FMO1 Is Involved in Excess Light Stress-Induced Signal Transduction and Cell Death Signaling
Abstract
1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Plasmid Construction
2.3. Stable Transformation of Arabidopsis
2.4. Excess Light (EL) Treatment
2.5. RNA Extraction and cDNA Synthesis
2.6. Relative Gene Expression Measurement by Real-Time qPCR
2.7. ROS Imaging
2.8. Cell Death Quantification by Electrolyte Leakage Analysis
2.9. Trypan Blue Staining and Micro-Lesions Counting
3. Results
3.1. FMO1 Expression Is Induced in Both Local EL-Treated and Systemic Leaves
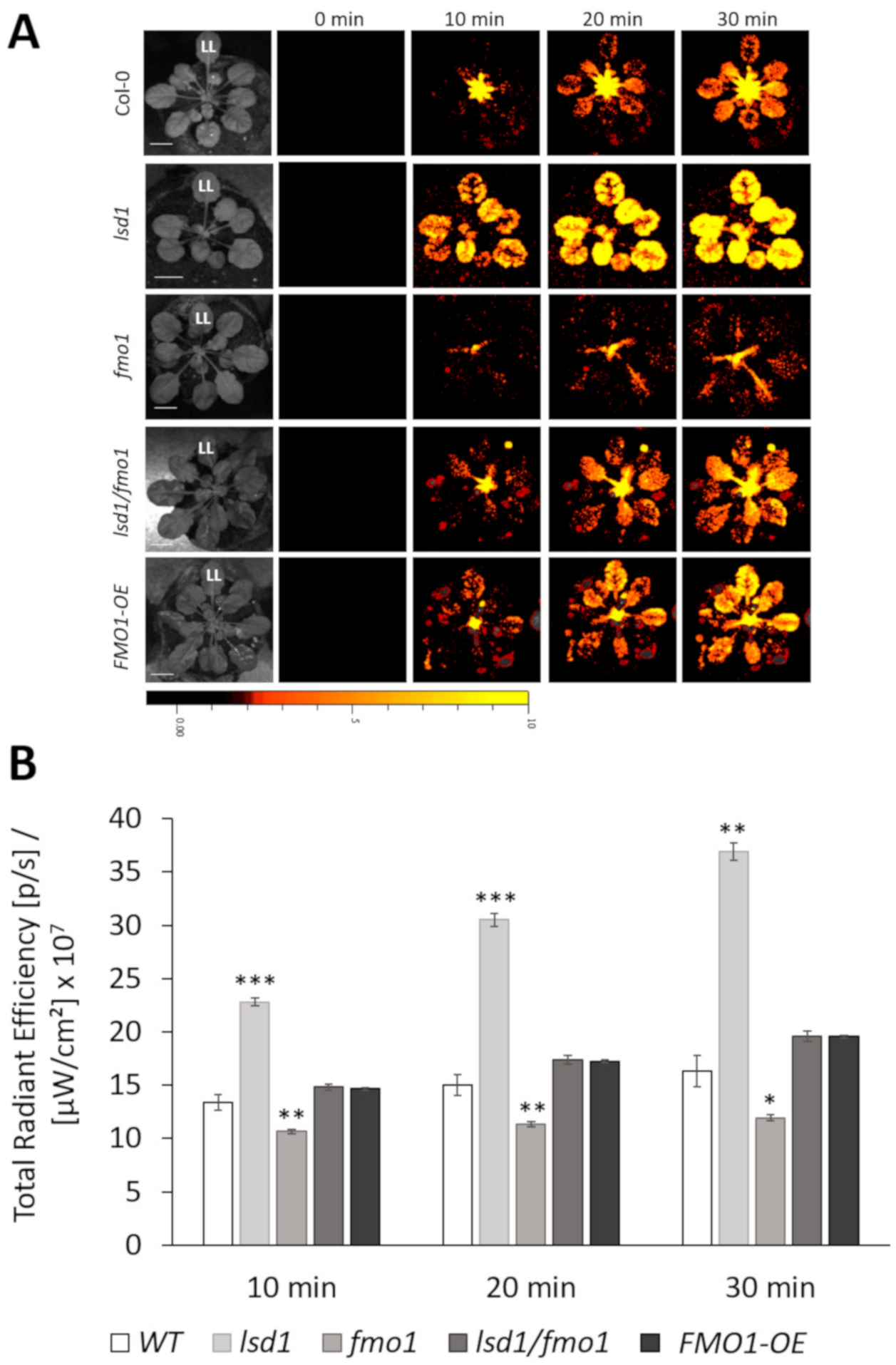
3.2. FMO1, Together with LSD1, Is Required for the Spread of the ROS Systemic Signal
3.3. Functional FMO1 Is Required for the Systemic Induction of APX2 and ZAT10 Expression upon Local EL Treatment
3.4. FMO1 Is Involved in LSD1-Dependent Spread of Systemic Cell Death
3.5. Proposed Model of FMO1 Involvement in ROS Signaling, SAAR and Cell Death
4. Discussion
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Gruner, K.; Griebel, T.; Návarová, H.; Attaran, E.; Zeier, J. Reprogramming of plants during systemic acquired resistance. Front. Plant Sci. 2013, 4, 252. [Google Scholar] [CrossRef] [PubMed]
- El-Shetehy, M.; Wang, C.; Shine, M.B.; Yu, K.; Kachroo, A.; Kachroo, P. Nitric oxide and reactive oxygen species are required for systemic acquired resistance in plants. Plant Signal. Behav. 2015, 10, e998544. [Google Scholar] [CrossRef] [PubMed]
- Cameron, R.K.; Paiva, N.L.; Lamb, C.J.; Dixon, R.A. Accumulation of salicylic acid and PR-1 gene transcripts in relation to the systemic acquired resistance (SAR) response induced by Pseudomonas syringae pv. tomato in Arabidopsis. Physiol. Mol. Plant Pathol. 1999, 55, 121–130. [Google Scholar] [CrossRef]
- Balint-Kurti, P. The plant hypersensitive response: Concepts, control and consequences. Mol. Plant Pathol. 2019, 20, 1163–1178. [Google Scholar] [CrossRef]
- Karpinski, S.; Reynolds, H.; Karpinska, B.; Wingsle, G.; Creissen, G.; Mullineaux, P. Systemic signaling and acclimation in response to excess excitation energy in Arabidopsis. Science 1999, 284, 654–657. [Google Scholar] [CrossRef]
- Zandalinas, S.I.; Sengupta, S.; Burks, D.; Azad, R.K.; Mittler, R. Identification and characterization of a core set of ROS wave-associated transcripts involved in the systemic acquired acclimation response of Arabidopsis to excess light. Plant J. 2019, 98, 126–141. [Google Scholar] [CrossRef]
- Mittler, R.; Blumwald, E. The Roles of ROS and ABA in Systemic Acquired Acclimation[OPEN]. Plant Cell 2015, 27, 64–70. [Google Scholar] [CrossRef]
