Notch Signaling in Breast Cancer: A Role in Drug Resistance
Abstract
1. Breast Cancer and Drug Resistance
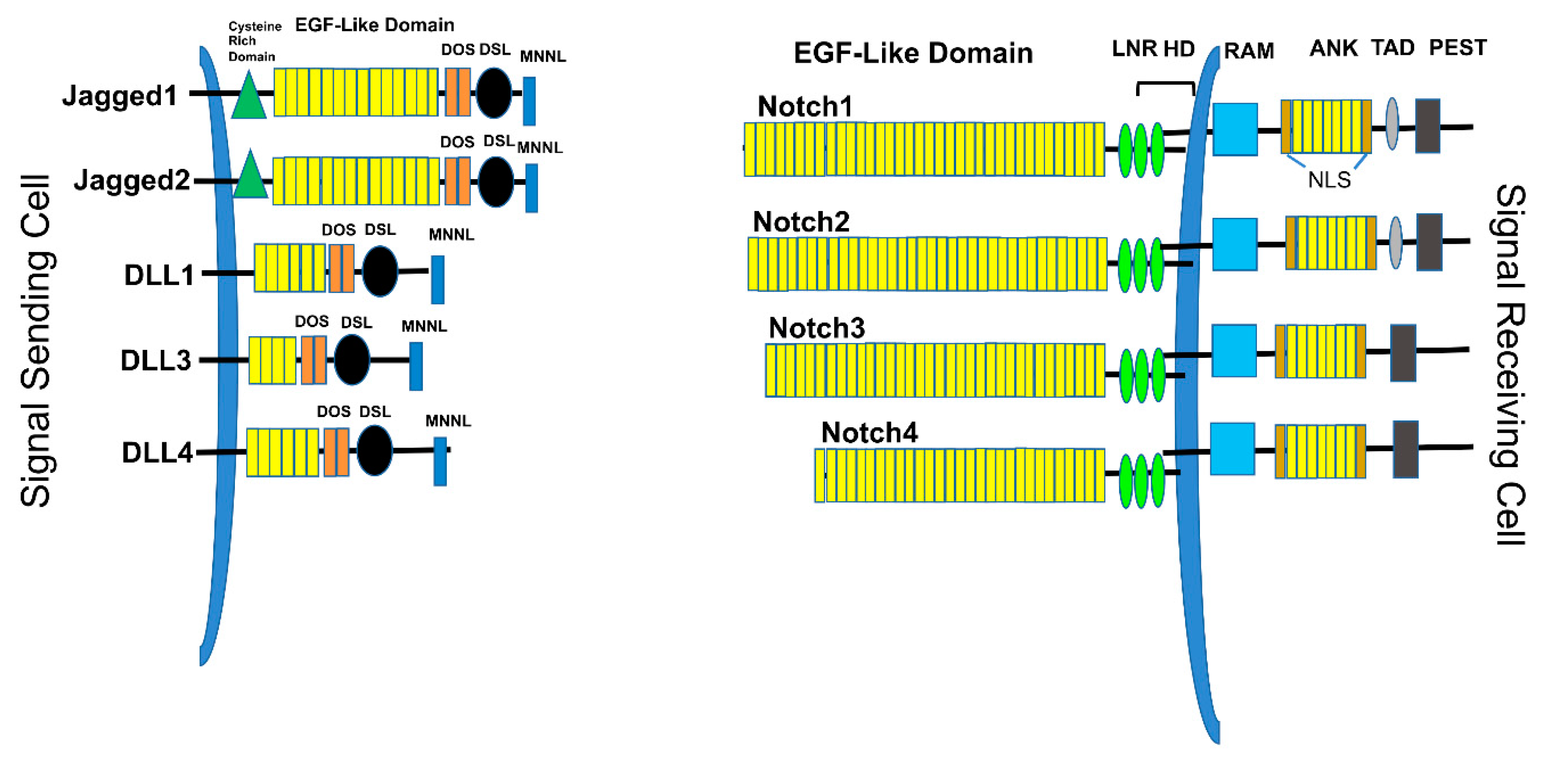
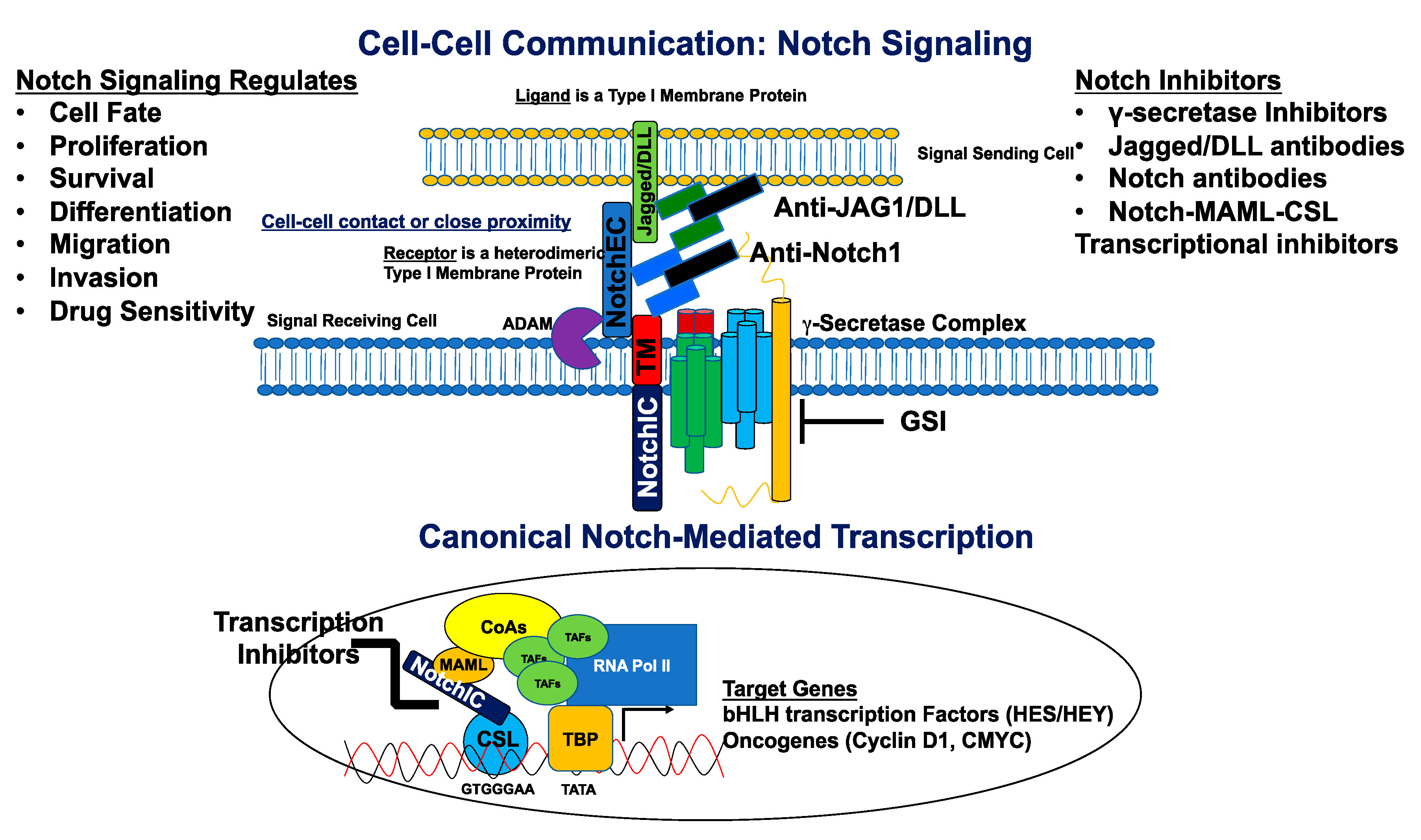
2. Overview of the Notch Signaling Pathway
3. A Role for Notch in Breast Cancer
3.1. Notch as a Breast Oncogene
3.2. Notch as a Prognostic Biomarker
3.3. Notch and Epithelial-to-Mesenchymal Transition
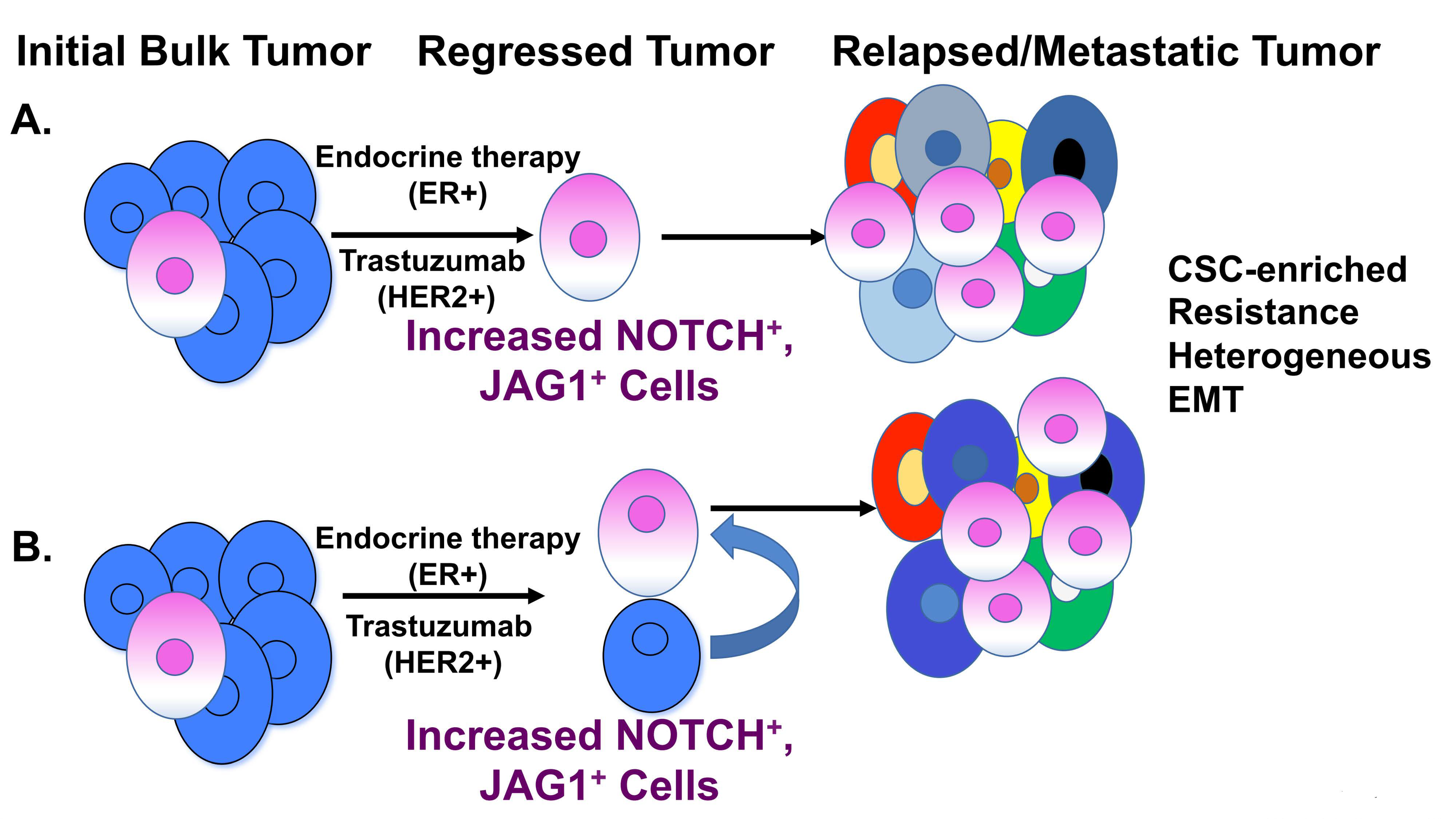
4. Notch and Breast Cancer Stem Cells (BCSCs)
4.1. Stem Cell Markers
4.2. Mammosphere Forming Efficiency
4.3. Extreme Limiting Dilution Assay
4.4. BCSCs and Drug Resistance
5. Notch-Mediated Resistance to Breast Cancer Treatment
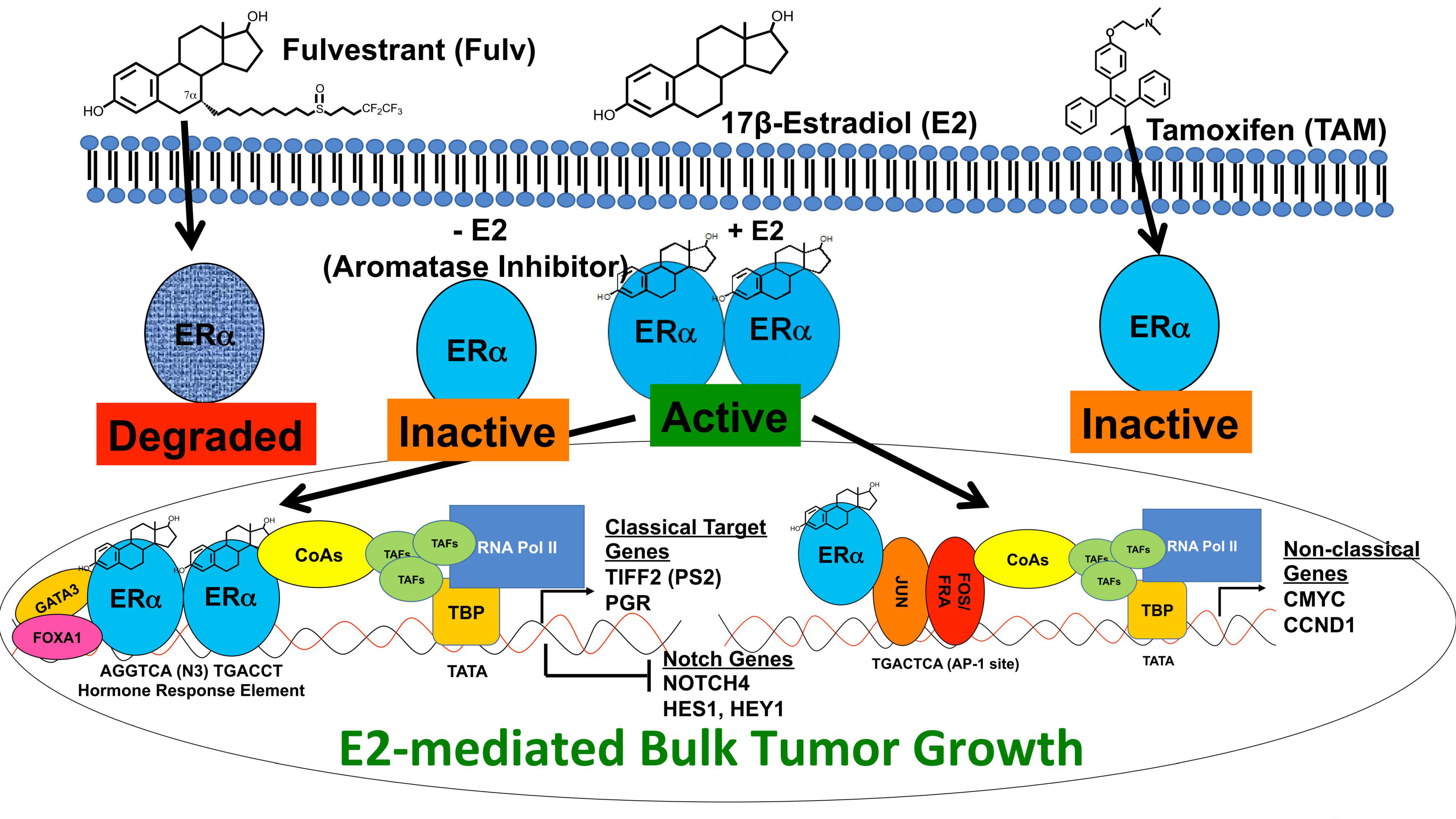
5.1. Estrogen Regulation of Notch Signaling
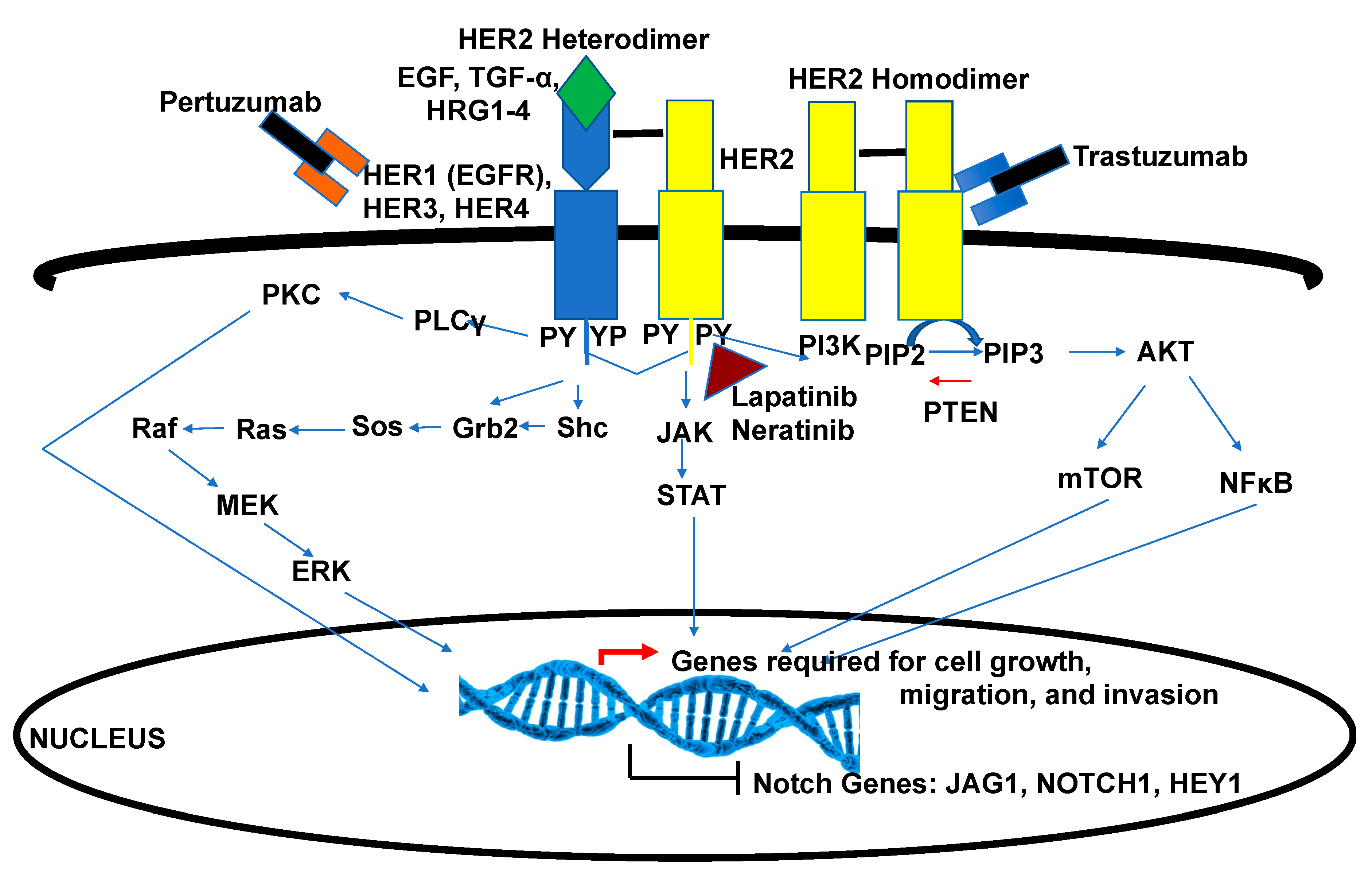
5.2. Crosstalk of Notch and HER2 Signaling
6. Notch Inhibition as a Therapeutic Approach
6.1. γ-Secretase Inhibitors
6.2. Notch Signaling Biologics
6.3. Notch Transcriptional Inhibitors
7. Future Ideas
8. Concluding Thoughts
Funding
Conflicts of Interest
References
- Siegel, R.L.; Mph, K.D.M.; Jemal, A. Cancer statistics, 2020. CA Cancer J. Clin. 2020, 70, 7–30. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Xiang, L.; Li, T.; Bai, Z. Cancer hallmarks, biomarkers and breast cancer molecular subtypes. J. Cancer 2016, 7, 1281–1294. [Google Scholar] [CrossRef] [PubMed]
- Hussain, S.A.; Williams, S.; Stevens, A.; Rea, D.W. Endocrine therapy for early breast cancer. Expert Rev. Anticancer Ther. 2004, 4, 877–888. [Google Scholar] [CrossRef] [PubMed]
- Tremont, A.; Lu, J.; Cole, J.T. Endocrine therapy for early breast cancer: Updated review. Ochsner J. 2017, 17, 405–411. [Google Scholar]
- Boccardo, F.; Rubagotti, A.; Amoroso, D.; Mesiti, M.; Pacini, P.; Gallo, L.; Sismondi, P.; Giai, M.; Genta, F.; Mustacchi, G.; et al. Italian breast cancer adjuvant chemo-hormone therapy cooperative group trials. Adv. Struct. Saf. Stud. 1998, 152, 453–470. [Google Scholar] [CrossRef]
- Higgins, M.; Liedke, P.E.; Goss, P.E. Extended adjuvant endocrine therapy in hormone dependent breast cancer: The paradigm of the NCIC-CTG MA.17/BIG 1–97 trial. Crit. Rev. Oncol. 2013, 86, 23–32. [Google Scholar] [CrossRef]
- Colleoni, M.; Munzone, E. Picking the optimal endocrine adjuvant treatment for pre-menopausal women. Breast 2015, 24, S11–S14. [Google Scholar] [CrossRef]
- Gianni, L.; Pienkowski, T.; Im, Y.-H.; Tseng, L.-M.; Liu, M.-C.; Lluch, A.; Starosławska, E.; De La Haba-Rodriguez, J.; Im, S.-A.; Pedrini, J.L.; et al. 5-year analysis of neoadjuvant pertuzumab and trastuzumab in patients with locally advanced, inflammatory, or early-stage HER2-positive breast cancer (NeoSphere): A multicentre, open-label, phase 2 randomised trial. Lancet Oncol. 2016, 17, 791–800. [Google Scholar] [CrossRef]
- Howie, L.; Scher, N.S.; Amiri-Kordestani, L.; Zhang, L.; King-Kallimanis, B.L.; Choudhry, Y.; Schroeder, J.; Goldberg, K.B.; Kluetz, P.G.; Ibrahim, A.; et al. FDA approval summary: Pertuzumab for adjuvant treatment of HER2-positive early breast cancer. Clin. Cancer Res. 2018, 25, 2949–2955. [Google Scholar] [CrossRef]
- Hudis, C.A. Trastuzumab—Mechanism of action and use in clinical practice. N. Engl. J. Med. 2007, 357, 39–51. [Google Scholar] [CrossRef]
- Jones, J.; Takeda, A.; Picot, J.; Von Keyserlingk, C.; Clegg, A. Lapatinib for the treatment of HER2-overexpressing breast cancer. Health Technol. Assess. 2009, 13, 1–6. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Martin, M.; Holmes, F.A.; Ejlertsen, B.; Delaloge, S.; Moy, B.; Iwata, H.; Von Minckwitz, G.; Chia, S.K.L.; Mansi, J.; Barrios, C.H.; et al. Neratinib after trastuzumab-based adjuvant therapy in HER2-positive breast cancer (ExteNET): 5-year analysis of a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet Oncol. 2017, 18, 1688–1700. [Google Scholar] [CrossRef]
- Miles, J.; White, Y. Neratinib for the treatment of early-stage HER2-positive breast cancer. J. Adv. Pract. Oncol. 2018, 9, 750–754. [Google Scholar] [CrossRef] [PubMed]
- Singh, H.; Walker, A.J.; Amiri-Kordestani, L.; Cheng, J.; Tang, S.; Balcazar, P.; Barnett-Ringgold, K.; Palmby, T.R.; Cao, X.; Zheng, N.; et al. U.S. Food and drug administration approval: Neratinib for the extended adjuvant treatment of early-stage HER2-positive breast cancer. Clin. Cancer Res. 2018, 24, 3486–3491. [Google Scholar] [CrossRef]
