Urine-Derived Epithelial Cell Lines: A New Tool to Model Fragile X Syndrome (FXS)
Abstract
1. Introduction
2. Materials and Methods
2.1. Study Participants
2.2. Collection of Urine
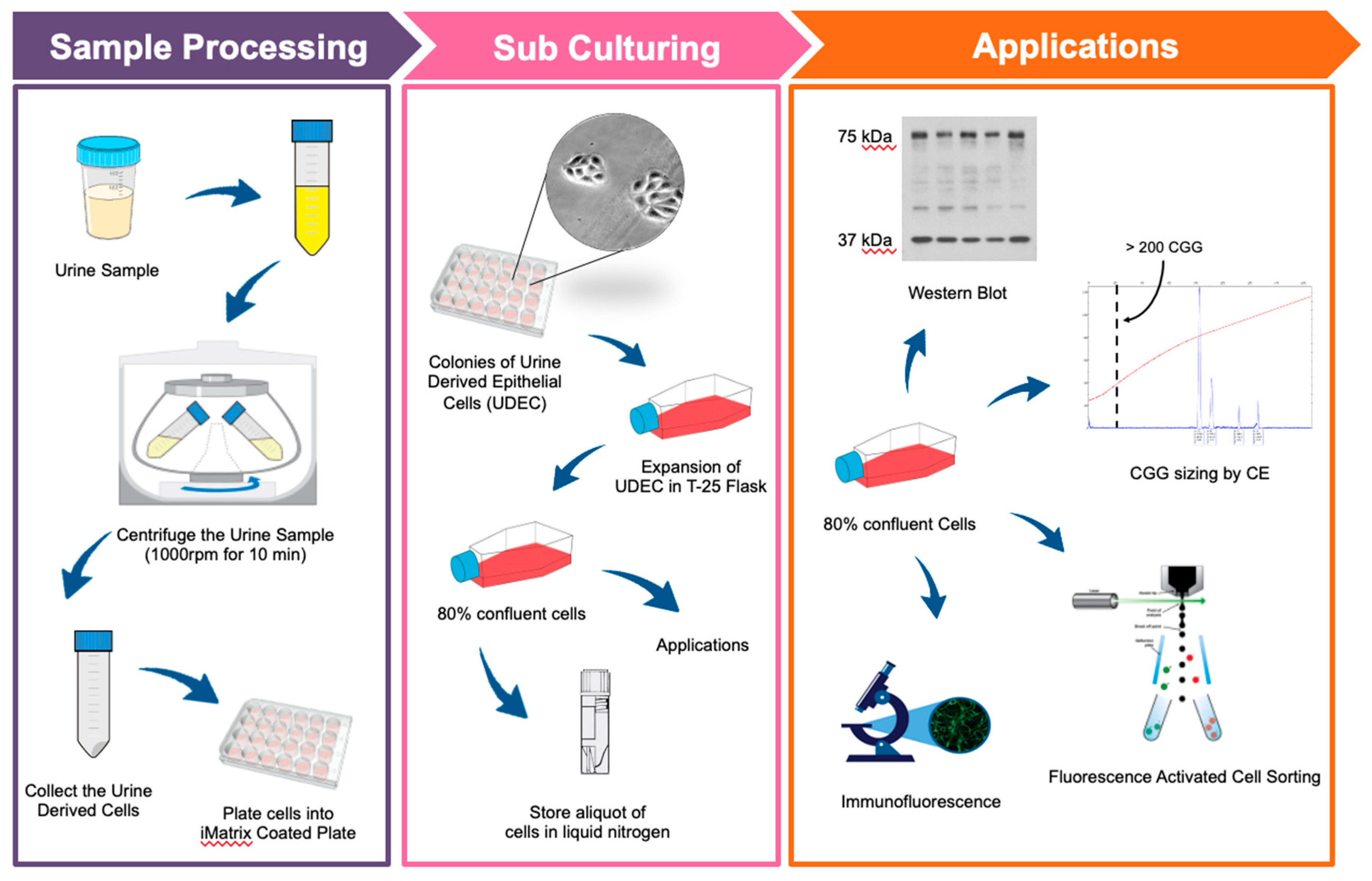
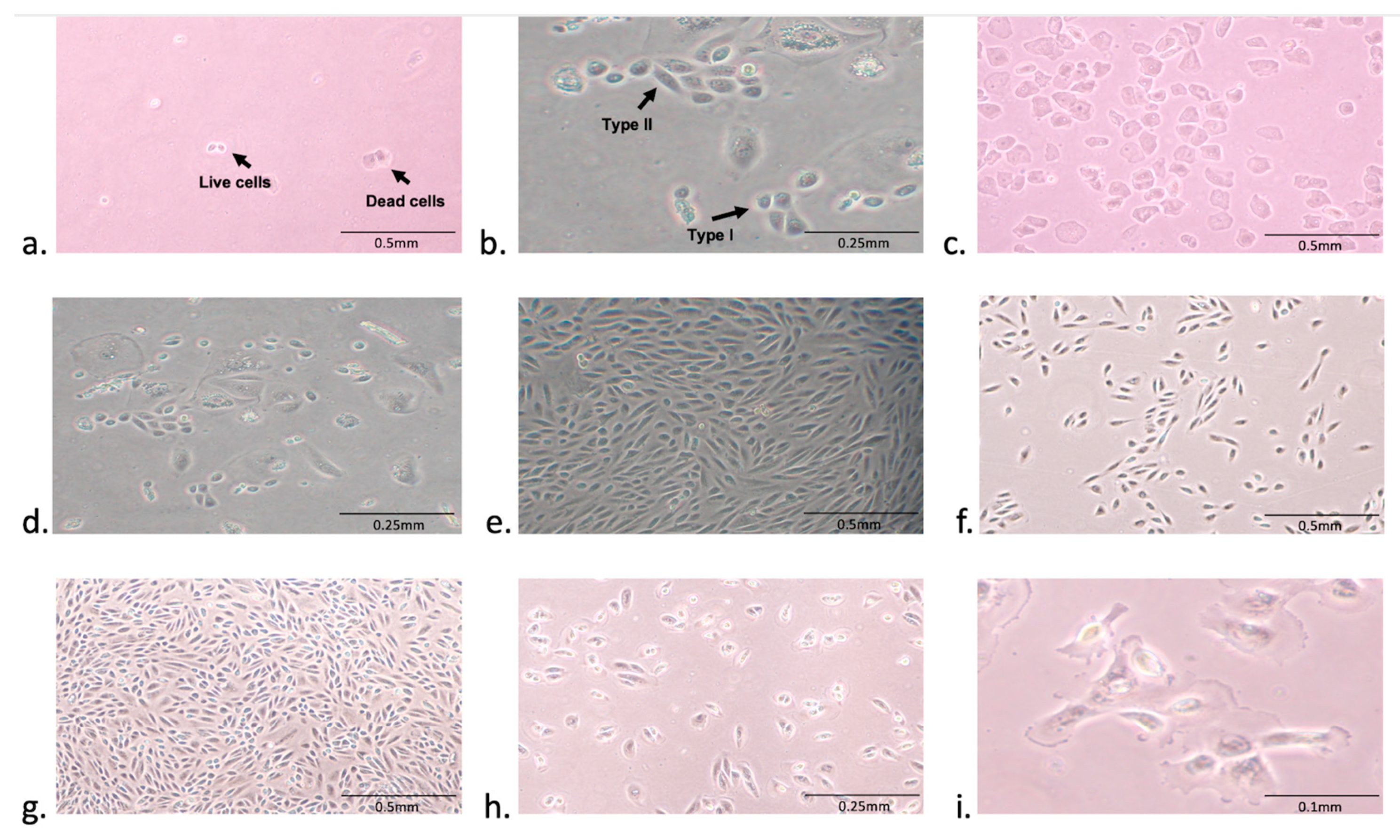
2.3. Primary Culture of Urine-Derived Epithelial Cells
2.4. Proliferation of Urine-Derived Epithelial Cells
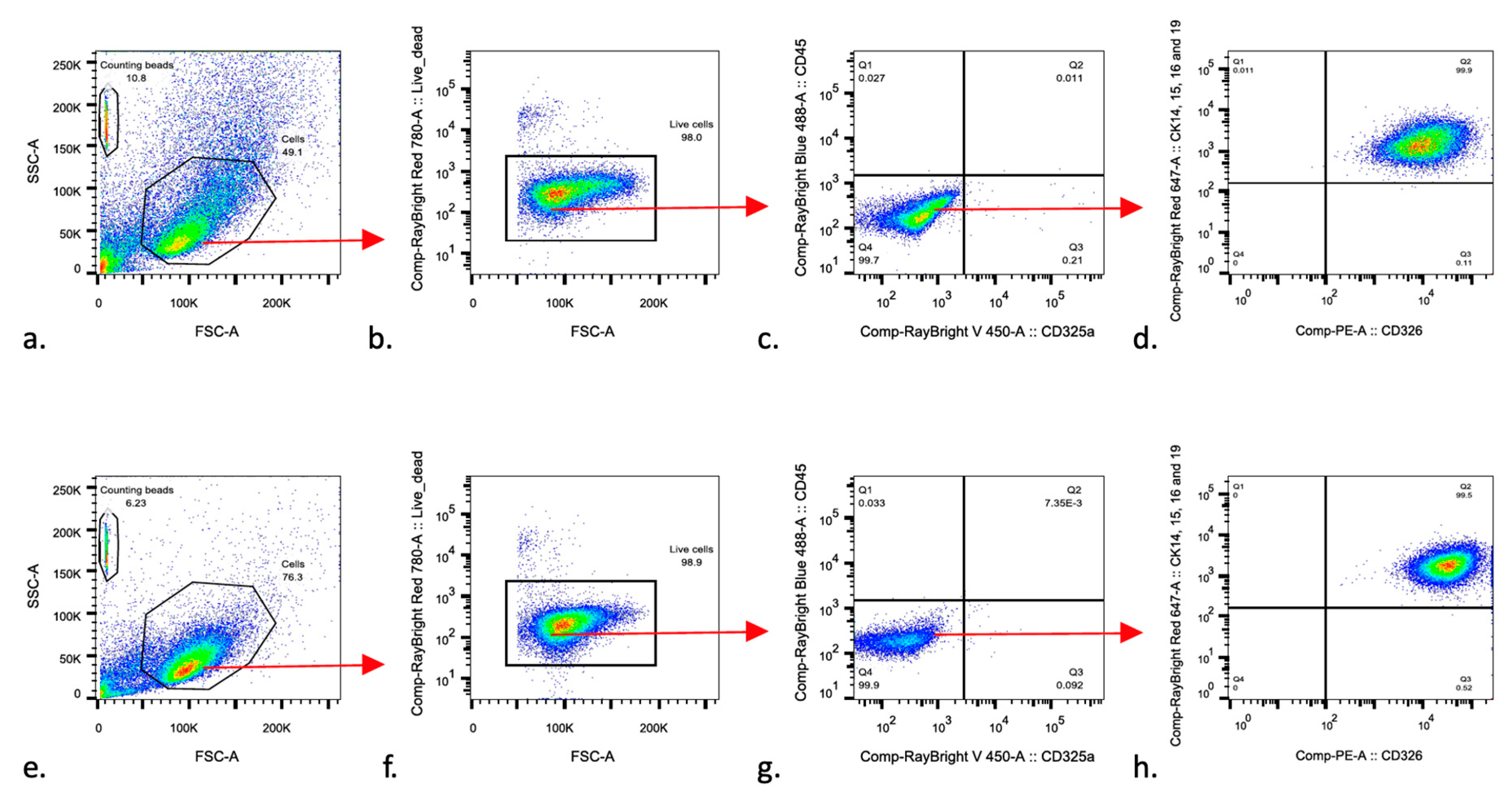
2.5. Fluorescence-Activated Cell Sorting (FACS)
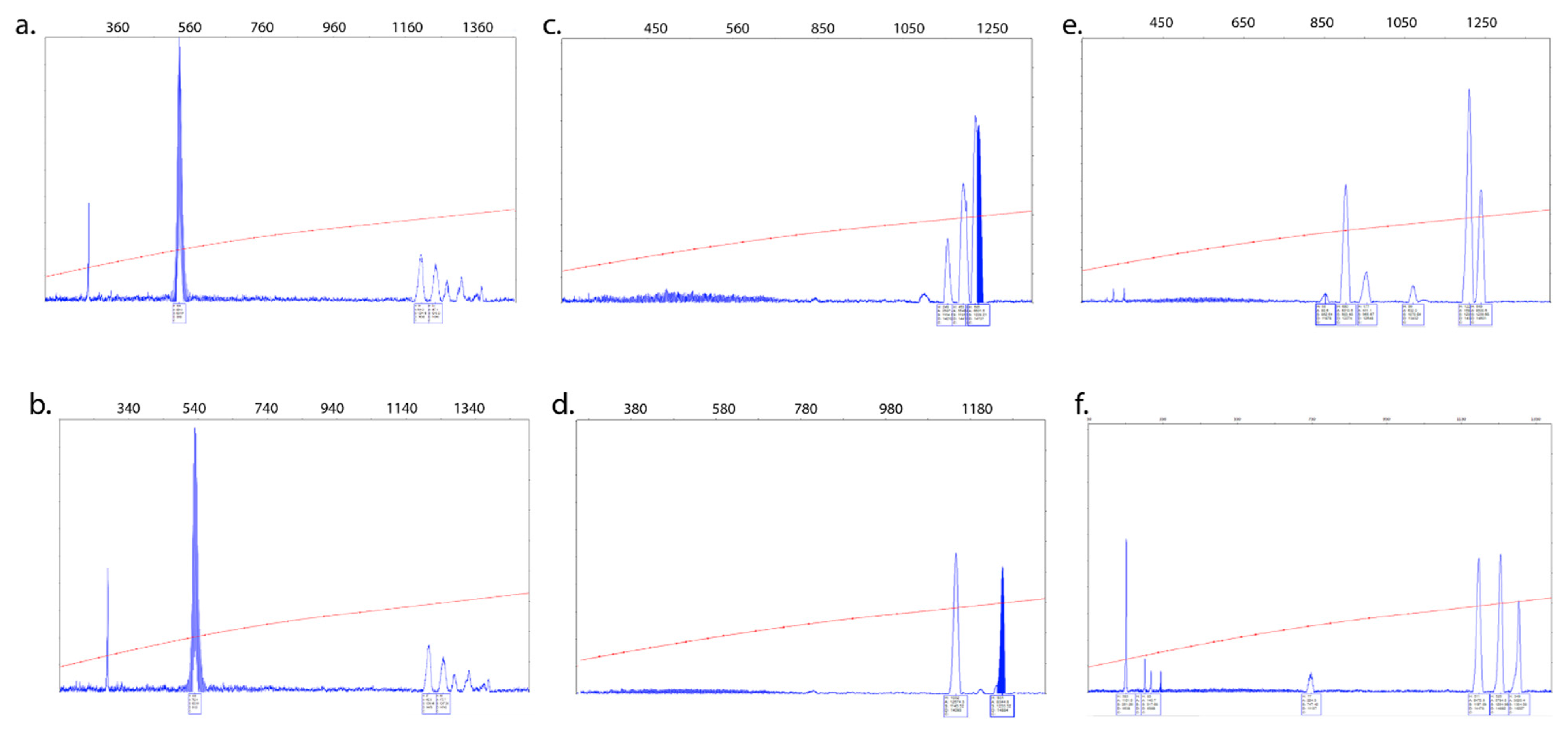
2.6. CGG Repeat Allele Size and Methylation Status
2.7. mRNA Expression Levels
2.8. Western Blot Analysis
2.9. Immunofluorescence Staining
3. Results
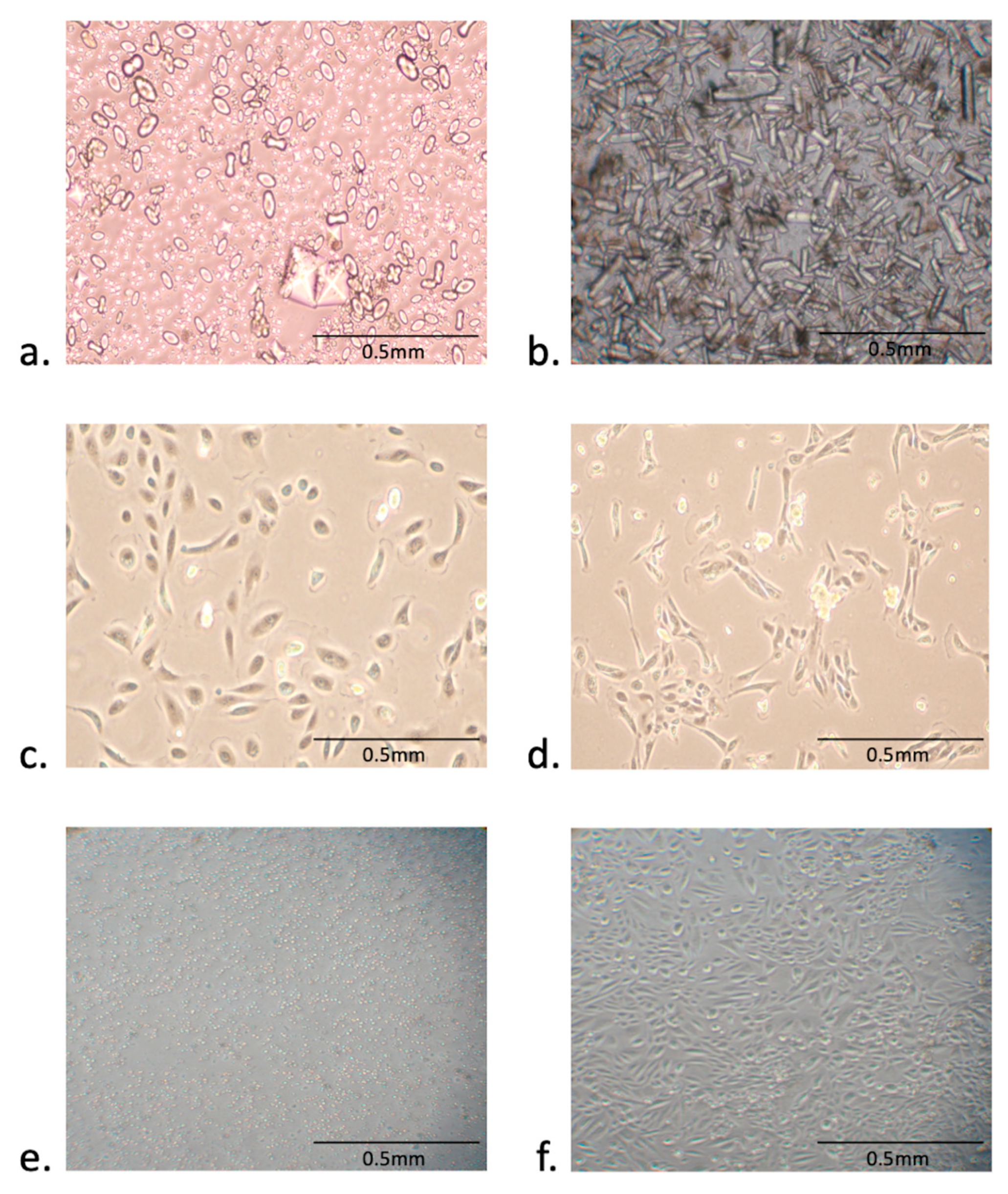
3.1. Isolation and Expansion of Urine-Derived Epithelial Cells
3.2. Urine-Derived Cells Expressed Epithelial Cell Surface Markers
3.3. Intra- and Inter-Tissue Mosaicism Detected in PBMCs and Urine-Derived Epithelial Cells
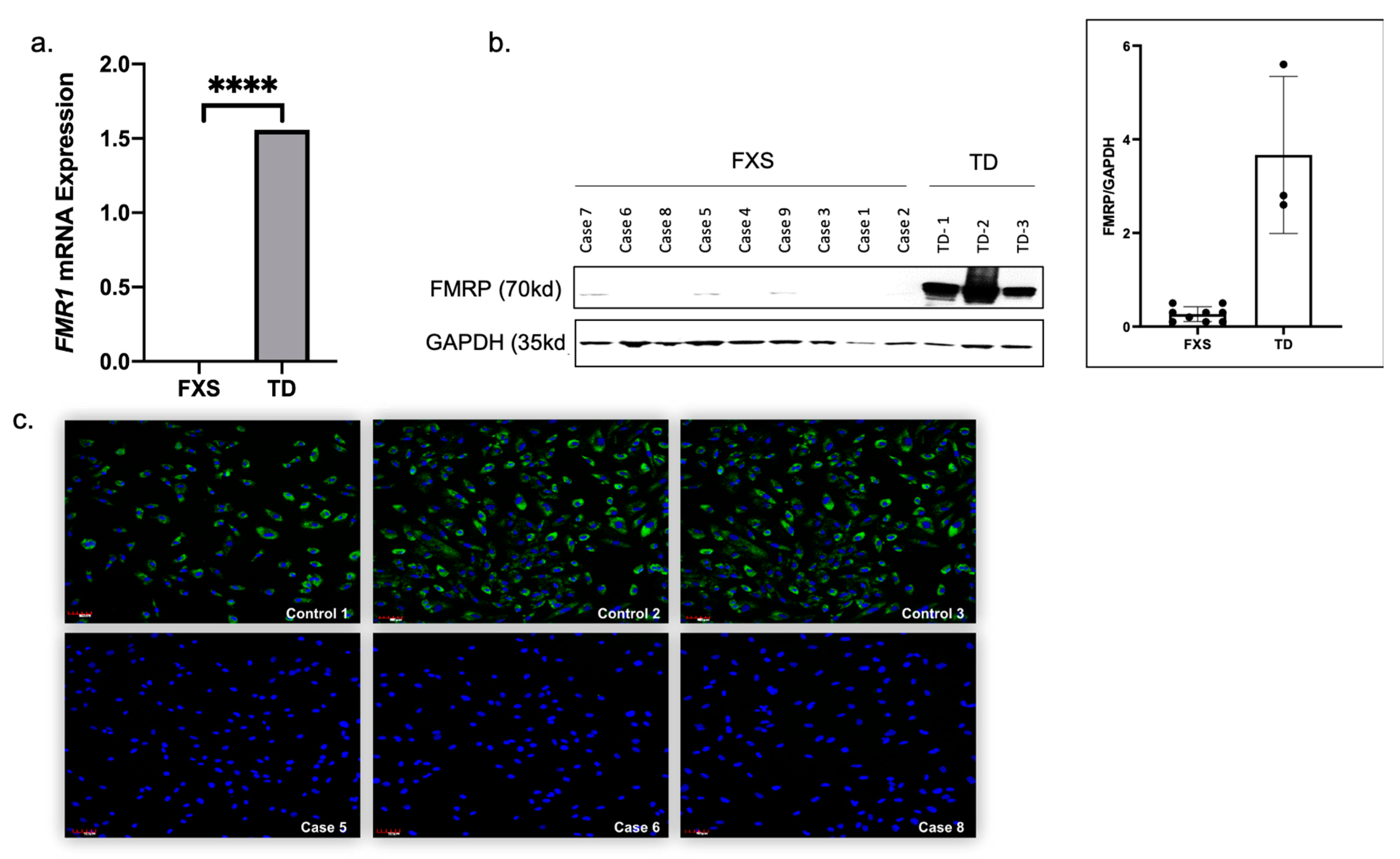
3.4. Urine-Derived Epithelial Cells Express FMR1 mRNA and FMRP Protein
3.5. Factors Affecting the Establishment of Urine-Derived Epithelial Cells
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Harris, J.C. New classification for neurodevelopmental disorders in DSM-5. Curr. Opin. Psychiatry 2014, 27, 95–97. [Google Scholar] [CrossRef]
