Tuberculosis-Associated MicroRNAs: From Pathogenesis to Disease Biomarkers
Abstract
:1. Introduction
2. MicroRNAs in Tuberculosis Pathogenesis
3. Modulation of Innate and Adaptive Immunity
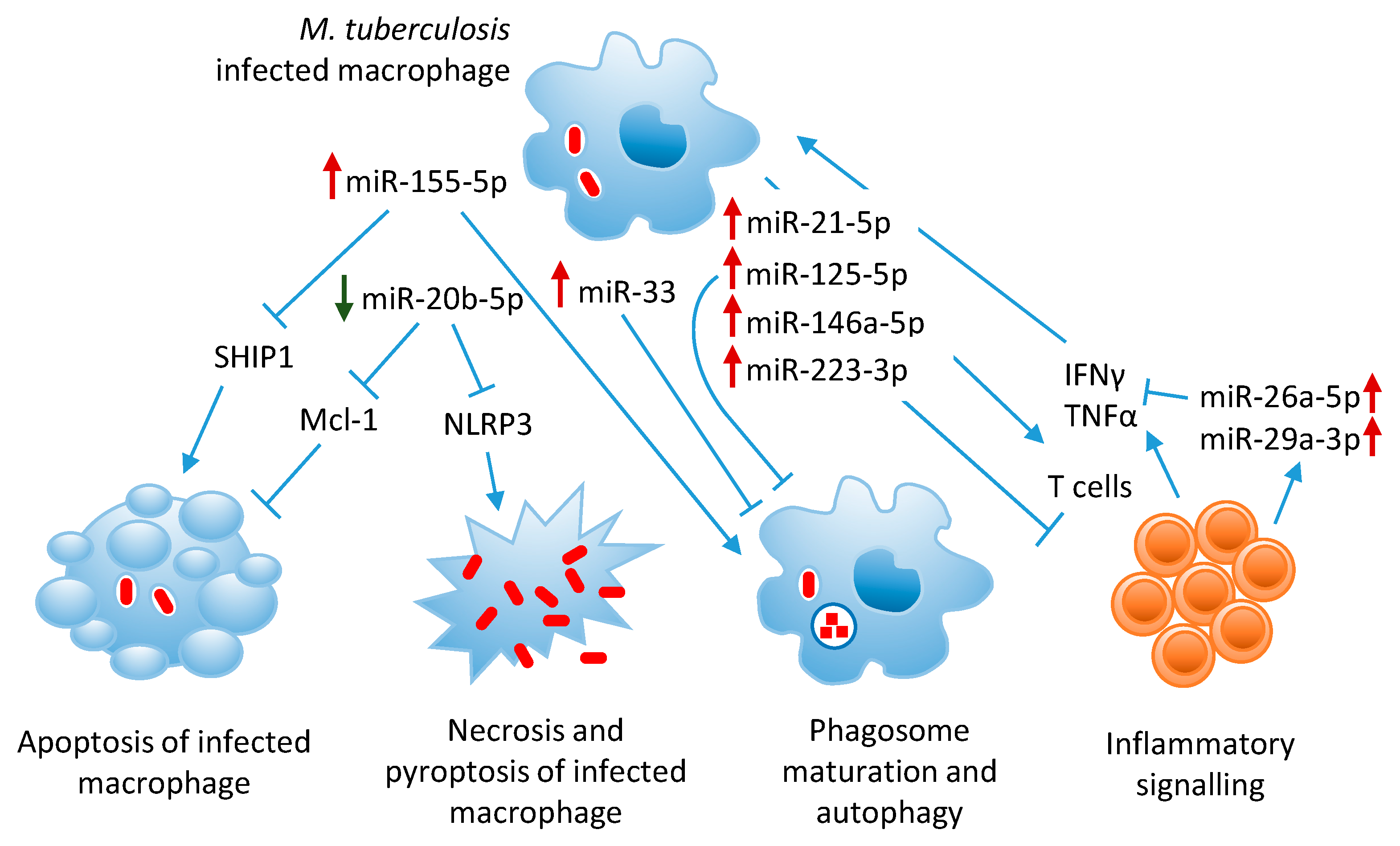
3.1. MiR-155-5p Inhibits Host Innate Immunity and Promotes M. tuberculosis Clearance
3.2. MiR-29a-3p Targets IFNγ and Is Downregulated in Experimental Mycobacterial Infection
3.3. Targeting IFNγ Signaling by MiR-26a-5p
4. Suppression of Inflammatory Signaling Pathways
4.1. MiR-21-5p, An Anti-Inflammatory MiRNA Upregulated in Mycobacterial Infections
4.2. Let-7 Family, Anti-Inflammatory MiRNAs Downregulated in TB
4.3. MiR-125-5p Suppresses TNFα Production and Autophagy Activation
4.4. MiR-146a-5p, Upregulated by Mycobacteria, Targets TRAF6 and Decreases NO Production
4.5. MiR-223-3p, Overexpressed in TB, Mitigates Excessive Inflammation
4.6. MiR-27b-3p and MiR-99b-5p Prevent Excessive Inflammation in TB
4.7. MiR-142-3p, An Anti-Inflammatory MiRNA Downregulated in TB
5. Inhibition of Phagosome Maturation and Autophagy
5.1. MiR-33 Locus MiRNAs Target Autophagy Effectors
5.2. MiR-27a-5p Downregulates Culcium Signaling and Autophagosome Formation
5.3. MiR-144-5p Inhibits Phagosome Maturation and T Cell Function
5.4. MiR-155-5p Promotes M. tuberculosis Killing Through Autophagy
5.5. MiR-889-5p, Overexpressed in LTBI, Inhibits Autophagy
6. Subversion of Macrophage Death Pathways
6.1. MiR-20b-5p, Downregulated in M. tuberculosis Infection, Inhibits Inflammasome Activation and Promotes Apoptosis
6.2. MiR-325-3p, Upregulated in M. tuberculosis Infection, Inhibits Apoptosis
6.3. MiR-155-5p Modulates Apoptosis
7. Biomarker Discovery Studies
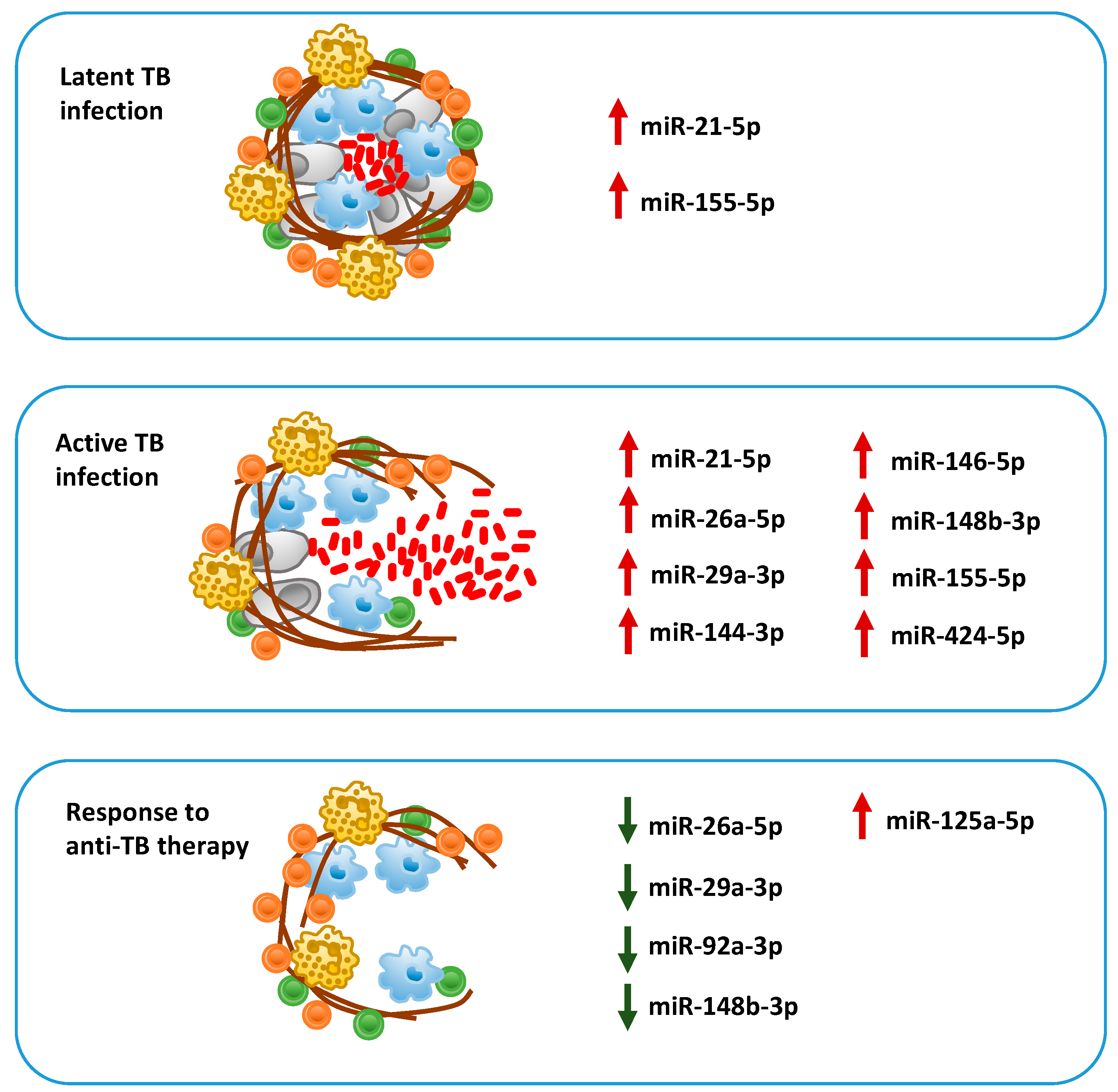
7.1. Biomarkers of Active Tuberculosis
7.2. MiRNAs in Latent vs. Active Tuberculosis
7.3. Prognostic Biomarkers of Risk of Progression to Tuberculosis and Response to Therapy
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Dye, C.; Williams, B.G. The population dynamics and control of tuberculosis. Science 2010, 328, 856–861. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- WHO. Global Tuberculosis Report 2019; Licence: CC BY-NC-SA 3.0 IGO; World Health Organization: Geneva, Switzerland, 2019. [Google Scholar]
- Pai, M.; Behr, M.A.; Dowdy, D.; Dheda, K.; Divangahi, M.; Boehme, C.C.; Ginsberg, A.; Swaminathan, S.; Spigelman, M.; Getahun, H.; et al. Tuberculosis. Nat. Rev. Dis. Primers 2016, 2, 16076. [Google Scholar] [CrossRef] [PubMed]
- Simmons, J.D.; Stein, C.M.; Seshadri, C.; Campo, M.; Alter, G.; Fortune, S.; Schurr, E.; Wallis, R.S.; Churchyard, G.; Mayanja-Kizza, H.; et al. Immunological mechanisms of human resistance to persistent Mycobacterium tuberculosis infection. Nat. Immun. 1996, 64, 683–690. [Google Scholar] [CrossRef] [PubMed]
- Smith, I. Mycobacterium tuberculosis pathogenesis and molecular determinants of virulence. Clin. Microbiol. Rev. 2003, 16, 463–496. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Natarajan, K.; Kundu, M.; Sharma, P.; Basu, J. Innate immune responses to M. tuberculosis infection. Tuberculosis 2011, 91, 427–431. [Google Scholar] [CrossRef]
- Ahmad, S. Pathogenesis, immunology, and diagnosis of latent Mycobacterium tuberculosis infection. Clin. Dev. Immunol. 2011, 814943. [Google Scholar] [CrossRef] [Green Version]
- Das, K.; Garnica, O.; Dhandayuthapani, S. Modulation of host miRNAs by intracellular bacterial pathogens. Front. Cell. Infect. Microbiol. 2016, 6, 79. [Google Scholar] [CrossRef]
- Maudet, C.; Mano, M.; Eulalio, A. MicroRNAs in the interaction between host and bacterial pathogens. FEBS Lett. 2014, 588, 4140–4147. [Google Scholar] [CrossRef] [Green Version]
- Pillai, R.S.; Bhattacharyya, S.N.; Filipowicz, W. Repression of protein synthesis by miRNAs: How many mechanisms? Trends Cell. Biol. 2007, 17, 118–126. [Google Scholar] [CrossRef]
- Hammond, S.M. An overview of microRNAs. Adv. Drug Deliv. Rev. 2015, 87, 3–14. [Google Scholar] [CrossRef] [Green Version]
- Tribolet, L.; Kerr, E.; Cowled, C.; Bean, A.G.D.; Stewart, C.R.; Dearnley, M.; Farr, R.J. MicroRNA biomarkers for infectious diseases: From basic research to biosensing. Front. Microbiol. 2020, 11, 1197. [Google Scholar] [CrossRef] [PubMed]
- Walzl, G.; McNerney, R.; du Plessis, N.; Bates, M.; McHugh, T.D.; Chegou, N.N.; Zumla, A. Tuberculosis: Advances and challenges in development of new diagnostics and biomarkers. Lancet Infect. Dis. 2018, 18, e199–e210. [Google Scholar] [CrossRef]
- Lalvani, A.; Berrocal-Almanza, L.C.; Halliday, A. Predicting progression to active tuberculosis: A rate-limiting step on the path to elimination. PLoS Med. 2019, 16, e1002814. [Google Scholar] [CrossRef] [Green Version]
- Yang, T.; Ge, B. miRNAs in immune responses to Mycobacterium tuberculosis infection. Cancer Lett. 2018, 431, 22–30. [Google Scholar] [CrossRef]
- Behrouzi, A.; Alimohammadi, M.; Nafari, A.H.; Yousefi, M.-H.; Riazi Rad, F.; Vaziri, F.; Siadat, S.D. The role of host miRNAs on Mycobacterium tuberculosis. ExRNA 2019, 1, 40. [Google Scholar] [CrossRef] [Green Version]