- Czarnocka, W.; Karpiński, S. Friend or foe? Reactive oxygen species production, scavenging and signaling in plant response to environmental stresses. Free Radic. Biol. Med. 2018, 122, 4–20. [Google Scholar] [CrossRef]
- Gilroy, S.; Białasek, M.; Suzuki, N.; Górecka, M.; Devireddy, A.R.; Karpiński, S.; Mittler, R. ROS, Calcium, and Electric Signals: Key Mediators of Rapid Systemic Signaling in Plants1[OPEN]. Plant Physiol. 2016, 171, 1606–1615. [Google Scholar] [CrossRef]
- Szechyńska-Hebda, M.; Kruk, J.; Górecka, M.; Karpińska, B.; Karpiński, S. Evidence for Light Wavelength-Specific Photoelectrophysiological Signaling and Memory of Excess Light Episodes in Arabidopsis[W][OA]. Plant Cell 2010, 22, 2201–2218. [Google Scholar] [CrossRef]
- Ciszak, K.; Kulasek, M.; Barczak, A.; Grzelak, J.; Maćkowski, S.; Karpiński, S. PsbS is required for systemic acquired acclimation and post-excess-light-stress optimization of chlorophyll fluorescence decay times in Arabidopsis. Plant Signal. Behav. 2015, 10, e982018. [Google Scholar] [CrossRef] [PubMed]
- Białasek, M.; Górecka, M.; Mittler, R.; Karpiński, S. Evidence for the Involvement of Electrical, Calcium and ROS Signaling in the Systemic Regulation of Non-Photochemical Quenching and Photosynthesis. Plant Cell Physiol. 2017, 58, 207–215. [Google Scholar] [CrossRef] [PubMed]
- Carmody, M.; Crisp, P.A.; d’Alessandro, S.; Ganguly, D.; Gordon, M.; Havaux, M.; Albrecht-Borth, V.; Pogson, B.J. Uncoupling High Light Responses from Singlet Oxygen Retrograde Signaling and Spatial-Temporal Systemic Acquired Acclimation1[OPEN]. Plant Physiol. 2016, 171, 1734–1749. [Google Scholar] [CrossRef] [PubMed]
- Karpinski, S.; Szechynska-Hebda, M. Secret life of plants. Plant Signal. Behav. 2010, 5, 1391–1394. [Google Scholar] [CrossRef]
- Rossel, J.B.; Wilson, P.B.; Hussain, D.; Woo, N.S.; Gordon, M.J.; Mewett, O.P.; Howell, K.A.; Whelan, J.; Kazan, K.; Pogson, B.J. Systemic and Intracellular Responses to Photooxidative Stress in Arabidopsis. Plant Cell 2007, 19, 4091–4110. [Google Scholar] [CrossRef] [PubMed]
- Fryer, M.J.; Ball, L.; Oxborough, K.; Karpinski, S.; Mullineaux, P.M.; Baker, N.R. Control of Ascorbate Peroxidase 2 expression by hydrogen peroxide and leaf water status during excess light stress reveals a functional organisation of Arabidopsis leaves. Plant J. 2003, 33, 691–705. [Google Scholar] [CrossRef]
- Galvez-Valdivieso, G.; Fryer, M.J.; Lawson, T.; Slattery, K.; Truman, W.; Smirnoff, N.; Asami, T.; Davies, W.J.; Jones, A.M.; Baker, N.R.; et al. The High Light Response in Arabidopsis Involves ABA Signaling between Vascular and Bundle Sheath Cells. Plant Cell 2009, 21, 2143–2162. [Google Scholar] [CrossRef]