- Pretto, D.; Yrigollen, C.M.; Tang, H.-T.; Williamson, J.; Espinal, G.; Iwahashi, C.K.; Durbin-Johnson, B.; Hagerman, R.J.; Hagerman, P.J.; Tassone, F. Clinical and molecular implications of mosaicism in FMR1 full mutations. Front. Genet. 2014, 5, 318. [Google Scholar] [CrossRef]
- Nolin, S.L.; Glicksman, A.; Ersalesi, N.; Dobkin, C.; Ted Brown, W.; Cao, R.; Blatt, E.; Sah, S.; Latham, G.J.; Hadd, A.G. Fragile X full mutation expansions are inhibited by one or more AGG interruptions in premutation carriers. Genet. Med. 2015, 17, 358–364. [Google Scholar] [CrossRef]
- Chen, X.; Wang, J.; Xie, H.; Zhou, W.; Wu, Y.; Wang, J.; Qin, J.; Guo, J.; Gu, Q.; Zhang, X.; et al. Fragile X syndrome screening in Chinese children with unknown intellectual developmental disorder. BMC Pediatr. 2015, 15, 77. [Google Scholar] [CrossRef]
- Kim, E.Y.; Page, P.; Dellefave-Castillo, L.M.; McNally, E.M.; Wyatt, E.J. Direct reprogramming of urine-derived cells with inducible MyoD for modeling human muscle disease. Skeletal Muscle 2016, 6, 32. [Google Scholar] [CrossRef]
- Lazzeri, E.; Ronconi, E.; Angelotti, M.L.; Peired, A.; Mazzinghi, B.; Becherucci, F.; Conti, S.; Sansavini, G.; Sisti, A.; Ravaglia, F.; et al. Human Urine-Derived Renal Progenitors for Personalized Modeling of Genetic Kidney Disorders. J. Am. Soc. Nephrol. 2015, 26, 1961–1974. [Google Scholar] [CrossRef] [PubMed]
