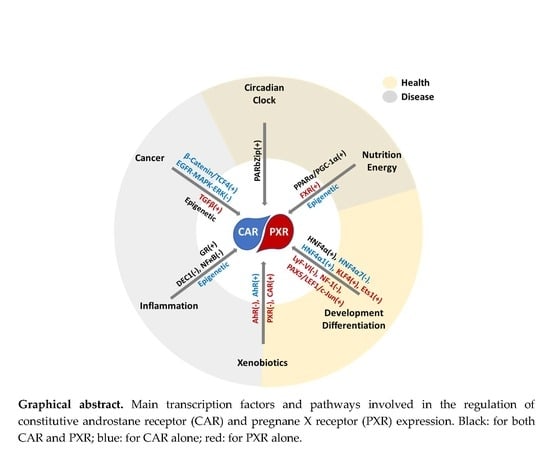
Regulation of CAR and PXR Expression in Health and Disease
Abstract
:1. Introduction
2. CAR and PXR Expression during Development
3. PXR and CAR Tissue Distribution
4. Splicing Variants
5. Circadian Clock
6. Hepatic Functional Zonation
7. Nutritional Status and Microbiome Metabolites
8. Transcriptional Regulation by Transcription Factors
8.1. Hepatic Nuclear Factor 4α
8.2. Glucocorticoid Receptor
8.3. Aryl Hydrocarbon Receptor
8.4. Farnesoid X Receptor
8.5. Thyroid Receptor
8.6. Other Transcription Factors
9. Epigenetic and Post-Transcriptional Regulations
9.1. Histone Modifications
9.2. DNA Methylation
9.3. microRNAs (miRNAs)
9.4. Long Non-Coding RNAs (lncRNAs)
9.5. RNA Editing
10. Post-Translational Regulation
11. Pathologies
11.1. Inflammation
11.1.1. Liver
11.1.2. Intestine
11.2. Chronic Diseases
11.2.1. Diabetes and Obesity
11.2.2. Fibrosis and Cirrhosis
11.2.3. Cholestasis
11.3. Cancer
12. Xenobiotics
12.1. Nanoparticles
12.2. Environmental Pollutants and Food Contaminants
12.3. Drugs
12.4. Food Components
13. Conclusions
Funding
Conflicts of Interest
References
- Yan, J.; Xie, W. A brief history of the discovery of PXR and CAR as xenobiotic receptors. Acta Pharm. Sin. B 2016, 6, 450–452. [Google Scholar] [CrossRef] [Green Version]
- Oladimeji, P.O.; Chen, T. PXR: More than just a master xenobiotic receptor. Mol. Pharmacol. 2017, 93, 119–127. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, M.; Robbins, D.; Chen, T. Targeting xenobiotic receptors PXR and CAR in human diseases. Drug Discov. Today 2015, 20, 618–628. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mackowiak, B.; Hodge, J.; Stern, S.; Wang, H. The roles of xenobiotic receptors: Beyond chemical disposition. Drug Metab. Dispos. 2018, 46, 1361–1371. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xiao, L.; Nickbarg, E.; Wang, W.; Thomas, A.; Ziebell, M.; Prosise, W.W.; Lesburg, C.A.; Taremi, S.S.; Gerlach, V.L.; Le, H.V.; et al. Evaluation of in vitro PXR-based assays and in silico modeling approaches for understanding the binding of a structurally diverse set of drugs to PXR. Biochem. Pharmacol. 2011, 81, 669–679. [Google Scholar] [CrossRef]
- Yang, H.; Wang, H. Signaling control of the constitutive androstane receptor (CAR). Protein Cell 2014, 5, 113–123. [Google Scholar] [CrossRef] [Green Version]
- Balasubramaniyan, N.; Shahid, M.; Suchy, F.J.; Ananthanarayanan, M. Multiple mechanisms of ontogenic regulation of nuclear receptors during rat liver development. Am. J. Physiol. Liver Physiol. 2005, 288, G251–G260. [Google Scholar] [CrossRef]
- Xu, S.-F.; Hu, A.-L.; Xie, L.; Liu, J.-J.; Wu, Q.; Liu, J. Age-associated changes of cytochrome P450 and related phase-2 gene/proteins in livers of rats. PeerJ 2019, 7, e7429. [Google Scholar] [CrossRef]
- Chen, S.; He, N.; Yu, J.; Li, L.; Hu, Y.; Deng, R.; Zhong, S.; Shen, L. Post-transcriptional regulation by miR-137 underlies the low abundance of CAR and low rate of bilirubin clearance in neonatal mice. Life Sci. 2014, 107, 8–13. [Google Scholar] [CrossRef]
- Greger, D.; Philipona, C.; Blum, J. Ontogeny of mRNA abundance of nuclear receptors and nuclear receptor target genes in young cattle. Domest. Anim. Endocrinol. 2006, 31, 76–87. [Google Scholar] [CrossRef]
- Marques, C.; Roberto, V.P.; Granadeiro, L.; Trindade, M.; Gavaia, P.J.; Laizé, V.; Cancela, M.L.; Fernández, I. The xenobiotic sensor PXR in a marine flatfish species (Solea senegalensis): Gene expression patterns and its regulation under different physiological conditions. Mar. Environ. Res. 2017, 130, 187–199. [Google Scholar] [CrossRef]
