Interplay between the Ubiquitin Proteasome System and Ubiquitin-Mediated Autophagy in Plants
Abstract
1. Introduction
2. Ubiquitin Proteasome System
2.1. Ubiquitin-Proteasome System
2.2. Enzymes of the UPS
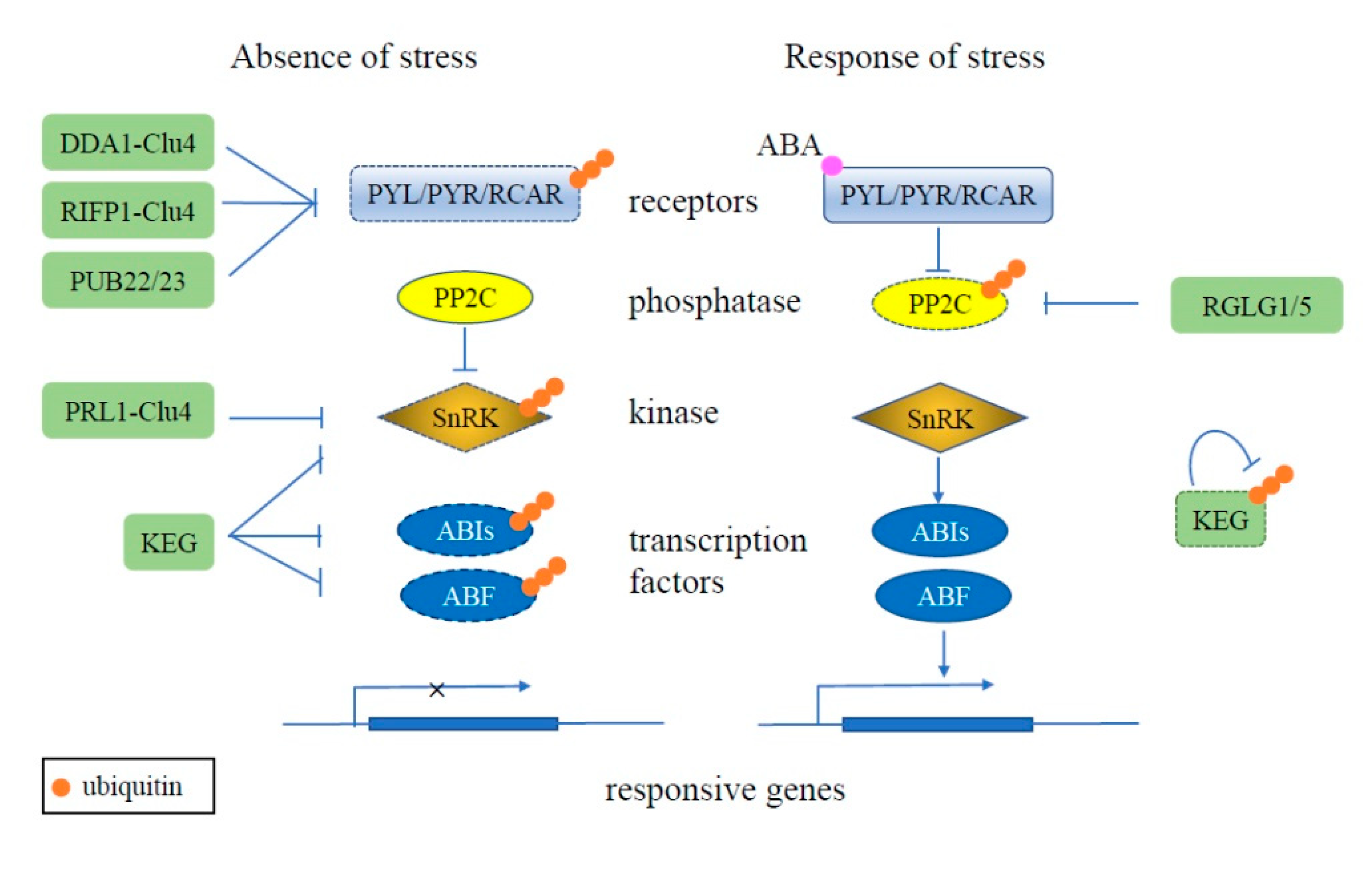
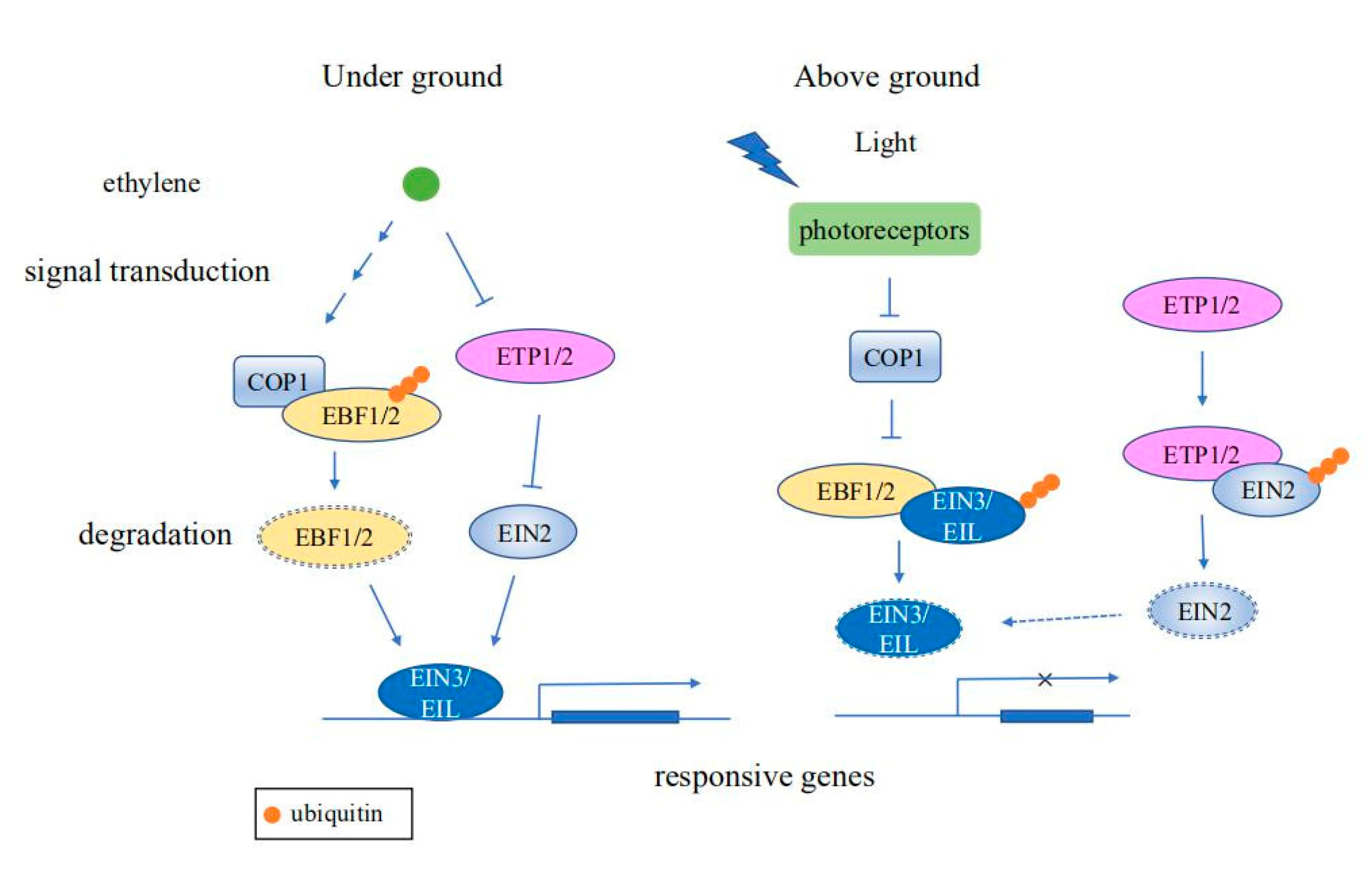
3. The Role of the UPS in Plant Stress Response
4. Ubiquitin-Mediated Regulation of Autophagy in Plants
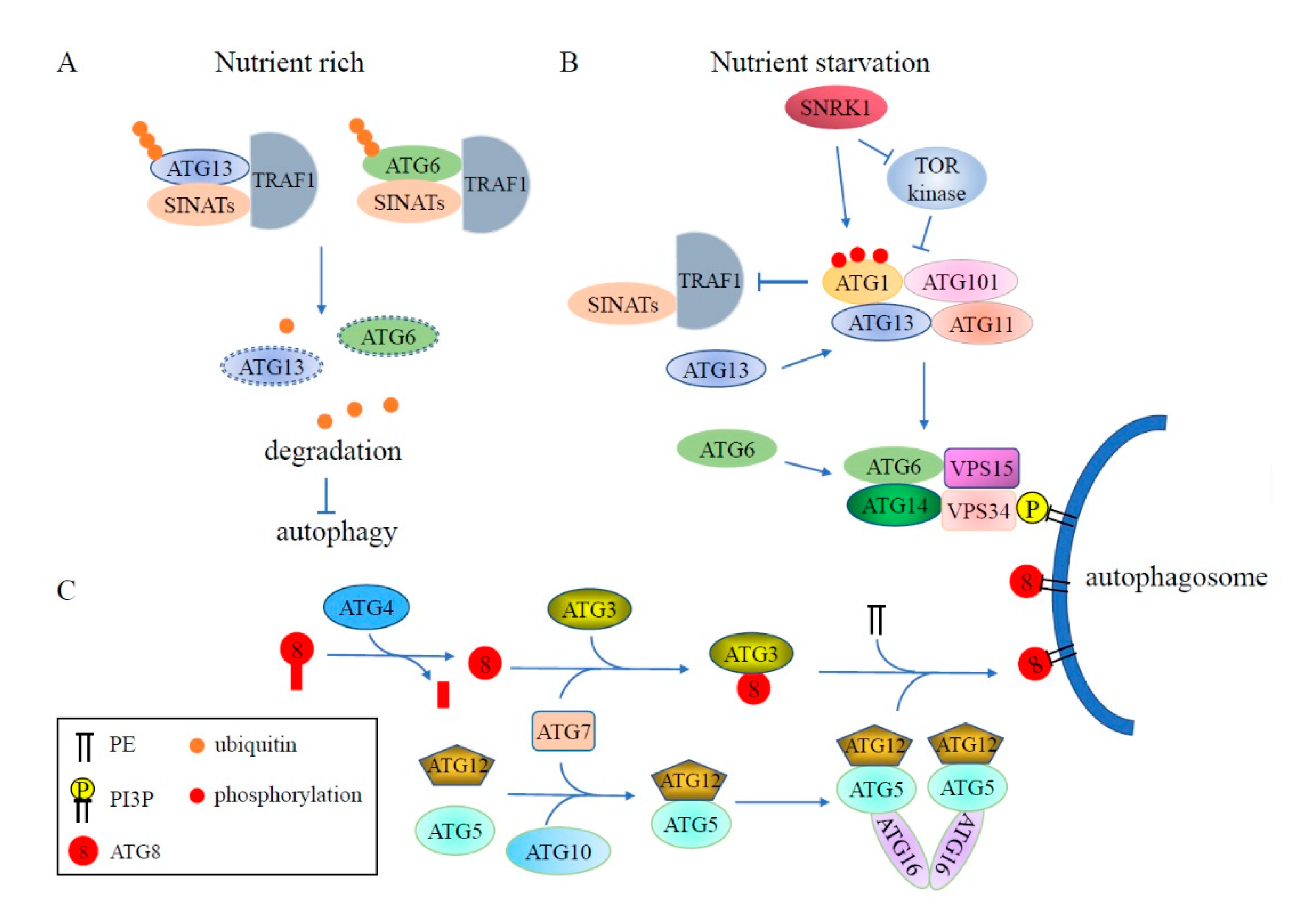
4.1. Regulation of Autophagy by Ubiquitination
4.2. Ubiquitin-Like Systems in Autophagy
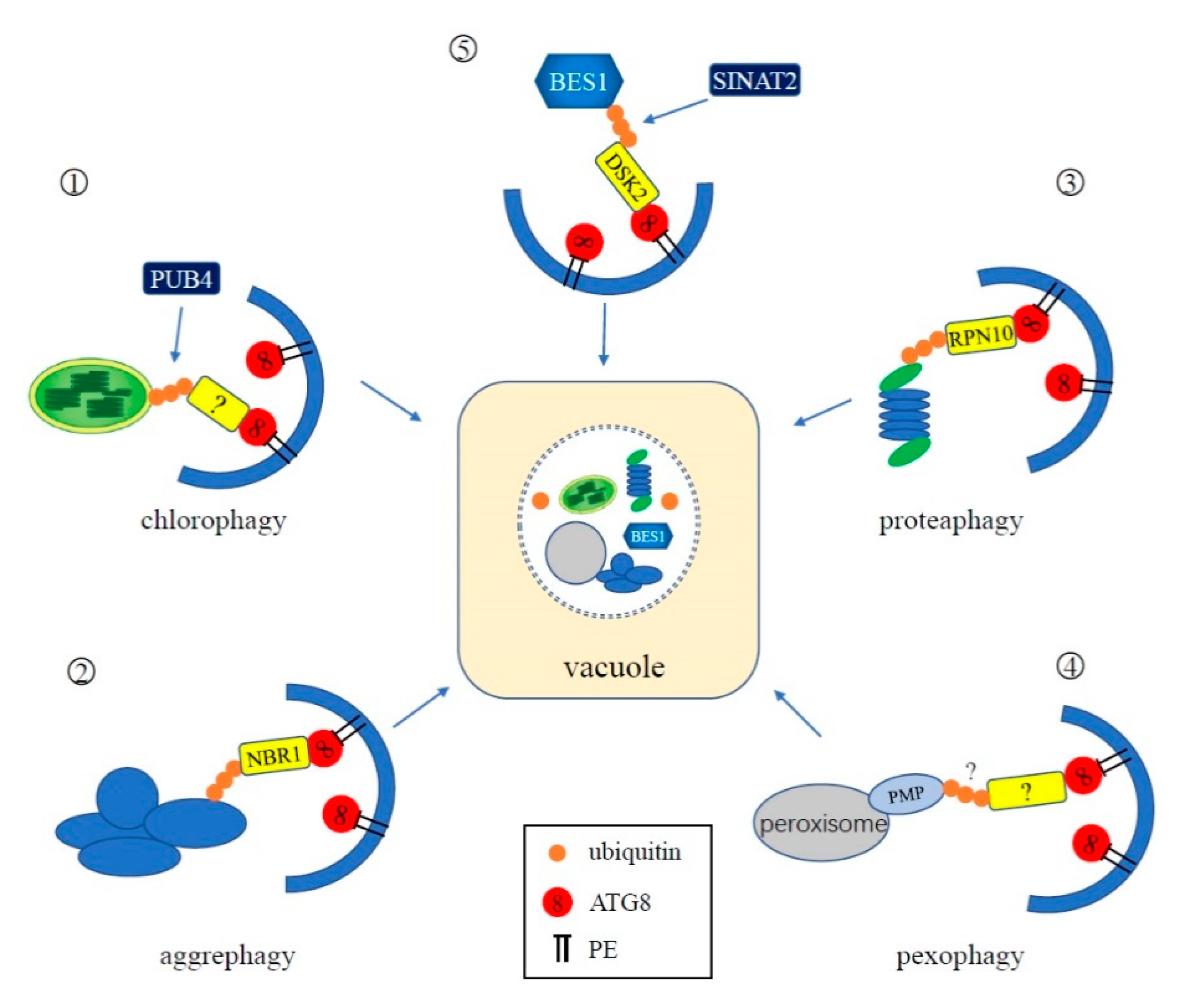
4.3. Ubiquitination in Selective Autophagy
4.3.1. Chlorophagy
4.3.2. Aggrephagy
4.3.3. Proteaphagy
4.3.4. Pexophagy
4.3.5. Others
5. Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Smalle, J.; Vierstra, R.D. The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 2004, 55, 555–590. [Google Scholar] [CrossRef] [PubMed]
- Balchin, D.; Hayer-Hartl, M.; Hartl, F.U. In vivo aspects of protein folding and quality control. Science 2016, 353, aac4354. [Google Scholar] [CrossRef] [PubMed]
- Xu, F.Q.; Xue, H.W. The ubiquitin-proteasome system in plant responses to environments. Plant Cell Environ. 2019, 42, 2931–2944. [Google Scholar] [CrossRef] [PubMed]
- Zientara-Rytter, K.; Subramani, S. The roles of ubiquitin-binding protein shuttles in the degradative fate of ubiquitinated proteins in the ubiquitin-proteasome system and autophagy. Cells 2019, 8, 40. [Google Scholar] [CrossRef] [PubMed]
- Dikic, I. Proteasomal and autophagic degradation systems. Annu. Rev. Biochem. 2017, 86, 193–224. [Google Scholar] [CrossRef] [PubMed]
- Avin-Wittenberg, T. Autophagy and its role in plant abiotic stress management. Plant Cell Environ. 2019, 42, 1045–1053. [Google Scholar] [CrossRef]
- Rabinowitz, J.D.; White, E. Autophagy and metabolism. Science 2010, 330, 1344–1348. [Google Scholar] [CrossRef]
- Marshall, R.S.; Vierstra, R.D. Autophagy: The master of bulk and selective recycling. Annu. Rev. Plant Biol. 2018, 69, 173–208. [Google Scholar] [CrossRef]
- Stone, S.L. Role of the ubiquitin proteasome system in plant response to abiotic stress. Int. Rev. Cell Mol. Biol. 2019, 343, 65–110. [Google Scholar]
- Ciechanover, A.; Heller, H.; Elias, S.; Haas, A.L.; Hershko, A. ATP-dependent conjugation of reticulocyte proteins with the polypeptide required for protein degradation. Proc. Natl. Acad. Sci. USA 1980, 77, 1365–1368. [Google Scholar] [CrossRef]
- Hershko, A.; Ciechanover, A.; Heller, H.; Haas, A.L.; Rose, I.A. Proposed role of ATP in protein breakdown: Conjugation of protein with multiple chains of the polypeptide of ATP-dependent proteolysis. Proc. Natl. Acad. Sci. USA 1980, 77, 1783–1786. [Google Scholar] [CrossRef] [PubMed]
- Kwon, Y.T.; Ciechanover, A. The ubiquitin code in the ubiquitin-proteasome system and autophagy. Trends Biochem. Sci. 2017, 42, 873–886. [Google Scholar] [CrossRef] [PubMed]