- Afzal, M.Z.; Strande, J.L. Generation of Induced Pluripotent Stem Cells from Muscular Dystrophy Patients: Efficient Integration-free Reprogramming of Urine Derived Cells. J. Vis. Exp. 2015, 95, e52032. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-Z.; Li, H.-F.; Ma, L.-X.; Qian, W.-J.; Wang, Z.-F.; Wu, Z.-Y. Urine-derived induced pluripotent stem cells as a modeling tool for paroxysmal kinesigenic dyskinesia. Biol. Open 2015, 4, 1744–1752. [Google Scholar] [CrossRef] [PubMed]
- Zhang, D.; Wei, G.; Li, P.; Zhou, X.; Zhang, Y. Urine-derived stem cells: A novel and versatile progenitor source for cell-based therapy and regenerative medicine. Genes Dis. 2014, 1, 8–17. [Google Scholar] [CrossRef]
- Filipovic-Sadic, S.; Sah, S.; Chen, L.; Krosting, J.; Sekinger, E.; Zhang, W.; Hagerman, P.J.; Stenzel, T.T.; Hadd, A.G.; Latham, G.J.; et al. A novel FMR1 PCR method for the routine detection of low abundance expanded alleles and full mutations in fragile X syndrome. Clin. Chem. 2010, 56, 399–408. [Google Scholar] [CrossRef]
