Adaptation of Proteasomes and Lysosomes to Cellular Environments
Abstract
:1. Background
2. Proteasome Plasticity
3. Recognition of Proteins by the Proteasome
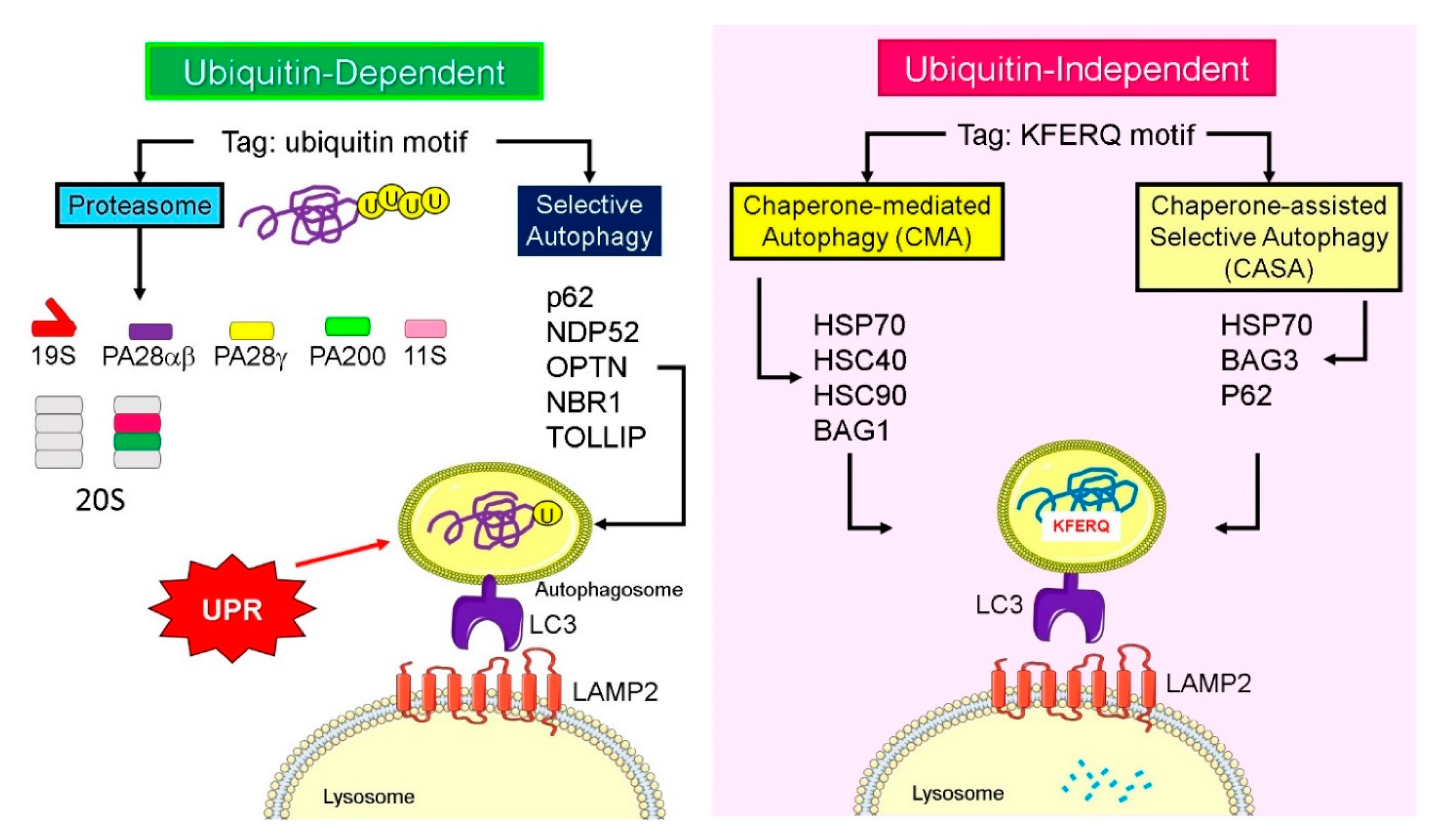
4. Lysosomal Degradation and Recognition of Ubiquitylated Proteins
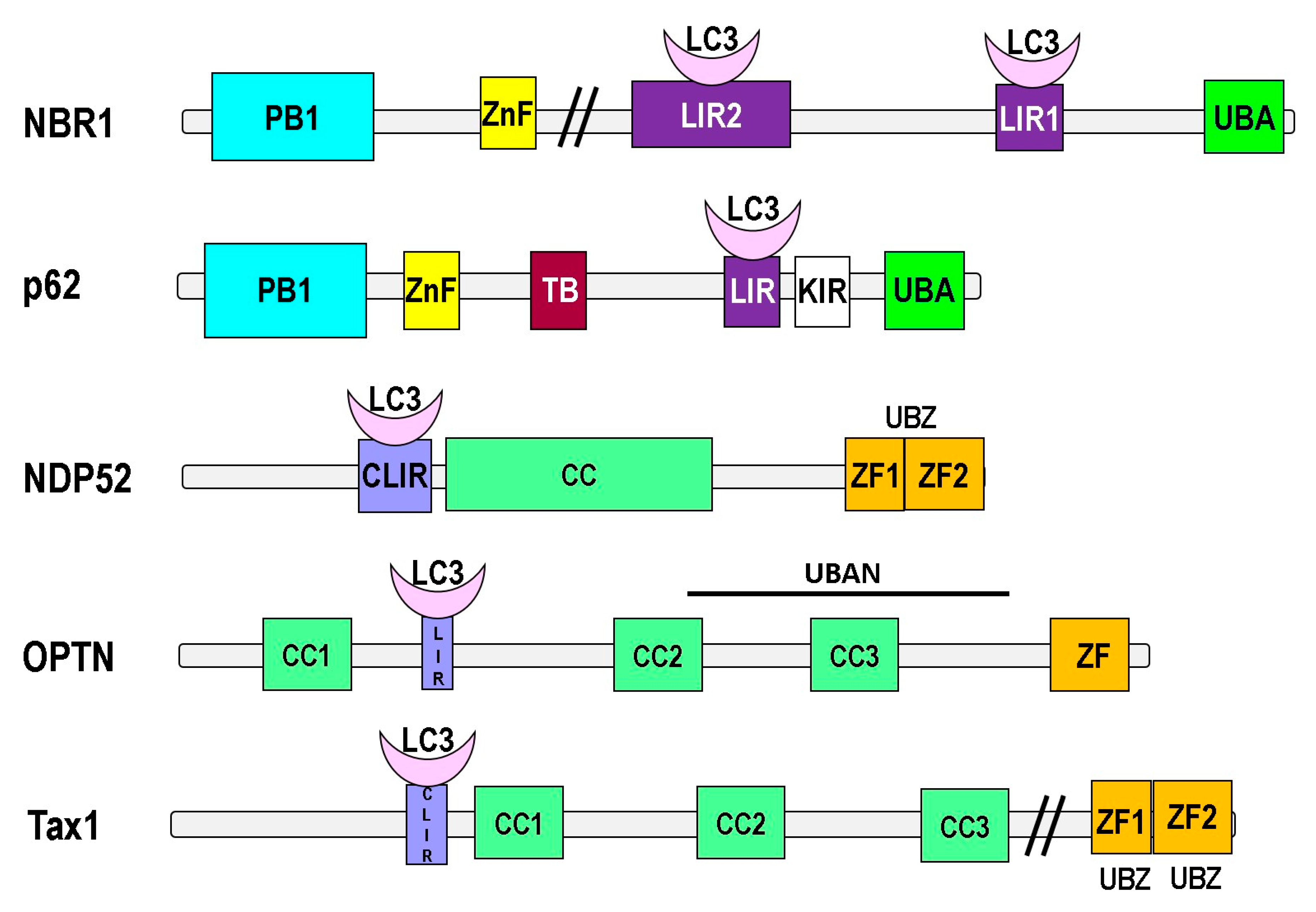
5. Ubiquitin-Like Proteins and Adaptors for Selective Autophagy
6. Signals That Determine Proteasomal and Lysosomal Degradation
7. Concluding Remarks
Funding
Conflicts of Interest
References
- Dong, Z.; Cui, H. The Autophagy-Lysosomal Pathways and Their Emerging Roles in Modulating Proteostasis in Tumors. Cells 2018, 8, 4. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clague, M.J.; Urbé, S. Ubiquitin: Same Molecule, Different Degradation Pathways. Cell 2010, 143, 682–685. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, E.; Park, S.; Lee, J.H.; Mun, J.Y.; Choi, W.H.; Yun, Y.; Lee, J.; Kim, J.H.; Kang, M.-J.; Lee, M.J. Dual Function of USP14 Deubiquitinase in Cellular Proteasomal Activity and Autophagic Flux. Cell Rep. 2018, 24, 732–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gan, J.; Leestemaker, Y.; Sapmaz, A.; Ovaa, H. Highlighting the Proteasome: Using Fluorescence to Visualize Proteasome Activity and Distribution. Front. Mol. Biosci. 2019, 6, 14. [Google Scholar] [CrossRef] [Green Version]
- Morozov, A.V.; Karpov, V.L. Proteasomes and Several Aspects of Their Heterogeneity Relevant to Cancer. Front. Oncol. 2019, 9, 761. [Google Scholar] [CrossRef] [Green Version]
- Tanaka, K. The proteasome: Overview of structure and functions. Proc. Jpn. Acad. Ser. B 2009, 85, 12–36. [Google Scholar] [CrossRef] [Green Version]
- Livneh, I.; Cohen-Kaplan, V.; Cohen-Rosenzweig, C.; Avni, N.; Ciechanover, A. The life cycle of the 26S proteasome: From birth, through regulation and function, and onto its death. Cell Res. 2016, 26, 869–885. [Google Scholar] [CrossRef] [Green Version]
- Saeki, Y. Ubiquitin recognition by the proteasome. J. Biochem. 2017, 161, 113–124. [Google Scholar] [CrossRef]
- Hoeller, D.; Dikic, I. How the proteasome is degraded. Proc. Natl. Acad. Sci. USA 2016, 113, 13266–13268. [Google Scholar] [CrossRef] [Green Version]
- Koulich, E.; Li, X.; DeMartino, G.N. Relative Structural and Functional Roles of Multiple Deubiquitylating Proteins Associated with Mammalian 26S Proteasome. Mol. Boil. Cell 2008, 19, 1072–1082. [Google Scholar] [CrossRef] [Green Version]
- Finley, D. Recognition and processing of ubiquitin-protein conjugates by the proteasome. Annu. Rev. Biochem. 2009, 78, 477–513. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krzywda, S.; Brzozowski, A.M.; Higashitsuji, H.; Fujita, J.; Welchman, R.; Dawson, S.; Mayer, R.J.; Wilkinson, A.J. The Crystal Structure of Gankyrin, an Oncoprotein Found in Complexes with Cyclin-dependent Kinase 4, a 19 S Proteasomal ATPase Regulator, and the Tumor Suppressors Rb and p53. J. Boil. Chem. 2003, 279, 1541–1545. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaneko, T.; Hamazaki, J.; Iemura, S.-I.; Sasaki, K.; Furuyama, K.; Natsume, T.; Tanaka, K.; Murata, S. Assembly Pathway of the Mammalian Proteasome Base Subcomplex Is Mediated by Multiple Specific Chaperones. Cell 2009, 137, 914–925. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, Y.; Wu, J.; Dong, Y.; Chen, S.; Sun, S.; Ma, Y.-B.; Ouyang, Q.; Finley, D.; Kirschner, M.W.; Mao, Y.J. Conformational Landscape of the p28-Bound Human Proteasome Regulatory Particle. Mol. Cell 2017, 67, 322–333.e6. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Livnat-Levanon, N.; Kevei, E.; Kleifeld, O.; Krutauz, D.; Segref, A.; Rinaldi, T.; Erpapazoglou, Z.; Cohen, M.M.; Reis, N.; Hoppe, T.; et al. Reversible 26S Proteasome Disassembly upon Mitochondrial Stress. Cell Rep. 2014, 7, 1371–1380. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Chemmama, I.E.; Yu, C.; Huszagh, A.; Xu, Y.; Viner, R.; Block, S.A.; Cimermancic, P.; Rychnovsky, S.D.; Ye, Y.; et al. The proteasome-interacting Ecm29 protein disassembles the 26S proteasome in response to oxidative stress. J. Boil. Chem. 2017, 292, 16310–16320. [Google Scholar] [CrossRef] [Green Version]
- Pickering, A.M.; Davies, K.J. Differential roles of proteasome and immunoproteasome regulators Pa28alphabeta, Pa28gamma and Pa200 in the degradation of oxidized proteins. Arch. Biochem. Biophys. 2012, 523, 181–190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yewdell, J.W. Immunoproteasomes: Regulating the regulator. Proc. Natl. Acad. Sci. USA 2005, 102, 9089–9090. [Google Scholar] [CrossRef] [Green Version]
- Ferrington, D.A.; Gregerson, D.S. Immunoproteasomes: Structure, function, and antigen presentation. Prog. Mol. Biol. Transl. Sci. 2012, 109, 75–112. [Google Scholar]
- Murata, S.; Takahama, Y.; Tanaka, K. Thymoproteasome: Probable role in generating positively selecting peptides. Curr. Opin. Immunol. 2008, 20, 192–196. [Google Scholar] [CrossRef]
- Wendler, P.; Enenkel, C. Nuclear Transport of Yeast Proteasomes. Front. Mol. Biosci. 2019, 6, 34. [Google Scholar] [CrossRef] [PubMed]
- Palmer, A.; Mason, G.G.; Paramio, J.M.; Knecht, E.; Rivett, A.J. Changes in proteasome localization during the cell cycle. Eur. J. Cell Boil. 1994, 64, 163–175. [Google Scholar]
- Albert, S.; Schaffer, M.; Beck, F.; Mosalaganti, S.; Asano, S.; Thomas, H.F.; Plitzko, J.M.; Beck, M.; Baumeister, W.; Engel, B.D. Proteasomes tether to two distinct sites at the nuclear pore complex. Proc. Natl. Acad. Sci. USA 2017, 114, 13726–13731. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koch, B.; Yu, H.-G. Regulation of inner nuclear membrane associated protein degradation. Nucleus 2019, 10, 169–180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Niepel, M.; Molloy, K.R.; Williams, R.; Farr, J.C.; Meinema, A.C.; Vecchietti, N.; Cristea, I.M.; Chait, B.T.; Rout, M.P.; Strambio-De-Castillia, C. The nuclear basket proteins Mlp1p and Mlp2p are part of a dynamic interactome including Esc1p and the proteasome. Mol. Boil. Cell 2013, 24, 3920–3938. [Google Scholar] [CrossRef]
