Metabolism of Reactive Oxygen Species in Osteosarcoma and Potential Treatment Applications
Abstract
:1. Introduction
2. Materials and Methods
2.1. Microarray Data
2.2. Osteosarcoma Specimens
2.3. Immunohistochemistry
2.4. Cell Lines and Culture Conditions
2.5. Quantitative Real-Time Polymerase Chain Reaction (qRT-PCR)
2.6. Western Blot Analysis
2.7. Plasmid Construction and Cell Transfection
2.8. Cell Proliferation Assay
2.9. Xenograft Transplantation
2.10. Statistical Analysis
3. Results
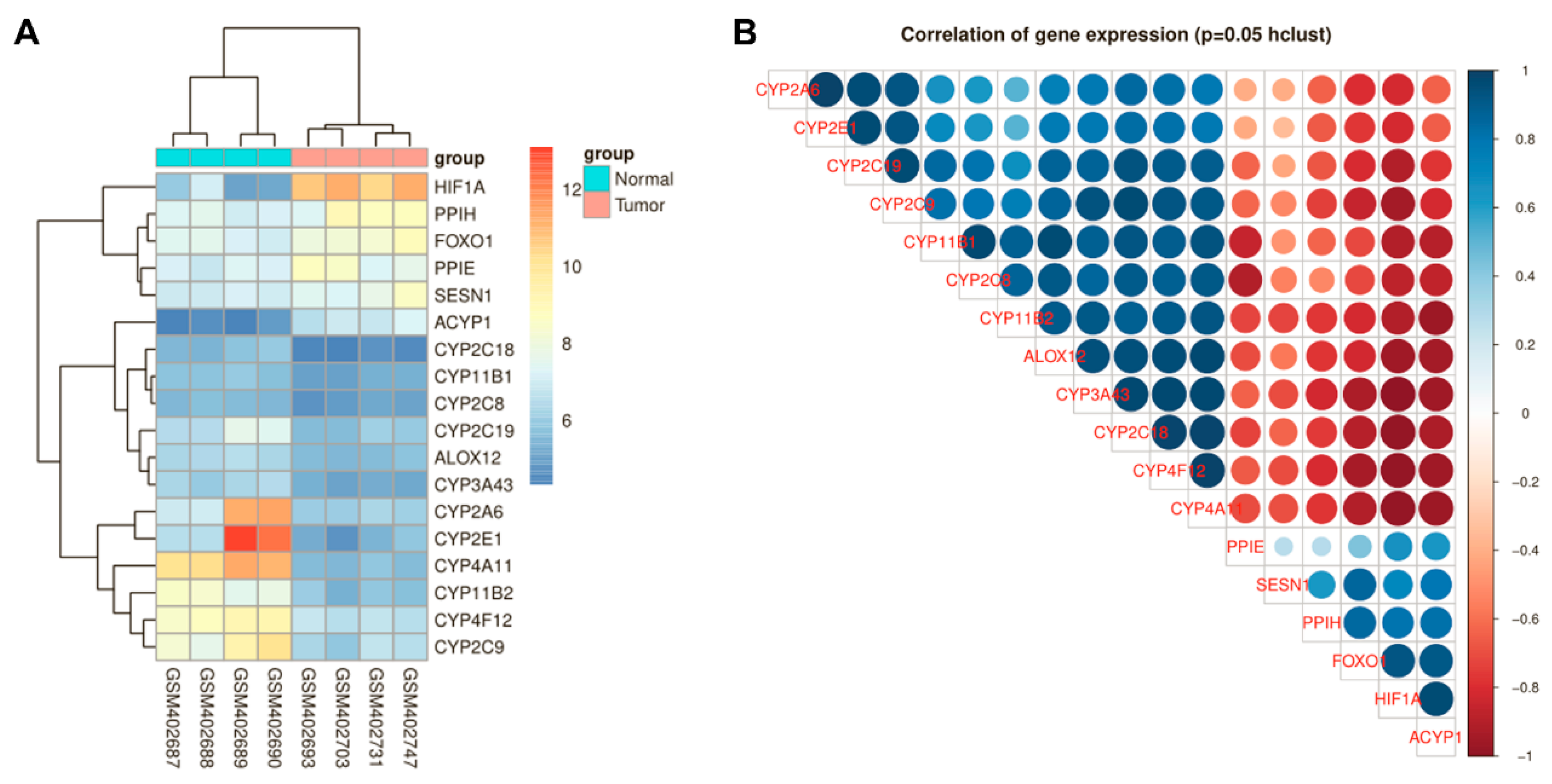
3.1. HIF-1α and FoxO1 Expression was Increased in Human Bone Cancer Tissues
3.2. ROS Production in OS Cells Was Related to HIF-1α Expression
3.3. HIF-1α Targeted FoxO1 Directly to Promote Its Expression in OS Cells
3.4. HIF-1α Expression Was Positively Correlated with FoxO1 and The Antioxidant Proteins MnSOD, catalase and Sesn3
3.5. The ROS Metabolism Regulation Pathway Is Involved in HIF-1α-Silenced OS Cells
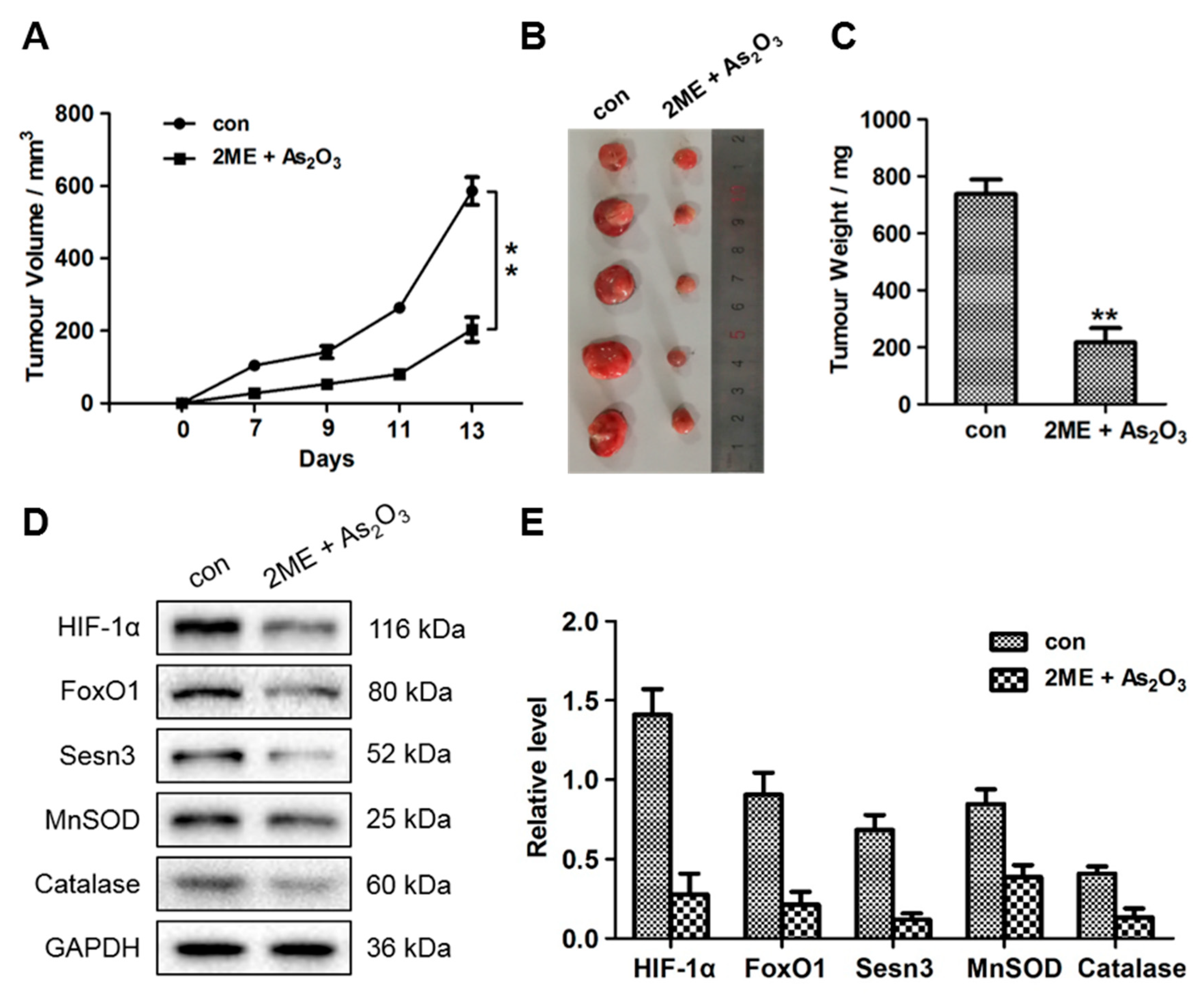
3.6. ME + As2O3 Treatment Attenuates Tumour Growth
3.7. ME + As2O3 Induce Bone Cancer Cell Death and Inhibit Migration
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Mirabello, L.; Troisi, R.J.; Savage, S.A. Osteosarcoma incidence and survival rates from 1973 to 2004: Data from the surveillance, epidemiology, and end results program. Cancer 2009, 115, 1531–1543. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Biazzo, A.; De Paolis, M. Multidisciplinary approach to osteosarcoma. Acta Orthop. Belg. 2016, 82, 690–698. [Google Scholar] [PubMed]
- Lagmay, J.P.; Krailo, M.D.; Dang, H.; Kim, A.; Hawkins, D.S.; Beaty, O., III.; Widemann, B.C.; Zwerdling, T.; Bomgaars, L.; Langevin, A.M.; et al. Outcome of patients with recurrent osteosarcoma enrolled in seven phase II trials through children’s cancer group, pediatric oncology group, and children’s oncology group: Learning from the past to move forward. J. Clin. Oncol. 2016, 34, 3031–3038. [Google Scholar] [CrossRef] [PubMed]
- Amant, F.; Moerman, P.; Neven, P.; Timmerman, D.; Van Limbergen, E.; Vergote, I. Endometrial cancer. Lancet 2005, 366, 491–505. [Google Scholar] [CrossRef]