- Tassone, F.; Pan, R.; Amiri, K.; Taylor, A.K.; Hagerman, P.J. A Rapid Polymerase Chain Reaction-Based Screening Method for Identification of All Expanded Alleles of the Fragile X (FMR1) Gene in Newborn and High-Risk Populations. J. Mol. Diagn. 2008, 10, 43–49. [Google Scholar] [CrossRef] [PubMed]
- Tassone, F.; Hagerman, R.J.; Chamberlain, W.D.; Hagerman, P.J. Transcription of the FMR1 gene in individuals with fragile X syndrome. Am. J. Med. Genet. 2000, 97, 195–203. [Google Scholar] [CrossRef]
- Zhou, T.; Benda, C.; Dunzinger, S.; Huang, Y.; Ho, J.C.; Yang, J.; Wang, Y.; Zhang, Y.; Zhuang, Q.; Li, Y.; et al. Generation of human induced pluripotent stem cells from urine samples. Nat. Protoc. 2012, 7, 2080–2089. [Google Scholar] [CrossRef] [PubMed]
- Sauer, V.; Tchaikovskaya, T.; Wang, X.; Li, Y.; Zhang, W.; Tar, K.; Polgar, Z.; Ding, J.; Guha, C.; Fox, I.J.; et al. Human Urinary Epithelial Cells as a Source of Engraftable Hepatocyte-Like Cells Using Stem Cell Technology. Cell Transpl. 2016, 25, 2221–2243. [Google Scholar] [CrossRef]
- Loesch, D.Z.; Sherwell, S.; Kinsella, G.; Tassone, F.; Taylor, A.; Amor, D.; Sung, S.; Evans, A. Fragile X-associated tremor/ataxia phenotype in a male carrier of unmethylated full mutation in the FMR1 gene. Clin. Genet. 2012, 82, 88–92. [Google Scholar] [CrossRef]