- Braten, O.; Livneh, I.; Ziv, T.; Admon, A.; Kehat, I.; Caspi, L.H.; Gonen, H.; Bercovich, B.; Godzik, A.; Jahandideh, S.; et al. Numerous proteins with unique characteristics are degraded by the 26S proteasome following monoubiquitination. Proc. Natl. Acad. Sci. USA 2016, 113, E4639–E4647. [Google Scholar] [CrossRef] [PubMed]
- Kim, D.Y.; Scalf, M.; Smith, L.M.; Vierstra, R.D. Advanced proteomic analyses yield a deep catalog of ubiquitylation targets in Arabidopsis. Plant Cell 2013, 25, 1523–1540. [Google Scholar] [CrossRef] [PubMed]
- Maor, R.; Jones, A.; Nühse, T.S.; Studholme, D.J.; Peck, S.C.; Shirasu, K. Multidimensional protein identification technology (MudPIT) analysis of ubiquitinated proteins in plants. Mol. Cell Proteom. 2007, 6, 601–610. [Google Scholar] [CrossRef] [PubMed]
- Komander, D. Mechanism, specificity and structure of the deubiquitinases. Subcell Biochem. 2010, 54, 69–87. [Google Scholar]
- Hatfield, P.M.; Gosink, M.M.; Carpenter, T.B.; Vierstra, R.D. The ubiquitin-activating enzyme (E1) gene family in Arabidopsis thaliana. Plant J. 1997, 11, 213–226. [Google Scholar] [CrossRef]
- Kraft, E.; Stone, S.L.; Ma, L.; Su, N.; Gao, Y.; Lau, O.S.; Deng, X.W.; Callis, J. Genome analysis and functional characterization of the E2 and RING-type E3 ligase ubiquitination enzymes of Arabidopsis. Plant Physiol. 2005, 139, 1597–1611. [Google Scholar] [CrossRef]
- Goritschnig, S.; Zhang, Y.; Li, X. The ubiquitin pathway is required for innate immunity in Arabidopsis. Plant J. 2007, 49, 540–551. [Google Scholar] [CrossRef]
- Wenzel, D.M.; Stoll, K.E.; Klevit, R.E. E2s: Structurally economical and functionally replete. Biochem. J. 2011, 433, 31–42. [Google Scholar] [CrossRef]
- Yanagawa, Y.; Sullivan, J.A.; Komatsu, S.; Gusmaroli, G.; Suzuki, G.; Yin, J.; Ishibashi, T.; Saijo, Y.; Rubio, V.; Kimura, S.; et al. Arabidopsis COP10 forms a complex with DDB1 and DET1 in vivo and enhances the activity of ubiquitin conjugating enzymes. Genes Dev. 2004, 18, 2172–2181. [Google Scholar] [CrossRef] [PubMed]
- Stone, S.L.; Hauksdóttir, H.; Troy, A.; Herschleb, J.; Kraft, E.; Callis, J. Functional analysis of the RING-type ubiquitin ligase family of Arabidopsis. Plant Physiol. 2005, 137, 13–30. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Kim, W.T. Regulation of abiotic stress signal transduction by E3 ubiquitin ligases in Arabidopsis. Mol. Cells 2011, 31, 201–208. [Google Scholar] [CrossRef]
- Mizoi, J.; Kanazawa, N.; Kidokoro, S.; Takahashi, F.; Qin, F.; Morimoto, K.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Heat-induced inhibition of phosphorylation of the stress-protective transcription factor DREB2A promotes thermotolerance of Arabidopsis thaliana. J. Biol. Chem. 2019, 294, 902–917. [Google Scholar] [CrossRef]
- Qin, F.; Sakuma, Y.; Tran, L.S.; Maruyama, K.; Kidokoro, S.; Fujita, Y.; Fujita, M.; Umezawa, T.; Sawano, Y.; Miyazono, K.; et al. Arabidopsis DREB2A-interacting proteins function as RING E3 ligases and negatively regulate plant drought stress-responsive gene expression. Plant Cell 2008, 20, 1693–1707. [Google Scholar] [CrossRef] [PubMed]