- Sabir, N.; Hussain, T.; Shah, S.Z.A.; Peramo, A.; Zhao, D.; Zhou, X. miRNAs in tuberculosis: New avenues for diagnosis and host-directed therapy. Front. Microbiol. 2018, 9, 602. [Google Scholar] [CrossRef] [Green Version]
- Ruiz-Tagle, C.; Naves, R.; Balcells, M.E. Unraveling the role of microRNAs in Mycobacterium tuberculosis infection and disease: Advances and pitfalls. Infect. Immun. 2020, 88, e00649-19. [Google Scholar] [CrossRef]
- Harapan, H.; Fitra, F.; Ichsan, I.; Mulyadi, M.; Miotto, P.; Hasan, N.A.; Calado, M.; Cirillo, D.M. The roles of microRNAs on tuberculosis infection: Meaning or myth? Tuberculosis 2013, 93, 596–605. [Google Scholar] [CrossRef] [Green Version]
- Philips, J.A.; Ernst, J.D. Tuberculosis pathogenesis and immunity. Annu. Rev. Pathol. 2012, 7, 353–384. [Google Scholar] [CrossRef]
- Kim, J.K.; Kim, T.S.; Basu, J.; Jo, E.K. MicroRNA in innate immunity and autophagy during mycobacterial infection. Cell. Microbiol. 2017, 19. [Google Scholar] [CrossRef] [Green Version]
- Liu, F.; Chen, J.; Wang, P.; Li, H.; Zhou, Y.; Liu, H.; Liu, Z.; Zheng, R.; Wang, L.; Yang, H.; et al. MicroRNA-27a controls the intracellular survival of Mycobacterium tuberculosis by regulating calcium-associated autophagy. Nat. Commun. 2018, 9, 4295. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ouimet, M.; Koster, S.; Sakowski, E.; Ramkhelawon, B.; van Solingen, C.; Oldebeken, S.; Karunakaran, D.; Portal-Celhay, C.; Sheedy, F.J.; Dutta Ray, T.; et al. Mycobacterium tuberculosis induces the miR-33 locus to reprogram autophagy and host lipid metabolism. Nat. Immunol. 2016, 17, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Yuk, J.M.; Kim, S.Y.; Kim, T.S.; Jin, H.S.; Yang, C.S.; Jo, E.K. MicroRNA-125a inhibits autophagy activation and antimicrobial responses during mycobacterial infection. J. Immunol. 2015, 194, 5355–5365. [Google Scholar] [CrossRef] [Green Version]
- Rajaram, M.V.; Ni, B.; Morris, J.D.; Brooks, M.N.; Carlson, T.K.; Bakthavachalu, B.; Schoenberg, D.R.; Torrelles, J.B.; Schlesinger, L.S. Mycobacterium tuberculosis lipomannan blocks TNF biosynthesis by regulating macrophage MAPK-activated protein kinase 2 (MK2) and microRNA miR-125b. Proc. Natl. Acad. Sci. USA 2011, 108, 17408–17413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, B.; Xue, W.; Zhang, H.; Zhang, R.; Feldman, K.; Zhao, Q.; Zhang, S.; Shi, L.; Pavani, K.C.; Nian, W.; et al. MicroRNA-325-3p facilitates immune escape of Mycobacterium tuberculosis through targeting LNX1 via NEK6 accumulation to promote anti-apoptotic STAT3 signaling. mBio 2020, 11, e00557-20. [Google Scholar] [CrossRef] [PubMed]
- Lou, J.; Wang, Y.; Zhang, Z.; Qiu, W. MiR-20b inhibits Mycobacterium tuberculosis induced inflammation in the lung of mice through targeting NLRP3. Exp. Cell Res. 2017, 358, 120–128. [Google Scholar] [CrossRef]
- Iwai, H.; Funatogawa, K.; Matsumura, K.; Kato-Miyazawa, M.; Kirikae, F.; Kiga, K.; Sasakawa, C.; Miyoshi-Akiyama, T.; Kirikae, T. MicroRNA-155 knockout mice are susceptible to M. tuberculosis infection. Tuberculosis 2015, 95, 246–250. [Google Scholar] [CrossRef]
- Rothchild, A.C.; Sissons, J.R.; Shafiani, S.; Plaisier, C.; Min, D.; Mai, D.; Gilchrist, M.; Peschon, J.; Larson, R.P.; Bergthaler, A.; et al. MiR-155–regulated molecular network orchestrates cell fate in the innate and adaptive immune response to Mycobacterium tuberculosis. Proc. Natl. Acad. Sci. USA 2016, 113, E6172–E6181. [Google Scholar] [CrossRef] [Green Version]
- Etna, M.P.; Sinigaglia, A.; Grassi, A.; Giacomini, E.; Romagnoli, A.; Pardini, M.; Severa, M.; Cruciani, M.; Rizzo, F.; Anastasiadou, E.; et al. Mycobacterium tuberculosis-induced miR-155 subverts autophagy by targeting ATG3 in human dendritic cells. PLoS Pathog. 2018, 14, e1006790. [Google Scholar] [CrossRef]
- Orme, I.M.; Robinson, R.T.; Cooper, A.M. The balance between protective and pathogenic immune responses in the TB-infected lung. Nat. Immunol. 2015, 16, 57–63. [Google Scholar] [CrossRef]
- Ernst, A.D. The immunological life cycle of tuberculosis. Nat. Rev. Immunol. 2012, 12, 581–591. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Yang, K.; Zhou, L.; Wu, M.; Wu, Y.; Zhu, M.; Lai, X.; Chen, T.; Feng, L.; Li, M.; et al. MicroRNA-155 promotes autophagy to eliminate intracellular mycobacteria by targeting Rheb. PLoS Pathog. 2013, 9, e1003697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kumar, R.; Halder, P.; Sahu, S.K.; Kumar, M.; Kumari, M.; Jana, K.; Ghosh, Z.; Sharma, P.; Kundu, M.; Basu, J. Identification of a novel role of ESAT-6-dependent miR-155 induction during infection of macrophages with Mycobacterium tuberculosis. Cell Microbiol. 2012, 14, 1620–1631. [Google Scholar] [CrossRef] [PubMed]
- Ghorpade, D.S.; Leyland, R.; Kurowska-Stolarska, M.; Patil, S.A.; Balaji, K.N. MicroRNA-155 is required for Mycobacterium bovis BCG-mediated apoptosis of macrophages. Mol. Cell. Biol. 2012, 32, 2239–2253. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wu, M.; Wen, J.; Yang, K.; Li, M.; Zhan, X.; Feng, L.; Li, M.; Huang, X. MicroRNA-155 induction by Mycobacterium bovis BCG enhances ROS production through targeting SHIP1. Mol. Immunol. 2014, 62, 29–36. [Google Scholar] [CrossRef] [PubMed]
