Use of a Hybrid Adeno-Associated Viral Vector Transposon System to Deliver the Insulin Gene to Diabetic NOD Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Vector Construction and Production
2.1.1. AAV Vectors
2.1.2. HIV/MSCV Lentiviral Vector
2.2. Transduction of Liver Tissue
2.3. Functional Analysis
2.4. Microscopic Analysis
2.5. Vector Copy Number Analysis
2.6. Reverse Transcriptase Polymerase Chain Reaction (RT-PCR) Analysis
2.7. Statistical Analysis
3. Results
3.1. Microscopic Analysis
3.2. Delivery of AAV8 Expressing INS-FUR ± Pdx1 Fails to Reverse Diabetes
3.3. Reversal of Autoimmune Diabetes Using the AAV8/piggyBac-LSP-INS-FUR Vector System and FFO Surgery
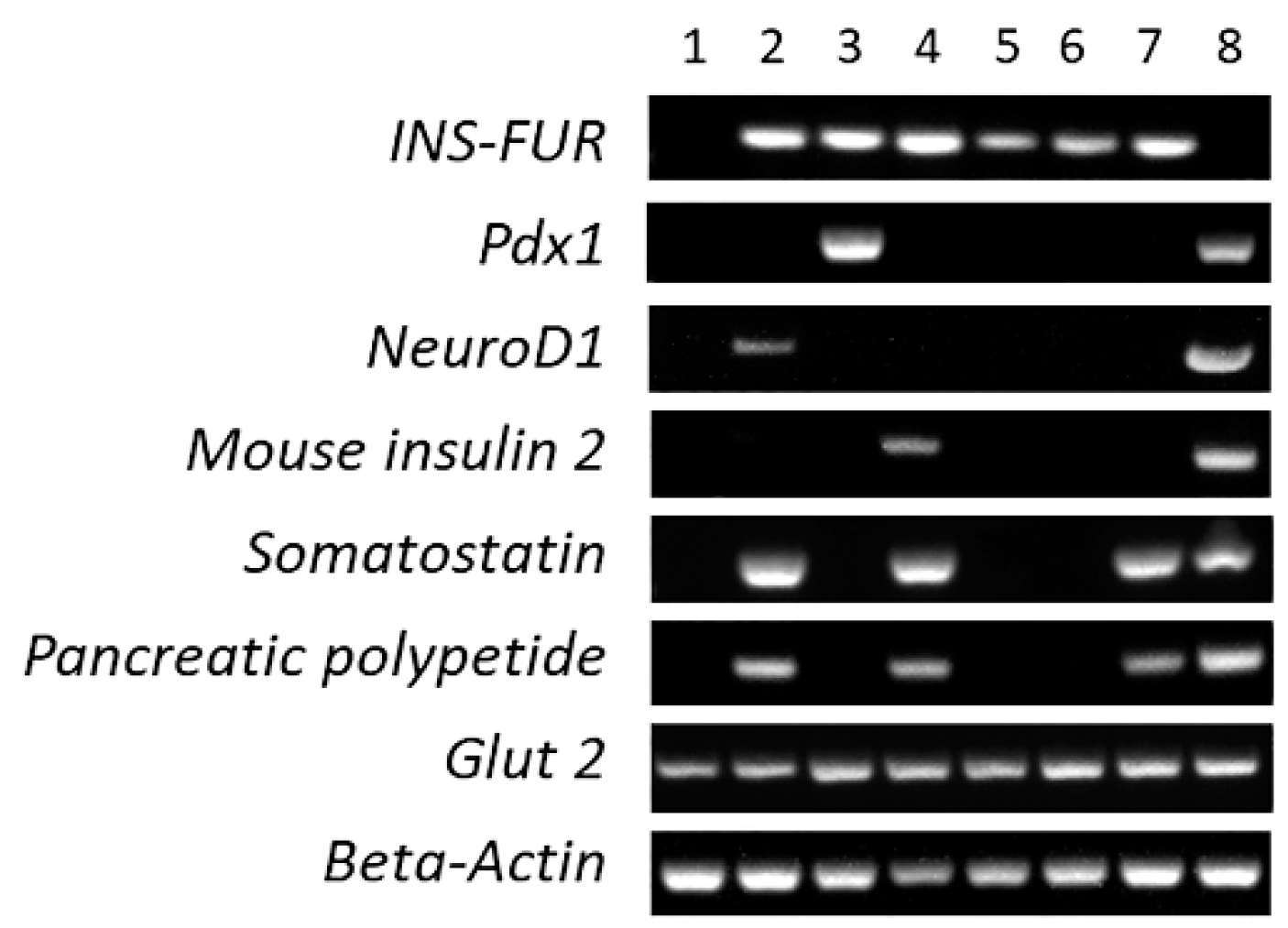
3.4. RT-PCR Analysis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Alberti, K.G.; Zimmet, P.G. Definition, diagnosis and classification of diabetes mellitus and its complication. Part 1: Diagnosis and classification of diabetes mellitus provisional report of a WHO consultation. Diabet. Med. 1998, 15, 539–553. [Google Scholar] [CrossRef]
- Yeh, H.C.; Brown, T.T.; Maruthur, N.; Ranasinghe, P.; Berger, Z.; Suh, Y.D.; Wilson, L.M.; Haberl, E.B.; Brick, J.; Bass, E.B.; et al. Comparative effectiveness and safety of methods of insulin delivery and glucose monitoring for diabetes mellitus: A systematic review and met-analysis. Ann. Intern. Med. 2012, 157, 336–347. [Google Scholar] [CrossRef] [PubMed]
- Franek, E.; Haluzik, M.; Varzic, S.C.; Sargin, M.; Macura, S.; Zacho, J.; Christiansen, J.S. Twice-daily insulin degludec/insulin aspart provides superior fasting plasma glucose control and a reduced rate of hypoglycaemia compared with biphasic insulin aspart 30 in insulin-naïve adults with type 2 diabetes. Diabet. Med. 2016, 33, 497–505. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Belle, T.L.; Coppieters, K.T.; von Herrath, M.G. Type 1 diabetes: Etiology, immunology, and therapeutic strategies. Physiol. Rev. 2011, 91, 79–118. [Google Scholar] [CrossRef] [PubMed]
- Weisman, A.; Bai, J.W.; Cardinez m Kramer, C.K.; Perkins, B.A. Effect of artificial pancreas systems on glycaemic control in patients with Type 1 duabetes: A systematic review and meta-analysis of outpatient randomised control trials. Lancet Diabetes Endocrinol. 2017, 5, 501–512. [Google Scholar] [CrossRef]
- Bekiari, E.; Kitsios, K.; Thabit, H.; Tauschmann, M.; Athanaasiadou, E.; Karagiannis, T.; Haidich, A.-B.; Hovorka, R.; Tsapas, A. Artificial pancreas treatment for outpatients with type 1 diabetes: Systematic review and meta-analysis. BMJ 2018, 361, k1310. [Google Scholar] [CrossRef] [Green Version]
