey, K.J.; et al. Hmgb1 release and redox regulates autophagy and apoptosis in cancer cells. Oncogene 2010, 29, 5299–5310. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Salim, H.; Akbar, N.S.; Zong, D.; Vaculova, A.H.; Lewensohn, R.; Moshfegh, A.; Viktorsson, K.; Zhivotovsky, B. Mirna-214 modulates radiotherapy response of non-small cell lung cancer cells through regulation of p38mapk, apoptosis and senescence. Br. J. Cancer 2012, 107, 1361–1373. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betin, V.M.; Lane, J.D. Caspase cleavage of atg4d stimulates gabarap-l1 processing and triggers mitochondrial targeting and apoptosis. J. Cell Sci. 2009, 122, 2554–2566. [Google Scholar] [CrossRef] [Green Version]
- Tan, J.Y.; Jia, L.Q.; Shi, W.H.; He, Q.; Zhu, L.; Yu, B. Rab5amediated autophagy regulates the phenotype and behavior of vascular smooth muscle cells. Mol. Med. Rep. 2016, 14, 4445–4453. [Google Scholar] [CrossRef] [Green Version]
- Ravikumar, B.; Imarisio, S.; Sarkar, S.; O’Kane, C.J.; Rubinsztein, D.C. Rab5 modulates aggregation and toxicity of mutant huntingtin through macroautophagy in cell and fly models of huntington disease. J. Cell Sci. 2008, 121, 1649–1660. [Google Scholar] [CrossRef] [Green Version]
- Borah, A.; Raveendran, S.; Rochani, A.; Maekawa, T.; Kumar, D.S. Targeting self-renewal pathways in cancer stem cells: Clinical implications for cancer therapy. Oncogenesis 2015, 4, e177. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rinkenbaugh, A.L.; Baldwin, A.S. The nf-kappab pathway and cancer stem cells. Cells 2016, 5, 16. [Google Scholar] [CrossRef] [PubMed]
- Housman, G.; Byler, S.; Heerboth, S.; Lapinska, K.; Longacre, M.; Snyder, N.; Sarkar, S. Drug resistance in cancer: An overview. Cancers 2014, 6, 1769–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- El-Sahli, S.; Xie, Y.; Wang, L.; Liu, S. Wnt signaling in cancer metabolism and immunity. Cancers 2019, 11, 904. [Google Scholar] [CrossRef] [Green Version]
- Zhang, K.; Guo, Y.; Wang, X.; Zhao, H.; Ji, Z.; Cheng, C.; Li, L.; Fang, Y.; Xu, D.; Zhu, H.H.; et al. Wnt/beta-catenin directs self-renewal symmetric cell division of htert(high) prostate cancer stem cells. Cancer Res. 2017, 77, 2534–2547. [Google Scholar] [CrossRef] [Green Version]
- Li, H.Y.; Liang, J.L.; Kuo, Y.L.; Lee, H.H.; Calkins, M.J.; Chang, H.T.; Lin, F.C.; Chen, Y.C.; Hsu, T.I.; Hsiao, M.; et al. Mir-105/93-3p promotes chemoresistance and circulating mir-105/93-3p acts as a diagnostic biomarker for triple negative breast cancer. Breast Cancer Res. 2017, 19, 133. [Google Scholar] [CrossRef]
- Lin, S.S.; Peng, C.Y.; Liao, Y.W.; Chou, M.Y.; Hsieh, P.L.; Yu, C.C. Mir-1246 targets ccng2 to enhance cancer stemness and chemoresistance in oral carcinomas. Cancers 2018, 10, 272. [Google Scholar] [CrossRef] [Green Version]
- Bernaudo, S.; Salem, M.; Qi, X.; Zhou, W.; Zhang, C.; Yang, W.; Rosman, D.; Deng, Z.; Ye, G.; Yang, B.B.; et al. Cyclin g2 inhibits epithelial-to-mesenchymal transition by disrupting wnt/beta-catenin signalling. Oncogene 2016, 35, 4828. [Google Scholar] [CrossRef] [Green Version]
- Wu, N.; Liu, C.; Bai, C.; Han, Y.P.; Cho, W.C.; Li, Q. Over-expression of deubiquitinating enzyme usp14 in lung adenocarcinoma promotes proliferation through the accumulation of beta-catenin. Int. J. Mol. Sci. 2013, 14, 10749–10760. [Google Scholar] [CrossRef]
- Yu, F.; Liu, J.B.; Wu, Z.J.; Xie, W.T.; Zhong, X.J.; Hou, L.K.; Wu, W.; Lu, H.M.; Jiang, X.H.; Jiang, J.J.; et al. Tumor suppressive microrna-124a inhibits stemness and enhances gefitinib sensitivity of non-small cell lung cancer cells by targeting ubiquitin-specific protease 14. Cancer Lett. 2018, 427, 74–84. [Google Scholar] [CrossRef]
- Xiao, W.; Gao, Z.; Duan, Y.; Yuan, W.; Ke, Y. Notch signaling plays a crucial role in cancer stem-like cells maintaining stemness and mediating chemotaxis in renal cell carcinoma. J. Exp. Clin. Cancer Res. 2017, 36, 41. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, E.Y.; Chang, E.; Lee, E.J.; Lee, H.W.; Kang, H.G.; Chun, K.H.; Woo, Y.M.; Kong, H.K.; Ko, J.Y.; Suzuki, H.; et al. Targeting of mir34a-notch1 axis reduced breast cancer stemness and chemoresistance. Cancer Res. 2014, 74, 7573–7582. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xu, K.; Shen, K.; Liang, X.; Li, Y.; Nagao, N.; Li, J.; Liu, J.; Yin, P. Mir-139-5p reverses cd44+/cd133+-associated multidrug resistance by downregulating notch1 in colorectal carcinoma cells. Oncotarget 2016, 7, 75118–75129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeong, J.Y.; Kang, H.; Kim, T.H.; Kim, G.; Heo, J.H.; Kwon, A.Y.; Kim, S.; Jung, S.G.; An, H.J. Microrna-136 inhibits cancer stem cell activity and enhances the anti-tumor effect of paclitaxel against chemoresistant ovarian cancer cells by targeting notch3. Cancer Lett. 2017, 386, 168–178. [Google Scholar] [CrossRef]