- Husnjak, K.; Dikic, I. Ubiquitin-Binding Proteins: Decoders of Ubiquitin-Mediated Cellular Functions. Annu. Rev. Biochem. 2012, 81, 291–322. [Google Scholar] [CrossRef]
- Marblestone, J.G.; Butt, S.; McKelvey, D.M.; Sterner, D.E.; Mattern, M.R.; Nicholson, B.; Eddins, M.J. Comprehensive Ubiquitin E2 Profiling of Ten Ubiquitin E3 Ligases. Cell Biophys. 2013, 67, 161–167. [Google Scholar] [CrossRef]
- Collins, G.A.; Goldberg, A.L. The Logic of the 26S Proteasome. Cell 2017, 169, 792–806. [Google Scholar] [CrossRef] [Green Version]
- Shoji, S.; Hanada, K.; Ohsawa, N.; Shirouzu, M. Central catalytic domain of BRAP (RNF52) recognizes the types of ubiquitin chains and utilizes oligo-ubiquitin for ubiquitylation. Biochem. J. 2017, 474, 3207–3226. [Google Scholar] [CrossRef] [Green Version]
- Hurley, J.H.; Lee, S.; Prag, G. Ubiquitin-binding domains. Biochem. J. 2006, 399, 361–372. [Google Scholar] [CrossRef]
- Grice, G.L.; Nathan, J.A. The recognition of ubiquitinated proteins by the proteasome. Cell. Mol. Life Sci. 2016, 73, 3497–3506. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asher, G.; Lotem, J.; Sachs, L.; Kahana, C.; Shaul, Y. Mdm-2 and ubiquitin-independent p53 proteasomal degradation regulated by NQO1. Proc. Natl. Acad. Sci. USA 2002, 99, 13125–13130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wiggins, C.M.; Tsvetkov, P.; Johnson, M.; Joyce, C.L.; Lamb, C.A.; Bryant, N.J.; Komander, D.; Shaul, Y.; Cook, S.J. BIM(EL), an intrinsically disordered protein, is degraded by 20S proteasomes in the absence of poly-ubiquitylation. J. Cell Sci. 2011, 124, 969–977. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Craxton, A.; Butterworth, M.; Harper, N.; Fairall, L.; Schwabe, J.; Ciechanover, A.; Cohen, G.M. NOXA, a sensor of proteasome integrity, is degraded by 26S proteasomes by an ubiquitin-independent pathway that is blocked by MCL-1. Cell Death Differ. 2012, 19, 1424–1434. [Google Scholar] [CrossRef]
- Prakash, S.; Tian, L.; Ratliff, K.S.; E Lehotzky, R.; Matouschek, A. An unstructured initiation site is required for efficient proteasome-mediated degradation. Nat. Struct. Mol. Boil. 2004, 11, 830–837. [Google Scholar] [CrossRef]
- Krappmann, D.; Wulczyn, F.G.; Scheidereit, C. Different mechanisms control signal-induced degradation and basal turnover of the NF-kappaB inhibitor IkappaB alpha in vivo. EMBO J. 1996, 15, 6716–6726. [Google Scholar] [CrossRef]
- Saftig, P.; Klumperman, J. Lysosome biogenesis and lysosomal membrane proteins: Trafficking meets function. Nat. Rev. Mol. Cell Boil. 2009, 10, 623–635. [Google Scholar] [CrossRef]