- Hatzis, C.; Symmans, W.F.; Zhang, Y.; Gould, R.E.; Moulder, S.L.; Hunt, K.K.; Abu-Khalaf, M.; Hofstatter, E.; Lannin, D.R.; Chagpar, A.B.; et al. Relationship between complete pathologic response to neoadjuvant chemotherapy and survival in triple-negative breast cancer. Clin. Cancer Res. 2015, 22, 26–33. [Google Scholar] [CrossRef]
- Ishikawa, T.; Shimizu, D.; Yamada, A.; Sasaki, T.; Morita, S.; Tanabe, M.; Kawachi, K.; Nozawa, A.; Chishima, T.; Kimura, M.; et al. Impacts and predictors of cytotoxic anticancer agents in different breast cancer subtypes. Oncol. Res. 2012, 20, 71–79. [Google Scholar] [CrossRef]
- Pal, S.K.; Childs, B.H.; Pegram, M. Triple negative breast cancer: Unmet medical needs. Breast Cancer Res. Treat. 2010, 125, 627–636. [Google Scholar] [CrossRef]
- Prat, A.; Fan, C.; Fernández-Martínez, A.; Hoadley, K.A.; Martinello, R.; Vidal, M.; Viladot, M.; Pineda, E.; Arance, A.; Muñoz, M.; et al. Response and survival of breast cancer intrinsic subtypes following multi-agent neoadjuvant chemotherapy. BMC Med. 2015, 13, 303. [Google Scholar] [CrossRef]
- Chumsri, S.; Li, Z.; Serie, D.J.; Mashadi-Hossein, A.; Colon-Otero, G.; Song, N.; Pogue-Geile, K.L.; Gavin, P.G.; Paik, S.; Moreno-Aspitia, A.; et al. Incidence of late relapses in patients with HER2-positive breast cancer receiving adjuvant trastuzumab: Combined analysis of NCCTG N9831 (Alliance) and NRG Oncology/NSABP B-31. J. Clin. Oncol. 2019, 37, 3425–3435. [Google Scholar] [CrossRef]
- Peterson, D.J.; Truong, P.T.; Sadek, B.; Alexander, C.S.; Wiksyk, B.; Shenouda, M.; Raad, R.A.; Taghian, A.G. Locoregional recurrence and survival outcomes by type of local therapy and trastuzumab use among women with node-negative, HER2-positive breast cancer. Ann. Surg. Oncol. 2014, 21, 3490–3496. [Google Scholar] [CrossRef]
- Rodriguez, D.; Ramkairsingh, M.; Lin, X.; Kapoor, A.; Major, P.; Tang, D. The central contributions of breast cancer stem cells in developing resistance to endocrine therapy in Estrogen Receptor (ER)-positive breast cancer. Cancers 2019, 11, 1028. [Google Scholar] [CrossRef] [PubMed]
- Patel, S.A.; DeMichele, A.M. Adding Adjuvant systemic treatment after neoadjuvant therapy in breast cancer: Review of the data. Curr. Oncol. Rep. 2017, 19, 56. [Google Scholar] [CrossRef] [PubMed]
- Artavanis-Tsakonas, S.; Matsuno, K.; Fortini, M. Notch signaling. Science 1995, 268, 225–232. [Google Scholar] [CrossRef]
- Lai, E.C. Notch signaling: Control of cell communication and cell fate. Development 2004, 131, 965–973. [Google Scholar] [CrossRef]
- Chiba, S. Concise review: Notch signaling in stem cell systems. Stem Cells 2006, 24, 2437–2447. [Google Scholar] [CrossRef] [PubMed]
- Miele, L.; Espinoza, I.; Pochampally, R.R.; Watabe, K.; Xing, F. Notch signaling: Targeting cancer stem cells and epithelial-to-mesenchymal transition. Onco Targets Ther. 2013, 6, 1249–1259. [Google Scholar] [CrossRef]
- Andersson, E.R.; Sandberg, R.; Lendahl, U. Notch signaling: Simplicity in design, versatility in function. Development 2011, 138, 3593–3612. [Google Scholar] [CrossRef] [PubMed]
- Kopan, R.; Ilagan, M.X.G.; Ilagan, M.X.G. The canonical notch signaling pathway: Unfolding the activation mechanism. Cell 2009, 137, 216–233. [Google Scholar] [CrossRef] [PubMed]
- Borggrefe, T.; Oswald, F. The Notch signaling pathway: Transcriptional regulation at Notch target genes. Cell. Mol. Life Sci. 2009, 66, 1631–1646. [Google Scholar] [CrossRef] [PubMed]
- Fischer, A.; Schumacher, N.; Maier, M.; Sendtner, M.; Gessler, M. The Notch target genes Hey1 and Hey2 are required for embryonic vascular development. Genes Dev. 2004, 18, 901–911. [Google Scholar] [CrossRef]
- Klinakis, A.; Szabolcs, M.; Politi, K.; Kiaris, H.; Artavanis-Tsakonas, S.; Efstratiadis, A. Myc is a Notch1 transcriptional target and a requisite for Notch1-induced mammary tumorigenesis in mice. Proc. Natl. Acad. Sci. USA 2006, 103, 9262–9267. [Google Scholar] [CrossRef] [PubMed]
- Cohen, B.; Shimizu, M.; Izrailit, J.; Ng, N.F.L.; Buchman, Y.; Pan, J.G.; Dering, J.; Reedijk, M. Cyclin D1 is a direct target of JAG1-mediated Notch signaling in breast cancer. Breast Cancer Res. Treat. 2009, 123, 113–124. [Google Scholar] [CrossRef]
- Bolόs, V.; Grego-Bessa, J.; De La Pompa, J.L. Notch signaling in development and cancer. Endocr. Rev. 2007, 28, 339–363. [Google Scholar] [CrossRef]