- Castel, D.; Mourikis, P.; Bartels, S.J.; Brinkman, A.B.; Tajbakhsh, S.; Stunnenberg, H.G. Dynamic binding of rbpj is determined by notch signaling status. Genes Dev. 2013, 27, 1059–1071. [Google Scholar] [CrossRef] [Green Version]
- Jin, Y.; Wang, M.; Hu, H.; Huang, Q.; Chen, Y.; Wang, G. Overcoming stemness and chemoresistance in colorectal cancer through mir-195-5p-modulated inhibition of notch signaling. Int. J. Biol. Macromol. 2018, 117, 445–453. [Google Scholar] [CrossRef]
- Dolatabadi, S.; Jonasson, E.; Linden, M.; Fereydouni, B.; Backsten, K.; Nilsson, M.; Martner, A.; Forootan, A.; Fagman, H.; Landberg, G.; et al. Jak-stat signalling controls cancer stem cell properties including chemotherapy resistance in myxoid liposarcoma. Int. J. Cancer 2019, 145, 435–449. [Google Scholar] [CrossRef]
- Wang, T.; Fahrmann, J.F.; Lee, H.; Li, Y.J.; Tripathi, S.C.; Yue, C.; Zhang, C.; Lifshitz, V.; Song, J.; Yuan, Y.; et al. Jak/stat3-regulated fatty acid beta-oxidation is critical for breast cancer stem cell self-renewal and chemoresistance. Cell Metab. 2018, 27, 136–150. [Google Scholar] [CrossRef] [Green Version]
- Ren, D.; Lin, B.; Zhang, X.; Peng, Y.; Ye, Z.; Ma, Y.; Liang, Y.; Cao, L.; Li, X.; Li, R.; et al. Maintenance of cancer stemness by mir-196b-5p contributes to chemoresistance of colorectal cancer cells via activating stat3 signaling pathway. Oncotarget 2017, 8, 49807–49823. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Morales, L.D.; Jang, I.S.; Cho, Y.Y.; Kim, D.J. Protein tyrosine phosphatases as potential regulators of stat3 signaling. Int. J. Mol. Sci. 2018, 19, 2708. [Google Scholar] [CrossRef] [Green Version]
- Long, J.; Jiang, C.; Liu, B.; Dai, Q.; Hua, R.; Chen, C.; Zhang, B.; Li, H. Maintenance of stemness by mir-589-5p in hepatocellular carcinoma cells promotes chemoresistance via stat3 signaling. Cancer Lett. 2018, 423, 113–126. [Google Scholar] [CrossRef] [PubMed]
- Cochrane, C.R.; Szczepny, A.; Watkins, D.N.; Cain, J.E. Hedgehog signaling in the maintenance of cancer stem cells. Cancers 2015, 7, 1554–1585. [Google Scholar] [CrossRef] [PubMed]
- Sari, I.N.; Phi, L.T.H.; Jun, N.; Wijaya, Y.T.; Lee, S.; Kwon, H.Y. Hedgehog signaling in cancer: A prospective therapeutic target for eradicating cancer stem cells. Cells 2018, 7, 208. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Santini, R.; Vinci, M.C.; Pandolfi, S.; Penachioni, J.Y.; Montagnani, V.; Olivito, B.; Gattai, R.; Pimpinelli, N.; Gerlini, G.; Borgognoni, L.; et al. Hedgehog-gli signaling drives self-renewal and tumorigenicity of human melanoma-initiating cells. Stem Cells 2012, 30, 1808–1818. [Google Scholar] [CrossRef] [PubMed]
- Tang, B.; Xu, A.; Xu, J.; Huang, H.; Chen, L.; Su, Y.; Zhang, L.; Li, J.; Fan, F.; Deng, J.; et al. Microrna-324-5p regulates stemness, pathogenesis and sensitivity to bortezomib in multiple myeloma cells by targeting hedgehog signaling. Int. J. Cancer 2018, 142, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Park, M.H.; Hong, J.T. Roles of nf-kappab in cancer and inflammatory diseases and their therapeutic approaches. Cells 2016, 5, 15. [Google Scholar] [CrossRef]
- Li, S.; Zeng, A.; Hu, Q.; Yan, W.; Liu, Y.; You, Y. Mir-423-5p contributes to a malignant phenotype and temozolomide chemoresistance in glioblastomas. Neuro Oncol. 2017, 19, 55–65. [Google Scholar] [CrossRef] [Green Version]
- Sun, G.; Ding, X.; Bi, N.; Wu, L.; Wang, J.; Zhang, W.; Dong, X.; Lv, N.; Song, Y.; Zhan, Q.; et al. Mir-423-5p in brain metastasis: Potential role in diagnostics and molecular biology. Cell Death Dis. 2018, 9, 936. [Google Scholar] [CrossRef] [Green Version]
- Yang, H.; Fu, H.; Wang, B.; Zhang, X.; Mao, J.; Li, X.; Wang, M.; Sun, Z.; Qian, H.; Xu, W. Exosomal mir-423-5p targets sufu to promote cancer growth and metastasis and serves as a novel marker for gastric cancer. Mol. Carcinog. 2018, 57, 1223–1236. [Google Scholar] [CrossRef] [PubMed]
- Wei, F.; Zhang, T.; Deng, S.C.; Wei, J.C.; Yang, P.; Wang, Q.; Chen, Z.P.; Li, W.L.; Chen, H.C.; Hu, H.; et al. Pd-l1 promotes colorectal cancer stem cell expansion by activating hmga1-dependent signaling pathways. Cancer Lett. 2019, 450, 1–13. [Google Scholar] [CrossRef]
- Hsu, J.M.; Xia, W.; Hsu, Y.H.; Chan, L.C.; Yu, W.H.; Cha, J.H.; Chen, C.T.; Liao, H.W.; Kuo, C.W.; Khoo, K.H.; et al. Stt3-dependent pd-l1 accumulation on cancer stem cells promotes immune evasion. Nat. Commun. 2018, 9, 1908. [Google Scholar] [CrossRef] [PubMed]