- Urbanek, P.; Klotz, L.O. Posttranscriptional regulation of FOXO expression: MicroRNAs and beyond. Br. J. Pharmacol. 2017, 174, 1514–1532. [Google Scholar] [CrossRef] [Green Version]
- Kenyon, C. The first long-lived mutants: Discovery of the insulin/IGF-1 pathway for ageing. Philos. Trans. R. Soc. B Biol. Sci. 2011, 366, 9–16. [Google Scholar] [CrossRef] [Green Version]
- Chatterton, D.E.; Nguyen, D.N.; Bering, S.B.; Sangild, P.T. Anti-inflammatory mechanisms of bioactive milk proteins in the intestine of newborns. Int. J. Biochem. Cell Biol. 2013, 45, 1730–1747. [Google Scholar] [CrossRef]
- Kim, S.Y.; Ko, Y.S.; Park, J.; Choi, Y.; Park, J.W.; Kim, Y.; Pyo, J.S.; Yoo, Y.B.; Lee, J.S.; Lee, B.L. Forkhead transcription factor FOXO1 inhibits angiogenesis in gastric cancer in relation to SIRT1. Cancer Res. Treat. 2016, 48, 345–354. [Google Scholar] [CrossRef]
- Gao, Z.; Liu, R.; Ye, N.; Liu, C.; Li, X.; Guo, X.; Zhang, Z.; Li, X.; Yao, Y.; Jiang, X. FOXO1 inhibits tumor cell migration via regulating cell surface morphology in non-small cell lung cancer cells. Cell. Physiol. Biochem. 2018, 48, 138–148. [Google Scholar] [CrossRef]
- Zhang, B.; Gui, L.; Zhu, L.; Zhao, X.; Yang, Y.; Li, Q. Forkhead box protein O1 mediates apoptosis in a cancer cervical cell line treated with the antitumor agent tumor necrosis factor-alpha. Genet. Mol. Res. 2015, 14, 7446–7454. [Google Scholar] [CrossRef]
- Kim, T.H.; Jo, S.W.; Lee, Y.S.; Kim, Y.J.; Lee, S.C.; Kim, W.J.; Yun, S.J. Forkhead box O-class 1 and forkhead box G1 as prognostic markers for bladder cancer. J. Korean Med. Sci. 2009, 24, 468–473. [Google Scholar] [CrossRef] [Green Version]
- Carbajo-Pescador, S.; Mauriz, J.L.; Garcia-Palomo, A.; Gonzalez-Gallego, J. FoxO proteins: Regulation and molecular targets in liver cancer. Curr. Med. Chem. 2014, 21, 1231–1246. [Google Scholar] [CrossRef] [PubMed]
- Reagan-Shaw, S.; Ahmad, N. RNA interference-mediated depletion of phosphoinositide 3-kinase activates forkhead box class O transcription factors and induces cell cycle arrest and apoptosis in breast carcinoma cells. Cancer Res. 2006, 66, 1062–1069. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferber, E.C.; Peck, B.; Delpuech, O.; Bell, G.P.; East, P.; Schulze, A. FOXO3a regulates reactive oxygen metabolism by inhibiting mitochondrial gene expression. Cell Death Differ. 2012, 19, 968–979. [Google Scholar] [CrossRef] [PubMed]