- Ernst, J.D. Mechanisms of M. tuberculosis immune evasion as challenges to TB vaccine design. Cell Host Microbe 2018, 24, 34–42. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, F.; Xu, S.; Liu, X.; Zhang, Q.; Xu, X.; Liu, M.; Hua, M.; Li, N.; Yao, H.; Cao, X. The microRNA miR-29 controls innate and adaptive immune responses to intracellular bacterial infection by targeting interferon-γ. Nat. Immunol. 2011, 12, 861–869. [Google Scholar] [CrossRef]
- Kleinsteuber, K.; Heesch, K.; Schattling, S.; Kohns, M.; Sander-Jülch, C.; Walzl, G.; Hesseling, A.; Mayatepek, E.; Fleischer, B.; Marx, F.M.; et al. Decreased expression of miR-21, miR-26a, miR-29a, and miR-142-3p in CD4+ T cells and peripheral blood from tuberculosis patients. PLoS ONE 2013, 8, e61609. [Google Scholar] [CrossRef] [Green Version]
- Afum-Adjei Awuah, A.; Ueberberg, B.; Owusu-Dabo, E.; Frempong, M.; Jacobsen, M. Dynamics of T-cell IFN-γ and miR-29a expression during active pulmonary tuberculosis. Int. Immunol. 2014, 26, 579–582. [Google Scholar] [CrossRef]
- Ni, B.; Rajaram, M.V.; Lafuse, W.P.; Landes, M.B.; Schlesinger, L.S. Mycobacterium tuberculosis decreases human macrophage IFN-γ responsiveness through miR-132 and miR-26a. J. Immunol. 2014, 193, 4537–4547. [Google Scholar] [CrossRef] [Green Version]
- Sahu, S.K.; Kumar, M.; Chakraborty, S.; Banerjee, S.K.; Kumar, R.; Gupta, P.; Jana, K.; Gupta, U.D.; Ghosh, Z.; Kundu, M.; et al. MicroRNA26a (miR-26a)/KLF4 and CREB-C/EBPβ-regulate innate immune signaling, the polarization of macrophages and the trafficking of Mycobacterium tuberculosis to lysosomes during infection. PLoS Pathog. 2017, 13, e1006410. [Google Scholar] [CrossRef] [PubMed]
- Cardona, P.; Cardona, P.J. Regulatory T cells in Mycobacterium tuberculosis infection. Front. Immunol. 2019, 10, 2139. [Google Scholar] [CrossRef] [PubMed]
- Yamamura, M.; Uyemura, K.; Deans, R.J.; Weinberg, T.; Rea, H.; Bloom, B.R.; Modlin, R.L. Defining protective responses to pathogens: Cytokine profiles in leprosy lesions. Science 1991, 254, 277–279. [Google Scholar] [CrossRef] [PubMed]
- Montoya, D.; Cruz, D.; Teles, R.M.; Lee, D.J.; Ochoa, M.T.; Krutzik, S.R.; Chun, R.; Schenk, M.; Zhang, X.; Ferguson, B.G.; et al. Divergence of macrophage phagocytic and antimicrobial programs in leprosy. Cell Host Microbe 2009, 6, 343–353. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, P.T.; Wheelwright, M.; Teles, R.; Komisopoulou, E.; Edfeldt, K.; Ferguson, B.; Mehta, M.D.; Vazirnia, A.; Rea, T.H.; Sarno, E.N.; et al. MicroRNA-21 targets the vitamin D–dependent antimicrobial pathway in leprosy. Nat. Med. 2012, 18, 267–273. [Google Scholar] [CrossRef]
- Wu, Z.; Lu, H.; Sheng, J.; Li, L. Inductive microRNA-21 impairs anti-mycobacterial responses by targeting IL-12 and Bcl-2. FEBS Lett. 2012, 586, 2459–2467. [Google Scholar] [CrossRef]
- Hackett, E.E.; Charles-Messance, H.; O’Leary, S.M.; Gleeson, L.E.; Muñoz-Wolf, N.; Case, S.; Wedderburn, A.; Johnston, D.G.W.; Williams, M.A.; Smyth, A.; et al. Mycobacterium tuberculosis limits host glycolysis and IL-1β by restriction of PFK-M via microRNA-21. Cell Rep. 2020, 30, 124–136.e4. [Google Scholar] [CrossRef]
- Kumar, M.; Kumar Sahu, S.; Kumar, R.; Subuddhi, A.; Kumar Maji, R.; Jana, K.; Gupta, P.; Raffetseder, J.; Lerm, M.; Ghosh, Z.; et al. MicroRNA let-7 modulates the immune response to Mycobacterium tuberculosis infection via control of A20, an inhibitor of the NF-kB pathway. Cell Host Microbe 2015, 17, 345–356. [Google Scholar] [CrossRef] [Green Version]
- Vereecke, L.; Beyaert, R.; van Loo, G. The ubiquitin-editing enzyme A20 (TNFAIP3) is a central regulator of immunopathology. Trends Immunol. 2009, 30, 383–391. [Google Scholar] [CrossRef]
- Yang, S.; Li, F.; Jia, S.; Zhang, K.; Jiang, W.; Shang, Y.; Chang, K.; Deng, S.; Chen, M. Early secreted antigen ESAT-6 of Mycobacterium tuberculosis promotes apoptosis of macrophages via targeting the microRNA155–SOCS1 interaction. Cell. Physiol. Biochem. 2015, 35, 1276–1288. [Google Scholar] [CrossRef]
- Li, M.; Wang, J.; Fang, Y.; Gong, S.; Li, M.; Wu, M.; Lai, X.; Zeng, G.; Wang, Y.; Yang, K.; et al. microRNA-146a promotes mycobacterial survival in macrophages through suppressing nitric oxide production. Sci. Rep. 2016, 6, 23351. [Google Scholar] [CrossRef] [Green Version]
- Johnnidis, J.B.; Harris, M.H.; Wheeler, R.T.; Stehling-Sun, S.; Lam, M.H.; Kirak, O.; Brummelkamp, T.R.; Fleming, M.D.; Camargo, F.D. Regulation of progenitor cell proliferation and granulocyte function by microRNA-223. Nature 2008, 451, 1125–1129. [Google Scholar] [CrossRef] [PubMed]
- Dorhoi, A.; Iannaccone, M.; Farinacci, M.; Faé, K.C.; Schreiber, J.; Moura-Alves, P.; Nouailles, G.; Mollenkopf, H.J.; Oberbeck-Müller, D.; Jörg, S.; et al. MicroRNA-223 controls susceptibility to tuberculosis by regulating lung neutrophil recruitment. J. Clin. Investig. 2013, 123, 4836–4848. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Morgan, M.J.; Choksi, S.; Zhang, Y.; Kim, Y.S.; Liu, Z.G. MicroRNAs modulate the noncanonical transcription factor NF-kappaB pathway by regulating expression of the kinase IKKalpha during macrophage differentiation. Nat. Immunol. 2010, 11, 799–805. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wang, R.; Jiang, J.; Yang, B.; Cao, Z.; Cheng, X. MiR-223 is upregulated in monocytes from patients with tuberculosis and regulates function of monocyte-derived macrophages. Mol. Immunol. 2015, 67, 475–481. [Google Scholar] [CrossRef] [PubMed]