- Gao, L.; Guo, Q.; Li, X.; Yang, X.; Ni, H.; Wang, T.; Zhao, Q.; Liu, H.; Xing, Y.; Xi, T.; et al. Mir-873/pd-l1 axis regulates the stemness of breast cancer cells. EBioMedicine 2019, 41, 395–407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, Y.; Jiang, Z.; Guan, X.; Chen, Y.; Tang, Q.; Wang, G.; Wang, X. Mir-450b-5p suppresses stemness and the development of chemoresistance by targeting sox2 in colorectal cancer. DNA Cell Biol. 2016, 35, 249–256. [Google Scholar] [CrossRef] [PubMed]
- Zeng, J.F.; Ma, X.Q.; Wang, L.P.; Wang, W. Microrna-145 exerts tumor-suppressive and chemo-resistance lowering effects by targeting cd44 in gastric cancer. World J. Gastroenterol. 2017, 23, 2337–2345. [Google Scholar] [CrossRef]
- Zhu, Y.; Wang, C.; Becker, S.A.; Hurst, K.; Nogueira, L.M.; Findlay, V.J.; Camp, E.R. Mir-145 antagonizes snai1-mediated stemness and radiation resistance in colorectal cancer. Mol. Ther. 2018, 26, 744–754. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dimitrova, N.; Gocheva, V.; Bhutkar, A.; Resnick, R.; Jong, R.M.; Miller, K.M.; Bendor, J.; Jacks, T. Stromal expression of mir-143/145 promotes neoangiogenesis in lung cancer development. Cancer Discov. 2016, 6, 188–201. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Q.; Zhuang, J.; Deng, Y.; Yang, L.; Cao, W.; Chen, W.; Lin, T.; Lv, X.; Yu, H.; Xue, Y.; et al. Mir34a/golph3 axis abrogates urothelial bladder cancer chemoresistance via reduced cancer stemness. Theranostics 2017, 7, 4777–4790. [Google Scholar] [CrossRef] [PubMed]
- Jia, B.; Liu, W.; Gu, J.; Wang, J.; Lv, W.; Zhang, W.; Hao, Q.; Pang, Z.; Mu, N.; Zhang, W.; et al. Mir-7-5p suppresses stemness and enhances temozolomide sensitivity of drug-resistant glioblastoma cells by targeting yin yang 1. Exp. Cell Res. 2019, 375, 73–81. [Google Scholar] [CrossRef]
- Li, J.; Song, J.; Guo, F. Mir-186 reverses cisplatin resistance and inhibits the formation of the glioblastoma-initiating cell phenotype by degrading yin yang 1 in glioblastoma. Int. J. Mol. Med. 2019, 43, 517–524. [Google Scholar] [CrossRef] [Green Version]
- Wang, Z.; Yang, J.; Xu, G.; Wang, W.; Liu, C.; Yang, H.; Yu, Z.; Lei, Q.; Xiao, L.; Xiong, J.; et al. Targeting mir-381-nefl axis sensitizes glioblastoma cells to temozolomide by regulating stemness factors and multidrug resistance factors. Oncotarget 2015, 6, 3147–3164. [Google Scholar] [CrossRef]
- Chanmee, T.; Ontong, P.; Mochizuki, N.; Kongtawelert, P.; Konno, K.; Itano, N. Excessive hyaluronan production promotes acquisition of cancer stem cell signatures through the coordinated regulation of twist and the transforming growth factor beta (tgf-beta)-snail signaling axis. J. Biol. Chem. 2014, 289, 26038–26056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Leenders, G.J.; Sookhlall, R.; Teubel, W.J.; de Ridder, C.M.; Reneman, S.; Sacchetti, A.; Vissers, K.J.; van Weerden, W.; Jenster, G. Activation of c-met induces a stem-like phenotype in human prostate cancer. PLoS ONE 2011, 6, e26753. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hiramoto, H.; Muramatsu, T.; Ichikawa, D.; Tanimoto, K.; Yasukawa, S.; Otsuji, E.; Inazawa, J. Mir-509-5p and mir-1243 increase the sensitivity to gemcitabine by inhibiting epithelial-mesenchymal transition in pancreatic cancer. Sci. Rep. 2017, 7, 4002. [Google Scholar] [CrossRef] [PubMed]
- Yue, J.; Lv, D.; Wang, C.; Li, L.; Zhao, Q.; Chen, H.; Xu, L. Epigenetic silencing of mir-483-3p promotes acquired gefitinib resistance and emt in egfr-mutant nsclc by targeting integrin beta3. Oncogene 2018, 37, 4300–4312. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sese, M.; Fuentes, P.; Esteve-Codina, A.; Bejar, E.; McGrail, K.; Thomas, G.; Aasen, T.; Ramon, Y.C.S. Hypoxia-mediated translational activation of itgb3 in breast cancer cells enhances tgf-beta signaling and malignant features in vitro and in vivo. Oncotarget 2017, 8, 114856–114876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raab-Westphal, S.; Marshall, J.F.; Goodman, S.L. Integrins as therapeutic targets: Successes and cancers. Cancers 2017, 9, 110. [Google Scholar] [CrossRef] [PubMed]
- Seguin, L.; Kato, S.; Franovic, A.; Camargo, M.F.; Lesperance, J.; Elliott, K.C.; Yebra, M.; Mielgo, A.; Lowy, A.M.; Husain, H.; et al. An integrin beta (3)-kras-ralb complex drives tumour stemness and resistance to egfr inhibition. Nat. Cell Biol. 2014, 16, 457–468. [Google Scholar] [CrossRef] [Green Version]
- Jiao, D.; Chen, J.; Li, Y.; Tang, X.; Wang, J.; Xu, W.; Song, J.; Li, Y.; Tao, H.; Chen, Q. Mir-1-3p and mir-206 sensitizes hgf-induced gefitinib-resistant human lung cancer cells through inhibition of c-met signalling and emt. J. Cell Mol. Med. 2018, 22, 3526–3536. [Google Scholar] [CrossRef]
- Garofalo, M.; Romano, G.; Di Leva, G.; Nuovo, G.; Jeon, Y.J.; Ngankeu, A.; Sun, J.; Lovat, F.; Alder, H.; Condorelli, G.; et al. Egfr and met receptor tyrosine kinase-altered microrna expression induces tumorigenesis and gefitinib resistance in lung cancers. Nat. Med. 2011, 18, 74–82. [Google Scholar] [CrossRef]