- Sakuma, Y.; Maruyama, K.; Qin, F.; Osakabe, Y.; Shinozaki, K.; Yamaguchi-Shinozaki, K. Dual function of an Arabidopsis transcription factor DREB2A in water-stress-responsive and heat-stress-responsive gene expression. Proc. Natl. Acad. Sci. USA 2006, 103, 18822–18827. [Google Scholar] [CrossRef] [PubMed]
- Kang, M.; Fokar, M.; Abdelmageed, H.; Allen, R.D. Arabidopsis SAP5 functions as a positive regulator of stress responses and exhibits E3 ubiquitin ligase activity. Plant Mol. Biol. 2011, 75, 451–466. [Google Scholar] [CrossRef]
- Chinnusamy, V.; Ohta, M.; Kanrar, S.; Lee, B.H.; Hong, X.; Agarwal, M.; Zhu, J.K. ICE1: A regulator of cold-induced transcriptome and freezing tolerance in Arabidopsis. Genes Dev. 2003, 17, 1043–1054. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Z.; Xie, S.; Si, T.; Li, Y.; Zhu, J.K. Mutational evidence for the critical role of CBF transcription factors in cold acclimation in Arabidopsis. Plant Physiol. 2016, 171, 2744–2759. [Google Scholar]
- Ding, Y.; Li, H.; Zhang, X.; Xie, Q.; Gong, Z.; Yang, S. OST1 kinase modulates freezing tolerance by enhancing ICE1 stability in Arabidopsis. Dev. Cell 2015, 32, 278–289. [Google Scholar] [CrossRef]
- Cutler, S.R.; Rodriguez, P.L.; Finkelstein, R.R.; Abrams, S.R. Abscisic acid: Emergence of a core signaling network. Annu. Rev. Plant Biol. 2010, 61, 651–679. [Google Scholar] [CrossRef] [PubMed]
- Irigoyen, M.L.; Iniesto, E.; Rodriguez, L.; Puga, M.I.; Yanagawa, Y.; Pick, E.; Strickland, E.; Paz-Ares, J.; Wei, N.; De Jaeger, G.; et al. Targeted degradation of abscisic acid receptors is mediated by the ubiquitin ligase substrate adaptor DDA1 in Arabidopsis. Plant Cell 2014, 26, 712–728. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Zhang, L.; Li, D.; Liu, Z.; Wang, J.; Li, X.; Yang, Y. The Arabidopsis F-box E3 ligase RIFP1 plays a negative role in abscisic acid signalling by facilitating ABA receptor RCAR3 degradation. Plant Cell Environ. 2016, 39, 571–582. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Zhao, L.; Zhang, M.; Zafar, S.A.; Fang, J.; Li, M.; Zhang, W.; Li, X. Arabidopsis E3 ubiquitin ligases PUB22 and PUB23 negatively regulate drought tolerance by targeting ABA receptor PYL9 for degradation. Int. J. Mol. Sci. 2017, 18, 1841. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.H.; Terzaghi, W.; Gusmaroli, G.; Charron, J.B.; Yoon, H.J.; Chen, H.; He, Y.J.; Xiong, Y.; Deng, X.W. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell 2008, 20, 152–167. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Liu, H.; Schofield, A.; Muise-Hennessey, A.; Stone, S.L. Arabidopsis CIPK26 interacts with KEG, components of the ABA signalling network and is degraded by the ubiquitin-proteasome system. J. Exp. Bot. 2013, 64, 2779–2791. [Google Scholar] [CrossRef] [PubMed]
- Liu, H.; Stone, S.L. Cytoplasmic degradation of the Arabidopsis transcription factor abscisic acid insensitive 5 is mediated by the RING-type E3 ligase KEEP ON GOING. J. Biol. Chem. 2013, 288, 20267–20279. [Google Scholar] [CrossRef]