- Mittler, R.; Kim, Y.; Song, L.; Coutu, J.; Coutu, A.; Ciftci-Yilmaz, S.; Lee, H.; Stevenson, B.; Zhu, J.-K. Gain- and loss-of-function mutations in Zat10 enhance the tolerance of plants to abiotic stress. FEBS Lett. 2006, 580, 6537–6542. [Google Scholar] [CrossRef]
- Bechtold, U.; Richard, O.; Zamboni, A.; Gapper, C.; Geisler, M.; Pogson, B.; Karpinski, S.; Mullineaux, P.M. Impact of chloroplastic- and extracellular-sourced ROS on high light-responsive gene expression in Arabidopsis. J. Exp. Bot. 2008, 59, 121–133. [Google Scholar] [CrossRef]
- Miller, G.; Shulaev, V.; Mittler, R. Reactive oxygen signaling and abiotic stress. Physiol. Plant. 2008, 133, 481–489. [Google Scholar] [CrossRef]
- Baxter, A.; Mittler, R.; Suzuki, N. ROS as key players in plant stress signalling. J. Exp. Bot. 2014, 65, 1229–1240. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Apel, K.; Kim, C. Singlet oxygen-mediated and EXECUTER-dependent signalling and acclimation of Arabidopsis thaliana exposed to light stress. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369. [Google Scholar] [CrossRef] [PubMed]
- Zeier, J.; Pink, B.; Mueller, M.J.; Berger, S. Light conditions influence specific defence responses in incompatible plant-pathogen interactions: Uncoupling systemic resistance from salicylic acid and PR-1 accumulation. Planta 2004, 219, 673–683. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Karpiński, S. Programmed Cell Death as a Response to High Light, UV and Drought Stress in Plants. Abiotic Stress Plant Responses Appl. Agric. 2013, 207–246. [Google Scholar] [CrossRef]
- Rustérucci, C.; Aviv, D.H.; Holt, B.F.; Dangl, J.L.; Parker, J.E. The Disease Resistance Signaling Components EDS1 and PAD4 Are Essential Regulators of the Cell Death Pathway Controlled by LSD1 in Arabidopsis. Plant Cell 2001, 13, 2211–2224. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Szechyńska-Hebda, M.; Sobczak, M.; Rusaczonek, A.; Kozłowska-Makulska, A.; Witoń, D.; Karpiński, S. Lesion simulating disease 1 and enhanced disease susceptibility 1 differentially regulate UV-C-induced photooxidative stress signalling and programmed cell death in Arabidopsis thaliana. Plant Cell Environ. 2015, 38, 315–330. [Google Scholar] [CrossRef]
- Mateo, A.; Mühlenbock, P.; Rustérucci, C.; Chang, C.C.-C.; Miszalski, Z.; Karpinska, B.; Parker, J.E.; Mullineaux, P.M.; Karpinski, S. LESION SIMULATING DISEASE 1 Is Required for Acclimation to Conditions That Promote Excess Excitation Energy. Plant Physiol. 2004, 136, 2818–2830. [Google Scholar] [CrossRef]
- Mühlenbock, P.; Plaszczyca, M.; Plaszczyca, M.; Mellerowicz, E.; Karpinski, S. Lysigenous aerenchyma formation in Arabidopsis is controlled by LESION SIMULATING DISEASE1. Plant Cell 2007, 19, 3819–3830. [Google Scholar] [CrossRef]