- Diévart, A.; Beaulieu, N.; Jolicoeur, P. Involvement of Notch1 in the development of mouse mammary tumors. Oncogene 1999, 18, 5973–5981. [Google Scholar] [CrossRef] [PubMed]
- Hu, C.; Diévart, A.; Lupien, M.; Calvo, E.; Tremblay, G.; Jolicoeur, P. Overexpression of activated murine Notch1 and Notch3 in transgenic mice blocks mammary gland development and induces mammary tumors. Am. J. Pathol. 2006, 168, 973–990. [Google Scholar] [CrossRef] [PubMed]
- Jhappan, C.; Gallahan, D.; Stahle, C.; Chu, E.; Smith, G.H.; Merlino, G.; Callahan, R. Expression of an activated Notch-related int-3 transgene interferes with cell differentiation and induces neoplastic transformation in mammary and salivary glands. Genes Dev. 1992, 6, 345–355. [Google Scholar] [CrossRef]
- Raafat, A.; Bargo, S.; Anver, M.R.; Callahan, R. Mammary development and tumorigenesis in mice expressing a truncated human Notch4/Int3 intracellular domain (h-Int3sh). Oncogene 2004, 23, 9401–9407. [Google Scholar] [CrossRef][Green Version]
- Dickson, B.C.; Mulligan, A.M.; Zhang, H.; Lockwood, G.; O’Malley, F.P.; Egan, S.E.; Reedijk, M. High-level JAG1 mRNA and protein predict poor outcome in breast cancer. Mod. Pathol. 2007, 20, 685–693. [Google Scholar] [CrossRef]
- Reedijk, M. High-level coexpression of JAG1 and NOTCH1 Is observed in human breast cancer and is associated with poor overall survival. Cancer Res. 2005, 65, 8530–8537. [Google Scholar] [CrossRef]
- Pece, S.; Serresi, M.; Santolini, E.; Capra, M.; Hulleman, E.; Galimberti, V.; Zurrida, S.; Maisonneuve, P.; Viale, G.; Di Fiore, P.P. Loss of negative regulation by Numb over Notch is relevant to human breast carcinogenesis. J. Cell Biol. 2004, 167, 215–221. [Google Scholar] [CrossRef]
- Stylianou, S.; Clarke, R.; Brennan, K. Aberrant activation of Notch signaling in human breast cancer. Cancer Res. 2006, 66, 1517–1525. [Google Scholar] [CrossRef] [PubMed]
- Yao, K.; Rizzo, P.; Rajan, P.; Albain, K.; Rychlik, K.; Shah, S.; Miele, L. Notch-1 and Notch-4 receptors as prognostic markers in breast cancer. Int. J. Surg. Pathol. 2010, 19, 607–613. [Google Scholar] [CrossRef] [PubMed]
- Speiser, J.J.; Foreman, K.; Drinka, E.; Godellas, C.; Perez, C.; Salhadar, A.; Erşahin, Ç.; Rajan, P. Notch-1 and Notch-4 biomarker expression in triple-negative breast cancer. Int. J. Surg. Pathol. 2011, 20, 137–143. [Google Scholar] [CrossRef] [PubMed]
- Reedijk, M.; Pinnaduwage, D.; Dickson, B.C.; Mulligan, A.M.; Zhang, H.; Bull, S.B.; O’Malley, F.P.; Egan, S.E.; Andrulis, I. JAG1 expression is associated with a basal phenotype and recurrence in lymph node-negative breast cancer. Breast Cancer Res. Treat. 2007, 111, 439–448. [Google Scholar] [CrossRef]
- Xing, F.; Okuda, H.; Watabe, M.; Kobayashi, A.; Pai, S.K.; Liu, W.; Pandey, P.R.; Fukuda, K.; Hirota, S.; Sugai, T.; et al. Hypoxia-induced Jagged2 promotes breast cancer metastasis and self-renewal of cancer stem-like cells. Oncogene 2011, 30, 4075–4086. [Google Scholar] [CrossRef]
- Speiser, J.J.; Erşahin, Ç.; Osipo, C. The functional role of Notch signaling in triple-negative breast cancer. Pancreat. Beta Cell 2013, 93, 277–306. [Google Scholar] [CrossRef]
- Pandya, K.; Wyatt, D.; Gallagher, B.; Shah, D.; Baker, A.; Bloodworth, J.; Zlobin, A.; Pannuti, A.; Green, A.; Ellis, I.; et al. PKCα attenuates Jagged-1-mediated Notch signaling in ErbB-2-positive breast cancer to reverse trastuzumab resistance. Clin. Cancer Res. 2015, 22, 175–186. [Google Scholar] [CrossRef]
- Yang, S.; Liu, Y.; Xia, B.; Deng, J.; Liu, T.; Li, Q.; Yang, Y.; Wang, Y.; Ning, X.; Zhang, Y.; et al. DLL4 as a predictor of pelvic lymph node metastasis and a novel prognostic biomarker in patients with early-stage cervical cancer. Tumor Biol. 2015, 37, 5063–5074. [Google Scholar] [CrossRef]
- Strati, T.-M.; Kotoula, V.; Kostopoulos, I.; Manousou, K.; Papadimitriou, C.; Lazaridis, G.; Lakis, S.; Pentheroudakis, G.; Pectasides, D.; Pazarli, E.; et al. Prognostic subcellular Notch2, Notch3 and Jagged1 localization patterns in early triple-negative breast cancer. Anticancer Res. 2017, 37, 2334. [Google Scholar] [CrossRef]
- Elzamly, S.; Badri, N.; Padilla, O.; Dwivedi, A.K.; Alvarado, L.A.; Hamilton, M.; Diab, N.; Rock, C.; Elfar, A.; Teleb, M.; et al. Epithelial-mesenchymal transition markers in breast cancer and pathological responseafter neoadjuvant chemotherapy. Breast Cancer Basic Clin. Res. 2018, 12, 1178223418788074. [Google Scholar] [CrossRef]