- Zorov, D.B.; Juhaszova, M.; Sollott, S.J. Mitochondrial reactive oxygen species (ROS) and ROS-induced ROS release. Physiol. Rev. 2014, 94, 909–950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Breau, M.; Houssaini, A.; Lipskaia, L.; Abid, S.; Born, E.; Marcos, E.; Czibik, G.; Attwe, A.; Beaulieu, D.; Palazzo, A.; et al. The antioxidant N-acetylcysteine protects from lung emphysema but induces lung adenocarcinoma in mice. JCI Insight 2019, 4, e127647. [Google Scholar] [CrossRef] [PubMed]
- Hagenbuchner, J.; Kuznetsov, A.; Hermann, M.; Hausott, B.; Obexer, P.; Ausserlechner, M.J. FOXO3-induced reactive oxygen species are regulated by BCL2L11 (Bim) and SESN3. J. Cell Sci. 2012, 125, 1191–1203. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vyas, S.; Zaganjor, E.; Haigis, M.C. Mitochondria and cancer. Cell 2016, 166, 555–566. [Google Scholar] [CrossRef]
- Allen, J.F. Why chloroplasts and mitochondria retain their own genomes and genetic systems: Colocation for redox regulation of gene expression. Proc. Natl. Acad. Sci. USA 2015, 112, 10231–10238. [Google Scholar] [CrossRef] [Green Version]
- Gleyzer, N.; Scarpulla, R.C. Concerted action of PGC-1-related Coactivator (PRC) and c-MYC in the stress response to mitochondrial dysfunction. J. Biol. Chem. 2016, 291, 25529–25541. [Google Scholar] [CrossRef] [Green Version]
- Gleyzer, N.; Vercauteren, K.; Scarpulla, R.C. Control of mitochondrial transcription specificity factors (TFB1M and TFB2M) by nuclear respiratory factors (NRF-1 and NRF-2) and PGC-1 family coactivators. Mol. Cell. Biol. 2005, 25, 1354–1366. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ji, H.; Wu, L.; Ma, X.; Ma, X.; Qin, G. The effect of resveratrol on the expression of AdipoR1 in kidneys of diabetic nephropathy. Mol. Biol. Rep. 2014, 41, 2151–2159. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, X.; Su, X.; Liu, T. HIF-2alpha affects proliferation and apoptosis of MG-63 osteosarcoma cells through MAPK signaling. Mol. Med. Rep. 2017, 15, 2174–2178. [Google Scholar] [CrossRef] [Green Version]