- Funakoshi, N.; Duret, C.; Pascussi, J.-M.; Blanc, P.; Maurel, P.; Daujat-Chavanieu, M.; Gerbal-Chaloin, S. Comparison of hepatic-like cell production from human embryonic stem cells and adult liver progenitor cells: CAR transduction activates a battery of detoxification genes. Stem Cell Rev. Rep. 2011, 7, 518–531. [Google Scholar] [CrossRef] [Green Version]
- Betts, S.; Björkhem-Bergman, L.; Rane, A.; Ekström, L. Expression of CYP3A4 and CYP3A7 in human foetal tissues and its correlation with nuclear receptors. Basic Clin. Pharmacol. Toxicol. 2015, 117, 261–266. [Google Scholar] [CrossRef] [PubMed]
- Chen, F.; Zamule, S.M.; Coslo, D.M.; Chen, T.; Omiecinski, C.J. The human constitutive androstane receptor promotes the differentiation and maturation of hepatic-like cells. Dev. Biol. 2013, 384, 155–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Miki, Y.; Suzuki, T.; Tazawa, C.; Blumberg, B.; Sasano, H. Steroid and xenobiotic receptor (SXR), cytochrome P450 3A4 and multidrug resistance gene 1 in human adult and fetal tissues. Mol. Cell. Endocrinol. 2005, 231, 75–85. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Soto-Gutierrez, A.; Baptista, P.M.; Spee, B. Biotechnology challenges to in vitro maturation of hepatic stem cells. Gastroenterology 2018, 154, 1258–1272. [Google Scholar] [CrossRef] [Green Version]
- Godoy, P.; Schmidt-Heck, W.; Natarajan, K.; Lucendo-Villarin, B.; Szkolnicka, D.; Asplund, A.; Björquist, P.; Widera, A.; Stöber, R.; Campos, G.; et al. Gene networks and transcription factor motifs defining the differentiation of stem cells into hepatocyte-like cells. J. Hepatol. 2015, 63, 934–942. [Google Scholar] [CrossRef] [Green Version]
- Xie, W.; Barwick, J.L.; Downes, M.; Blumberg, B.; Simon, C.M.; Nelson, M.C.; Neuschwander-Tetri, B.A.; Brunt, E.M.; Guzelian, P.S.; Evans, R.M. Humanized xenobiotic response in mice expressing nuclear receptor SXR. Nat. Cell Biol. 2000, 406, 435–439. [Google Scholar] [CrossRef]
- Wei, P.; Zhang, J.; Egan-Hafley, M.; Liang, S.; Moore, D.D. The nuclear receptor CAR mediates specific xenobiotic induction of drug metabolism. Nat. Cell Biol. 2000, 407, 920–923. [Google Scholar] [CrossRef]
- Waxman, D.J.; Holloway, M.G. Sex differences in the expression of hepatic drug metabolizing enzymes. Mol. Pharmacol. 2009, 76, 215–228. [Google Scholar] [CrossRef] [Green Version]
- Lu, Y.-F.; Jin, T.; Xu, Y.; Zhang, D.; Wu, Q.; Zhang, Y.-K.J.; Liu, J. Sex differences in the circadian variation of cytochrome P450 genes and corresponding nuclear receptors in mouse liver. Chronobiol. Int. 2013, 30, 1135–1143. [Google Scholar] [CrossRef]
- Kennedy, M.J. Hormonal regulation of hepatic drug-metabolizing enzyme activity during adolescence. Clin. Pharmacol. Ther. 2008, 84, 662–673. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, H.; LeCulyse, E.; Liu, L.; Hu, M.; Matoney, L.; Zhu, W.; Yan, B. Rat pregnane X receptor: Molecular cloning, tissue distribution, and xenobiotic regulation. Arch. Biochem. Biophys. 1999, 368, 14–22. [Google Scholar] [CrossRef] [PubMed]
- Nishimura, M.; Naito, S.; Yokoi, T. Tissue-specific mRNA expression profiles of human nuclear receptor subfamilies. Drug Metab. Pharmacokinet. 2004, 19, 135–149. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Wang, Y.; Chen, Z.; Ma, Z.-C.; You, Q.; Zhang, X.; Zhou, T.; Xiao, Y.; Liang, Q.; Tan, H.; et al. Artemisinin protects against dextran sulfate-sodium-induced inflammatory bowel disease, which is associated with activation of the pregnane X receptor. Eur. J. Pharmacol. 2014, 738, 273–284. [Google Scholar] [CrossRef]