- Shafiee, A.; Patel, J.; Lee, J.S.; Hutmacher, D.W.; Fisk, N.M.; Khosrotehrani, K. Mesenchymal stem/stromal cells enhance engraftment, vasculogenic and pro-angiogenic activities of endothelial colony forming cells in immunocompetent hosts. Sci. Rep. 2017, 7, 13558. [Google Scholar] [CrossRef] [PubMed]
- Pathak, V.; Pathak, N.M.; O’neill, C.L.; Guduric-Fuchs, J.; Medina, R.J. Therapies for type 1 diabetes: Current scenario and future perspectives. Clin. Med. Insights 2019, 12, 1–13. [Google Scholar] [CrossRef]
- Ren, B.; O’Brien, B.A.; Swan, M.A.; Kiona, M.E.; Nassif, N.T.; Wei, M.Q.; Simpson, A.M. Long-term correction of diabetes in rats following lentiviral hepatic insulin gene therapy. Diabetologia 2007, 50, 1910–1920. [Google Scholar] [CrossRef] [Green Version]
- Ren, B.; O’Brien, B.A.; Byrne, M.R.; Ch’ng, E.; Gatt, P.N.; Swan, M.A.; Nassif, N.T.; Wei, M.Q.; Gijsbers, R.; Debyser, Z.; et al. Long term reversal of diabetes in non obese diabetic mice by liver-directed gene therapy. J. Gene Med. 2013, 15, 28–41. [Google Scholar] [CrossRef]
- Gerace, D.; Ren, B.; Hawthorne, W.J.; Byrne, M.R.; Phillips, P.M.; O’Brien, B.A.; Nassif, N.T.; Alexander, I.E.; Simpson, A.M. Pancreatic transdifferentiation in porcine liver following lentiviral delivery of human furin-cleavable insulin. Trans. Proc. 2013, 45, 1869–1874. [Google Scholar] [CrossRef] [PubMed]
- Elsner, M.; Terbish, T.; Jorns, A.; Naujok, O.; Wedekind, D.; Hedrich, H.J.; Lenzen, S. Reversal of diabetes through gene therapy of diabetic rats by hepatic insulin expression via lentiviral transduction. Mol. Ther. 2012, 20, 918–926. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ber, I.; Shternhall, K.; Perl, S.; Ohanuna, Z.; Goldberg, I.; Barshack, I.; Benvenisti-Zarum, L.; Meivar-Levy, I.; Ferber, S. Functional, Persistent, and Extended liver to pancreas transdifferentiation. J. Biol. Chem. 2003, 278, 31950–31957. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Li, X.; Lam, K.S.L.; Tam, S.; Xiao, W.; Xu, R. Adeno-associated virus-mediated pancreatic and duodenal homeobox gene-1 expression enhanced differentiation of hepatic oval stem cells to insulin-producing cells in diabetic rats. J. Biomed. Sci. 2008, 15, 487–497. [Google Scholar] [CrossRef] [PubMed]
- Tuch, B.E.; Szymanska, B.; Yao, M.; Tabiin, M.T.; Gross, D.J.; Holman, S.; Swan, M.A.; Humphrey, R.K.B.; Marshall, G.M.; Simpson, A.M. Function of a genetically modified human liver cell line that stores, processes and secretes insulin. Gene Ther. 2003, 10, 490–503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferber, S.; Cohen, H.; Ber, I.; Einav, Y.; Goldberg, I.; Barshack, I.; Seijffers, R.; Kopolovic, J.; Kaiser, N.; Karasik, A. Pancreatic and duodenal homeobox gene 1 induces expression of insulin genes in liver and ameliorates streptozotocin-induced hyperglycaemia. Nat. Med. 2000, 6, 568–572. [Google Scholar] [CrossRef] [PubMed]