- Leong, K.G.; Niessen, K.; Kulic, I.; Raouf, A.; Eaves, C.; Pollet, I.; Karsan, A. Jagged1-mediated Notch activation induces epithelial-to-mesenchymal transition through Slug-induced repression of E-cadherin. J. Exp. Med. 2007, 204, 2935–2948. [Google Scholar] [CrossRef] [PubMed]
- Shao, S.; Zhao, X.; Zhang, X.; Luo, M.; Zuo, X.; Huang, S.; Wang, Y.; Gu, S.; Zhao, X. Notch1 signaling regulates the epithelial-mesenchymal transition and invasion of breast cancer in a Slug-dependent manner. Mol. Cancer 2015, 14, 28. [Google Scholar] [CrossRef]
- Sethi, N.; Dai, X.; Winter, C.G.; Kang, Y. Tumor-derived Jagged1 promotes osteolytic bone metastasis of breast cancer by engaging Notch signaling in bone cells. Cancer Cell 2011, 19, 192–205. [Google Scholar] [CrossRef]
- Tao, J.; Erez, A.; Lee, B.H. One NOTCH further: Jagged 1 in bone metastasis. Cancer Cell 2011, 19, 159–161. [Google Scholar] [CrossRef] [PubMed]
- Sahlgren, C.; Gustafsson, M.V.; Jin, S.; Poellinger, L.; Lendahl, U. Notch signaling mediates hypoxia-induced tumor cell migration and invasion. Proc. Natl. Acad. Sci. USA 2008, 105, 6392–6397. [Google Scholar] [CrossRef] [PubMed]
- Chen, J.; Imanaka, N.; Griffin, J.D. Hypoxia potentiates Notch signaling in breast cancer leading to decreased E-cadherin expression and increased cell migration and invasion. Br. J. Cancer 2009, 102, 351–360. [Google Scholar] [CrossRef] [PubMed]
- Mani, S.A.; Guo, W.; Liao, M.-J.; Eaton, E.N.; Ayyanan, A.; Zhou, A.Y.; Brooks, M.; Reinhard, F.; Zhang, C.C.; Shipitsin, M.; et al. The Epithelial-mesenchymal transition generates cells with properties of stem cells. Cell 2008, 133, 704–715. [Google Scholar] [CrossRef]
- Morel, A.-P.; Lièvre, M.; Thomas, C.; Hinkal, G.; Ansieau, S.; Puisieux, A. Generation of breast cancer stem cells through epithelial-mesenchymal transition. PLoS ONE 2008, 3, e2888. [Google Scholar] [CrossRef]
- Inan, S.; Hayran, M. Cell Signaling pathways related to epithelial mesenchymal transition in cancer metastasis. Crit. Rev. Oncog. 2019, 24, 47–54. [Google Scholar] [CrossRef]
- Liu, S.; Cong, Y.; Wang, N.; Sun, Y.; Deng, L.; Liu, Y.; Martin-Trevino, R.; Shang, L.; McDermott, S.P.; Landis, M.D.; et al. Breast cancer stem cells transition between epithelial and mesenchymal states reflective of their normal counterparts. Stem Cell Rep. 2013, 2, 78–91. [Google Scholar] [CrossRef]
- Luo, M.; Brooks, M.; Wicha, M.S.; Brooke, M. Epithelial-mesenchymal plasticity of breast cancer stem cells: Implications for metastasis and therapeutic resistance. Curr. Pharm. Des. 2015, 21, 1301–1310. [Google Scholar] [CrossRef] [PubMed]
- May, C.D.; Sphyris, N.; Evans, K.W.; Werden, S.J.; Guo, W.; Mani, S.A. Epithelial-mesenchymal transition and cancer stem cells: A dangerously dynamic duo in breast cancer progression. Breast Cancer Res. 2011, 13, 202. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.; Steeg, P.S.; Price, J.E.; Krishnamurthy, S.; Mani, S.A.; Reuben, J.; Cristofanilli, M.; Dontu, G.; Bidaut, L.M.; Valero, V.; et al. Breast cancer metastasis: Challenges and opportunities. Cancer Res. 2009, 69, 4951–4953. [Google Scholar] [CrossRef] [PubMed]
- Pannuti, A.; Foreman, K.; Rizzo, P.; Osipo, C.; Golde, T.; Osborne, B.; Miele, L. Targeting Notch to target cancer stem cells. Clin. Cancer Res. 2010, 16, 3141–3152. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Zhang, H.; Sun, X.-F.; Guo, X.-H.; He, Y.-N.; Cui, S.-D.; Fan, Q.-X. Tamoxifen-resistant breast cancer cells possess cancer stem-like cell properties. Chin. Med. J. 2013, 126, 3030–3034. [Google Scholar] [PubMed]
- Suman, S.; Das, T.P.; Damodaran, C. Silencing NOTCH signaling causes growth arrest in both breast cancer stem cells and breast cancer cells. Br. J. Cancer 2013, 109, 2587–2596. [Google Scholar] [CrossRef]
- Al-Hajj, M.; Wicha, M.S.; Benito-Hernandez, A.; Morrison, S.J.; Clarke, M.F. Prospective identification of tumorigenic breast cancer cells. Proc. Natl. Acad. Sci. USA 2003, 100, 3983–3988. [Google Scholar] [CrossRef]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Tarpin, C.; Diebel, M.; Esterni, B.; Houvenaeghel, G.; Extra, J.-M.; Bertucci, F.; Jacquemier, J.; et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin. Cancer Res. 2009, 16, 45–55. [Google Scholar] [CrossRef]