- Pollock, C.B.; Rogatcheva, M.B.; Schook, L.B. Comparative genomics of xenobiotic metabolism: A porcine-human PXR gene comparison. Mamm. Genome 2007, 18, 210–219. [Google Scholar] [CrossRef]
- Wassmur, B.; Gräns, J.; Kling, P.; Celander, M.C. Interactions of pharmaceuticals and other xenobiotics on hepatic pregnane X receptor and cytochrome P450 3A signaling pathway in rainbow trout (Oncorhynchus mykiss). Aquat. Toxicol. 2010, 100, 91–100. [Google Scholar] [CrossRef] [Green Version]
- Xing, Y.; Yan, J.; Niu, Y. PXR: A center of transcriptional regulation in cancer. Acta Pharm. Sin. B 2019, 10, 197–206. [Google Scholar] [CrossRef]
- Hudson, G.M.; Flannigan, K.L.; Erickson, S.L.; Vicentini, F.A.; Zamponi, A.; Hirota, C.L.; Alston, L.; Altier, C.; Ghosh, S.; Rioux, K.P.; et al. Constitutive androstane receptor regulates the intestinal mucosal response to injury. Br. J. Pharmacol. 2017, 174, 1857–1871. [Google Scholar] [CrossRef]
- Chirulli, V.; Longo, V.; Marini, S.; Mazzaccaro, A.; Fiorio, R.; Gervasi, P.G. CAR and PXR expression and inducibility of CYP2B and CYP3A activities in rat and rabbit lungs. Life Sci. 2005, 76, 2535–2546. [Google Scholar] [CrossRef]
- Baes, M.; Gulick, T.; Choi, H.S.; Martinoli, M.G.; Simha, D.; Moore, D.D. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol. Cell. Biol. 1994, 14, 1544–1552. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Choi, H.-S.; Chung, M.; Tzameli, I.; Simha, D.; Lee, Y.-K.; Seol, W.; Moore, D.D. Differential transactivation by two isoforms of the orphan nuclear hormone receptor CAR. J. Biol. Chem. 1997, 272, 23565–23571. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, Z.; Kruijt, J.K.; Van Der Sluis, R.J.; Van Berkel, T.J.C.; Hoekstra, M.; Der Sluis, R.J.V.; Berkel, T.J.C.V. Nuclear receptor atlas of female mouse liver parenchymal, endothelial, and Kupffer cells. Physiol. Genom. 2013, 45, 268–275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lu, P.; Prost, S.; Caldwell, H.; Tugwood, J.D.; Betton, G.R.; Harrison, D.J. Microarray analysis of gene expression of mouse hepatocytes of different ploidy. Mamm. Genome 2007, 18, 617–626. [Google Scholar] [CrossRef]
- Wallace, K.; Cowie, D.E.; Konstantinou, D.K.; Hill, S.J.; Tjelle, T.E.; Axon, A.; Koruth, M.; White, S.A.; Carlsen, H.; Mann, D.A.; et al. The PXR is a drug target for chronic inflammatory liver disease. J. Steroid Biochem. Mol. Biol. 2010, 120, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Haughton, E.L.; Tucker, S.J.; Marek, C.J.; Durward, E.; Leel, V.; Bascal, Z.; Monaghan, T.M.; Koruth, M.; Collie–Duguid, E.; Mann, D.A.; et al. Pregnane X receptor activators inhibit human hepatic stellate cell transdifferentiation in vitro. Gastroenterology 2006, 131, 194–209. [Google Scholar] [CrossRef]
- Renga, B.; Daniela, F.; D’Amore, C.; Schiaroli, E.; Carino, A.; Baldelli, F.; Fiorucci, S. HIV-1 infection is associated with changes in nuclear receptor transcriptome, pro-inflammatory and lipid profile of monocytes. BMC Infect. Dis. 2012, 12, 274. [Google Scholar] [CrossRef] [Green Version]
- Schote, A.B.; Turner, J.D.; Schiltz, J.; Muller, C.P. Nuclear receptors in human immune cells: Expression and correlations. Mol. Immunol. 2007, 44, 1436–1445. [Google Scholar] [CrossRef] [PubMed]