- Kojima, H.; Matsummura, K.; Younan, P.; Imaeda, H.; Maeda, M.; Chan, L. NeuroD-betacellulin gene therapy induces islet neogenesis in the liver and reverses diabetes in mice. Nat. Med. 2003, 9, 596–603. [Google Scholar] [CrossRef]
- Tang, D.-Q.; Shun, L.; Koya, V.; Sun, Y.; Wang, Q.; Wang, H.; Li, S.-W.; Sun, Y.; Purich, D.L.; Zhang, C.; et al. Genetically reprogrammed, liver-derived insulin-producing cells are glucose-responsive, but susceptible to autoimmune destruction in settings of murine model of type 1 diabetes. Am. J. Transl. Res. 2013, 5, 184–199. [Google Scholar] [PubMed]
- Wang, A.Y.; Ehrhardt, A.; Xu, H.; Kay, M.A. Adenovirus transduction is required for the correction of diabetes using Pdx1 of Neurogenin 3 in the liver. Am. Soc. Ggene Ther. 2007, 15, 255–263. [Google Scholar] [CrossRef]
- Ren, B.; Tao, C.; Swan, M.A.; Joachim, N.; Martiniello-Wilks, R.; Nassif, N.T.; O’Brien, B.A.; Simpson, A.M. Pancreatic transdiffereniation and glucose-regulated production of human insulin in the H4 IIE rat liver cell line. Int. J. Mol. Sci. 2016, 17, 534. [Google Scholar] [CrossRef] [Green Version]
- Alam, T.; Wai, P.; Held, D.; Vakill, S.T.T.; Forsberg, E.; Sollinger, H. Correction of diabetic hyperglycaemia and amelioration of metabolic anomalies by minicircle DNA mediated glucose-dependent hepatic insulin production. PLoS ONE 2013, 8, e67515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hashimoto, H.; Mizushima, T.; Ogura, T.; Kagawa, T.; Tomiyama, K.; Takahashi, R.; Yagoto, M.; Kawai, K.; Chijiwa, T.; Nakamuar, M.; et al. Study on AAV-mediated gene therapy for diabetes in humanized liver mouse to predict efficacy in humans. Biochem. Biophys. Res. Commun. 2016, 478, 1254–1260. [Google Scholar] [CrossRef] [Green Version]
- Gan, S.U.; Fu, Z.; Sia, K.C.; Kon, O.L.; Calne, R.; Lee, K.O. development of a liver-specific Tet-off AAV8 vector for improved safety of isnulin gene therapy for diabetes. J. Gene Med. 2018, 21, e3067. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Recino, A.; Gan, S.U.; Sia, K.C.; Sawyer, Y.; Trendell, J.; Kay, R.; Gribble, F.M.; Reimann, F.; Foale, R.; Notaridou, M. Immunosuppression overcomes insulin- and vector-specific immune responses that limit efficacy of AAV2/ 8-mediated insulin gene therapy in NOD mice. Gene Ther. 2019, 26, 40–56. [Google Scholar] [CrossRef] [PubMed]
- Ren, B.; La, Q.T.; O’Brien, B.A.; Nassif, N.T.; Yan, Y.; Gerace, D.; Martiniello-Wilks, R.; Torpy, F.; Dane, A.P.; Alexander, I.E.; et al. Partial pancreatic transdifferentiation of primary human hepatocytes in the livers of an humanized mouse model. J. Gene Med. 2018, 20, e3017. [Google Scholar] [CrossRef]