- Wu, Y.; Guo, Z.; Yao, K.; Miao, Y.; Liang, S.; Liu, F.; Wang, Y.; Zhang, Y. The transcriptional foundations of Sp110-mediated macrophage (RAW264.7) resistance to Mycobacterium tuberculosis H37Ra. Sci. Rep. 2016, 6, 22041. [Google Scholar] [CrossRef]
- Liang, S.; Song, Z.; Wu, Y.; Gao, Y.; Gao, M.; Liu, F.; Wang, F.; Zhang, Y. MicroRNA-27b modulates inflammatory response and apoptosis during Mycobacterium tuberculosis infection. J. Immunol. 2018, 200, 3506–3518. [Google Scholar] [CrossRef] [Green Version]
- Singh, Y.; Kaul, V.; Mehra, A.; Chatterjee, S.; Tousif, S.; Dwivedi, V.P.; Suar, M.; Van Kaer, L.; Bishai, W.R.; Das, G. Mycobacterium tuberculosis controls microRNA-99b (miR-99b) expression in infected murine dendritic cells to modulate host immunity. J. Biol. Chem. 2013, 288, 5056–5061. [Google Scholar] [CrossRef] [Green Version]
- Xu, G.; Zhang, Z.; Wei, J.; Zhang, Y.; Zhang, Y.; Guo, L.; Liu, X. MicroR-142-3p down-regulates IRAK-1 in response to Mycobacterium bovis BCG infection in macrophages. Tuberculosis 2013, 93, 606–611. [Google Scholar] [CrossRef]
- Zhai, W.; Wu, F.; Zhang, Y.; Fu, Y.; Liu, Z. The immune escape mechanisms of Mycobacterium tuberculosis. Int. J. Mol. Sci. 2019, 20, 340. [Google Scholar] [CrossRef] [Green Version]
- Najafi-Shoushtari, S.H.; Kristo, F.; Li, Y.; Shioda, T.; Cohen, D.E.; Gerszten, R.E.; Näär, A.M. MicroRNA-33 and the SREBP host genes cooperate to control cholesterol homeostasis. Science 2010, 328, 1566–1569. [Google Scholar] [CrossRef] [Green Version]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 contributes to the regulation of cholesterol homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lv, Y.; Guo, S.; Li, X.G.; Chi, J.Y.; Qu, Y.Q.; Zhong, H.L. Sputum and serum microRNA-144 levels in patients with tuberculosis before and after treatment. Int. J. Infect. Dis. 2016, 43, 68–73. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, Y.; Wang, X.; Jiang, J.; Cao, Z.; Yang, B.; Cheng, X. Modulation of T cell cytokine production by miR-144* with elevated expression in patients with pulmonary tuberculosis. Mol. Immunol. 2011, 48, 1084–1090. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.K.; Lee, H.M.; Park, K.S.; Shin, D.M.; Kim, T.S.; Kim, Y.S.; Suh, H.W.; Kim, S.Y.; Kim, I.S.; Kim, J.M.; et al. MIR144* inhibits antimicrobial responses against Mycobacterium tuberculosis in human monocytes and macrophages by targeting the autophagy protein DRAM2. Autophagy 2017, 13, 423–441. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.-Y.; Chen, Y.-M.; Lin, C.-F.; Lo, C.-M.; Liu, H.-J.; Liao, T.-L. MicroRNA-889 inhibits autophagy to maintain mycobacterial survival in patients with latent tuberculosis infection by targeting TWEAK. mBio 2020, 11, e03045-19. [Google Scholar] [CrossRef] [Green Version]
- Behar, S.M.; Divangahi, M.; Remold, H.G. Evasion of innate immunity by M. tuberculosis: Is death an exit strategy? Nat. Rev. Microbiol. 2010, 8, 668–674. [Google Scholar] [CrossRef] [Green Version]
- Beckwith, K.S.; Beckwith, M.S.; Ullmann, S.; Sætra, R.S.; Kim, H.; Marstad, A.; Åsberg, S.E.; Strand, T.A.; Haug, M.; Niederweis, M.; et al. Plasma membrane damage causes NLRP3 activation and pyroptosis during Mycobacterium tuberculosis infection. Nat. Commun. 2020, 11, 2270. [Google Scholar] [CrossRef]
- Zhang, D.; Yi, Z.; Fu, Y. Downregulation of miR-20b-5p facilitates Mycobacterium tuberculosis survival in RAW 264.7 macrophages via attenuating the cell apoptosis by Mcl-1 upregulation. J. Cell. Biochem. 2019, 120, 5889–5896. [Google Scholar] [CrossRef]
- Huang, J.; Jiao, J.; Xu, W.; Zhao, H.; Zhang, C.; Shi, Y.; Xiao, Z. MiR-155 is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO3. Mol. Med. Rep. 2015, 12, 7102–7108. [Google Scholar] [CrossRef] [Green Version]
- Fiore-Gartland, A.; Carpp, L.N.; Naidoo, K.; Thompson, E.; Zak, D.E.; Self, S.; Churchyard, G.; Walzl, G.; Penn-Nicholson, A.; Scriba, T.J.; et al. Considerations for biomarker-targeted intervention strategies for tuberculosis disease prevention. Tuberculosis 2018, 109, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Levin, M.; Kaforou, M. Predicting active tuberculosis progression by RNA analysis. Lancet 2016, 387, 2268–2270. [Google Scholar] [CrossRef]