- Jolly, M.K.; Boareto, M.; Debeb, B.G.; Aceto, N.; Farach-Carson, M.C.; Woodward, W.A.; Levine, H. Inflammatory breast cancer: A model for investigating cluster-based dissemination. NPJ Breast Cancer 2017, 3, 21. [Google Scholar] [CrossRef]
- Vassalli, G. Aldehyde dehydrogenases: Not just markers, but functional regulators of stem cells. Stem Cells Int. 2019, 2019, 1–15. [Google Scholar] [CrossRef]
- Armstrong, L.; Stojkovic, M.; Dimmick, I.; Ahmad, S.; Stojković, P.; Hole, N.; Lako, M. Phenotypic characterization of murine primitive hematopoietic progenitor cells isolated on basis of aldehyde dehydrogenase activity. Stem Cells 2004, 22, 1142–1151. [Google Scholar] [CrossRef] [PubMed]
- Chute, J.P.; Muramoto, G.G.; Whitesides, J.; Colvin, M.; Safi, R.; Chao, N.J.; McDonnell, N.P. Inhibition of aldehyde dehydrogenase and retinoid signaling induces the expansion of human hematopoietic stem cells. Proc. Natl. Acad. Sci. USA 2006, 103, 11707–11712. [Google Scholar] [CrossRef] [PubMed]
- Corti, S.; Locatelli, F.; Papadimitriou, D.; Donadoni, C.; Salani, S.; Del Bo, R.; Strazzer, S.; Bresolin, N.; Comi, G.P. Identification of a primitive brain-derived neural stem cell population based on aldehyde dehydrogenase activity. Stem Cells 2006, 24, 975–985. [Google Scholar] [CrossRef] [PubMed]
- Storms, R.W.; Trujillo, A.P.; Springer, J.B.; Shah, L.; Colvin, O.M.; Ludeman, S.M.; Smith, C. Isolation of primitive human hematopoietic progenitors on the basis of aldehyde dehydrogenase activity. Proc. Natl. Acad. Sci. USA 1999, 96, 9118–9123. [Google Scholar] [CrossRef]
- Cheung, A.M.S.; Wan, T.S.K.; Leung, J.C.K.; Chan, L.Y.Y.; Huang, H.; Kwong, Y.L.; Liang, R.; Leung, A.Y.H. Aldehyde dehydrogenase activity in leukemic blasts defines a subgroup of acute myeloid leukemia with adverse prognosis and superior NOD/SCID engrafting potential. Leukemia 2007, 21, 1423–1430. [Google Scholar] [CrossRef]
- Matsui, W.; Wang, Q.; Barber, J.P.; Brennan, S.; Smith, B.D.; Borrello, I.; Mcniece, I.; Lin, L.; Ambinder, R.F.; Peacock, C.; et al. Clonogenic multiple myeloma progenitors, stem cell properties, and drug resistance. Cancer Res. 2008, 68, 190–197. [Google Scholar] [CrossRef]
- Ginestier, C.; Hur, M.H.; Charafe-Jauffret, E.; Monville, F.; Dutcher, J.; Brown, M.; Jacquemier, J.; Viens, P.; Kleer, C.G.; Liu, S.; et al. ALDH1 is a marker of normal and malignant human mammary stem cells and a predictor of poor clinical outcome. Cell Stem Cell 2007, 1, 555–567. [Google Scholar] [CrossRef]
- Balicki, D. Moving forward in human mammary stem cell biology and breast cancer prognostication using ALDH1. Cell Stem Cell 2007, 1, 485–487. [Google Scholar] [CrossRef]
- Vesuna, F.; Lisok, A.; Kimble, B.; Raman, V. Twist modulates breast cancer stem cells by transcriptional regulation of CD24 expression. Neoplasia 2009, 11, 1318–1328. [Google Scholar] [CrossRef]
- Yang, M.-H.; Hsu, D.S.-S.; Wang, H.-W.; Wang, H.-J.; Lan, H.-Y.; Yang, W.-H.; Huang, C.-H.; Kao, S.-Y.; Tzeng, C.-H.; Tai, S.-K.; et al. Bmi1 is essential in Twist1-induced epithelial–mesenchymal transition. Nat. Cell Biol. 2010, 12, 982–992. [Google Scholar] [CrossRef]
- Evdokimova, V.; Tognon, C.; Ng, T.; Ruzanov, P.; Melnyk, N.; Fink, D.; Sorokin, A.; Ovchinnikov, L.P.; Davicioni, E.; Triche, T.J.; et al. Translational activation of snail1 and other developmentally regulated transcription factors by YB-1 promotes an epithelial-mesenchymal transition. Cancer Cell 2009, 15, 402–415. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Ware, K.E.; Gilja, S.; Somarelli, J.A.; Levine, H. EMT and MET: Necessary or permissive for metastasis? Mol. Oncol. 2017, 11, 755–769. [Google Scholar] [CrossRef] [PubMed]
- Jolly, M.K.; Celia-Terrassa, T. Dynamics of phenotypic heterogeneity associated with EMT and stemness during cancer progression. J. Clin. Med. 2019, 8, 1542. [Google Scholar] [CrossRef] [PubMed]
- Dontu, G.; Al-Hajj, M.; Abdallah, W.M.; Clarke, M.F.; Wicha, M.S. Stem cells in normal breast development and breast cancer. Cell Prolif. 2003, 36, 59–72. [Google Scholar] [CrossRef] [PubMed]
- Reynolds, B.A.; Weiss, S. Clonal and population analyses demonstrate that an EGF-responsive mammalian embryonic CNS precursor is a stem cell. Dev. Biol. 1996, 175, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Dontu, G.; Jackson, K.W.; McNicholas, E.; Kawamura, M.J.; Abdallah, W.M.; Wicha, M.S. Role of Notch signaling in cell-fate determination of human mammary stem/progenitor cells. Breast Cancer Res. 2004, 6, 1–11. [Google Scholar] [CrossRef]