- Stone, S.L.; Williams, L.A.; Farmer, L.M.; Vierstra, R.D.; Callis, J. KEEP ON GOING, a RING E3 ligase essential for Arabidopsis growth and development, is involved in abscisic acid signaling. Plant Cell 2006, 18, 3415–3428. [Google Scholar] [CrossRef]
- Chen, Y.T.; Liu, H.; Stone, S.; Callis, J. ABA and the ubiquitin E3 ligase KEEP ON GOING affect proteolysis of the Arabidopsis thaliana transcription factors ABF1 and ABF3. Plant J. 2013, 75, 965–976. [Google Scholar] [CrossRef]
- Ma, Y.; Szostkiewicz, I.; Korte, A.; Moes, D.; Yang, Y.; Christmann, A.; Grill, E. Regulators of PP2C phosphatase activity function as abscisic acid sensors. Science 2009, 324, 1064–1068. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, X.; Peirats-Llobet, M.; Belda-Palazon, B.; Wang, X.; Cui, S.; Yu, X.; Rodriguez, P.L.; An, C. Ubiquitin ligases RGLG1 and RGLG5 regulate abscisic acid signaling by controlling the turnover of phosphatase PP2CA. Plant Cell 2016, 28, 2178–2196. [Google Scholar] [CrossRef] [PubMed]
- Lyzenga, W.J.; Sullivan, V.; Liu, H.; Stone, S.L. The kinase activity of calcineurin B-like interacting protein kinase 26 (CIPK26) influences its own stability and that of the ABA-regulated ubiquitin ligase, Keep on Going (KEG). Front. Plant Sci. 2017, 8, 502. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.; Lu, X.; Ma, B.; Chen, S.-Y.; Zhang, J.-S. Ethylene signaling in rice and Arabidopsis: Conserved and diverged aspects. Mol. Plant 2015, 8, 495–505. [Google Scholar] [CrossRef] [PubMed]
- Dolgikh, V.A.; Pukhovaya, E.M.; Zemlyanskaya, E.V. Shaping ethylene response: The role of EIN3/EIL1 transcription factors. Front. Plant Sci. 2019, 10, 1030. [Google Scholar] [CrossRef]
- An, F.; Zhao, Q.; Ji, Y.; Li, W.; Jiang, Z.; Yu, X.; Zhang, C.; Han, Y.; He, W.; Liu, Y.; et al. Ethylene-induced stabilization of ETHYLENE INSENSITIVE3 and EIN3-LIKE1 is mediated by proteasomal degradation of EIN3 binding F-box 1 and 2 that requires EIN2 in Arabidopsis. Plant Cell 2010, 22, 2384–2401. [Google Scholar] [CrossRef]
- Shi, H.; Liu, R.; Xue, C.; Shen, X.; Wei, N.; Deng, X.W.; Zhong, S. Seedlings transduce the depth and mechanical pressure of covering soil using COP1 and ethylene to regulate EBF1/EBF2 for soil emergence. Curr. Biol. 2016, 26, 139–149. [Google Scholar] [CrossRef]
- Podolec, R.; Ulm, R. Photoreceptor-mediated regulation of the COP1/SPA E3 ubiquitin ligase. Curr. Opin. Plant Biol. 2018, 45, 18–25. [Google Scholar] [CrossRef]
- Qiao, H.; Chang, K.N.; Yazaki, J.; Ecker, J.R. Interplay between ethylene, ETP1/ETP2 F-box proteins, and degradation of EIN2 triggers ethylene responses in Arabidopsis. Genes Dev. 2009, 23, 512–521. [Google Scholar] [CrossRef]
- Vain, T.; Raggi, S.; Ferro, N.; Barange, D.K.; Kieffer, M.; Ma, Q.; Doyle, S.M.; Thelander, M.; Pařízková, B.; Novák, O.; et al. Selective auxin agonists induce specific AUX/IAA protein degradation to modulate plant development. Proc. Natl. Acad. Sci. USA 2019, 116, 6463–6472. [Google Scholar] [CrossRef]