- Tassone, F.; Hagerman, R.J.; Loesch, D.Z.; Lachiewicz, A.; Taylor, A.K.; Hagerman, P.J. Fragile X males with unmethylated, full mutation trinucleotide repeat expansions have elevated levels ofFMR1 messenger RNA. Am. J. Med. Genet. 2000, 94, 232–236. [Google Scholar] [CrossRef]
- Pretto, D.I.; Mendoza-Morales, G.; Lo, J.; Cao, R.; Hadd, A.; Latham, G.J.; Durbin-Johnson, B.; Hagerman, R.; Tassone, F. CGG allele size somatic mosaicism and methylation inFMR1premutation alleles. J. Med. Genet. 2014, 51, 309–318. [Google Scholar] [CrossRef]
- Hadd, A.G.; Filipovic-Sadic, S.; Zhou, L.; Williams, A.; Latham, G.J.; Berry-Kravis, E.; Hall, D.A. A methylation PCR method determines FMR1 activation ratios and differentiates premutation allele mosaicism in carrier siblings. Clin. Epigenetics 2016, 8, 130. [Google Scholar] [CrossRef]
- Mailick, M.R.; Movaghar, A.; Hong, J.; Greenberg, J.S.; DaWalt, L.S.; Zhou, L.; Jackson, J.; Rathouz, P.J.; Baker, M.W.; Brilliant, M.; et al. Health Profiles of Mosaic Versus Non-mosaic FMR1 Premutation Carrier Mothers of Children With Fragile X Syndrome. Front. Genet. 2018, 9, 173. [Google Scholar] [CrossRef]
- Tassone, F.; Longshore, J.; Zunich, J.; Steinbach, P.; Salat, U.; Taylor, A.K. Tissue-specific methylation differences in a fragile X premutation carrier. Clin. Genet. 1999, 55, 346–352. [Google Scholar] [CrossRef]