- Hudson, G.M.; Flannigan, K.L.; Venu, V.K.P.; Alston, L.; Sandall, C.F.; Macdonald, J.A.; Muruve, D.A.; Chang, T.K.; Mani, S.; A Hirota, S. Pregnane X receptor activation triggers rapid atp release in primed macrophages that mediates NLRP3 inflammasome activation. J. Pharmacol. Exp. Ther. 2019, 370, 44–53. [Google Scholar] [CrossRef]
- Mencarelli, A.; Distrutti, E.; Renga, B.; D’Amore, C.; Cipriani, S.; Palladino, G.; Donini, A.; Ricci, P.; Fiorucci, S. Probiotics modulate intestinal expression of nuclear receptor and provide counter-regulatory signals to inflammation-driven adipose tissue activation. PLoS ONE 2011, 6, e22978. [Google Scholar] [CrossRef] [Green Version]
- Takeuchi, Y.; Kito, A.; Itoh, S.; Naruse, H.; Fujikawa, J.; Sadek, K.M.; Akiyama, S.; Yamashiro, T.; Wakisaka, S.; Abe, M. Kruppel-like factor 4 represses osteoblast differentiation via ciliary Hedgehog signaling. Exp. Cell Res. 2018, 371, 417–425. [Google Scholar] [CrossRef]
- Pavek, P.; Smutný, T. Nuclear receptors in regulation of biotransformation enzymes and drug transporters in the placental barrier. Drug Metab. Rev. 2013, 46, 19–32. [Google Scholar] [CrossRef]
- Torres-Vergara, P.; Ho, Y.S.; Espinoza, F.; Nualart, F.; Escudero, C.; Penny, J. The constitutive androstane receptor and pregnane X receptor in the brain. Br. J. Pharmacol. 2020, 177, 2666–2682. [Google Scholar] [CrossRef] [PubMed]
- Auerbach, S.S.; Ramsden, R.; Stoner, M.A.; Verlinde, C.; Hassett, C.; Omiecinski, C.J. Alternatively spliced isoforms of the human constitutive androstane receptor. Nucleic Acids Res. 2003, 31, 3194–3207. [Google Scholar] [CrossRef] [Green Version]
- Jinno, H.; Tanaka-Kagawa, T.; Hanioka, N.; Ishida, S.; Saeki, M.; Soyama, A.; Itoda, M.; Nishimura, T.; Saito, Y.; Ozawa, S.; et al. Identification of novel alternative splice variants of human constitutive androstane receptor and characterization of their expression in the liver. Mol. Pharmacol. 2004, 65, 496–502. [Google Scholar] [CrossRef] [Green Version]
- Lamba, J.K.; Lamba, V.; Yasuda, K.; Lin, Y.S.; Assem, M.; Thompson, E.; Strom, S.C.; Schuetz, E. Expression of constitutive androstane receptor splice variants in human tissues and their functional consequences. J. Pharmacol. Exp. Ther. 2004, 311, 811–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- DeKeyser, J.G.; Laurenzana, E.M.; Peterson, E.C.; Chen, T.; Omiecinski, C.J. Selective phthalate activation of naturally occurring human constitutive androstane receptor splice variants and the pregnane X receptor. Toxicol. Sci. 2011, 120, 381–391. [Google Scholar] [CrossRef] [PubMed]
- DeKeyser, J.G.; Stagliano, M.C.; Auerbach, S.S.; Prabhu, K.S.; Jones, A.D.; Omiecinski, C.J. Di(2-ethylhexyl) phthalate is a highly potent agonist for the human constitutive androstane receptor splice variant CAR2. Mol. Pharmacol. 2009, 75, 1005–1013. [Google Scholar] [CrossRef] [Green Version]
- Rondini, E.A.; Duniec-Dmuchowski, Z.; Kocarek, T.A. Nonsterol isoprenoids activate human constitutive androstane receptor in an isoform-selective manner in primary cultured mouse hepatocytes. Drug Metab. Dispos. 2016, 44, 595–604. [Google Scholar] [CrossRef] [Green Version]
- Sharma, A.; Yun, H.; Jyotsana, N.; Chaturvedi, A.; Schwarzer, A.; Yung, E.; Lai, C.K.; Kuchenbauer, F.; Argiropoulos, B.; Görlich, K.; et al. Constitutive IRF8 expression inhibits AML by activation of repressed immune response signaling. Leukemia 2014, 29, 157–168. [Google Scholar] [CrossRef]