- Su, T.; Li, X.; Yang, M.; Shao, Q.; Zhao, Y.; Ma, C.; Wang, P. Autophagy: An intracellular degradation pathway regulating plant survival and stress response. Front. Plant Sci. 2020, 11, 164. [Google Scholar] [CrossRef]
- van Doorn, W.G.; Papini, A. Ultrastructure of autophagy in plant cells. Autophagy 2013, 9, 1922–1936. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Abeliovich, H.; Agostinis, P.; Agrawal, D.K.; Aliev, G.; Askew, D.S.; Baba, M.; Baehrecke, E.H.; Bahr, B.A.; Ballabio, A.; et al. Guidelines for the use and interpretation of assays for monitoring autophagy in higher eukaryotes. Autophagy 2008, 4, 151–175. [Google Scholar] [CrossRef] [PubMed]
- Korolchuk, V.I.; Menzies, F.M.; Rubinsztein, D.C. Mechanisms of cross-talk between the ubiquitin-proteasome and autophagy-lysosome systems. FEBS Lett. 2010, 584, 1393–1398. [Google Scholar] [CrossRef]
- Streich, F.C., Jr.; Lima, C.D. Structural and functional insights to ubiquitin-like protein conjugation. Annu. Rev. Biophys. 2014, 43, 357–379. [Google Scholar] [CrossRef] [PubMed]
- Suttangkakul, A.; Li, F.; Chung, T.; Vierstra, R.D. The ATG1/ATG13 protein kinase complex is both a regulator and a target of autophagic recycling in Arabidopsis. Plant Cell 2011, 23, 3761–3779. [Google Scholar] [CrossRef] [PubMed]
- Qi, H.; Li, J.; Xia, F.N.; Chen, J.Y.; Lei, X.; Han, M.Q.; Xie, L.J.; Zhou, Q.M.; Xiao, S. Arabidopsis SINAT proteins control autophagy by mediating ubiquitylation and degradation of ATG13. Plant Cell 2020, 32, 263–284. [Google Scholar] [CrossRef]
- Qi, H.; Xia, F.-N.; Xiao, S. Autophagy in plants: Physiological roles and post-translational regulation. J. Integr. Plant Biol. 2020. [Google Scholar] [CrossRef]
- Qi, H.; Xia, F.N.; Xie, L.J.; Yu, L.J.; Chen, Q.F.; Zhuang, X.H.; Wang, Q.; Li, F.; Jiang, L.; Xie, Q.; et al. TRAF family proteins regulate autophagy dynamics by modulating AUTOPHAGY PROTEIN6 stability in Arabidopsis. Plant Cell 2017, 29, 890–911. [Google Scholar] [CrossRef]
- Xia, F.N.; Zeng, B.; Liu, H.S.; Qi, H.; Xie, L.J.; Yu, L.J.; Chen, Q.F.; Li, J.F.; Chen, Y.Q.; Jiang, L.; et al. SINAT E3 ubiquitin ligases mediate FREE1 and VPS23A degradation to modulate abscisic acid signalling. Plant Cell 2020. [Google Scholar] [CrossRef]
- Xiao, Z.; Yang, C.; Liu, C.; Yang, L.; Yang, S.; Zhou, J.; Li, F.; Jiang, L.; Xiao, S.; Gao, C.; et al. SINAT E3 ligases regulate the stability of the ESCRT component FREE1 in response to iron deficiency in plants. J. Integr. Plant Biol. 2020, 62, 1399–1417. [Google Scholar] [CrossRef]
- Ohsumi, Y. Molecular dissection of autophagy: Two ubiquitin-like systems. Nat. Rev. Mol. Cell Biol. 2001, 2, 211–216. [Google Scholar] [CrossRef] [PubMed]
- Chung, T.; Phillips, A.R.; Vierstra, R.D. ATG8 lipidation and ATG8-mediated autophagy in Arabidopsis require ATG12 expressed from the differentially controlled ATG12A and ATG12B loci. Plant J. 2010, 62, 483–493. [Google Scholar] [CrossRef] [PubMed]