- Hu, X.; Miao, J.; Zhang, M.; Wang, X.; Wang, Z.; Han, J.; Tong, D.; Huang, C. Mirna-103a-3p promotes human gastric cancer cell proliferation by targeting and suppressing atf7 in vitro. Mol. Cells 2018, 41, 390–400. [Google Scholar]
- Wu, D.M.; Hong, X.W.; Wang, L.L.; Cui, X.F.; Lu, J.; Chen, G.Q.; Zheng, Y.L. Microrna-17 inhibition overcomes chemoresistance and suppresses epithelial-mesenchymal transition through a dedd-dependent mechanism in gastric cancer. Int. J. Biochem. Cell Biol. 2018, 102, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Li, Y.; Yu, J.; Pei, H.; Luo, P.; Zhang, J. Mir-128 modulates chemosensitivity and invasion of prostate cancer cells through targeting zeb1. Jpn. J. Clin. Oncol. 2015, 45, 474–482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fischer, K.R.; Durrans, A.; Lee, S.; Sheng, J.; Li, F.; Wong, S.T.; Choi, H.; El Rayes, T.; Ryu, S.; Troeger, J.; et al. Epithelial-to-mesenchymal transition is not required for lung metastasis but contributes to chemoresistance. Nature 2015, 527, 472–476. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Men, X.; Zhao, R.; Han, J.; Fan, Z.; Wang, Y.; Lv, Y.; Zuo, J.; Zhao, L.; Sang, M.; et al. Mir-200c inhibits tgf-beta-induced-emt to restore trastuzumab sensitivity by targeting zeb1 and zeb2 in gastric cancer. Cancer Gene Ther. 2018, 25, 68–76. [Google Scholar] [CrossRef] [Green Version]
- Wu, G.; Wang, J.; Chen, G.; Zhao, X. Microrna-204 modulates chemosensitivity and apoptosis of prostate cancer cells by targeting zinc-finger e-box-binding homeobox 1 (zeb1). Am. J. Transl. Res. 2017, 9, 3599–3610. [Google Scholar]
- Cao, L.; Wan, Q.; Li, F.; Tang, C.E. Mir-363 inhibits cisplatin chemoresistance of epithelial ovarian cancer by regulating snail-induced epithelial-mesenchymal transition. BMB Rep. 2018, 51, 456–461. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Zhang, R.; Zhang, S.; Xu, R.; Yang, Q. Microrna-574-3p regulates epithelial mesenchymal transition and cisplatin resistance via targeting zeb1 in human gastric carcinoma cells. Gene 2019, 700, 110–119. [Google Scholar] [CrossRef]
- Lee, J.W.; Guan, W.; Han, S.; Hong, D.K.; Kim, L.S.; Kim, H. Microrna-708-3p mediates metastasis and chemoresistance through inhibition of epithelial-to-mesenchymal transition in breast cancer. Cancer Sci. 2018, 109, 1404–1413. [Google Scholar] [CrossRef] [Green Version]
- Wang, G.; Dong, Y.; Liu, H.; Ji, N.; Cao, J.; Liu, A.; Tang, X.; Ren, Y. Loss of mir-873 contributes to gemcitabine resistance in triple-negative breast cancer via targeting zeb1. Oncol. Lett. 2019, 18, 3837–3844. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Gibbons, D.L.; Goswami, S.; Cortez, M.A.; Ahn, Y.H.; Byers, L.A.; Zhang, X.; Yi, X.; Dwyer, D.; Lin, W.; et al. Metastasis is regulated via microrna-200/zeb1 axis control of tumour cell pd-l1 expression and intratumoral immunosuppression. Nat. Commun. 2014, 5, 5241. [Google Scholar] [CrossRef]
- Lv, Q.; Wang, W.; Xue, J.; Hua, F.; Mu, R.; Lin, H.; Yan, J.; Lv, X.; Chen, X.; Hu, Z.W. Dedd interacts with pi3kc3 to activate autophagy and attenuate epithelial-mesenchymal transition in human breast cancer. Cancer Res. 2012, 72, 3238–3250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Raposo, G.; Stoorvogel, W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol. 2013, 200, 373–383. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jabalee, J.; Towle, R.; Garnis, C. The role of extracellular vesicles in cancer: Cargo, function, and therapeutic implications. Cells 2018, 7, 93. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zonneveld, M.I.; Keulers, T.G.H.; Rouschop, K.M.A. Extracellular vesicles as transmitters of hypoxia tolerance in solid cancers. Cancers 2019, 11, 154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sansone, P.; Savini, C.; Kurelac, I.; Chang, Q.; Amato, L.B.; Strillacci, A.; Stepanova, A.; Iommarini, L.; Mastroleo, C.; Daly, L.; et al. Packaging and transfer of mitochondrial DNA via exosomes regulate escape from dormancy in hormonal therapy-resistant breast cancer. Proc. Natl. Acad. Sci. USA 2017, 114, E9066–E9075. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- O’Neill, C.P.; Gilligan, K.E.; Dwyer, R.M. Role of extracellular vesicles (evs) in cell stress response and resistance to cancer therapy. Cancers 2019, 11, 136. [Google Scholar] [CrossRef] [Green Version]
- Kreger, B.T.; Johansen, E.R.; Cerione, R.A.; Antonyak, M.A. The enrichment of survivin in exosomes from breast cancer cells treated with paclitaxel promotes cell survival and chemoresistance. Cancers 2016, 8, 111. [Google Scholar] [CrossRef] [Green Version]