- Sigal, G.B.; Segal, M.R.; Mathew, A.; Jarlsberg, L.; Wang, M.; Barbero, S.; Small, N.; Haynesworth, K.; Davis, J.L.; Weiner, M.; et al. Biomarkers of tuberculosis severity and treatment effect: A directed screen of 70 host markers in a randomized clinical trial. EBioMedicine 2017, 25, 112–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anderson, S.T.; Kaforou, M.; Brent, A.J.; Wright, V.J.; Banwell, C.M.; Chagaluka, G.; Crampin, A.C.; Dockrell, H.M.; French, N.; Hamilton, M.S.; et al. Diagnosis of childhood tuberculosis and host RNA expression in Africa. N. Engl. J. Med. 2014, 370, 1712–1723. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leong, S.; Zhao, Y.; Joseph, N.M.; Hochberg, N.S.; Sarkar, S.; Pleskunas, J.; Hom, D.; Lakshminarayanan, S.; Horsburgh, C.R., Jr.; Roy, G.; et al. Existing blood transcriptional classifiers accurately discriminate active tuberculosis from latent infection in individuals from south India. Tuberculosis 2018, 109, 41–51. [Google Scholar] [CrossRef] [PubMed]
- Maertzdorf, J.; McEwen, G.; Weiner, J., 3rd; Tian, S.; Lader, E.; Schriek, U.; Mayanja-Kizza, H.; Ota, M.; Kenneth, J.; Kaufmann, S.H. Concise gene signature for point-of-care classification of tuberculosis. EMBO Mol. Med. 2016, 8, 86–95. [Google Scholar] [CrossRef]
- Sambarey, A.; Devaprasad, A.; Mohan, A.; Ahmed, A.; Nayak, S.; Swaminathan, S.; D’Souza, G.; Jesuraj, A.; Dhar, C.; Babu, S.; et al. Unbiased identification of blood-based biomarkers for pulmonary tuberculosis by modeling and mining molecular interaction networks. EBioMedicine 2017, 15, 112–126. [Google Scholar] [CrossRef] [Green Version]
- Suliman, S.; Thompson, E.; Sutherland, J.; Weiner, J., 3rd; Ota, M.O.C.; Shankar, S.; Penn-Nicholson, A.; Thiel, B.; Erasmus, M.; Maertzdorf, J.; et al. Four-Gene Pan-African Blood Signature Predicts Progression to Tuberculosis. Am. J. Respir. Crit. Care Med. 2018. [Google Scholar] [CrossRef]
- Sweeney, T.E.; Braviak, L.; Tato, C.M.; Khatri, P. Genome-wide expression for diagnosis of pulmonary tuberculosis: A multicohort analysis. Lancet Respir. Med. 2016, 4, 213–224. [Google Scholar] [CrossRef] [Green Version]
- Zak, D.E.; Penn-Nicholson, A.; Scriba, T.J.; Thompson, E.; Suliman, S.; Amon, L.M.; Mahomed, H.; Erasmus, M.; Whatney, W.; Hussey, G.D.; et al. A blood RNA signature for tuberculosis disease risk: A prospective cohort study. Lancet 2016, 387, 2312–2322. [Google Scholar] [CrossRef] [Green Version]
- Thompson, E.G.; Du, Y.; Malherbe, S.T.; Shankar, S.; Braun, J.; Valvo, J.; Ronacher, K.; Tromp, G.; Tabb, D.L.; Alland, D.; et al. Host blood RNA signatures predict the outcome of tuberculosis treatment. Tuberculosis 2017, 107, 48–58. [Google Scholar] [CrossRef]
- Pedersen, J.L.; Bokil, N.J.; Saunders, B.M. Developing new TB biomarkers, are miRNA the answer? Tuberculosis 2019, 118, 101860. [Google Scholar] [CrossRef] [PubMed]
- de Araujo, L.S.; Ribeiro-Alves, M.; Leal-Calvo, T.; Leung, J.; Durán, V.; Samir, M.; Talbot, S.; Tallam, A.; Mello, F.C.Q.; Geffers, R.; et al. Reprogramming of small noncoding RNA populations in peripheral blood reveals host biomarkers for latent and active Mycobacterium tuberculosis infection. mBio 2019, 10, e01037-19. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lyu, L.; Zhang, X.; Li, C.; Yang, T.; Wang, J.; Pan, L.; Jia, H.; Li, Z.; Sun, Q.; Yue, L.; et al. Small RNA profiles of serum exosomes derived from individuals with latent and active tuberculosis. Front. Microbiol. 2019, 10, 1174. [Google Scholar] [CrossRef] [PubMed]
- Ndzi, E.N.; Nkenfou, C.N.; Mekue, L.M.; Zentilin, L.; Tamgue, O.; Pefura, E.W.Y.; Kuiaté, J.R.; Giacca, M.; Ndjolo, A. MicroRNA hsa-miR-29a-3p is a plasma biomarker for the differential diagnosis and monitoring of tuberculosis. Tuberculosis 2019, 114, 69–76. [Google Scholar] [CrossRef]
- Chakrabarty, S.; Kumar, A.; Raviprasad, K.; Mallya, S.; Satyamoorthy, K.; Chawla, K. Host and MTB genome encoded miRNA markers for diagnosis of tuberculosis. Tuberculosis 2019, 116, 37–43. [Google Scholar] [CrossRef]
- Hu, X.; Liao, S.; Bai, H.; Wu, L.; Wang, M.; Wu, Q.; Zhou, J.; Jiao, L.; Chen, X.; Zhou, Y.; et al. Integrating exosomal microRNAs and electronic health data improved tuberculosis diagnosis. EBioMedicine 2019, 40, 564–573. [Google Scholar] [CrossRef] [Green Version]
- Alipoor, S.D.; Tabarsi, P.; Varahram, M.; Movassaghi, M.; Dizaji, M.K.; Folkerts, G.; Garssen, J.; Adcock, I.M.; Mortaz, E. Serum exosomal miRNAs are associated with active pulmonary tuberculosis. Dis. Markers 2019, 2019, 1907426. [Google Scholar] [CrossRef] [Green Version]
- Barry, S.E.; Ellis, M.; Yang, Y.; Guan, G.; Wang, X.; Britton, W.J.; Saunders, B.M. Identification of a plasma microRNA profile in untreated pulmonary tuberculosis patients that is modulated by anti-mycobacterial therapy. J. Infect. 2018, 77, 341–348. [Google Scholar] [CrossRef] [Green Version]
- Duffy, F.J.; Thompson, E.; Downing, K.; Suliman, S.; Mayanja-Kizza, H.; Boom, W.H.; Thiel, B.; Weiner, J., III; Kaufmann, S.H.E.; Dover, D.; et al. A serum circulating miRNA signature for short-term risk of progression to active tuberculosis among household contacts. Front. Immunol. 2018, 9, 661. [Google Scholar] [CrossRef] [Green Version]
- Wang, C.; Yang, S.; Liu, C.M.; Jiang, T.T.; Chen, Z.L.; Tu, H.H.; Mao, L.G.; Li, Z.J.; Li, J.C. Screening and identification of four serum miRNAs as novel potential biomarkers for cured pulmonary tuberculosis. Tuberculosis 2018, 108, 26–34. [Google Scholar] [CrossRef] [PubMed]