- Kloet, D.E.; Polderman, P.E.; Eijkelenboom, A.; Smits, L.M.; van Triest, M.H.; van den Berg, M.C.; Groot Koerkamp, M.J.; van Leenen, D.; Lijnzaad, P.; Holstege, F.C.; et al. FOXO target gene CTDSP2 regulates cell cycle progression through Ras and p21(Cip1/Waf1). Biochem. J. 2015, 469, 289–298. [Google Scholar] [CrossRef] [Green Version]
- Ames, K.; Da Cunha, D.S.; Gonzalez, B.; Konta, M.; Lin, F.; Shechter, G.; Starikov, L.; Wong, S.; Bulow, H.E.; Melendez, A. A Non-cell-autonomous role of BEC-1/BECN1/Beclin1 in coordinating cell-cycle progression and stem cell proliferation during germline development. Curr. Biol. 2017, 27, 905–913. [Google Scholar] [CrossRef] [Green Version]
- Akasaki, Y.; Alvarez-Garcia, O.; Saito, M.; Carames, B.; Iwamoto, Y.; Lotz, M.K. FoxO transcription factors support oxidative stress resistance in human chondrocytes. Arthritis Rheumatol. 2014, 66, 3349–3358. [Google Scholar] [CrossRef] [Green Version]
- Salminen, A.; Kaarniranta, K.; Kauppinen, A. AMPK and HIF signaling pathways regulate both longevity and cancer growth: The good news and the bad news about survival mechanisms. Biogerontology 2016, 17, 655–680. [Google Scholar] [CrossRef]
- Forstermann, U.; Xia, N.; Li, H. Roles of Vascular Oxidative Stress and Nitric Oxide in the Pathogenesis of Atherosclerosis. Circ. Res. 2017, 120, 713–735. [Google Scholar] [CrossRef]
- Tian, T.; Wang, Z.; Zhang, J. Pathomechanisms of oxidative stress in inflammatory bowel disease and potential antioxidant therapies. Oxid. Med. Cell. Longev. 2017, 2017, 4535194. [Google Scholar] [CrossRef]
- Seidlmayer, L.K.; Juettner, V.V.; Kettlewell, S.; Pavlov, E.V.; Blatter, L.A.; Dedkova, E.N. Distinct mPTP activation mechanisms in ischaemia-reperfusion: Contributions of Ca2+, ROS, pH, and inorganic polyphosphate. Cardiovasc. Res. 2015, 106, 237–248. [Google Scholar] [CrossRef] [Green Version]