- Ross, J.; Plummer, S.M.; Rode, A.; Scheer, N.; Bower, C.C.; Vogel, O.; Henderson, C.J.; Wolf, C.R.; Elcombe, C. Human Constitutive Androstane Receptor (CAR) and Pregnane X Receptor (PXR) support the hypertrophic but not the hyperplastic response to the murine nongenotoxic hepatocarcinogens phenobarbital and chlordane in vivo. Toxicol. Sci. 2010, 116, 452–466. [Google Scholar] [CrossRef] [Green Version]
- Arnold, K.; Eichelbaum, M.; Burk, O. Alternative splicing affects the function and tissue-specific expression of the human constitutive androstane receptor. Nucl. Recept. 2004, 2, 1. [Google Scholar] [CrossRef] [Green Version]
- Gray, M.A.; Peacock, J.N.; Squires, E.J. Characterization of the porcine constitutive androstane receptor (CAR) and its splice variants. Xenobiotica 2009, 39, 915–930. [Google Scholar] [CrossRef]
- Fukuen, S.; Fukuda, T.; Matsuda, H.; Sumida, A.; Yamamoto, I.; Inaba, T.; Azuma, J. Identification of the novel splicing variants for the hPXR in human livers. Biochem. Biophys. Res. Commun. 2002, 298, 433–438. [Google Scholar] [CrossRef]
- Lamba, J.K. PXR (NR1I2): Splice variants in human tissues, including brain, and identification of neurosteroids and nicotine as PXR activators. Toxicol. Appl. Pharmacol. 2004, 199, 251–265. [Google Scholar] [CrossRef]
- Gardner-Stephen, D.; Heydel, J.-M.; Goyal, A.; Lu, Y.; Xie, W.; Lindblom, T.; MacKenzie, P.I.; Radominska-Pandya, A. Human PXR variants and their differential effects on the regulation of human udp-glucuronosyltransferase gene expression. Drug Metab. Dispos. 2004, 32, 340–347. [Google Scholar] [CrossRef] [Green Version]
- Kliewer, S.; Moore, J.T.; Wade, L.; Staudinger, J.L.; Watson, M.; Jones, S.; McKee, D.D.; Oliver, B.B.; Willson, T.M.; Zetterström, R.H.; et al. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell 1998, 92, 73–82. [Google Scholar] [CrossRef] [Green Version]
- Brewer, C.T.; Chen, T. PXR variants: The impact on drug metabolism and therapeutic responses. Acta Pharm. Sin. B 2016, 6, 441–449. [Google Scholar] [CrossRef] [Green Version]
- Breuker, C.; Planque, C.; Rajabi, F.; Nault, J.-C.; Couchy, G.; Zucman-Rossi, J.; Evrard, A.; Kantar, J.; Chevet, E.; Bioulac-Sage, P.; et al. Characterization of a novel PXR isoform with potential dominant-negative properties. J. Hepatol. 2014, 61, 609–616. [Google Scholar] [CrossRef]
- Mensah-Osman, E.J.; Thomas, D.G.; Tabb, M.M.; Larios, J.M.; Hughes, D.P.; Giordano, T.J.; Ms, M.L.L.; Rae, J.M.; Blumberg, B.; Hollenberg, P.F.; et al. Expression levels and activation of a PXR variant are directly related to drug resistance in osteosarcoma cell lines. Cancer 2007, 109, 957–965. [Google Scholar] [CrossRef]
- Liu, Y.; Ji, W.; Yin, Y.; Fan, L.; Zhang, J.; Yun, H.; Wang, N.; Li, Q.; Wei, Z.; Ouyang, D.; et al. The effects of splicing variant of PXR PAR-2 on CYP3A4 and MDR1 mRNA expressions. Clin. Chim. Acta 2009, 403, 142–144. [Google Scholar] [CrossRef] [PubMed]
- Matic, M.; Corradin, A.P.; Tsoli, M.; Clarke, S.J.; Polly, P.; Robertson, G.R. The alternatively spliced murine pregnane X receptor isoform, mPXRΔ171–211 exhibits a repressive action. Int. J. Biochem. Cell Biol. 2010, 42, 672–682. [Google Scholar] [CrossRef] [PubMed]
- Gray, M.; Pollock, C.B.; Schook, L.B.; Squires, E.J. Characterization of porcine pregnane X receptor, farnesoid X receptor and their splice variants. Exp. Biol. Med. 2010, 235, 718–736. [Google Scholar] [CrossRef]
- Burris, T.P. Nuclear hormone receptors for heme: REV-ERBα and REV-ERBβ are ligand-regulated components of the mammalian clock. Mol. Endocrinol. 2008, 22, 1509–1520. [Google Scholar] [CrossRef] [Green Version]