- Cui, J.Y.; Liang, H.W.; Pan, X.L.; Li, D.; Jiao, N.; Liu, Y.H.; Fu, J.; He, X.Y.; Sun, G.X.; Zhang, C.L.; et al. Characterization of a novel panel of plasma microRNAs that discriminates between Mycobacterium tuberculosis infection and healthy individuals. PLoS ONE 2017, 12, e0184113. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Liu, C.M.; Wei, L.L.; Shi, L.Y.; Pan, Z.F.; Mao, L.G.; Wan, X.C.; Ping, Z.P.; Jiang, T.T.; Chen, Z.L.; et al. A Group of novel serum diagnostic biomarkers for multidrug-resistant tuberculosis by iTRAQ-2D LC-MS/MS and Solexa sequencing. Int. J. Biol. Sci. 2016, 12, 246–256. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagh, V.; Urhekar, A.; Modi, D. Levels of microRNA miR-16 and miR-155 are altered in serum of patients with tuberculosis and associate with responses to therapy. Tuberculosis 2017, 102, 24–30. [Google Scholar] [CrossRef] [PubMed]
- Zheng, M.L.; Zhou, N.K.; Luo, C.H. MiRNA-155 and miRNA-132 as potential diagnostic biomarkers for pulmonary tuberculosis: A preliminary study. Microb. Pathog. 2016, 100, 78–83. [Google Scholar] [CrossRef]
- Xin, H.; Yang, Y.; Liu, J.; Li, X.; Li, M.; Feng, B.; Li, Z.; Zhang, H.; Li, H.; Shen, F.; et al. Association between tuberculosis and circulating microRNA hsa-let-7b and hsa-miR-30b: A pilot study in a Chinese population. Tuberculosis 2016, 99, 63–69. [Google Scholar] [CrossRef]
- Xu, Z.; Zhou, A.; Ni, J.; Zhang, Q.; Wang, Y.; Lu, J.; Wu, W.; Karakousis, P.C.; Lu, S.; Yao, Y. Differential expression of miRNAs and their relation to active tuberculosis. Tuberculosis 2015, 95, 395–403. [Google Scholar] [CrossRef]
- Honeyborne, I.; Lipman, M.C.; Eckold, C.; Evangelopoulos, D.; Gillespie, S.H.; Pym, A.; McHugh, T.D. Effective anti-tuberculosis therapy correlates with plasma small RNA. Eur. Respir. J. 2015, 45, 1741–1744. [Google Scholar] [CrossRef]
- Latorre, I.; Leidinger, P.; Backes, C.; Domínguez, J.; de Souza-Galvão, M.L.; Maldonado, J.; Prat, C.; Ruiz-Manzano, J.; Sánchez, F.; Casas, I.; et al. A novel whole-blood miRNA signature for a rapid diagnosis of pulmonary tuberculosis. Eur. Respir. J. 2015, 45, 1173–1176. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Wang, Q.; Xi, X.; Jiao, J.; Xu, W.; Huang, J.; Lai, Z. High serum miR-183 level is associated with the bioactivity of macrophage derived from tuberculosis patients. Int. J. Clin. Exp. Pathol. 2015, 8, 655–659. [Google Scholar]
- Zhang, H.; Sun, Z.; Wei, W.; Liu, Z.; Fleming, J.; Zhang, S.; Lin, N.; Wang, M.; Chen, M.; Xu, Y.; et al. Identification of Serum microRNA Biomarkers for Tuberculosis Using RNA-seq. PLoS ONE 2014, 9, e88909. [Google Scholar] [CrossRef] [Green Version]
- Abd-El-Fattah, A.A.; Sadik, N.A.; Shaker, O.G.; Aboulftouh, M.L. Differential microRNAs expression in serum of patients with lung cancer, pulmonary tuberculosis, and pneumonia. Cell Biochem. Biophys. 2013, 67, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Guo, J.; Fan, S.; Li, Y.; Wei, L.; Yang, X.; Jiang, T.; Chen, Z.; Wang, C.; Liu, J.; et al. Screening and identification of six serum microRNAs as novel potential combination biomarkers for pulmonary tuberculosis diagnosis. PLoS ONE 2013, 8, e81076. [Google Scholar] [CrossRef] [Green Version]
- Miotto, P.; Mwangoka, G.; Valente, I.C.; Norbis, L.; Sotgiu, G.; Bosu, R.; Ambrosi, A.; Codecasa, L.R.; Goletti, D.; Matteelli, A.; et al. MiRNA signatures in sera of patients with active pulmonary tuberculosis. PLoS ONE 2013, 8, e80149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maertzdorf, J.; Weiner, J.; Mollenkopf, H.J.; TBornotTB Network; Bauer, T.; Prasse, A.; Müller-Quernheim, J.; Kaufmann, S.H. Common patterns and disease-related signatures in tuberculosis and sarcoidosis. Proc. Natl. Acad. Sci. USA 2012, 109, 7853–7858. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Cui, L.; Ge, Y.; Shi, Z.; Zhao, K.; Guo, X.; Yang, D.; Yu, H.; Cui, L.; Shan, Y.; et al. Altered serum microRNAs as biomarkers for the early diagnosis of pulmonary tuberculosis infection. BMC Infect. Dis. 2012, 12, 384. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Yi, Z.; Wu, X.; Li, J.; Xu, F. Circulating microRNAs in patients with active pulmonary tuberculosis. J. Clin. Microbiol. 2011, 49, 4246–4251. [Google Scholar] [CrossRef] [Green Version]
- Lin, Y.; Zhang, Y.; Yu, H.; Tian, R.; Wang, G.; Li, F. Identification of unique key genes and miRNAs in latent tuberculosis infection by network analysis. Mol. Immunol. 2019, 112, 103–114. [Google Scholar] [CrossRef]
- Zhang, X.; Zhu, M.; Hu, X. Integrated miRNA and mRNA expression profiling to identify mRNA targets of dysregulated miRNAs in pulmonary tuberculosis. Epigenomics 2018, 10, 1051–1069. [Google Scholar] [CrossRef] [Green Version]
- van Rensburg, I.C.; du Toit, L.; Walzl, G.; du Plessis, N.; Loxton, A.G. Decreased neutrophil–associated miRNA and increased B-cell associated miRNA expression during tuberculosis. Gene 2018, 655, 35–41. [Google Scholar] [CrossRef]
- Corral-Fernández, N.E.; Cortes-García, J.D.; Bruno, R.S.; Romano-Moreno, S.; Medellín-Garibay, S.E.; Magaña-Aquino, M.; Salazar-González, R.A.; González-Amaro, R.; Portales-Pérez, D.P. Analysis of transcription factors, microRNAs and cytokines involved in T lymphocyte differentiation in patients with tuberculosis after directly observed treatment short-course. Tuberculosis 2017, 105, 1–8. [Google Scholar] [CrossRef]