- Kowara, R.; Moraleja, K.L.; Chakravarthy, B. Involvement of nitric oxide synthase and ROS-mediated activation of L-type voltage-gated Ca2+ channels in NMDA-induced DPYSL3 degradation. Brain Res. 2006, 1119, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Wang, Y.; Zhou, Y.; He, Q.Y. Jolkinolide B induces apoptosis of colorectal carcinoma through ROS-ER stress-Ca2+-mitochondria dependent pathway. Oncotarget 2017, 8, 91223–91237. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.J.; Lee, S.B.; Park, J.K.; Yoo, Y.D. TNF-alpha-induced ROS production triggering apoptosis is directly linked to Romo1 and Bcl-X(L). Cell Death Differ. 2010, 17, 1420–1434. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feng, H.; Gu, J.; Gou, F.; Huang, W.; Gao, C.; Chen, G.; Long, Y.; Zhou, X.; Yang, M.; Liu, S.; et al. High glucose and lipopolysaccharide prime NLRP3 inflammasome via ROS/TXNIP pathway in mesangial cells. J. Diabetes Res. 2016, 2016, 6973175. [Google Scholar] [CrossRef] [Green Version]
- Koziel, K.; Smigelskaite, J.; Drasche, A.; Enthammer, M.; Ashraf, M.I.; Khalid, S.; Troppmair, J. RAF and antioxidants prevent cell death induction after growth factor abrogation through regulation of Bcl-2 proteins. Exp. Cell Res. 2013, 319, 2728–2738. [Google Scholar] [CrossRef] [Green Version]
- He, L.; He, T.; Farrar, S.; Ji, L.; Liu, T.; Ma, X. Antioxidants maintain cellular redox homeostasis by elimination of reactive oxygen species. Cell. Physiol. Biochem. 2017, 44, 532–553. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, W.; Qiu, E. Protection of oxidative stress induced apoptosis in osteosarcoma cells by dihydromyricetin through down-regulation of caspase activation and up-regulation of BcL-2. Saudi J. Biol. Sci. 2017, 24, 837–842. [Google Scholar] [CrossRef]
- Coomans de Brachene, A.; Demoulin, J.B. FOXO transcription factors in cancer development and therapy. Cell. Mol. Life Sci. 2016, 73, 1159–1172. [Google Scholar] [CrossRef]
- Kim, S.; Koh, H. Role of FOXO transcription factors in crosstalk between mitochondria and the nucleus. J. Bioenergy Biomembr. 2017, 49, 335–341. [Google Scholar] [CrossRef]