- Zheng, P.; Chen, L.; Yuan, X.; Luo, Q.; Liu, Y.; Xie, G.; Ma, Y.; Shen, L. Exosomal transfer of tumor-associated macrophage-derived mir-21 confers cisplatin resistance in gastric cancer cells. J. Exp. Clin. Cancer Res. 2017, 36, 53. [Google Scholar] [CrossRef] [Green Version]
- Qin, X.; Guo, H.; Wang, X.; Zhu, X.; Yan, M.; Wang, X.; Xu, Q.; Shi, J.; Lu, E.; Chen, W.; et al. Exosomal mir-196a derived from cancer-associated fibroblasts confers cisplatin resistance in head and neck cancer through targeting cdkn1b and ing5. Genome Biol. 2019, 20, 12. [Google Scholar] [CrossRef]
- Fu, X.; Liu, M.; Qu, S.; Ma, J.; Zhang, Y.; Shi, T.; Wen, H.; Yang, Y.; Wang, S.; Wang, J.; et al. Exosomal microrna-32-5p induces multidrug resistance in hepatocellular carcinoma via the pi3k/akt pathway. J. Exp. Clin. Cancer Res. 2018, 37, 52. [Google Scholar] [CrossRef] [Green Version]
- Wang, M.; Qiu, R.; Yu, S.; Xu, X.; Li, G.; Gu, R.; Tan, C.; Zhu, W.; Shen, B. Paclitaxelresistant gastric cancer mgc803 cells promote epithelialtomesenchymal transition and chemoresistance in paclitaxelsensitive cells via exosomal delivery of mir1555p. Int. J. Oncol. 2019, 54, 326–338. [Google Scholar] [PubMed]
- Santos, J.C.; Lima, N.D.S.; Sarian, L.O.; Matheu, A.; Ribeiro, M.L.; Derchain, S.F.M. Exosome-mediated breast cancer chemoresistance via mir-155 transfer. Sci. Rep. 2018, 8, 829. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, F.; Ma, C.; Zhou, T.; Dong, X.; Luo, Q.; Geng, L.; Ding, L.; Zhang, Y.; Zhang, L.; Li, N.; et al. Exosomes derived from gemcitabine-resistant cells transfer malignant phenotypic traits via delivery of mirna-222-3p. Mol. Cancer 2017, 16, 132. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Min, Q.H.; Wang, X.Z.; Zhang, J.; Chen, Q.G.; Li, S.Q.; Liu, X.Q.; Li, J.; Liu, J.; Yang, W.M.; Jiang, Y.H.; et al. Exosomes derived from imatinib-resistant chronic myeloid leukemia cells mediate a horizontal transfer of drug-resistant trait by delivering mir-365. Exp. Cell Res. 2018, 362, 386–393. [Google Scholar] [CrossRef]
- Cornell, L.; Wander, S.A.; Visal, T.; Wagle, N.; Shapiro, G.I. Microrna-mediated suppression of the tgf-beta pathway confers transmissible and reversible cdk4/6 inhibitor resistance. Cell Rep. 2019, 26, 2667–2680.e7. [Google Scholar] [CrossRef] [Green Version]
- Kwok, H.H.; Ning, Z.; Chong, P.W.; Wan, T.S.; Ng, M.H.; Ho, G.Y.F.; Ip, M.S.; Lam, D.C. Transfer of extracellular vesicle-associated-rnas induces drug resistance in alk-translocated lung adenocarcinoma. Cancers 2019, 11, 104. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Lu, Y.; Xu, Y.; Hou, S.; Huang, J.; Wang, B.; Zhao, J.; Xia, S.; Fan, S.; Yu, X.; et al. Exosomal transfer of mir-501 confers doxorubicin resistance and tumorigenesis via targeting of blid in gastric cancer. Cancer Lett. 2019, 459, 122–134. [Google Scholar] [CrossRef]
- Zeng, X.; Zhao, H.; Li, Y.; Fan, J.; Sun, Y.; Wang, S.; Wang, Z.; Song, P.; Ju, D. Targeting hedgehog signaling pathway and autophagy overcomes drug resistance of bcr-abl-positive chronic myeloid leukemia. Autophagy 2015, 11, 355–372. [Google Scholar] [CrossRef] [Green Version]
- Oddo, D.; Sennott, E.M.; Barault, L.; Valtorta, E.; Arena, S.; Cassingena, A.; Filiciotto, G.; Marzolla, G.; Elez, E.; van Geel, R.M.; et al. Molecular landscape of acquired resistance to targeted therapy combinations in braf-mutant colorectal cancer. Cancer Res. 2016, 76, 4504–4515. [Google Scholar] [CrossRef] [Green Version]
- Eskildsen, T.; Taipaleenmaki, H.; Stenvang, J.; Abdallah, B.M.; Ditzel, N.; Nossent, A.Y.; Bak, M.; Kauppinen, S.; Kassem, M. Microrna-138 regulates osteogenic differentiation of human stromal (mesenchymal) stem cells in vivo. Proc. Natl. Acad. Sci. USA 2011, 108, 6139–6144. [Google Scholar] [CrossRef] [Green Version]
- Ceppi, M.; Pereira, P.M.; Dunand-Sauthier, I.; Barras, E.; Reith, W.; Santos, M.A.; Pierre, P. Microrna-155 modulates the interleukin-1 signaling pathway in activated human monocyte-derived dendritic cells. Proc. Natl. Acad. Sci. USA 2009, 106, 2735–2740. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varshney, A.; Panda, J.J.; Singh, A.K.; Yadav, N.; Bihari, C.; Biswas, S.; Sarin, S.K.; Chauhan, V.S. Targeted delivery of microrna-199a-3p using self-assembled dipeptide nanoparticles efficiently reduces hepatocellular carcinoma in mice. Hepatology 2018, 67, 1392–1407. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Y.; Duo, Y.; Bi, J.; Zeng, X.; Mei, L.; Bao, S.; He, L.; Shan, A.; Zhang, Y.; Yu, X. Targeted delivery of anti-mir-155 by functionalized mesoporous silica nanoparticles for colorectal cancer therapy. Int. J. Nanomed. 2018, 13, 1241–1256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Q.; Huang, Z.; Guo, W.; Ni, S.; Xiao, X.; Wang, L.; Huang, D.; Tan, C.; Xu, Q.; Zha, R.; et al. Microrna-202-3p inhibits cell proliferation by targeting adp-ribosylation factor-like 5a in human colorectal carcinoma. Clin. Cancer Res. 2014, 20, 1146–1157. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, T.; Guo, J.; Zhang, X. Mir-202-5p/pten mediates doxorubicin-resistance of breast cancer cells via pi3k/akt signaling pathway. Cancer Biol. Ther. 2019, 20, 989–998. [Google Scholar] [CrossRef]