- Azuma, H.; Paulk, N.; Ranade, A.; Dorrell, C.; Al-Dhalimy, M.; Ellis, E.; Strom, S.; Kay, M.A.; Finegold, M.; Grompe, M. Robust expansion of human hepatocytes in Fah−/−/Rag2−/−/Ilrg−/− mice. Nat. Biotech. 2007, 25, 903–910. [Google Scholar] [CrossRef] [Green Version]
- Bissig, K.-D.; Le, T.; Verma, I.M. Repopulation of adult and neonatal mice with human hepatocytes: A chimeric animal model. Proc. Natl. Acad. Sci. USA 2007, 104, 20507–20511. [Google Scholar] [CrossRef] [Green Version]
- DePolo, N.J.; Reed, J.D.; Sheridan, P.L.; Townsend, K.; Sauter, S.L.; Jolly, D.J.; Dubensky, T.W. VSV-G pseudotyped lentiviral vector particles produced in human cells are inactivated by human serum. Mol. Ther. 2000, 2, 218–222. [Google Scholar] [CrossRef]
- Lisowski, L.; Dane, A.P.; Chu, K.; Cunningham, S.C.; Wilson, E.M.; Nygaard, S.; Grompe, M.; Alexander, I.E.; Kay, M.A. Selection and evaluation of clinically relevant AAV variants in a xenograft liver model. Nature 2014, 506, 382–386. [Google Scholar] [CrossRef] [Green Version]
- Cabanes-Cres, M.; Westhaus, A.; Navarro, R.G.; Baltazar, G.; Zhu, E.; Amaya, A.K.; Liao, S.H.Y.; Scott, S.; Sallard, E.; Dilworth, K.L.; et al. Attentuation of heparin sulfate proteoglycan binding enhances in vivo transduction of human primary hepatocytes with AAV2. Mol. Ther. Meth. Clin. Dev. 2020, 17, 1139–1154. [Google Scholar] [CrossRef]
- Nathwani, A.C.; Tuddenham, E.G.D.; Rangarajan, S.; McIntosh, J.; Linch, D.C.; Chir, B.; Chowdary, P.; Ridell, A.; Jaquilmac, A.; Harrington, C.; et al. Adenovirus-associated virus vector-mediated gene transfer in haemophilia B. N. Engl. J. Med. 2011, 365, 2357–2365. [Google Scholar] [CrossRef] [PubMed]
- Dane, A.P.; Wowro, S.J.; Cunningham, S.C.; Alexander, I.E. Comparison of gene transfer to the murine liver following intraperitoneal and intraportal delivery of hepatotrophic AAV pseudo-serotypes. Gene Ther. 2013, 20, 460–464. [Google Scholar] [CrossRef] [PubMed]
- Cary, L.C.; Goebel, M.; Corsaro, B.G.; Wang, H.-G.; Rosen, E.; Fraser, M.J. Transposon mutagenesis of baculoviruses: Analysis of Trichopulsia ni transposon IFP2 insertions within the FP-locus of nuclear polyhedrosis viruses. Virology 1989, 172, 156–169. [Google Scholar] [CrossRef]
- Cunningham, S.C.; Siew, S.M.; Hallwirth, C.V.; Bolitho, C.; Garg, G.; Michael, I.P.; Hetherington, N.A.; Carpenter, K.; de Alencastro, G.; Nagy, A.; et al. Modeling correction of severe urea cycle defects in the growing murine liver using a hybrid recombinant adeno-associated virus/piggyback transposase gene delivery system. Hepatology 2015, 62, 417–428. [Google Scholar] [CrossRef] [PubMed]