- Braulke, T.; Bonifacino, J.S. Sorting of lysosomal proteins. Biochim. Biophys. Acta (BBA)-Bioenerg. 2009, 1793, 605–614. [Google Scholar] [CrossRef] [Green Version]
- Bonam, S.R.; Wang, F.; Muller, S. Lysosomes as a therapeutic target. Nat. Rev. Drug Discov. 2019, 18, 923–948. [Google Scholar] [CrossRef] [Green Version]
- Griffiths, G.; Hoflack, B.; Simons, K.; Mellman, I.; Kornfeld, S. The mannose 6-phosphate receptor and the biogenesis of lysosomes. Cell 1988, 52, 329–341. [Google Scholar] [CrossRef]
- Jongsma, M.L.; Berlin, I.; Wijdeven, R.H.; Janssen, L.; Janssen, G.M.; Garstka, M.A.; Janssen, H.; Mensink, M.; Van Veelen, P.; Spaapen, R.M.; et al. An ER-Associated Pathway Defines Endosomal Architecture for Controlled Cargo Transport. Cell 2016, 166, 152–166. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ballabio, A.; Bonifacino, J.S. Lysosomes as dynamic regulators of cell and organismal homeostasis. Nat. Rev. Mol. Cell Boil. 2019, 21, 101–118. [Google Scholar] [CrossRef] [PubMed]
- Bonifacino, J.S.; Neefjes, J. Moving and positioning the endolysosomal system. Curr. Opin. Cell Boil. 2017, 47, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Dubnikov, T.; Ben-Gedalya, T.; Cohen, E. Protein Quality Control in Health and Disease. Cold Spring Harb. Perspect. Boil. 2016, 9, a023523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lecker, S.H.; Goldberg, A.L.; Mitch, W.E. Protein Degradation by the Ubiquitin–Proteasome Pathway in Normal and Disease States. J. Am. Soc. Nephrol. 2006, 17, 1807–1819. [Google Scholar] [CrossRef] [PubMed]
- Qin, H.; Shao, Q.; Igdoura, S.A.; Alaoui-Jamali, M.A.; Laird, D. Lysosomal and Proteasomal Degradation Play Distinct Roles in the Life Cycle of Cx43 in Gap Junctional Intercellular Communication-deficient and -competent Breast Tumor Cells. J. Boil. Chem. 2003, 278, 30005–30014. [Google Scholar] [CrossRef] [Green Version]
- Mizushima, N.; Yoshimori, T.; Ohsumi, Y. The Role of Atg Proteins in Autophagosome Formation. Annu. Rev. Cell Dev. Boil. 2011, 27, 107–132. [Google Scholar] [CrossRef]
- Galluzzi, L.; Baehrecke, E.H.; Ballabio, A.; Boya, P.; Pedro, J.M.B.-S.; Cecconi, F.; Choi, A.M.; Chu, C.T.; Codogno, P.; Colombo, M.I.; et al. Molecular definitions of autophagy and related processes. EMBO J. 2017, 36, 1811–1836. [Google Scholar] [CrossRef]
- Stolz, A.; Ernst, A.; Dikic, I. Cargo recognition and trafficking in selective autophagy. Nat. Cell Biol. 2014, 16, 495–501. [Google Scholar] [CrossRef]
- Krick, R.; Bremer, S.; Welter, E.; Schlotterhose, P.; Muehe, Y.; Eskelinen, E.L.; Thumm, M. Cdc48/p97 and Shp1/p47 regulate autophagosome biogenesis in concert with ubiquitin-like Atg8. J. Cell Biol. 2010, 190, 965–973. [Google Scholar] [CrossRef] [Green Version]
- Hara, T.; Nakamura, K.; Matsui, M.; Yamamoto, A.; Nakahara, Y.; Suzuki-Migishima, R.; Yokoyama, M.; Mishima, K.; Saito, I.; Okano, H.; et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature 2006, 441, 885–889. [Google Scholar] [CrossRef] [PubMed]