- Kozomara, A.; Birgaoanu, M.; Griffiths-Jones, S. Mirbase: From microrna sequences to function. Nucleic Acids Res. 2019, 47, D155–D162. [Google Scholar] [CrossRef]
- Na, Y.J.; Kim, J.H. Understanding cooperativity of micrornas via microrna association networks. BMC Genom. 2013, 14 (Suppl. 5), S17. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Luo, J.; Liu, C.; Niu, H.; Wang, J.; Liu, Q.; Zhao, Z.; Xu, H.; Ding, Y.; Sun, J.; et al. Investigating microrna and transcription factor co-regulatory networks in colorectal cancer. BMC Bioinform. 2017, 18, 388. [Google Scholar] [CrossRef] [Green Version]
| MiRNAs | Target Gene(s) | Cancer Type | Effect of MiRNAs | Ref. |
|---|---|---|---|---|
| miR-20a-5p | MAPK1 | Breast cancer | Over-expression of miR-20a-5p increases the overall cytotoxicity of several agents, such as vinorelbine, doxorubicin, and paclitaxel | [44] |
| miR-27-3p | FZD7 | Hepatocellular carcinoma | Over-expression of miR-27-3p enhances the sensitivity of multidrug-resistant cells to 5-fluorouracil | [33] |
| miR-122-5p | CTNNB1 | Hepatocellular carcinoma | Up-regulation of miR-122-5p raises the anti-cancer effect of oxaliplatin | [34] |
| miR-129-5p | ABCB1 | Gastric cancer | Up-regulation of miR-129-5p heightens cisplatin-induced cell death and caspase activation | [17] |
| miR-133a-3p | ABCC1 | Hepatocellular carcinoma | Over-expression of miR-133a-3p leads to elevated cytotoxicity of doxorubicin | [23] |
| miR-148a-3p miR-148b-3p miR-152-3p | SPIN1 | Breast cancer | Over-expression of these miRNAs re-sensitizes the drug-resistant cells to doxorubicin | [45] |
| miR-199a-3p | MTOR | Cholangiocarcinoma | Reconstitution of miR-199a-3p increases growth inhibition rate and apoptosis induced by cisplatin | [39] |
| miR-210-3p | ABCC5 | Pancreatic cancer | Elevated miR-210-3p levels improve the overall cytotoxicity of gemcitabine | [26] |
| miR-218-5p | PRKCE | Gallbladder cancer | Elevated miR-218-5p levels potentiate gemcitabine-mediated cell death and growth inhibition | [38] |
| miR-223-3p | ABCB1 | Hepatocellular carcinoma | Down-regulation of miR-223-3p confers resistance to doxorubicin | [18] |
| miR-326 | ABCC1 | Hepatocellular carcinoma | Over-expression of miR-326 leads to elevated cytotoxicity of doxorubicin | [23] |
| miR-328-3p | ABCG2 | Breast cancer | Over-expression of miR-328-3p augments the sensitivity of drug-resistant cells to mitoxantrone | [29] |
| miR-491-3p | ABCB1, SP3 | Hepatocellular carcinoma | Down-regulation of miR-491-3p decreases the sensitivity to doxorubicin and vinblastin | [19] |
| miR-495-3p | ABCB1, UBE2C | Ovarian cancer, Gastric cancer, Lung cancer | Up-regulation of miR-495-3p re-sensitizes drug-resistant ovarian and gastric cancer cells to doxorubicin/paclitaxel combination, and cisplatin resistance is reversed in miR-495-3p over-expressing lung cancer cells | [20,46] |
| miR-506-3p | CTNNB1 | Colorectal cancer | Over-expression of miR-506-3p re-sensitizes drug-resistant cells to oxaliplatin | [35] |
| miR-508-5p | ABCB1, ZNRD1 | Gastric cancer | Down-regulation of miR-508-5p confers resistance to cisplatin, doxorubicin, vincristine, and 5-fluorouracil | [21] |
| miR-514 | ABCA1, ABCA10, ABCF2 | Ovarian cancer | Up-regulation of miR-514 re-sensitizes drug-resistant cells to cisplatin | [28] |
| miR-595 | SLC19A1, ABCB1 | Acute lymphoblastic leukemia, Ovarian cancer | Over-expression of miR-595 can either decrease or increase the efficacy of methotrexate or cisplatin, respectively | [30,31] |
| miR-1268a | ABCC1 | Glioblastoma | Over-expression of miR-1268a augments temozolomide sensitivity | [24] |
| MiRNAs | Target Gene(s) | Cancer Type | Effect of MiRNAs | Ref. |
|---|---|---|---|---|
| miR-7-5p | PARP1 | Lung cancer | Over-expression of miR-7-5p increases the overall cytotoxicity of doxorubicin | [55] |
| miR-30-5p | FANCF, REV1 | Breast cancer | Over-expression of miR-30-5p raises the anti-cancer effect of doxorubicin | [56] |
| miR-138-5p | ERCC1, ERCC4 | Gastric cancer | Knockdown of miR-138-5p lowers the efficacy of cisplatin, thus enhancing cisplatin resistance | [58] |
| miR-182-5p | BRCA1, RAD51 | Breast cancer, Acute Myelogenous Leukemia | Silencing of miR-182-5p results in resistance to PARP1 inhibitors and CNDAC | [63,64] |