- Nakanishi, H.; Higuchi y Kawakami, S.; Yamashita, F.; Hashida, S. piggyBack transposon-mediated long-term gene expression in mice. Mol. Ther. 2010, 18, 707–714. [Google Scholar] [CrossRef] [Green Version]
- Cunningham, S.C.; Dane, A.P.; Spinoulos, A.; Alexander, I.E. Gene delivery to the juvenile mouse liver using AAV2/8 vectors. Mol. Ther. 2008, 16, 1081–1088. [Google Scholar] [CrossRef]
- Cunningham, S.C.; Kok, C.Y.; Dane, A.P.; Carpenter, K.; Kizana, E.; Kuchel, P.W.; Alexander, I.E. Induction and prevention of severe hyperammonemia in the spfash mouse model of ornithine transcarbamylase deficiency using shRNA and rAAV-mediated gene delivery. Mol. Ther. 2011, 19, 854–859. [Google Scholar] [CrossRef]
- Choi, J.K.; Hoang, N.; Vilardi, A.M.; Conrad, P.; Emerson, S.G.; Gewirtz, A.M. Hybrid HIV/MSCV LTR enhances transgene expression of lentiviral vectors in human CD34+ hematopoetic cells. Stem Cells 2001, 19, 236–246. [Google Scholar] [CrossRef]
- Cunningham, S.C.; Spinoulas, A.; Carpenter, K.H.; Wicken, B.; Kuchel, P.W.; Alexander, I.E. AAV2/8-mediated correction of OTC deficiency is robust in adult but not neonatal Spfash mice. Mol. Ther. 2009, 17, 1340–1346. [Google Scholar] [CrossRef]
- Andrikopoulos, S.; Blair, A.R.; Deluca, N.; Fam, B.C.; Proietto, J. Evaluating the glucose test in mice. Am. J. Physiol. Endocrinol. Metab. 2008, 295, E1323–E1332. [Google Scholar] [CrossRef] [Green Version]
- Chellappan, D.K.; Sivam, N.S.; Xiang, T.K.; Pan, L.W.; Fui, T.Z.; Kien, C.; Khoo, N.; Yi, F.J.; Chellian, J.; Cheng, L.L.; et al. Gene therapy for type 1 diabetes mellitus. Biomed. Pharmacother. 2018, 108, 1188–1200. [Google Scholar] [CrossRef] [PubMed]
- Volpers, C.; Kochanek, S. Adenoviral vectors for gene transfer and theropy. J. Gene Med. 2004, 6, S164–S171. [Google Scholar] [CrossRef] [PubMed]
- Cavazzana-Calvo, M.; Hacein-bey, S.; Basile, C.D.; Gross, F.; Yvon, E.; Nusbaum, P.; Selz, F.; Hue, C.; Certain, S.; Casanova, J.-L.; et al. Gene therapy of severe combined immunodeficiency (SCID)-XI disease. Science 2000, 288, 669–672. [Google Scholar] [CrossRef] [PubMed]
- White, M.; Whittaker, R.; Gandere, C.; Stoll, E.A. A guide to approaching regulatory considerations for lentiviral-mediated gene transfer. Hum. Gene Ther. 2007, 28, 136–176. [Google Scholar]
- Wang, L.; Wang, H.; Bell, P.; McMenamin, D.; Wilson, J.M. Hepatic gene transfer in neonatal mice by adeno-associated virus serotype 8 vector. Hum. Gene Ther. 2012, 23, 533–539. [Google Scholar] [CrossRef]