- Mohawk, J.A.; Green, C.B.; Takahashi, J.S. Central and peripheral circadian clocks in mammals. Annu. Rev. Neurosci. 2012, 35, 445–462. [Google Scholar] [CrossRef] [Green Version]
- Yang, X.; Downes, M.; Yu, R.T.; Bookout, A.L.; He, W.; Straume, M.; Mangelsdorf, D.J.; Evans, R.M. Nuclear receptor expression links the circadian clock to metabolism. Cell 2006, 126, 801–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.-K.J.; Yeager, R.L.; Klaassen, C.D. Circadian expression profiles of drug-processing genes and transcription factors in mouse liver. Drug Metab. Dispos. 2009, 37, 106–115. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chan, G.N.; Saldivia, V.; Yang, Y.; Pang, H.; Lannoy, L.; Bendayan, R. In vivo induction of P-glycoprotein expression at the mouse blood-brain barrier: An intracerebral microdialysis study. J. Neurochem. 2013, 127, 342–352. [Google Scholar] [CrossRef]
- Gachon, F.; Olela, F.F.; Schaad, O.; Descombes, P.; Schibler, U. The circadian PAR-domain basic leucine zipper transcription factors DBP, TEF, and HLF modulate basal and inducible xenobiotic detoxification. Cell Metab. 2006, 4, 25–36. [Google Scholar] [CrossRef] [PubMed]
- De Vries, E.M.; Oosterman, J.E.; Eggink, H.M.; De Goede, P.; Şen, Ş.; Foppen, E.; Boudzovitch-Surovtseva, O.; Boelen, A.; Romijn, J.A.; LaFleur, S.E.; et al. Effects of meal composition and meal timing on the expression of genes involved in hepatic drug metabolism in rats. PLoS ONE 2017, 12, e0185520. [Google Scholar] [CrossRef]
- Kanno, Y.; Otsuka, S.; Hiromasa, T.; Nakahama, T.; Inouye, Y. Diurnal difference in CAR mRNA expression. Nucl. Recept. 2004, 2, 6. [Google Scholar] [CrossRef] [Green Version]
- Patel, R.D.; Hollingshead, B.D.; Omiecinski, C.J.; Perdew, G.H. Aryl-hydrocarbon receptor activation regulates constitutive androstane receptor levels in murine and human liver. Hepatology 2007, 46, 209–218. [Google Scholar] [CrossRef] [Green Version]
- Richardson, V.M.; Santostefano, M.J.; Birnbaum, L.S. Daily cycle of bHLH-PAS proteins, Ah receptor and arnt, in multiple tissues of female sprague–dawley rats. Biochem. Biophys. Res. Commun. 1998, 252, 225–231. [Google Scholar] [CrossRef]
- Lin, Y.; Wang, S.; Zhou, Z.; Guo, L.; Yu, F.; Wu, B. Bmal1 regulates circadian expression of cytochrome P450 3a11 and drug metabolism in mice. Commun. Biol. 2019, 2, 378. [Google Scholar] [CrossRef]
- Kang, H.S.; Angers, M.; Beak, J.Y.; Wu, X.; Gimble, J.M.; Wada, T.; Xie, W.; Collins, J.B.; Grissom, S.F.; Jetten, A.M. Gene expression profiling reveals a regulatory role for RORα and RORγ in phase I and phase II metabolism. Physiol. Genom. 2007, 31, 281–294. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, C.; Li, S.; Liu, T.; Borjigin, J.; Lin, J.D. Transcriptional coactivator PGC-1α integrates the mammalian clock and energy metabolism. Nat. Cell Biol. 2007, 447, 477–481. [Google Scholar] [CrossRef]
- Lamia, K.A.; Papp, S.J.; Yu, R.T.; Barish, G.D.; Uhlenhaut, N.H.; Jonker, J.W.; Downes, M.; Evans, R.M. Cryptochromes mediate rhythmic repression of the glucocorticoid receptor. Nat. Cell Biol. 2011, 480, 552–556. [Google Scholar] [CrossRef]
- Kettner, N.M.; Voicu, H.; Finegold, M.J.; Coarfa, C.; Sreekumar, A.; Putluri, N.; Katchy, C.A.; Lee, C.; Moore, D.D.; Fu, L. Circadian homeostasis of liver metabolism suppresses hepatocarcinogenesis. Cancer Cell 2016, 30, 909–924. [Google Scholar] [CrossRef]