- Olszak, B.; Malinovsky, F.G.; Brodersen, P.; Grell, M.; Giese, H.; Petersen, M.; Mundy, J. A putative flavin-containing mono-oxygenase as a marker for certain defense and cell death pathways. Plant Sci. 2006, 170, 614–623. [Google Scholar] [CrossRef]
- Schlaich, N.L. Flavin-containing monooxygenases in plants: Looking beyond detox. Trends Plant Sci. 2007, 12, 412–418. [Google Scholar] [CrossRef]
- Rossner, R.; Kaeberlein, M.; Leiser, S.F. Flavin-containing monooxygenases in aging and disease: Emerging roles for ancient enzymes. J. Biol. Chem. 2017, 292, 11138–11146. [Google Scholar] [CrossRef] [PubMed]
- Bartsch, M.; Gobbato, E.; Bednarek, P.; Debey, S.; Schultze, J.L.; Bautor, J.; Parker, J.E. Salicylic Acid–Independent ENHANCED DISEASE SUSCEPTIBILITY1 Signaling in Arabidopsis Immunity and Cell Death Is Regulated by the Monooxygenase FMO1 and the Nudix Hydrolase NUDT7. Plant Cell 2006, 18, 1038–1051. [Google Scholar] [CrossRef] [PubMed]
- Koch, M.; Vorwerk, S.; Masur, C.; Sharifi-Sirchi, G.; Olivieri, N.; Schlaich, N.L. A role for a flavin-containing mono-oxygenase in resistance against microbial pathogens in Arabidopsis. Plant J. Cell Mol. Biol. 2006, 47, 629–639. [Google Scholar] [CrossRef] [PubMed]
- Mishina, T.E.; Zeier, J. The Arabidopsis flavin-dependent monooxygenase FMO1 is an essential component of biologically induced systemic acquired resistance. Plant Physiol. 2006, 141, 1666–1675. [Google Scholar] [CrossRef]
- Hilfiker, O.; Groux, R.; Bruessow, F.; Kiefer, K.; Zeier, J.; Reymond, P. Insect eggs induce a systemic acquired resistance in Arabidopsis. Plant J. Cell Mol. Biol. 2014, 80, 1085–1094. [Google Scholar] [CrossRef]
- Orlovskis, Z.; Reymond, P. Pieris brassicae eggs trigger interplant systemic acquired resistance against a foliar pathogen in Arabidopsis. New Phytol. 2020. [Google Scholar] [CrossRef]
- Bernsdorff, F.; Döring, A.-C.; Gruner, K.; Schuck, S.; Bräutigam, A.; Zeier, J. Pipecolic Acid Orchestrates Plant Systemic Acquired Resistance and Defense Priming via Salicylic Acid-Dependent and -Independent Pathways. Plant Cell 2016, 28, 102–129. [Google Scholar] [CrossRef]
- Ding, P.; Rekhter, D.; Ding, Y.; Feussner, K.; Busta, L.; Haroth, S.; Xu, S.; Li, X.; Jetter, R.; Feussner, I.; et al. Characterization of a Pipecolic Acid Biosynthesis Pathway Required for Systemic Acquired Resistance. Plant Cell 2016, 28, 2603–2615. [Google Scholar] [CrossRef]
- Yang, C.-Y.; Hsu, F.-C.; Li, J.-P.; Wang, N.-N.; Shih, M.-C. The AP2/ERF transcription factor AtERF73/HRE1 modulates ethylene responses during hypoxia in Arabidopsis. Plant Physiol. 2011, 156, 202–212. [Google Scholar] [CrossRef]