- Spinelli, S.V.; Fernández, R.D.V.; Zoff, L.; Bongiovanni, B.; Díaz, A.; D’Attilio, L.; Santucci, N.; Alvarez, T.; Marchesini, M.M.; Bogue, C.; et al. MiR-30c is specifically repressed in patients with active pulmonary tuberculosis. Tuberculosis 2017, 105, 73–79. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Pan, M.; Xu, Y.M. Mir-29a expressions in peripheral blood mononuclear cell and cerebrospinal fluid: Diagnostic value in patients with pediatric tuberculous meningitis. Brain Res. Bull. 2017, 130, 231–235. [Google Scholar] [CrossRef] [PubMed]
- Zhou, M.; Yu, G.; Yang, X.; Zhu, C.; Zhang, Z.; Zhan, X. Circulating microRNAs as biomarkers for the early diagnosis of childhood tuberculosis infection. Mol. Med. Rep. 2016, 13, 4620–4626. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zheng, L.; Leung, E.; Lee, N.; Lui, G.; To, K.F.; Chan, R.C.; Ip, M. Differential microRNA expression in human macrophages with Mycobacterium tuberculosis infection of Beijing/W and Non-Beijing/W strain types. PLoS ONE 2015, 10, e0126018. [Google Scholar] [CrossRef]
- Wang, J.X.; Xu, J.; Han, Y.F.; Zhu, Y.B.; Zhang, W.J. Diagnostic values of microRNA-31 in peripheral blood mononuclear cells for pediatric pulmonary tuberculosis in Chinese patients. Genet. Mol. Res. 2015, 14, 17235–17243. [Google Scholar] [CrossRef]
- Fu, Y.; Yi, Z.; Li, J.; Li, R. Deregulated microRNAs in CD4+ T cells from individuals with latent tuberculosis versus active tuberculosis. J. Cell. Mol. Med. 2014, 18, 503–513. [Google Scholar] [CrossRef]
- Spinelli, S.V.; Diaz, A.; D’Attilio, L.; Marchesini, M.M.; Bogue, C.; Bay, M.L.; Bottasso, O.A. Altered microRNA expression levels in mononuclear cells of patients with pulmonary and pleural tuberculosis and their relation with components of the immune response. Mol. Immunol. 2013, 53, 265–269. [Google Scholar] [CrossRef]
- Wang, C.; Yang, S.; Sun, G.; Tang, X.; Lu, S.; Neyrolles, O.; Gao, Q. Comparative miRNA expression profiles in individuals with latent and active tuberculosis. PLoS ONE 2011, 6, e25832. [Google Scholar] [CrossRef]

| Function | Upregulated in TB | Dowregulated in TB |
|---|---|---|
| Inhibition of innate immunity | miR-26-5p, miR-132-3p, miR-155-5p | miR-29-3p |
| Suppression of inflammation | miR-21-5p, miR-27b-3p, miR-99b-5p, miR-125-5p, miR-146a-5p, miR-223-3p | let-7f, miR-20b-5p miR-142-3p |
| Inhibition of phagosome maturation and autophagy | miR-33 locus, miR-27a-5p, miR-144-5p, miR-889-5p | |
| Apoptosis inhibition | miR-155-5p, miR-325-3p |
| Cases No. (Category) | Controls No. (Category) | Country | Samples | Method for Screening | Method for Validation | Up-Regulated in TB | Down-Regulated in TB | Ref. | Year |
|---|---|---|---|---|---|---|---|---|---|
| 8 (TB), 21 (LTBI) | 6 treated TB, 14 HC | Brazil | blood | RNA-Seq (Illumina) | qRT-PCR | miR-589-5p | miR-196b-5p, let-7a-5p | [84] | 2020 |
| 60 (TB), 60 (LTBI) | 60 HC | China | exosomes from serum | RNA-Seq (Illumina) | qRT-PCR | miR-1246, miR-2110, miR-370-3p, miR-28-3p, miR-193b-5p | miR-3675-5p | [85] | 2019 |
| 84 (TB), 35 (LTBI) | 42 HC | Cameroon | plasma | literature (miRNA selection) | qRT-PCR | miR-29a-3p, miR-361-5p (vs LTBI); miR-155-5p (vs HC) | [86] | 2019 | |
| 15 (TB), 22 (extra-pulmonary TB) | 15 HC | India | serum | RNA-Seq (Ion Torrent) | qRT-PCR | miR-146a-5p (TB), miR-125b-5p (EPTB) | [87] | 2019 | |
| 246 (TB) | 105 HC | China | exosomes from plasma | Microarray (Affymetrix) | qRT-PCR | miR-20a-5p, miR-20b-5p, miR-26a-5p, miR-106a-5p, miR-191-5p, miR-486-5p | [88] | 2019 | |
| 25 (TB) | 25 HC | Iran | exosomes from serum | literature (miRNA selection) | qRT-PCR | miR-484, miR-425-5p, miR-96-3p | [89] | 2019 | |
| 100 (TB) | 89 treated TB, 100 HC | China | plasma | miRNA PCR panel (Exiqon) | qRT-PCR | miR-29a-3p, miR-99b-5p (vs HC), miR-29a-3p, miR-99b-5p, miR-26a-5p (vs treated) | miR-21-5p, miR-146a-5p, miR-652-5p | [90] | 2018 |
| 54 (TB) | 54 HC | South Africa, Uganda | serum | qRT-PCR | qRT-PCR | miR-21-5p, miR-484 | miR-148b-3p | [91] | 2018 |
| 53 (TB) | 53 treated TB, 53 HC | China | serum | RNA-Seq (Illumina) | qRT-PCR | miR-21-5p, miR-92a-3p, miR-148b-3p (vs treated) | miR-125a-5p (vs treated) | [92] | 2017 |
| 178 (TB) | 95 HC | China | plasma | RNA-Seq (Illumina) | qRT-PCR | miR-22-3p, miR-320a-5p, miR-769-5p | [93] | 2017 | |
| 60 (TB), 32 (MDR-TB) | 60 HC | China | serum | RNA-Seq (Illumina) | qRT-PCR | miR-424-5p, miR-4433b-5p (MDR vs. DS); miR-199b-5p, miR-424-5p (vs HC) | [94] | 2016 | |
| 124 (TB) | 117 HC | China | serum and sputum | literature (miRNA selection) | qRT-PCR | miR-144-3p | [64] | 2016 | |
| 30 (TB), 19 (MDR-TB) | 10 treated TB, 30 HC | India | serum | literature (miRNA selection) | qRT-PCR | miR-16-5p, miR-155-5p | [95] | 2016 | |
| 73 (TB) | 69 HC | China | blood | available microarray dataset | none | miR-132-3p, miR-155-5p | [96] | 2016 | |