- Ben-Moshe, S.; Itzkovitz, S. Spatial heterogeneity in the mammalian liver. Nat. Rev. Gastroenterol. Hepatol. 2019, 16, 395–410. [Google Scholar] [CrossRef]
- Benhamouche, S.; Decaens, T.; Godard, C.; Chambrey, R.; Rickman, D.S.; Moinard, C.; Vasseur-Cognet, M.; Kuo, C.J.; Kahn, A.; Perret, C.; et al. Apc tumor suppressor gene is the “Zonation-Keeper” of mouse liver. Dev. Cell 2006, 10, 759–770. [Google Scholar] [CrossRef] [Green Version]
- Rocha, A.S.; Vidal, V.; Mertz, M.; Kendall, T.J.; Charlet, A.; Okamoto, H.; Schedl, A. The angiocrine factor Rspondin3 is a key determinant of liver zonation. Cell Rep. 2015, 13, 1757–1764. [Google Scholar] [CrossRef] [Green Version]
- Braeuning, A.; Ittrich, C.; Köhle, C.; Hailfinger, S.; Bonin, M.; Buchmann, A.; Schwarz, M. Differential gene expression in periportal and perivenous mouse hepatocytes. FEBS J. 2006, 273, 5051–5061. [Google Scholar] [CrossRef] [PubMed]
- Braeuning, A.; Sanna, R.; Huelsken, J.; Schwarz, M. Inducibility of drug-metabolizing enzymes by xenobiotics in mice with liver-specific knockout of Ctnnb1. Drug Metab. Dispos. 2009, 37, 1138–1145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gougelet, A.; Torre, C.; Veber, P.; Sartor, C.; Bachelot, L.; Denechaud, P.-D.; Godard, C.; Moldes, M.; Burnol, A.; Dubuquoy, C.; et al. T-cell factor 4 and beta-catenin chromatin occupancies pattern zonal liver metabolism in mice. Hepatology 2014, 59, 2344–2357. [Google Scholar] [CrossRef] [PubMed]
- Kumari, S.; Mukhopadhyay, G.; Tyagi, R.K. Transcriptional regulation of mouse PXR gene: An interplay of transregulatory factors. PLoS ONE 2012, 7, e44126. [Google Scholar] [CrossRef] [Green Version]
- Gerbal-Chaloin, S.; Dumé, A.-S.; Briolotti, P.; Klieber, S.; Raulet, E.; Duret, C.; Fabre, J.-M.; Ramos, J.; Maurel, P.; Daujat-Chavanieu, M. The WNT/β-Catenin pathway is a transcriptional regulator of CYP2E1,CYP1A2, and aryl hydrocarbon receptor gene expression in primary human hepatocytes. Mol. Pharmacol. 2014, 86, 624–634. [Google Scholar] [CrossRef] [Green Version]
- Thomas, M.; Bayha, C.; Vetter, S.; Hofmann, U.; Schwarz, M.; Zanger, U.M.; Braeuning, A. Activating and inhibitory functions of WNT/beta-Catenin in the induction of cytochromes P450 by nuclear receptors in HepaRG cells. Mol. Pharmacol. 2015, 87, 1013–1020. [Google Scholar] [CrossRef] [Green Version]
- Jaworski, M.; Buchmann, A.; Bauer, P.; Riess, O.; Schwarz, M. B-Raf and Ha-ras mutations in chemically induced mouse liver tumors. Oncogene 2004, 24, 1290–1295. [Google Scholar] [CrossRef] [Green Version]
- Hailfinger, S.; Jaworski, M.; Braeuning, A.; Buchmann, A.; Schwarz, M. Zonal gene expression in murine liver: Lessons from tumors. Hepatology 2006, 43, 407–414. [Google Scholar] [CrossRef]
- Stahl, S.; Ittrich, C.; Marx-Stoelting, P.; Köhle, C.; Altug-Teber, Ö.; Riess, O.; Bonin, M.; Jobst, J.; Kaiser, S.; Buchmann, A.; et al. Genotype-phenotype relationships in hepatocellular tumors from mice and man. Hepatology 2005, 42, 353–361. [Google Scholar] [CrossRef]
- Braeuning, A.; Ittrich, C.; Köhle, C.; Buchmann, A.; Schwarz, M. Zonal gene expression in mouse liver resembles expression patterns of Ha-ras and beta-catenin mutated hepatomas. Drug Metab. Dispos. 2007, 35, 503–507. [Google Scholar] [CrossRef] [Green Version]
- Braeuning, A.; Kollotzek, F.; Zeller, E.; Knorpp, T.; Templin, M.F.; Schwarz, M. Mouse hepatomas with Ha-ras and B-raf mutations differ in mitogen-activated protein kinase signaling and response to constitutive androstane receptor activation. Drug Metab. Dispos. 2018, 46, 1462–1465. [Google Scholar] [CrossRef] [Green Version]