- Wituszyńska, W.; Ślesak, I.; Vanderauwera, S.; Szechyńska-Hebda, M.; Kornaś, A.; Kelen, K.V.D.; Mühlenbock, P.; Karpińska, B.; Maćkowski, S.; Breusegem, F.V.; et al. LESION SIMULATING DISEASE1, ENHANCED DISEASE SUSCEPTIBILITY1, and PHYTOALEXIN DEFICIENT4 Conditionally Regulate Cellular Signaling Homeostasis, Photosynthesis, Water Use Efficiency, and Seed Yield in Arabidopsis. Plant Physiol. 2013, 161, 1795–1805. [Google Scholar] [CrossRef]
- Joglekar, S.; Suliman, M.; Bartsch, M.; Halder, V.; Maintz, J.; Bautor, J.; Zeier, J.; Parker, J.E.; Kombrink, E. Chemical Activation of EDS1/PAD4 Signaling Leading to Pathogen Resistance in Arabidopsis. Plant Cell Physiol. 2018, 59, 1592–1607. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Chen, L.; Mu, J.; Zuo, J. LESION SIMULATING DISEASE1 Interacts with Catalases to Regulate Hypersensitive Cell Death in Arabidopsis1[C][W]. Plant Physiol. 2013, 163, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.-C.; Holmes, E.C.; Rajniak, J.; Kim, J.-G.; Tang, S.; Fischer, C.R.; Mudgett, M.B.; Sattely, E.S. N-hydroxy-pipecolic acid is a mobile metabolite that induces systemic disease resistance in Arabidopsis. Proc. Natl. Acad. Sci. USA. 2018, 115, E4920–E4929. [Google Scholar] [CrossRef] [PubMed]
- Hartmann, M.; Zeier, T.; Bernsdorff, F.; Reichel-Deland, V.; Kim, D.; Hohmann, M.; Scholten, N.; Schuck, S.; Bräutigam, A.; Hölzel, T.; et al. Flavin Monooxygenase-Generated N-Hydroxypipecolic Acid Is a Critical Element of Plant Systemic Immunity. Cell 2018, 173, 456–469. [Google Scholar] [CrossRef] [PubMed]
- Karimi, M.; Inzé, D.; Depicker, A. GATEWAY vectors for Agrobacterium-mediated plant transformation. Trends Plant Sci. 2002, 7, 193–195. [Google Scholar] [CrossRef]
- Clough, S.J.; Bent, A.F. Floral dip: A simplified method for Agrobacterium -mediated transformation of Arabidopsis thaliana. Plant J. 1998, 16, 735–743. [Google Scholar] [CrossRef]
- Czarnocka, W.; Van Der Kelen, K.; Willems, P.; Szechyńska-Hebda, M.; Shahnejat-Bushehri, S.; Balazadeh, S.; Rusaczonek, A.; Mueller-Roeber, B.; Van Breusegem, F.; Karpiński, S. The dual role of LESION SIMULATING DISEASE 1 as a condition-dependent scaffold protein and transcription regulator. Plant Cell Environ. 2017, 40, 2644–2662. [Google Scholar] [CrossRef]
- Hruz, T.; Laule, O.; Szabo, G.; Wessendorp, F.; Bleuler, S.; Oertle, L.; Widmayer, P.; Gruissem, W.; Zimmermann, P. Genevestigator V3: A Reference Expression Database for the Meta-Analysis of Transcriptomes. Available online: https://www.hindawi.com/journals/abi/2008/420747/ (accessed on 4 August 2020).