| 10 (TB), 13 (LTBI) | 11 HC | China | plasma | Microarray (Agilent) | qRT-PCR | let-7b-5p, miR-30b-5p | [97] | 2016 | |
| 11 (TB) | 10 HC | China | serum | available microarray dataset | miRNA PCR panel (TaqMan) | miR-1249-5p | list of 11 miRNAs | [98] | 2015 |
| 34 (TB, 17 HIV co-infected) | 30 treated TB (14 HIV co-infected) | South Africa | plasma | miRNA PCR panel (MIHS-106Z arrays) | qRT-PCR | miR-29a-3p, miR-17-3p, miR-133a | [99] | 2015 | |
| 17 (TB), 17 (LTBI) | 16 HC | Spain | blood | Microarray (Agilent) | qRT-PCR | miR-194-5p, miR-21-5p, miR-29c-3p (vs HC and LTBI) | miR-150-5p (vs HC and LTBI) | [100] | 2015 |
| 110 (TB) | 48 HC | China | serum | literature (miRNA selection) | qRT-PCR | miR-183-5p | [101] | 2015 | |
| 15 (TB), 14 (LTBI) | 68 HC | China | serum | RNA-Seq (Illumina) | qRT-PCR | miR-196b-5p, miR-376c-3p | [102] | 2014 | |
| 29 (TB) | 37 HC | Egypt | serum | literature (miRNA selection) | miRNA PCR Panel (miScript) | miR-197-3p | [103] | 2013 | |
| 108 (TB) | 88 HC | China | serum | RNA-Seq (Illumina) | qRT-PCR | miR-378a-5p, miR-483-5p, miR-22-3p, miR-29c-3p | miR-101-3p, miR-320b | [104] | 2013 |
| 269 (TB, 73 HIV co-infected), 109 (LTBI) | 105 HC | Italy, Tanzania, Uganda | serum | miRNA PCR panel (TaqMan), pooling of samples | qRT-PCR in a subset of individual samples | list of 12 miRNAs (e.g., miR-148a, miR-192, miR-193a-5p, miR-451, miR-590-5p, miR-885-5p) | let-7e-5p | [105] | 2013 |
| 8 (TB) | 8 HC | Germany | serum | Microarray (Agilent) | none | list of 17 miRNAs | miR-574-5p, miR-768-3p, miR-940 | [106] | 2012 |
| 30 (TB) | 65 HC | China | serum | miRNA PCR panels (TaqMan) | qRT-PCR | miR-361-5p, miR-889, miR-576-3p, miR-210, miR-26a-5p, miR-432-5p, miR-134 | [107] | 2012 | |
| 75 (TB) | 55 HC | China | serum | Microarray (Exiqon) | qRT-PCR | miR-93-3p, miR-29a-3p | miR-3125 | [108] | 2011 |
| Subjects No. (Category) | Controls No. (Category) | Country | Samples | Method for Screening | Method for Validation | Up-Regulated in TB | Down-Regulated in TB | Ref. | Year |
|---|---|---|---|---|---|---|---|---|---|
| 30 (TB), 35 (LTBI) | 35 HC | China | PBMCs | available microarray dataset | qRT-PCR | miR-212-3p | [109] | 2019 | |
| 3 (TB) | 3 HC | China | PBMCs | small RNA-seq | none | list of 18 miRNAs | list of 23 miRNAs | [110] | 2018 |
| 12 (TB) | 12 HC | South Africa | PBMCs | literature (miRNA selection) | qRT-PCR | miR-320a-3p, miR-204-5p, miR-331-3p, miR-147b, miR-210-3p | miR-197-3p, miR-99b-5p, miR-191-5p | [111] | 2018 |
| 21 (TB) | 21 treated TB | Mexico | PBMCs | literature (miRNA selection) | qRT-PCR | miR-29a-3p, miR-326 | [112] | 2017 | |
| 9 (TB) | 9 HC | Argentina | PBMCs | literature (miRNA selection) | qRT-PCR | miR-29a-3p, miR-30c-5p, miR-181a-5p, miR-181b-5p | [113] | 2017 | |
| 122 (TB) | 130 HC | China | PBMCs, CSF | literature (miRNA selection) | qRT-PCR | miR-29a-3p | [114] | 2017 | |
| 28 (TB) | 24 HC | China | PBMCs | Microarray (Agilent) | qRT-PCR | miR-29b-3p | miR-1-3p, miR-155-5p, miR-31-5p, miR-146a-5p, miR-10a-5p, miR-125b-5p, miR-150-5p | [115] | 2016 |
| 3 (TB), 4 (LTBI) | 3 HC | Hong Kong-China | Macrophages ex vivo | miRNA PCR panel (TaqMan) | none | miR-16-5p, miR-137, miR-140-3p, miR-193a-3p, miR-501-5p, miR-598 | miR-95 | [116] | 2015 |
| 65 (TB) | 60 HC | China | PBMCs | literature (miRNA selection) | qRT-PCR | miR-31 | [117] | 2015 | |
| 30 (TB), 28 (LTBI) | 30 HC | China | CD4 + T cells | Microarray (Exiqon) | qRT-PCR | miR-451a, miR-340-5p, miR-136-5p, miR-29b-3p | miR-4292 | [118] | 2013 |
| 22 (TB), 14 (LTBI) | 19 HC | Germany | CD4 + T cells | literature (miRNA selection) | qRT-PCR | miR-21-5p, miR-26a-5p, miR-29a-3p, miR-142-3p | [39] | 2013 | |
| 24 (TB) | 20 HC | Argentina | PBMCs | literature (miRNA selection) | qRT-PCR | miR-424-5p | miR-146a-5p | [119] | 2012 |
| 29 (TB), 29 (LTBI) | 18 HC | China | PBMCs | Microarray (Agilent) | qRT-PCR | miR-424-5p, miR-365a-3p (vs HC); miR-424-5p, miR-365a-3p, miR-144-3p, miR-223-3p, miR-451a (vs LTB) | [120] | 2011 | |
| 21 (TB) | 19 HC | China | PBMCs induced with PPD | Microarray (Agilent) | qRT-PCR | miR-155-5p, miR-155-3p | [112] | 2011 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sinigaglia, A.; Peta, E.; Riccetti, S.; Venkateswaran, S.; Manganelli, R.; Barzon, L. Tuberculosis-Associated MicroRNAs: From Pathogenesis to Disease Biomarkers. Cells 2020, 9, 2160. https://doi.org/10.3390/cells9102160
Sinigaglia A, Peta E, Riccetti S, Venkateswaran S, Manganelli R, Barzon L. Tuberculosis-Associated MicroRNAs: From Pathogenesis to Disease Biomarkers. Cells. 2020; 9(10):2160. https://doi.org/10.3390/cells9102160
Chicago/Turabian StyleSinigaglia, Alessandro, Elektra Peta, Silvia Riccetti, Seshasailam Venkateswaran, Riccardo Manganelli, and Luisa Barzon. 2020. "Tuberculosis-Associated MicroRNAs: From Pathogenesis to Disease Biomarkers" Cells 9, no. 10: 2160. https://doi.org/10.3390/cells9102160
APA StyleSinigaglia, A., Peta, E., Riccetti, S., Venkateswaran, S., Manganelli, R., & Barzon, L. (2020). Tuberculosis-Associated MicroRNAs: From Pathogenesis to Disease Biomarkers. Cells, 9(10), 2160. https://doi.org/10.3390/cells9102160