- Genc, B. Methylation mosaicism of 5′-(CGG)n-3’ repeats in fragile X, premutation and normal individuals. Nucleic Acids Res. 2000, 28, 2141–2152. [Google Scholar] [CrossRef] [PubMed]
- Bonarrigo, F.A.; Russo, S.; Vizziello, P.; Menni, F.; Cogliati, F.; Giorgini, V.; Monti, F.; Milani, D. Think About It. J. Child Neurol. 2014, 29, NP74–NP77. [Google Scholar] [CrossRef] [PubMed]
- Bharadwaj, S.; Liu, G.; Shi, Y.; Wu, R.; Yang, B.; He, T.; Fan, Y.; Lu, X.; Zhou, X.; Liu, H.; et al. Multipotential differentiation of human urine-derived stem cells: Potential for therapeutic applications in urology. Stem Cells 2013, 31, 1840–1856. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.-Z.; Ma, L.-X.; Qian, W.-J.; Li, H.-F.; Wang, Z.-F.; Wang, H.-X.; Wu, Z.-Y. Modeling Neurological Disease by Rapid Conversion of Human Urine Cells into Functional Neurons. Stem Cells Int. 2016, 2016, 2452985. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.; Wang, L.; Huang, W.; Su, H.; Xue, Y.; Su, Z.; Liao, B.; Wang, H.; Bao, X.; Qin, D.; et al. Generation of integration-free neural progenitor cells from cells in human urine. Nat. Methods 2013, 10, 84–89. [Google Scholar] [CrossRef]
- Cheng, L.; Hu, W.; Qiu, B.; Zhao, J.; Yu, Y.; Guan, W.; Wang, M.; Yang, W.; Pei, G. Generation of neural progenitor cells by chemical cocktails and hypoxia. Cell Res. 2014, 24, 665–679. [Google Scholar] [CrossRef]