- Ramakers, C.; Ruijter, J.M.; Deprez, R.H.L.; Moorman, A.F.M. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci. Lett. 2003, 339, 62–66. [Google Scholar] [CrossRef]
- Fichman, Y.; Miller, G.; Mittler, R. Whole-Plant Live Imaging of Reactive Oxygen Species. Mol. Plant 2019, 12, 1203–1210. [Google Scholar] [CrossRef]
- Fichman, Y.; Mittler, R. Noninvasive Live ROS Imaging of Whole Plants Grown in Soil. Trends Plant Sci. 2020, S1360–1385. [Google Scholar] [CrossRef]
- Czarnocka, W.; Rusaczonek, A.; Willems, P.; Sujkowska-Rybkowska, M.; Van Breusegem, F.; Karpiński, S. Novel Role of JAC1 in Influencing Photosynthesis, Stomatal Conductance, and Photooxidative Stress Signalling Pathway in Arabidopsis thaliana. Front. Plant Sci. 2020, 11, 1124. [Google Scholar] [CrossRef]
- Miller, G.; Schlauch, K.; Tam, R.; Cortes, D.; Torres, M.A.; Shulaev, V.; Dangl, J.L.; Mittler, R. The Plant NADPH Oxidase RBOHD Mediates Rapid Systemic Signaling in Response to Diverse Stimuli. Sci. Signal. 2009, 2, ra45. [Google Scholar] [CrossRef] [PubMed]
- Gadjev, I.; Vanderauwera, S.; Gechev, T.S.; Laloi, C.; Minkov, I.N.; Shulaev, V.; Apel, K.; Inzé, D.; Mittler, R.; Van Breusegem, F. Transcriptomic footprints disclose specificity of reactive oxygen species signaling in Arabidopsis. Plant Physiol. 2006, 141, 436–445. [Google Scholar] [CrossRef] [PubMed]
- Hahn, A.; Kilian, J.; Mohrholz, A.; Ladwig, F.; Peschke, F.; Dautel, R.; Harter, K.; Berendzen, K.W.; Wanke, D. Plant Core Environmental Stress Response Genes Are Systemically Coordinated during Abiotic Stresses. Int. J. Mol. Sci. 2013, 14, 7617–7641. [Google Scholar] [CrossRef] [PubMed]
- Mühlenbock, P.; Szechyńska-Hebda, M.; Płaszczyca, M.; Baudo, M.; Mateo, A.; Mullineaux, P.M.; Parker, J.E.; Karpińska, B.; Karpiński, S. Chloroplast Signaling and LESION SIMULATING DISEASE1 Regulate Crosstalk between Light Acclimation and Immunity in Arabidopsis. Plant Cell 2008, 20, 2339–2356. [Google Scholar] [CrossRef]
- Rusaczonek, A.; Czarnocka, W.; Kacprzak, S.; Witoń, D.; Ślesak, I.; Szechyńska-Hebda, M.; Gawroński, P.; Karpiński, S. Role of phytochromes A and B in the regulation of cell death and acclimatory responses to UV stress in Arabidopsis thaliana. J. Exp. Bot. 2015, 66, 6679–6695. [Google Scholar] [CrossRef]
- Alvarez, M.E.; Pennell, R.I.; Meijer, P.J.; Ishikawa, A.; Dixon, R.A.; Lamb, C. Reactive oxygen intermediates mediate a systemic signal network in the establishment of plant immunity. Cell 1998, 92, 773–784. [Google Scholar] [CrossRef]
- Clarke, J.D.; Aarts, N.; Feys, B.J.; Dong, X.; Parker, J.E. Constitutive disease resistance requires EDS1 in the Arabidopsis mutants cpr1 and cpr6 and is partially EDS1-dependent in cpr5. Plant J. Cell Mol. Biol. 2001, 26, 409–420. [Google Scholar] [CrossRef]
- Karpiński, S.; Szechyńska-Hebda, M.; Wituszyńska, W.; Burdiak, P. Light acclimation, retrograde signalling, cell death and immune defences in plants. Plant Cell Environ. 2013, 36, 736–744. [Google Scholar] [CrossRef]