- Juhasz, G.; Érdi, B.; Sass, M.; Neufeld, T.P. Atg7-dependent autophagy promotes neuronal health, stress tolerance, and longevity but is dispensable for metamorphosis in Drosophila. Genes Dev. 2007, 21, 3061–3066. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bjørkøy, G.; Lamark, T.; Brech, A.; Outzen, H.; Perander, M.; Øvervatn, A.; Stenmark, H.; Johansen, T. p62/SQSTM1 forms protein aggregates degraded by autophagy and has a protective effect on huntingtin-induced cell death. J. Cell Biol. 2005, 171, 603–614. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pankiv, S.; Clausen, T.H.; Lamark, T.; Brech, A.; Bruun, J.A.; Outzen, H.; Øvervatn, A.; Bjørkøy, G.; Johansen, T. p62/SQSTM1 binds directly to Atg8/LC3 to facilitate degradation of ubiquitinated protein aggregates by autophagy. J. Biol. Chem. 2007, 282, 24131–24145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Moscat, J.; Diaz-Meco, M.T.; Wooten, M.W. Signal integration and diversification through the p62 scaffold protein. Trends Biochem. Sci. 2007, 32, 95–100. [Google Scholar] [CrossRef]
- Sumimoto, H.; Kamakura, S.; Ito, T. Structure and Function of the PB1 Domain, a Protein Interaction Module Conserved in Animals, Fungi, Amoebas, and Plants. Sci. STKE 2007, 2007, re6. [Google Scholar] [CrossRef]
- Johansen, T.; Lamark, T. Selective autophagy mediated by autophagic adapter proteins. Autophagy 2011, 7, 279–296. [Google Scholar] [CrossRef]
- Komatsu, M.; Kurokawa, H.; Waguri, S.; Taguchi, K.; Kobayashi, A.; Ichimura, Y.; Sou, Y.-S.; Ueno, I.; Sakamoto, A.; Tong, K.I.; et al. The selective autophagy substrate p62 activates the stress responsive transcription factor Nrf2 through inactivation of Keap1. Nature 2010, 12, 213–223. [Google Scholar] [CrossRef]
- Rogov, V.; Dötsch, V.; Johansen, T.; Kirkin, V. Interactions between Autophagy Receptors and Ubiquitin-like Proteins Form the Molecular Basis for Selective Autophagy. Mol. Cell 2014, 53, 167–178. [Google Scholar] [CrossRef] [Green Version]
- Fujita, N.; Morita, E.; Itoh, T.; Tanaka, A.; Nakaoka, M.; Osada, Y.; Umemoto, T.; Saitoh, T.; Nakatogawa, H.; Kobayashi, S.; et al. Recruitment of the autophagic machinery to endosomes during infection is mediated by ubiquitin. J. Cell Boil. 2013, 203, 115–128. [Google Scholar] [CrossRef] [Green Version]
- Alemu, E.A.; Lamark, T.; Torgersen, K.M.; Birgisdottir, Å.B.; Larsen, K.B.; Jain, A.; Olsvik, H.; Øvervatn, A.; Kirkin, V.; Johansen, T. ATG8 Family Proteins Act as Scaffolds for Assembly of the ULK Complex. J. Boil. Chem. 2012, 287, 39275–39290. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamark, T.; Perander, M.; Outzen, H.; Kristiansen, K.; Øvervatn, A.; Michaelsen, E.; Bjørkøy, G.; Johansen, T. Interaction Codes within the Family of Mammalian Phox and Bem1p Domain-containing Proteins. J. Boil. Chem. 2003, 278, 34568–34581. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kirkin, V.; Lamark, T.; Sou, Y.-S.; Bjørkøy, G.; Nunn, J.L.; Bruun, J.A.; Shvets, E.; McEwan, D.G.; Clausen, T.H.; Wild, P.; et al. A Role for NBR1 in Autophagosomal Degradation of Ubiquitinated Substrates. Mol. Cell 2009, 33, 505–516. [Google Scholar] [CrossRef] [Green Version]