- Yoshimoto, K.; Hanaoka, H.; Sato, S.; Kato, T.; Tabata, S.; Noda, T.; Ohsumi, Y. Processing of ATG8s, ubiquitin-like proteins, and their deconjugation by ATG4s are essential for plant autophagy. Plant Cell 2004, 16, 2967–2983. [Google Scholar] [CrossRef] [PubMed]
- Doelling, J.H.; Walker, J.M.; Friedman, E.M.; Thompson, A.R.; Vierstra, R.D. The APG8/12-activating enzyme APG7 is required for proper nutrient recycling and senescence in Arabidopsis thaliana. J. Biol. Chem. 2002, 277, 33105–33114. [Google Scholar] [CrossRef] [PubMed]
- Ichimura, Y.; Kirisako, T.; Takao, T.; Satomi, Y.; Shimonishi, Y.; Ishihara, N.; Mizushima, N.; Tanida, I.; Kominami, E.; Ohsumi, M.; et al. A ubiquitin-like system mediates protein lipidation. Nature 2000, 408, 488–492. [Google Scholar] [CrossRef] [PubMed]
- Klionsky, D.J.; Schulman, B.A. Dynamic regulation of macroautophagy by distinctive ubiquitin-like proteins. Nat. Struct. Mol. Biol. 2014, 21, 336–345. [Google Scholar] [CrossRef]
- Phillips, A.R.; Suttangkakul, A.; Vierstra, R.D. The ATG12-conjugating enzyme ATG10 is essential for autophagic vesicle formation in Arabidopsis thaliana. Genetics 2008, 178, 1339–1353. [Google Scholar] [CrossRef]
- Thompson, A.R.; Doelling, J.H.; Suttangkakul, A.; Vierstra, R.D. Autophagic nutrient recycling in Arabidopsis directed by the ATG8 and ATG12 conjugation pathways. Plant Physiol. 2005, 138, 2097–2110. [Google Scholar] [CrossRef]
- Bu, F.; Yang, M.; Guo, X.; Huang, W.; Chen, L. Multiple functions of ATG8 family proteins in plant autophagy. Front. Cell Dev. Biol. 2020, 8, 466. [Google Scholar] [CrossRef]
- Jarvis, P.; López-Juez, E. Biogenesis and homeostasis of chloroplasts and other plastids. Nat. Rev. Mol. Cell Biol. 2013, 14, 787–802. [Google Scholar] [CrossRef]
- Izumi, M.; Ishida, H.; Nakamura, S.; Hidema, J. Entire photodamaged chloroplasts are transported to the central vacuole by autophagy. Plant Cell 2017, 29, 377–394. [Google Scholar] [CrossRef] [PubMed]
- Havé, M.; Marmagne, A.; Chardon, F.; Masclaux-Daubresse, C. Nitrogen remobilization during leaf senescence: Lessons from Arabidopsis to crops. J. Exp. Bot. 2017, 68, 2513–2529. [Google Scholar] [PubMed]
- Woodson, J.D.; Joens, M.S.; Sinson, A.B.; Gilkerson, J.; Salomé, P.A.; Weigel, D.; Fitzpatrick, J.A.; Chory, J. Ubiquitin facilitates a quality-control pathway that removes damaged chloroplasts. Science 2015, 350, 450–454. [Google Scholar] [CrossRef]
- Kikuchi, Y.; Nakamura, S.; Woodson, J.D.; Ishida, H.; Ling, Q.; Hidema, J.; Jarvis, R.P.; Hagihara, S.; Izumi, M. Chloroplast autophagy and ubiquitination combine to manage oxidative damage and starvation responses. Plant Physiol. 2020, 183, 1531–1544. [Google Scholar] [CrossRef] [PubMed]
- Ling, Q.; Huang, W.; Baldwin, A.; Jarvis, P. Chloroplast biogenesis is regulated by direct action of the ubiquitin-proteasome system. Science 2012, 338, 655–659. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296. [Google Scholar] [CrossRef] [PubMed]