- Farnie, G.; Clarke, R. Mammary stem cells and breast cancer—Role of Notch signalling. Stem Cell Rev. Rep. 2007, 3, 169–175. [Google Scholar] [CrossRef]
- Farnie, G.; Clarke, R.; Spence, K.; Pinnock, N.; Brennan, K.; Anderson, N.G.; Bundred, N.J. Novel cell culture technique for primary ductal carcinoma in situ: Role of Notch and epidermal growth factor receptor signaling pathways. J. Natl. Cancer Inst. 2007, 99, 616–627. [Google Scholar] [CrossRef]
- Grudzien, P.; Lo, S.; Albain, K.S.; Robinson, P.; Rajan, P.; Strack, P.R.; Golde, T.E.; Miele, L.; Foreman, K.E. Inhibition of Notch signaling reduces the stem-like population of breast cancer cells and prevents mammosphere formation. Anticancer Res. 2010, 30, 3853–3867. [Google Scholar]
- Charafe-Jauffret, E.; Ginestier, C.; Iovino, F.; Wicinski, J.; Cervera, N.; Finetti, P.; Hur, M.-H.; Diebel, M.E.; Monville, F.; Dutcher, J.; et al. Breast cancer cell lines contain functional cancer stem cells with metastatic capacity and a distinct molecular signature. Cancer Res. 2009, 69, 1302–1313. [Google Scholar] [CrossRef]
- Hu, Y.; Smyth, G.K. ELDA: Extreme limiting dilution analysis for comparing depleted and enriched populations in stem cell and other assays. J. Immunol. Methods 2009, 347, 70–78. [Google Scholar] [CrossRef] [PubMed]
- Harrison, H.; Farnie, G.; Brennan, K.R.; Clarke, R. Breast cancer stem cells: Something out of Notching? Cancer Res. 2010, 70, 8973–8976. [Google Scholar] [CrossRef] [PubMed]
- Wong, N.; Fuller, M.; Sung, S.; Wong, F.; Karsan, A. Heterogeneity of breast cancer stem cells as evidenced with Notch-dependent and Notch-independent populations. Cancer Med. 2012, 1, 105–113. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Lewis, M.T.; Huang, J.; Gutierrez, C.; Osborne, C.K.; Wu, M.-F.; Hilsenbeck, S.G.; Pavlick, A.; Zhang, X.; Chamness, G.C.; et al. Intrinsic resistance of tumorigenic breast cancer cells to chemotherapy. J. Natl. Cancer Inst. 2008, 100, 672–679. [Google Scholar] [CrossRef] [PubMed]
- Tanei, T.; Morimoto, K.; Shimazu, K.; Kim, S.J.; Tanji, Y.; Taguchi, T.; Tamaki, Y.; Noguchi, S. Association of breast cancer stem cells identified by aldehyde dehydrogenase 1 expression with resistance to sequential paclitaxel and epirubicin-based chemotherapy for breast cancers. Clin. Cancer Res. 2009, 15, 4234–4241. [Google Scholar] [CrossRef]
- Simões, B.M.; O’Brien, C.S.; Eyre, R.; Silva, A.; Yu, L.; Sarmiento-Castro, A.; Alférez, D.G.; Spence, K.; Santiago-Gómez, A.; Chemi, F.; et al. Anti-estrogen Resistance in human breast tumors is driven by JAG1-NOTCH4-dependent cancer stem cell activity. Cell Rep. 2015, 12, 1968–1977. [Google Scholar] [CrossRef]
- Shah, D.; Wyatt, D.; Baker, A.T.; Simms, P.; Peiffer, D.S.; Fernandez, M.; Rakha, E.; Green, A.; Filipovic, A.; Miele, L.; et al. Inhibition of HER2 increases JAGGED1-dependent breast cancer stem cells: Role for membrane JAGGED1. Clin. Cancer Res. 2018, 24, 4566–4578. [Google Scholar] [CrossRef]
- Baker, A.; Wyatt, D.; Bocchetta, M.; Li, J.; Filipovic, A.; Green, A.; Peiffer, D.S.; Fuqua, S.; Miele, L.; Albain, K.S.; et al. Notch-1-PTEN-ERK1/2 signaling axis promotes HER2+ breast cancer cell proliferation and stem cell survival. Oncogene 2018, 37, 4489–4504. [Google Scholar] [CrossRef]
- Jordan, V.C.; Brodie, A. Development and evolution of therapies targeted to the estrogen receptor for the treatment and prevention of breast cancer. Steroids 2007, 72, 7–25. [Google Scholar] [CrossRef]
- Pearce, S.T.; Jordan, V. The biological role of estrogen receptors α and β in cancer. Crit. Rev. Oncol. 2004, 50, 3–22. [Google Scholar] [CrossRef]
- Brodie, A.M. Aromatase inhibition and its pharmacologic implications. Biochem. Pharmacol. 1985, 34, 3213–3219. [Google Scholar] [CrossRef]
- Johnston, S.; MacLennan, K.; Sacks, N.; Salter, J.; Smith, I.; Dowsett, M. Modulation of Bcl-2 and Ki-67 expression in oestrogen receptor-positive human breast cancer by tamoxifen. Eur. J. Cancer 1994, 30, 1663–1669. [Google Scholar] [CrossRef]
- Sutepvarnon, A.; Warnnissorn, M.; Srimuninnimit, V. Predictive value of Ki67 for adjuvant chemotherapy in node-negative, hormone receptor-positive breast cancer. J. Med. Assoc. Thail. 2013, 96 (Suppl. S2), S60–S66. [Google Scholar]