| miR-205-5p | PRKCE, ZEB1 | Prostate cancer | Reconstitution of miR-205-5p escalates the efficiency of radiotherapy | [68] |
| miR-211-5p | POLH, TDP1, ATRX, MRPS11, ERCC6L2 | Ovarian cancer | Elevated miR-211-5p levels improve the overall cytotoxicity of carboplatin | [69] |
| miR-488-3p | EIF3A | Lung cancer | Elevated miR-488-3p levels impede cisplatin-mediated induction of cell death and growth inhibition | [73] |
| miR-493-5p | CHD4 | Ovarian cancer | Down-regulation of miR-493-5p levels leads to enhanced responsiveness to cisplatin and olaparib | [75] |
| miR-520g-3p, miR-520h | APEX1 | Multiple myeloma | Over-expression of both miR-520g-3p and miR-520h hampers the growth of bortezomib resistant multiple myeloma cells | [69] |
| miR-4429 | RAD51 | Cervical cancer | Over-expression of miR-4429 enhances radiosensitivity | [65] |
| MiRNAs | Target Gene(s) | Cancer Type | Effect of MiRNAs | Ref. |
|---|---|---|---|---|
| miR-23-3p | ATG12, HMGB2 | Gastric cancer | Over-expression of miR-23-3p leads to the enhanced efficacy of 5-fluorouracil, cisplatin, and vincristine in drug-resistant cells | [90] |
| miR-26-5p | ULK1 | Hepatocellular carcinoma | Over-expression of miR-26-5p promotes doxorubicin-induced apoptosis | [91] |
| miR-34-5p | HMGB1 | Retinoblastoma | Reconstitution of miR-34-5p enhances cell death following treatment of etoposide, vincristine, and carboplatin | [92] |
| miR-101-3p | RAB5A, STMN1, ATG4D | Hepatocellular carcinoma, Breast cancer | Up-regulation of miR-101-3p increases cisplatin- and 4-hydroxytamoxifen-induced cell death in hepatocellular carcinoma and breast cancer cells, respectively | [93,94] |
| miR-129-3p | MTOR | Hepatocellular carcinoma, Gastric cancer | Silencing of miR-129-3p escalates the efficiency of Trichostatin A | [87] |
| miR-137-3p | ATG5 | Pancreatic cancer | Elevated miR-137-3p levels enhances the effects of doxorubicin on growth inhibition and apoptosis | [95] |
| miR-140-5p | HMGN5 | Osteosarcoma | Reconstitution of miR-140-5p sensitizes cells to cisplatin, doxorubicin, and methotrexate | [96] |
| miR-142-3p | ATG5, ATG16L1 | Hepatocellular carcinoma | Reconstitution of miR-142-3p enhances the cytotoxicity of sorafenib | [97] |
| miR-148-3p | RAB12 | Gastric cancer | Up-regulation of miR-148-3p reverses cisplatin resistance | [89] |
| miR-152-3p | ATG14 | Ovarian cancer | Over-expression of miR-152-3p sensitizes cisplatin-resistant cells toward cisplatin via enhancing cell death and inhibiting cell growth | [98] |
| miR-214-3p | ATG12 | Colorectal cancer | Down-regulation of miR-214-3p induces radioresistance | [99] |
| miR-224-3p | ATG5 | Glioblastoma, Astrocytoma | Over-expression of miR-224-3p enhances the efficacy of temozolomide with increased apoptosis induction | [100] |
| miR-409-3p | BECLIN1 | Colorectal cancer | Replacement of miR-409-3p sensitizes resistant cancer cells to oxaliplatin | [101] |
| miR-410-3p | HMGB1 | Pancreatic cancer | Over-expression of miR-410-3p improves gemcitabine-induced cell death and growth inhibition in drug-resistant cells | [102] |
| miR-520-3p | ATG7 | Hepatocellular carcinoma | Replacement of miR-520-3p increases the sensitivity of drug-resistant cells to doxorubicin by enhancing cell death and growth inhibition | [103] |
| miR-874-3p | ATG16L1 | Gastric cancer | Restoration of miR-874-3p sensitizes cells to 5-fluorouracil and cisplatin | [104] |
| MiRNAs | Target Gene(s) | Cancer Type | Effect of MiRNAs | Ref. |
|---|---|---|---|---|
| miR-7-5p | YY1 | Glioblastoma | Treatment with miR-7-5p enhances the sensitivity of drug-resistant cells to temozolomide | [148] |
| miR-34-5p | NOTCH1, GOLPH3 | Breast cancer, Urothelial bladder cancer | Ectopic expression of miR-34-5p increases the sensitivity to doxorubicin by enhancing apoptosis induction in drug-resistant breast cancer cells. Down-regulation of miR-34-5p desensitizes bladder cancer cells to gemcitabine and cisplatin | [122,147] |
| miR-93-3p, miR-105-5p | SFRP1 | Breast cancer | Silencing of miR-93-3p and miR-105-5p enhances the sensitivity to cisplatin and chemoradiotherapy | [116] |
| miR-124-3p | USP14 | Lung cancer | Over-expression of miR-124-3p increases the effects of gefitinib on apoptosis and growth inhibition | [120] |
| miR-136-5p | NOTCH3 | Ovarian cancer | Over-expression of miR-136-5p escalates paclitaxel-induced cell death in drug-resistant cells | [124] |
| miR-139-5p | NOTCH1 | Colorectal cancer | Ectopic expression of miR-139-5p enhances the sensitivity of CD44+/CD133+ cells to oxaliplatin, vincristine, 5-fluorouracil, and mitomycin C | [123] |
| miR-145-5p | c-MYC, CD44, KLF4, OCT4 | Colorectal cancer, Gastric cancer | Over-expression of miR-145-5p sensitizes cells to cisplatin and 5-fluorouracil in gastric cancer. This miRNA also enhances the efficacy of radiation and oxaliplatin | [144,145] |