- Shaid, S.; Brandts, C.H.; Serve, H.; Dikic, I. Ubiquitination and selective autophagy. Cell Death Differ. 2013, 20, 21–30. [Google Scholar] [CrossRef]
- Svenning, S.; Lamark, T.; Krause, K.; Johansen, T. Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 2011, 7, 993–1010. [Google Scholar] [CrossRef]
- Jung, H.; Lee, H.N.; Marshall, R.S.; Lomax, A.W.; Yoon, M.J.; Kim, J.; Kim, J.H.; Vierstra, R.D.; Chung, T. Arabidopsis cargo receptor NBR1 mediates selective autophagy of defective proteins. J. Exp. Bot. 2020, 71, 73–89. [Google Scholar] [CrossRef]
- Zhou, J.; Zhang, Y.; Qi, J.; Chi, Y.; Fan, B.; Yu, J.Q.; Chen, Z. E3 ubiquitin ligase CHIP and NBR1-mediated selective autophagy protect additively against proteotoxicity in plant stress responses. PLoS Genet. 2014, 10, e1004116. [Google Scholar] [CrossRef]
- Tarnowski, L.; Rodriguez, M.C.; Brzywczy, J.; Piecho-Kabacik, M.; Krčkova, Z.; Martinec, J.; Wawrzynska, A.; Sirko, A. A selective autophagy cargo receptor NBR1 modulates abscisic acid signalling in Arabidopsis thaliana. Sci. Rep. 2020, 10, 7778. [Google Scholar] [CrossRef] [PubMed]
- Livneh, I.; Cohen-Kaplan, V.; Cohen-Rosenzweig, C.; Avni, N.; Ciechanover, A. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 2016, 26, 869–885. [Google Scholar] [CrossRef] [PubMed]
- Marshall, R.S.; McLoughlin, F.; Vierstra, R.D. Autophagic turnover of inactive 26S proteasomes in yeast is directed by the ubiquitin receptor Cue5 and the Hsp42 chaperone. Cell Rep. 2016, 16, 1717–1732. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Kaplan, V.; Livneh, I.; Avni, N.; Fabre, B.; Ziv, T.; Kwon, Y.T.; Ciechanover, A. p62- and ubiquitin-dependent stress-induced autophagy of the mammalian 26S proteasome. Proc. Natl. Acad. Sci. USA 2016, 113, E7490–E7499. [Google Scholar] [CrossRef]
- Marshall, R.S.; Li, F.; Gemperline, D.C.; Book, A.J.; Vierstra, R.D. Autophagic degradation of the 26S proteasome is mediated by the dual ATG8/ubiquitin receptor RPN10 in Arabidopsis. Mol. Cell 2015, 58, 1053–1066. [Google Scholar] [CrossRef]
- Pan, R.; Liu, J.; Hu, J. Peroxisomes in plant reproduction and seed-related development. Integr. Plant Biol. 2019, 61, 784–802. [Google Scholar] [CrossRef]
- Su, T.; Wang, P.; Li, H.; Zhao, Y.; Lu, Y.; Dai, P.; Ren, T.; Wang, X.; Li, X.; Shao, Q.; et al. The Arabidopsis catalase triple mutant reveals important roles of catalases and peroxisome-derived signaling in plant development. J. Integr. Plant Biol. 2018, 60, 591–607. [Google Scholar] [CrossRef]
- Corpas, F.J.; Del Río, L.A.; Palma, J.M. Plant peroxisomes at the crossroad of NO and H2O2 metabolism. J. Integr. Plant Biol. 2019, 61, 803–816. [Google Scholar]
- Su, T.; Li, W.; Wang, P.; Ma, C. Dynamics of peroxisome homeostasis and its role in stress response and signaling in plants. Front. Plant Sci. 2019, 10, 705. [Google Scholar] [CrossRef]
- Till, A.; Lakhani, R.; Burnett, S.F.; Subramani, S. Pexophagy: The selective degradation of peroxisomes. Int. J. Cell Biol. 2012, 2012, 512721. [Google Scholar] [CrossRef]