- Ellis, M.J.; Suman, V.J.; Hoog, J.; Goncalves, R.; Sanati, S.; Creighton, C.J.; DeSchryver, K.; Crouch, E.; Brink, A.; Watson, M.; et al. Ki67 proliferation index as a tool for chemotherapy decisions during and after neoadjuvant aromatase inhibitor treatment of breast cancer: Results from the american college of surgeons oncology group Z1031 trial (alliance). J. Clin. Oncol. 2017, 35, 1061–1069. [Google Scholar] [CrossRef]
- Haque, M.; Desai, K.V. Pathways to endocrine therapy resistance in breast cancer. Front. Endocrinol. 2019, 10, 573. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, C.; Patten, D.K.; Januszewski, A.; Zucchini, G.; Howell, S.J. Breast cancer: Current and future endocrine therapies. Mol. Cell. Endocrinol. 2014, 382, 695–723. [Google Scholar] [CrossRef] [PubMed]
- Rizzo, P.; Miao, H.; D’Souza, G.; Osipo, C.; Song, L.L.; Yun, J.; Zhao, H.; Mascarenhas, J.; Wyatt, D.; Antico, G.; et al. Cross-talk between notch and the estrogen receptor in breast cancer suggests novel therapeutic approaches. Cancer Res. 2008, 68, 5226–5235. [Google Scholar] [CrossRef] [PubMed]
- Acar, A.; Simões, B.M.; Clarke, R.; Brennan, K. A role for Notch signalling in breast cancer and endocrine resistance. Stem Cells Int. 2016, 2016, 1–6. [Google Scholar] [CrossRef]
- Fuqua, S.A.W.; Chamness, G.C.; McGuire, W.L. Estrogen receptor mutations in breast cancer. J. Cell. Biochem. 1993, 51, 135–139. [Google Scholar] [CrossRef]
- Gelsomino, L.; Panza, S.; Giordano, C.; Barone, I.; Gu, G.; Spina, E.; Catalano, S.; Fuqua, S.; Andò, S. Mutations in the estrogen receptor alpha hormone binding domain promote stem cell phenotype through notch activation in breast cancer cell lines. Cancer Lett. 2018, 428, 12–20. [Google Scholar] [CrossRef]
- Jelovac, D.; Wolff, A.C. The adjuvant treatment of HER2-positive breast cancer. Curr. Treat. Options Oncol. 2012, 13, 230–239. [Google Scholar] [CrossRef] [PubMed]
- Conte, B.; Fabi, A.; Poggio, F.; Blondeaux, E.; Dellepiane, C.; D’Alonzo, A.; Buono, G.; Arpino, G.; Magri, V.; Naso, G.; et al. T-DM1 efficacy in patients with HER2-positive metastatic breast cancer progressing after a taxane plus pertuzumab and trastuzumab: An Italian multicenter observational study. Clin. Breast Cancer 2020, 20, e181–e187. [Google Scholar] [CrossRef] [PubMed]
- Baselga, J. Mechanism of action of trastuzumab and scientific update. Semin. Oncol. 2001, 28, 4–11. [Google Scholar] [CrossRef]
- Xuhong, J.-C.; Qi, X.-W.; Zhang, Y.; Jiang, J. Mechanism, safety and efficacy of three tyrosine kinase inhibitors lapatinib, neratinib and pyrotinib in HER2-positive breast cancer. Am. J. Cancer Res. 2019, 9, 2103–2119. [Google Scholar] [PubMed]
- Pernas, S.; Tolaney, S.M. HER2-positive breast cancer: New therapeutic frontiers and overcoming resistance. Ther. Adv. Med. Oncol. 2019, 11, 1758835919833519. [Google Scholar] [CrossRef] [PubMed]
- Wong, A.L.A.; Lee, S.-C. Mechanisms of resistance to trastuzumab and novel therapeutic strategies in HER2-positive breast cancer. Int. J. Breast Cancer 2012, 2012, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Guo, S.; Liu, M.; Gonzalez-Perez, R.R. Role of Notch and its oncogenic signaling crosstalk in breast cancer. Biochim. Biophys. Acta 2011, 1815, 197–213. [Google Scholar] [CrossRef]
- Lindsay, J.; Jiao, X.; Sakamaki, T.; Casimiro, M.C.; Shirley, L.A.; Tran, T.H.; Ju, X.; Liu, M.; Li, Z.; Wang, C.; et al. ErbB2 induces Notch1 activity and function in breast cancer cells. Clin. Transl. Sci. 2008, 1, 107–115. [Google Scholar] [CrossRef]
- Farnie, G.; Willan, P.M.; Clarke, R.; Bundred, N.J. Combined inhibition of ErbB1/2 and Notch receptors effectively targets breast Ductal Carcinoma In Situ (DCIS) stem/progenitor cell activity regardless of ErbB2 status. PLoS ONE 2013, 8, e56840. [Google Scholar] [CrossRef]
- Osipo, C.; Patel, P.; Rizzo, P.; Clementz, A.G.; Hao, L.; Golde, T.E.; Miele, L. ErbB-2 inhibition activates Notch-1 and sensitizes breast cancer cells to a γ-secretase inhibitor. Oncogene 2008, 27, 5019–5032. [Google Scholar] [CrossRef]
- Magnifico, A.; Albano, L.; Campaner, S.; Delia, D.; Castiglioni, F.; Gasparini, P.; Sozzi, G.; Fontanella, E.; Ménard, S.; Tagliabue, E. Tumor-initiating cells of HER2-positive carcinoma cell lines express the highest oncoprotein levels and are sensitive to trastuzumab. Clin. Cancer Res. 2009, 15, 2010–2021. [Google Scholar] [CrossRef] [PubMed]