- Deosaran, E.; Larsen, K.B.; Hua, R.; Sargent, G.; Wang, Y.; Kim, S.; Lamark, T.; Jauregui, M.; Law, K.; Lippincott-Schwartz, J.; et al. NBR1 acts as an autophagy receptor for peroxisomes. J. Cell Sci. 2012, 126, 939–952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wild, P.; Farhan, H.; McEwan, D.G.; Wagner, S.; Rogov, V.V.; Brady, N.R.; Richter, B.; Korac, J.; Waidmann, O.; Choudhary, C.; et al. Phosphorylation of the Autophagy Receptor Optineurin Restricts Salmonella Growth. Science 2011, 333, 228–233. [Google Scholar] [CrossRef] [Green Version]
- Korac, J.; Schaeffer, V.; Kovacević, I.; Clement, A.M.; Jungblut, B.; Behl, C.; Terzic, J.; Dikic, I. Ubiquitin-independent function of optineurin in autophagic clearance of protein aggregates. J. Cell Sci. 2012, 126, 580–592. [Google Scholar] [CrossRef] [Green Version]
- Kachaner, D.; Génin, P.; Laplantine, E.; Weil, R. Toward an integrative view of Optineurin functions. Cell Cycle 2012, 11, 2808–2818. [Google Scholar] [CrossRef]
- Von Muhlinen, N.; Akutsu, M.; Ravenhill, B.J.; Foeglein, Á.; Bloor, S.; Rutherford, T.J.; Freund, S.M.; Komander, D.; Randow, F. LC3C, Bound Selectively by a Noncanonical LIR Motif in NDP52, Is Required for Antibacterial Autophagy. Mol. Cell 2012, 48, 329–342. [Google Scholar] [CrossRef] [Green Version]
- Newman, A.C.; Scholefield, C.L.; Kemp, A.J.; Newman, M.; McIver, E.G.; Kamal, A.; Wilkinson, S. TBK1 kinase addiction in lung cancer cells is mediated via autophagy of Tax1bp1/Ndp52 and non-canonical NF-κB signalling. PLoS ONE 2012, 7, e50672. [Google Scholar] [CrossRef] [Green Version]
- Thurston, T.L.M.; Ryzhakov, G.; Bloor, S.; Von Muhlinen, N.; Randow, F. The TBK1 adaptor and autophagy receptor NDP52 restricts the proliferation of ubiquitin-coated bacteria. Nat. Immunol. 2009, 10, 1215–1221. [Google Scholar] [CrossRef]
- Farré, J.-C.; Burkenroad, A.; Burnett, S.F.; Subramani, S. Phosphorylation of mitophagy and pexophagy receptors coordinates their interaction with Atg8 and Atg11. EMBO Rep. 2013, 14, 441–449. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schweers, R.L.; Zhang, J.; Randall, M.S.; Loyd, M.R.; Li, W.; Dorsey, F.C.; Kundu, M.; Opferman, J.T.; Cleveland, J.L.; Miller, J.L.; et al. NIX is required for programmed mitochondrial clearance during reticulocyte maturation. Proc. Natl. Acad. Sci. USA 2007, 104, 19500–19505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lampert, M.A.; Orogo, A.M.; Najor, R.H.; Hammerling, B.C.; Leon, L.J.; Wang, B.; Kim, T.; Sussman, M.A.; Gustafsson, Å.B. BNIP3L/NIX and FUNDC1-mediated mitophagy is required for mitochondrial network remodeling during cardiac progenitor cell differentiation. Autophagy 2019, 15, 1182–1198. [Google Scholar] [CrossRef] [PubMed]