- Griebel, T.; Zeier, J. Light regulation and daytime dependency of inducible plant defenses in Arabidopsis: Phytochrome signaling controls systemic acquired resistance rather than local defense. Plant Physiol. 2008, 147, 790–801. [Google Scholar] [CrossRef]
- Bienert, G.P.; Møller, A.L.B.; Kristiansen, K.A.; Schulz, A.; Møller, I.M.; Schjoerring, J.K.; Jahn, T.P. Specific aquaporins facilitate the diffusion of hydrogen peroxide across membranes. J. Biol. Chem. 2007, 282, 1183–1192. [Google Scholar] [CrossRef] [PubMed]
- Laloi, C.; Havaux, M. Key players of singlet oxygen-induced cell death in plants. Front. Plant Sci. 2015, 6, 39. [Google Scholar] [CrossRef] [PubMed]
- Chen, P.; Umeda, M. DNA double-strand breaks induce the expression of flavin-containing monooxygenase and reduce root meristem size in Arabidopsis thaliana. Genes Cells Dev. Mol. Cell. Mech. 2015, 20, 636–646. [Google Scholar] [CrossRef] [PubMed]
- Sun, T.; Zhang, Y.; Li, Y.; Zhang, Q.; Ding, Y.; Zhang, Y. ChIP-seq reveals broad roles of SARD1 and CBP60g in regulating plant immunity. Nat. Commun. 2015, 6, 10159. [Google Scholar] [CrossRef]
- Budimir, J.; Treffon, K.; Nair, A.; Thurow, C.; Gatz, C. Redox-active cysteines in TGACG-BINDING FACTOR 1 (TGA1) do not play a role in salicylic acid or pathogen-induced expression of TGA1-regulated target genes in Arabidopsis thaliana. New Phytol. 2020. [Google Scholar] [CrossRef]
- Brodersen, P.; Petersen, M.; Pike, H.M.; Olszak, B.; Skov, S.; Ødum, N.; Jørgensen, L.B.; Brown, R.E.; Mundy, J. Knockout of Arabidopsis ACCELERATED-CELL-DEATH11 encoding a sphingosine transfer protein causes activation of programmed cell death and defense. Genes Dev. 2002, 16, 490–502. [Google Scholar] [CrossRef]




© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Czarnocka, W.; Fichman, Y.; Bernacki, M.; Różańska, E.; Sańko-Sawczenko, I.; Mittler, R.; Karpiński, S. FMO1 Is Involved in Excess Light Stress-Induced Signal Transduction and Cell Death Signaling. Cells 2020, 9, 2163. https://doi.org/10.3390/cells9102163
Czarnocka W, Fichman Y, Bernacki M, Różańska E, Sańko-Sawczenko I, Mittler R, Karpiński S. FMO1 Is Involved in Excess Light Stress-Induced Signal Transduction and Cell Death Signaling. Cells. 2020; 9(10):2163. https://doi.org/10.3390/cells9102163
Chicago/Turabian StyleCzarnocka, Weronika, Yosef Fichman, Maciej Bernacki, Elżbieta Różańska, Izabela Sańko-Sawczenko, Ron Mittler, and Stanisław Karpiński. 2020. "FMO1 Is Involved in Excess Light Stress-Induced Signal Transduction and Cell Death Signaling" Cells 9, no. 10: 2163. https://doi.org/10.3390/cells9102163
APA StyleCzarnocka, W., Fichman, Y., Bernacki, M., Różańska, E., Sańko-Sawczenko, I., Mittler, R., & Karpiński, S. (2020). FMO1 Is Involved in Excess Light Stress-Induced Signal Transduction and Cell Death Signaling. Cells, 9(10), 2163. https://doi.org/10.3390/cells9102163