- Bento, G.; Shafigullina, A.K.; Rizvanov, A.A.; Sardão, V.A.; Macedo, M.P.; Oliveira, P.J. Urine-Derived Stem Cells: Applications in Regenerative and Predictive Medicine. Cells 2020, 9, 573. [Google Scholar] [CrossRef]
| Participants | Age | Gender | Peripheral Blood Mutation Category | Peripheral Blood CGG Repeat Number * | Peripheral Blood (%) Methylation | Mutation Category | Epithelial Cells CGG Repeat Number | Peripheral Blood FMR1 mRNA Level (StErr) | Epithelial Cells FMR1 mRNA Level |
|---|---|---|---|---|---|---|---|---|---|
| Case 1 | 14 | M | Full mutation | >200 | Full mutation | >200 | 0 | 0 | |
| Case 2 | 20 | M | Full mutation | >200 | Full mutation | >200 | 0 | ||
| Case 3 | 8 | M | Full mutation | >200 | Full mutation | >200 | 0 | ||
| Case 4 | 25 | M | Full mutation | >200 | Full mutation | >200 | 0 | ||
| Case 5 | 13 | M | Full mutation | >200 | Full mutation | >200 | 0.01 (0.002) | 0 | |
| Case 6 | 20 | M | Full mutation | >200 | Full mutation | >200 | 0.009 (0.001) | 0 | |
| Case 7 | 18 | M | Full mutation, Meth mosaic | >200 (30–200) ** | >95% | Full mutation, Meth mosaic | >200 (30–200) ** | 0.29 (0.03) | |