| miR-186-5p | YY1 | Glioblastoma | Ectopic expression of miR-186-5p improves the cisplatin cytotoxicity | [149] |
| miR-195-5p | NOTCH2, RBPJ | Colorectal cancer | Over-expression of miR-195-5p subdues resistance to 5-fluorouracil | [126] |
| miR-196-5p | SOCS1, SOCS3 | Colorectal cancer | Knockdown of miR-196-5p sensitizes cancer cells to 5-fluorouracil by augmenting apoptosis | [129] |
| miR-324-5p | SMO, GLI1 | Multiple myeloma | Over-expression of miR-324-5p heightens the efficacy of bortezomib in multiple myeloma cells | [135] |
| miR-381-3p | NEFL | Glioblastoma | Silencing of miR-381-3p increases the sensitivity of cells to temozolomide | [150] |
| miR-423-5p | ING4 | Glioblastoma | Over-expression of miR-423-5p significantly attenuates the chemosensitivity of glioma cells to temozolomide | [137] |
| miR-450b-5p | SOX2 | Colorectal cancer | Ectopic expression of miR-450b-5p sensitizes cells to 5-fluorouracil | [143] |
| miR-589-5p | SOCS2, SOCS5, PTPN1, PTPN11 | Hepatocellular carcinoma | Ectopic expression of miR-589-5p promotes the emergence of acquired resistance to doxorubicin | [131] |
| miR-873-5p | PD-L1 | Breast cancer | Over-expression of miR-873-5p attenuates therapeutic resistance to doxorubicin | [142] |
| miR-1246 | CCNG2 | Oral cancer | Knockdown of miR-1246 sensitizes cancer cells to cisplatin | [117] |
| MiRNAs | Target Gene(s) | Cancer Type | Effect of MiRNAs | Ref. |
|---|---|---|---|---|
| miR-1-3p | MET | Lung cancer | Over-expression of miR-1-3p increases the anti-proliferative effects of gefitinib | [158] |
| miR-17-5p | DEDD | Gastric cancer | Inhibition of miR-17-5p augments cisplatin- and 5-fluorouracil-induced apoptosis | [161] |
| miR-103-3p | PRKCE | Lung cancer | Enforced expression of miR-103-3p elevates the anti-proliferative effects of gefitinib along with caspase 3/7 activation | [159] |
| miR-128-3p | ZEB1 | Prostate cancer | Over-expression of miR-128-3p improves the effect of cisplatin on cell growth and invasion | [162] |
| miR-200 family | ZEB1, ZEB2 | Gastric cancer, Breast cancer | Enforced expression of miR-200 family restores trastuzumab and cyclophosphamide sensitivity in gastric and breast cancer, respectively | [163,164] |
| miR-203a-3p | SRC | Lung cancer | Enforced expression of miR-203a-3p elevates the anti-proliferative effects of gefitinib along with caspase 3/7 activation | [159] |
| miR-204-5p | TGFBR2, ZEB1 | Gastric cancer, Prostate cancer | Over-expression of miR-204-5p improves the efficacy of 5-fluorouracil in gastric cancer cells. In prostate cancer cells, miR-204-5p promotes docetaxel-mediated apoptosis | [154,165] |
| miR-206 | MET | Lung cancer | Over-expression of miR-206 increases the anti-proliferative effects of gefitinib | [158] |
| miR-363-3p | SNAI1 | Ovarian cancer | Silencing of miR-363-3p diminishes the anti-proliferative effects of cisplatin | [166] |
| miR-483-3p | ITGB3 | Lung cancer | Epigenetic silencing of miR-483-3p desensitizes cells to gefitinib | [154] |
| miR-509-5p | VIM, HMGA2 | Pancreatic cancer | Over-expression of miR-509-5p increases the anti-proliferative effects of gemcitabine | [153] |
| miR-574-3p | ZEB1 | Gastric cancer | Enforced expression of miR-574-3p elevates cisplatin-induced apoptosis | [167] |
| miR-708-3p | ZEB1, CDH2, VIM | Breast cancer | Over-expression of miR-708-3p augments doxorubicin-mediated apoptosis | [168] |
| miR-873-5p | ZEB1 | Breast cancer | Ectopic expression of miR-873-5p elevates the gemcitabine-induced cell growth arrest | [169] |
| miR-1243 | SMAD2, SMAD4 | Pancreatic cancer | Over-expression of miR-1243 increases the anti-proliferative effects of gemcitabine | [153] |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Seo, H.A.; Moeng, S.; Sim, S.; Kuh, H.J.; Choi, S.Y.; Park, J.K. MicroRNA-Based Combinatorial Cancer Therapy: Effects of MicroRNAs on the Efficacy of Anti-Cancer Therapies. Cells 2020, 9, 29. https://doi.org/10.3390/cells9010029
Seo HA, Moeng S, Sim S, Kuh HJ, Choi SY, Park JK. MicroRNA-Based Combinatorial Cancer Therapy: Effects of MicroRNAs on the Efficacy of Anti-Cancer Therapies. Cells. 2020; 9(1):29. https://doi.org/10.3390/cells9010029
Chicago/Turabian StyleSeo, Hyun Ah, Sokviseth Moeng, Seokmin Sim, Hyo Jeong Kuh, Soo Young Choi, and Jong Kook Park. 2020. "MicroRNA-Based Combinatorial Cancer Therapy: Effects of MicroRNAs on the Efficacy of Anti-Cancer Therapies" Cells 9, no. 1: 29. https://doi.org/10.3390/cells9010029
APA StyleSeo, H. A., Moeng, S., Sim, S., Kuh, H. J., Choi, S. Y., & Park, J. K. (2020). MicroRNA-Based Combinatorial Cancer Therapy: Effects of MicroRNAs on the Efficacy of Anti-Cancer Therapies. Cells, 9(1), 29. https://doi.org/10.3390/cells9010029