- Novak, I.; Kirkin, V.; McEwan, D.G.; Zhang, J.; Wild, P.; Rozenknop, A.; Rogov, V.; Löhr, F.; Popovic, D.; Occhipinti, A.; et al. Nix is a selective autophagy receptor for mitochondrial clearance. EMBO Rep. 2009, 11, 45–51. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Feng, D.; Chen, G.; Chen, M.; Zheng, Q.; Song, P.; Ma, Q.; Zhu, C.; Wang, R.; Qi, W.; et al. Mitochondrial outer-membrane protein FUNDC1 mediates hypoxia-induced mitophagy in mammalian cells. Nature 2012, 14, 177–185. [Google Scholar] [CrossRef]
- Lu, J.-H.; He, L.; Behrends, C.; Araki, M.; Araki, K.; Wang, Q.J.; Catanzaro, J.M.; Friedman, S.L.; Zong, W.-X.; Fiel, M.I.; et al. NRBF2 regulates autophagy and prevents liver injury by modulating Atg14L-linked phosphatidylinositol-3 kinase III activity. Nat. Commun. 2014, 5, 3920. [Google Scholar] [CrossRef]
- B’chir, W.; Maurin, A.C.; Carraro, V.; Averous, J.; Jousse, C.; Muranishi, Y.; Parry, L.; Stepien, G.; Fafournoux, P.; Bruhat, A. The eIF2alpha/ATF4 pathway is essential for stress-induced autophagy gene expression. Nucleic Acids Res. 2013, 41, 7683–7699. [Google Scholar] [CrossRef] [Green Version]
- Bao, Y.; Pu, Y.; Yu, X.; Gregory, B.D.; Srivastava, R.; Howell, S.H.; Bassham, D.C. IRE1B degrades RNAs encoding proteins that interfere with the induction of autophagy by ER stress in Arabidopsis thaliana. Autophagy 2018, 14, 1562–1573. [Google Scholar] [CrossRef] [Green Version]
- Deegan, S.; Koryga, I.; Glynn, S.A.; Gupta, S.; Gorman, A.M.; Samali, A. A close connection between the PERK and IRE arms of the UPR and the transcriptional regulation of autophagy. Biochem. Biophys. Res. Commun. 2015, 456, 305–311. [Google Scholar] [CrossRef]
- Guilbert, S.M.; Lambert, H.; Rodrigue, M.-A.; Fuchs, M.; Landry, J.; Lavoie, J.N. HSPB8 and BAG3 cooperate to promote spatial sequestration of ubiquitinated proteins and coordinate the cellular adaptive response to proteasome insufficiency. FASEB J. 2018, 32, 3518–3535. [Google Scholar] [CrossRef] [Green Version]
- Gao, Y.-S.; Hubbert, C.C.; Lu, J.; Lee, Y.-S.; Lee, J.Y.; Yao, T.-P. Histone Deacetylase 6 Regulates Growth Factor-Induced Actin Remodeling and Endocytosis. Mol. Cell. Boil. 2007, 27, 8637–8647. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, J.Y.; Koga, H.; Kawaguchi, Y.; Tang, W.; Wong, E.; Gao, Y.-S.; Pandey, U.B.; Kaushik, S.; Tresse, E.; Lu, J.; et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010, 29, 969–980. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, J.S.; Miller, S.E.; Hanson, P.I.; Weihl, C.C. Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J. Biol. Chem. 2008, 283, 30289–30299. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalveram, B.; Schmidtke, G.; Groettrup, M. The ubiquitin-like modifier FAT10 interacts with HDAC6 and localizes to aggresomes under proteasome inhibition. J. Cell Sci. 2008, 121, 4079–4088. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meriin, A.B.; Narayanan, A.; Meng, L.; Alexandrov, I.; Varelas, X.; Cissé, I.I.; Sherman, M.Y. Hsp70–Bag3 complex is a hub for proteotoxicity-induced signaling that controls protein aggregation. Proc. Natl. Acad. Sci. USA 2018, 115, E7043–E7052. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schneider, J.L.; Suh, Y.; Cuervo, A.M. Deficient Chaperone-Mediated Autophagy in Liver Leads to Metabolic Dysregulation. Cell Metab. 2014, 20, 417–432. [Google Scholar] [CrossRef] [Green Version]


| Ubiquitin Ligases | Number of Ligases | Deubiquitinases | Number of DUBs |
|---|---|---|---|
| Tag Proteins with Ubiquitin Residues | Remove a Ubiquitin Residue from Proteins. | ||
| Ubiquitin activating enzyme E1 Ligase | 2 | Cystein Proteases: i. Ubiquitin specific proteases (USPs) ii. Ubiquitin carboxy-terminal hydrolases (UCHs) iii. Ovarian-tumor proteases (OTUs) iv. Machado-Joseph disease protein domain proteases (MJDs) v. Monocyte chemotactic protein-induced proteins (MCPIPs) vi. Permuted papain fold peptidases of dsRNA viruses and eukaryotes (PPPDEs) | 62 USPs 4 UCHs 15 OTUs 4 MJDs 7 MCPIPs |
| Ubiquitin conjugating enzyme E2 Ligase | 40 | Zinc-dependent metalloproteinases: i. JAMMs/MPN+ proteases | 4 JAMMs |
| Ubiquitin ligating enzymeE3 Ligase | 600 |
| Adapter Protein | Interaction Domain | Selective Autophagy Involved in |
|---|---|---|
| p62/SQSTM1 | UBA | Aggrephagy, mitophagy, xenophagy, pexophagy and zymophagy |
| NBR1 | UBA | Aggrephagy, mitophagy, xenophagy |
| Optineurin | UBAN, UBZ | Aggrephagy, mitophagy, xenophagy |
| NDP52 | UBZ | Aggrephagy, mitophagy, xenophagy |
| BNIP3L/NIX | - | Mitophagy |
| FUNDC1 | - | Mitophagy |
| HDAC6 | - | Aggrephagy and mitophagy |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mebratu, Y.A.; Negasi, Z.H.; Dutta, S.; Rojas-Quintero, J.; Tesfaigzi, Y. Adaptation of Proteasomes and Lysosomes to Cellular Environments. Cells 2020, 9, 2221. https://doi.org/10.3390/cells9102221
Mebratu YA, Negasi ZH, Dutta S, Rojas-Quintero J, Tesfaigzi Y. Adaptation of Proteasomes and Lysosomes to Cellular Environments. Cells. 2020; 9(10):2221. https://doi.org/10.3390/cells9102221
Chicago/Turabian StyleMebratu, Yohannes Afework, Zerihun Hailemariam Negasi, Saugata Dutta, Joselyn Rojas-Quintero, and Yohannes Tesfaigzi. 2020. "Adaptation of Proteasomes and Lysosomes to Cellular Environments" Cells 9, no. 10: 2221. https://doi.org/10.3390/cells9102221
APA StyleMebratu, Y. A., Negasi, Z. H., Dutta, S., Rojas-Quintero, J., & Tesfaigzi, Y. (2020). Adaptation of Proteasomes and Lysosomes to Cellular Environments. Cells, 9(10), 2221. https://doi.org/10.3390/cells9102221