| Case 8 | 8 | M | Full mutation, Meth mosaic | >200 (240–350) ** | 85% | Full mutation | >200*** | 0.47 (0.01) | 0 |
| Case 9 | 8 | M | Full mutation | >200*** | Full mutation | >200 | 0.16 (0.004) | 0 | |
| Case 10 | 13 | M | Full mutation | >200 | Full mutation | >200 | 0 | ||
| Case 11 | 15 | M | Full mutation, Size mosaic | >200 (103) ** | 96% | Full mutation, Size mosaic | >200 (103) ** | 0.15 (0.06) | |
| Case 12 | 17 | M | Full mutation | >200 | Full mutation | >200 | 0 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zafarullah, M.; Jasoliya, M.; Tassone, F. Urine-Derived Epithelial Cell Lines: A New Tool to Model Fragile X Syndrome (FXS). Cells 2020, 9, 2240. https://doi.org/10.3390/cells9102240
Zafarullah M, Jasoliya M, Tassone F. Urine-Derived Epithelial Cell Lines: A New Tool to Model Fragile X Syndrome (FXS). Cells. 2020; 9(10):2240. https://doi.org/10.3390/cells9102240
Chicago/Turabian StyleZafarullah, Marwa, Mittal Jasoliya, and Flora Tassone. 2020. "Urine-Derived Epithelial Cell Lines: A New Tool to Model Fragile X Syndrome (FXS)" Cells 9, no. 10: 2240. https://doi.org/10.3390/cells9102240
APA StyleZafarullah, M., Jasoliya, M., & Tassone, F. (2020). Urine-Derived Epithelial Cell Lines: A New Tool to Model Fragile X Syndrome (FXS). Cells, 9(10), 2240. https://doi.org/10.3390/cells9102240