- Farré, J.C.; Manjithaya, R.; Mathewson, R.D.; Subramani, S. PpAtg30 tags peroxisomes for turnover by selective autophagy. Dev. Cell 2008, 14, 365–376. [Google Scholar] [CrossRef] [PubMed]
- Motley, A.M.; Nuttall, J.M.; Hettema, E.H. Atg36: The Saccharomyces cerevisiae receptor for pexophagy. Autophagy 2012, 8, 1680–1681. [Google Scholar] [CrossRef]
- Zientara-Rytter, K.; Sirko, A. To deliver or to degrade—An interplay of the ubiquitin-proteasome system, autophagy and vesicular transport in plants. FEBS J. 2016, 283, 3534–3555. [Google Scholar] [CrossRef] [PubMed]
- Yamashita, S.; Abe, K.; Tatemichi, Y.; Fujiki, Y. The membrane peroxin PEX3 induces peroxisome-ubiquitination-linked pexophagy. Autophagy 2014, 10, 1549–1564. [Google Scholar] [CrossRef]
- Sargent, G.; van Zutphen, T.; Shatseva, T.; Zhang, L.; Di Giovanni, V.; Bandsma, R.; Kim, P.K. PEX2 is the E3 ubiquitin ligase required for pexophagy during starvation. J. Cell Biol. 2016, 214, 677–690. [Google Scholar] [CrossRef]
- Zhang, J.; Tripathi, D.N.; Jing, J.; Alexander, A.; Kim, J.; Powell, R.T.; Dere, R.; Tait-Mulder, J.; Lee, J.H.; Paull, T.T.; et al. ATM functions at the peroxisome to induce pexophagy in response to ROS. Nat. Cell Biol. 2015, 17, 1259–1269. [Google Scholar] [CrossRef] [PubMed]
- Kaur, N.; Zhao, Q.; Xie, Q.; Hu, J. Arabidopsis RING peroxins are E3 ubiquitin ligases that interact with two homologous ubiquitin receptor proteins (F). J. Integr. Plant Biol. 2013, 55, 108–120. [Google Scholar] [CrossRef]
- Yin, Y.; Wang, Z.Y.; Mora-Garcia, S.; Li, J.; Yoshida, S.; Asami, T.; Chory, J. BES1 accumulates in the nucleus in response to brassinosteroids to regulate gene expression and promote stem elongation. Cell 2002, 109, 181–191. [Google Scholar] [CrossRef]
- Nolan, T.M.; Brennan, B.; Yang, M.; Chen, J.; Zhang, M.; Li, Z.; Wang, X.; Bassham, D.C.; Walley, J.; Yin, Y. Selective autophagy of BES1 mediated by DSK2 balances plant growth and survival. Dev. Cell 2017, 41, 33–46.e7. [Google Scholar] [CrossRef]
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Su, T.; Yang, M.; Wang, P.; Zhao, Y.; Ma, C. Interplay between the Ubiquitin Proteasome System and Ubiquitin-Mediated Autophagy in Plants. Cells 2020, 9, 2219. https://doi.org/10.3390/cells9102219
Su T, Yang M, Wang P, Zhao Y, Ma C. Interplay between the Ubiquitin Proteasome System and Ubiquitin-Mediated Autophagy in Plants. Cells. 2020; 9(10):2219. https://doi.org/10.3390/cells9102219
Chicago/Turabian StyleSu, Tong, Mingyue Yang, Pingping Wang, Yanxiu Zhao, and Changle Ma. 2020. "Interplay between the Ubiquitin Proteasome System and Ubiquitin-Mediated Autophagy in Plants" Cells 9, no. 10: 2219. https://doi.org/10.3390/cells9102219
APA StyleSu, T., Yang, M., Wang, P., Zhao, Y., & Ma, C. (2020). Interplay between the Ubiquitin Proteasome System and Ubiquitin-Mediated Autophagy in Plants. Cells, 9(10), 2219. https://doi.org/10.3390/cells9102219