- Shanmukhappa, K.; Mourya, R.; Sabla, G.E.; Degen, J.L.; Bezerra, J.A. Hepatic to pancreatic switch defines a role for hemostatic factors in cellular plasticity in mice. Proc. Natl. Acad. Sci. USA 2005, 102, 10182–10187. [Google Scholar] [CrossRef] [Green Version]
- Yang, L.; Shiwu, L.; Hatch, H.; Ahrens, K.; Cornelius, J.G.; Petersen, B.E.; Ammon, A.B. In vitro trans-differentiation of adult hepatic stem cells into pancreatic endocrine hormone-producing cells. Proc. Natl. Acad. Sci. USA 2002, 99, 8078–8083. [Google Scholar] [CrossRef] [Green Version]
- Cerda-Esteban, N.; Naumann, H.; Ruzittu, S.; Mah, N.; Pongrac, I.M.; Cozzitorto, C.; Hommel, A.; Andrade-Navarro, M.A.; Bonifacio, E.; Spagnoli, F.M. Stepwise reprogramming of liver cells to a pancreas progenitor state by the transcriptional regulator Tgif2. Nat. Commun. 2017, 8, 1–13. [Google Scholar] [CrossRef]
- Wang, Y.; Dorrell, C.; Naugler, W.E.; Heskett, M.; Spellman, P.; Li, B.; Gavivo, F.; Haft, A.; Wakefield, L.; Grompe, M. Long-term correction of diabetes in mice by in vivo reprogramming of pancreatic ducts. Mol. Ther. 2018, 26, 1327–1342. [Google Scholar] [CrossRef] [Green Version]
- Coleman, M.A.; Jessup, C.F.; Bridge, J.A.; Overgaard, N.H.; Penko, D.; Walters, S.; Borg, D.J.; Galea, R.; Forbes, J.M.; Thomas, R.; et al. Antigen-encoding bone marrow terminates islet-directed memory CD+ T-cell responses to alleviate islet transplant rejection. Diabetes 2016, 65, 1328–1340. [Google Scholar] [CrossRef] [Green Version]





© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
La, Q.T.; Ren, B.; Logan, G.J.; Cunningham, S.C.; Khandekar, N.; Nassif, N.T.; O’Brien, B.A.; Alexander, I.E.; Simpson, A.M. Use of a Hybrid Adeno-Associated Viral Vector Transposon System to Deliver the Insulin Gene to Diabetic NOD Mice. Cells 2020, 9, 2227. https://doi.org/10.3390/cells9102227
La QT, Ren B, Logan GJ, Cunningham SC, Khandekar N, Nassif NT, O’Brien BA, Alexander IE, Simpson AM. Use of a Hybrid Adeno-Associated Viral Vector Transposon System to Deliver the Insulin Gene to Diabetic NOD Mice. Cells. 2020; 9(10):2227. https://doi.org/10.3390/cells9102227
Chicago/Turabian StyleLa, Que T., Binhai Ren, Grant J. Logan, Sharon C. Cunningham, Neeta Khandekar, Najah T. Nassif, Bronwyn A. O’Brien, Ian E. Alexander, and Ann M. Simpson. 2020. "Use of a Hybrid Adeno-Associated Viral Vector Transposon System to Deliver the Insulin Gene to Diabetic NOD Mice" Cells 9, no. 10: 2227. https://doi.org/10.3390/cells9102227
APA StyleLa, Q. T., Ren, B., Logan, G. J., Cunningham, S. C., Khandekar, N., Nassif, N. T., O’Brien, B. A., Alexander, I. E., & Simpson, A. M. (2020). Use of a Hybrid Adeno-Associated Viral Vector Transposon System to Deliver the Insulin Gene to Diabetic NOD Mice. Cells, 9(10), 2227. https://doi.org/10.3390/cells9102227