- Storz, P. Forkhead homeobox type O transcription factors in the responses to oxidative stress. Antioxid. Redox Signal. 2011, 14, 593–605. [Google Scholar] [CrossRef] [Green Version]
- Hameedaldeen, A.; Liu, J.; Batres, A.; Graves, G.S.; Graves, D.T. FOXO1, TGF-beta regulation and wound healing. Int. J. Mol. Sci. 2014, 15, 16257–16269. [Google Scholar] [CrossRef] [Green Version]
- Siddiqui, W.A.; Ahad, A.; Ahsan, H. The mystery of BCL2 family: Bcl-2 proteins and apoptosis: An update. Arch. Toxicol. 2015, 89, 289–317. [Google Scholar] [CrossRef] [PubMed]
- Pihan, P.; Carreras-Sureda, A.; Hetz, C. BCL-2 family: Integrating stress responses at the ER to control cell demise. Cell Death Differ. 2017, 24, 1478–1487. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Herold, M.J.; Rohrbeck, L.; Lang, M.J.; Grumont, R.; Gerondakis, S.; Tai, L.; Bouillet, P.; Kaufmann, T.; Strasser, A. Foxo-mediated Bim transcription is dispensable for the apoptosis of hematopoietic cells that is mediated by this BH3-only protein. EMBO Rep. 2013, 14, 992–998. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Valis, K.; Prochazka, L.; Boura, E.; Chladova, J.; Obsil, T.; Rohlena, J.; Truksa, J.; Dong, L.F.; Ralph, S.J.; Neuzil, J. Hippo/Mst1 stimulates transcription of the proapoptotic mediator NOXA in a FoxO1-dependent manner. Cancer Res. 2011, 71, 946–954. [Google Scholar] [CrossRef] [PubMed] [Green Version]






© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Wang, B.; Qu, X.-L.; Zheng, B.-Q.; Huang, W.-D.; Sun, Z.-W.; Wang, C.-M.; Chen, Y. Metabolism of Reactive Oxygen Species in Osteosarcoma and Potential Treatment Applications. Cells 2020, 9, 87. https://doi.org/10.3390/cells9010087
Sun W, Wang B, Qu X-L, Zheng B-Q, Huang W-D, Sun Z-W, Wang C-M, Chen Y. Metabolism of Reactive Oxygen Species in Osteosarcoma and Potential Treatment Applications. Cells. 2020; 9(1):87. https://doi.org/10.3390/cells9010087
Chicago/Turabian StyleSun, Wei, Bing Wang, Xing-Long Qu, Bi-Qiang Zheng, Wen-Ding Huang, Zheng-Wang Sun, Chun-Meng Wang, and Yong Chen. 2020. "Metabolism of Reactive Oxygen Species in Osteosarcoma and Potential Treatment Applications" Cells 9, no. 1: 87. https://doi.org/10.3390/cells9010087