- Pandya, K.; Meeke, K.; Clementz, A.G.; Rogowski, A.; Roberts, J.; Miele, L.; Albain, K.S.; Osipo, C. Targeting both Notch and ErbB-2 signalling pathways is required for prevention of ErbB-2-positive breast tumour recurrence. Br. J. Cancer 2011, 105, 796–806. [Google Scholar] [CrossRef] [PubMed]
- Kondratyev, M.; Kreso, A.; Hallett, R.M.; Girgis-Gabardo, A.; Barcelon, M.E.; Ilieva, D.; Ware, C.; Majumder, P.K.; Hassell, J.A. Gamma-secretase inhibitors target tumor-initiating cells in a mouse model of ERBB2 breast cancer. Oncogene 2011, 31, 93–103. [Google Scholar] [CrossRef]
- Osipo, C.; Zlobin, A.; Olsauskas-Kuprys, R. Gamma secretase inhibitors of Notch signaling. OncoTargets Ther. 2013, 6, 943–955. [Google Scholar] [CrossRef]
- Takebe, N.; Nguyen, D.; Yang, S.X. Targeting Notch signaling pathway in cancer: Clinical development advances and challenges. Pharmacol. Ther. 2014, 141, 140–149. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Paranjape, A.N.; Rangarajan, A.; Dighe, R.R. A monoclonal antibody against human Notch1 ligand-binding domain depletes subpopulation of putative breast cancer stem-like cells. Mol. Cancer Ther. 2011, 11, 77–86. [Google Scholar] [CrossRef]
- Moellering, R.E.; Cornejo, M.; Davis, T.N.; Del Bianco, C.; Aster, J.C.; Blacklow, S.C.; Kung, A.L.; Gilliland, D.G.; Verdine, G.L.; Bradner, J.E. Direct inhibition of the NOTCH transcription factor complex. Nature 2009, 462, 182–188. [Google Scholar] [CrossRef]
- Astudillo, L.; Da Silva, T.G.; Wang, Z.; Han, X.; Jin, K.; VanWye, J.; Zhu, X.; Weaver, K.; Oashi, T.; Lopes, P.E.M.; et al. The small molecule IMR-1 inhibits the Notch transcriptional activation complex to suppress tumorigenesis. Cancer Res. 2016, 76, 3593–3603. [Google Scholar] [CrossRef]
- Yong, T.; Sun, A.; Henry, M.D.; Meyers, S.; Davis, J.N. Down regulation of CSL activity inhibits cell proliferation in prostate and breast cancer cells. J. Cell. Biochem. 2011, 112, 2340–2351. [Google Scholar] [CrossRef]
- Abdel-Rahman, N.; Martinez-Arias, A.; Blundell, T.L. Probing the druggability of protein–protein interactions: Targeting the Notch1 receptor ankyrin domain using a fragment-based approach. Biochem. Soc. Trans. 2011, 39, 1327–1333. [Google Scholar] [CrossRef][Green Version]
- Wilson, J.J.; Kovall, R.A. Crystal structure of the CSL-Notch-mastermind ternary complex bound to DNA. Cell 2006, 124, 985–996. [Google Scholar] [CrossRef] [PubMed]
- Albain, K.S.; Zlobin, A.Y.; Covington, K.R.; Gallahger, B.T.; Hilsenbeck, S.G.; Czerlanis, C.M.; Lo, S.; Robinson, P.A.; Gaynor, E.R.; Godellas, C.; et al. Abstract S4-03: Identification of a notch-driven breast cancer stem cell gene signature for anti-notch therapy in an ER+ presurgical window model. In Proceedings of the Thirty-Seventh Annual CTRC-AACR San Antonio Breast Cancer Symposium, San Antonio, TX, USA, 9–13 December 2014; Volume 75. [Google Scholar]
- Peiffer, D.S.; Wyatt, D.; Zlobin, A.; Piracha, A.; Ng, J.; Dingwall, A.K.; Albain, K.S.; Osipo, C.; Ng, J. DAXX suppresses tumor-initiating cells in estrogen receptor–positive breast cancer following endocrine therapy. Cancer Res. 2019, 79, 4965–4977. [Google Scholar] [CrossRef] [PubMed]
- Peiffer, D.S.; Ma, E.; Wyatt, D.; Albain, K.S.; Osipo, C. DAXX-inducing phytoestrogens inhibit ER+ tumor initiating cells and delay tumor development. NPJ Breast Cancer 2020, 6, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Trimble, M.S.; Xin, J.H.; Guy, C.T.; Muller, W.J.; Hassell, J.A. PEA3 is overexpressed in mouse metastatic mammary adenocarcinomas. Oncogene 1993, 8, 3037–3042. [Google Scholar] [PubMed]
- Clementz, A.G.; Rogowski, A.; Pandya, K.; Miele, L.; Osipo, C. NOTCH-1 and NOTCH-4 are novel gene targets of PEA3 in breast cancer: Novel therapeutic implications. Breast Cancer Res. 2011, 13, R63. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
BeLow, M.; Osipo, C. Notch Signaling in Breast Cancer: A Role in Drug Resistance. Cells 2020, 9, 2204. https://doi.org/10.3390/cells9102204
BeLow M, Osipo C. Notch Signaling in Breast Cancer: A Role in Drug Resistance. Cells. 2020; 9(10):2204. https://doi.org/10.3390/cells9102204
Chicago/Turabian StyleBeLow, McKenna, and Clodia Osipo. 2020. "Notch Signaling in Breast Cancer: A Role in Drug Resistance" Cells 9, no. 10: 2204. https://doi.org/10.3390/cells9102204
APA StyleBeLow, M., & Osipo, C. (2020). Notch Signaling in Breast Cancer: A Role in Drug Resistance. Cells, 9(10), 2204. https://doi.org/10.3390/cells9102204