- Braeuning, A.; Menzel, M.; Kleinschnitz, E.-M.; Harada, N.; Tamai, Y.; Köhle, C.; Buchmann, A.; Schwarz, M. Serum components and activated Ha-ras antagonize expression of perivenous marker genes stimulated by beta-catenin signaling in mouse hepatocytes. FEBS J. 2007, 274, 4766–4777. [Google Scholar] [CrossRef]
- Bachleda, P.; Vrza, R.; Dvorak, Z. Activation of MAPKs influences the expression of drug-metabolizing enzymes in primary human hepatocytes. Gen. Physiol. Biophys. 2009, 28, 316–320. [Google Scholar] [CrossRef]
- De Boussac, H.; Gondeau, C.; Briolotti, P.; Duret, C.; Treindl, F.; Römer, M.; Fabre, J.-M.; Herrero, A.; Ramos, J.; Maurel, P.; et al. Epidermal growth factor represses constitutive androstane receptor expression in primary human hepatocytes and favors regulation by pregnane X receptor. Drug Metab. Dispos. 2017, 46, 223–236. [Google Scholar] [CrossRef] [PubMed]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Tóth, B.; Lemze, D.; Golan, M.; Massasa, E.E.; Baydatch, S.; Landen, S.; Moor, A.E.; et al. Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nat. Cell Biol. 2017, 542, 352–356. [Google Scholar] [CrossRef] [PubMed]
- Halpern, K.B.; Shenhav, R.; Matcovitch-Natan, O.; Tóth, B.; Lemze, D.; Golan, M.; Massasa, E.; Baydatch, S.; Landen, S.; Moor, A.; et al. Erratum: Single-cell spatial reconstruction reveals global division of labour in the mammalian liver. Nature 2017, 543, 742. [Google Scholar] [CrossRef] [Green Version]
- Yan, J.; Chen, B.; Lu, J.; Xie, W. Deciphering the roles of the constitutive androstane receptor in energy metabolism. Acta Pharmacol. Sin. 2014, 36, 62–70. [Google Scholar] [CrossRef] [Green Version]
- Zhao, L.-Y.; Xu, J.-Y.; Shi, Z.; Englert, N.A.; Zhang, S.-Y. Pregnane X receptor (PXR) deficiency improves high fat diet-induced obesity via induction of fibroblast growth factor 15 (FGF15) expression. Biochem. Pharmacol. 2017, 142, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Sookoian, S.; Castaño, G.O.; Burgueño, A.L.; Gianotti, T.F.; Rosselli, M.S.; Pirola, C.J. The nuclear receptor PXR gene variants are associated with liver injury in nonalcoholic fatty liver disease. Pharm. Genom. 2010, 20, 1–8. [Google Scholar] [CrossRef]
- Maglich, J.M.; Watson, J.; McMillen, P.J.; Goodwin, B.; Willson, T.M.; Moore, J.T. The nuclear receptor CAR is a regulator of thyroid hormone metabolism during caloric restriction. J. Biol. Chem. 2004, 279, 19832–19838. [Google Scholar] [CrossRef] [Green Version]
- Ding, X.; Lichti, K.; Kim, I.; Gonzalez, F.J.; Staudinger, J.L. Regulation of constitutive androstane receptor and its target genes by fasting, cAMP, hepatocyte nuclear factor alpha, and the coactivator peroxisome proliferator-activated receptor gamma coactivator-1alpha. J. Biol. Chem. 2006, 281, 26540–26551. [Google Scholar] [CrossRef] [Green Version]
- Wieneke, N.; Hirsch-Ernst, K.I.; Kuna, M.; Kersten, S.; Puschel, G.P. PPARalpha-dependent induction of the energy homeostasis-regulating nuclear receptor NR1i3 (CAR) in rat hepatocytes: Potential role in starvation adaptation. FEBS Lett. 2007, 581, 5617–5626. [Google Scholar] [CrossRef] [Green Version]
- Bougarne, N.; Weyers, B.; Desmet, S.J.; Deckers, J.; Ray, D.W.; Staels, B.; De Bosscher, K. Molecular actions of PPARalpha in Lipid metabolism and inflammation. Endocr. Rev. 2018, 39, 760–802. [Google Scholar] [CrossRef] [Green Version]
- Saito, K.; Kobayashi, K.; Mizuno, Y.; Fukuchi, Y.; Furihata, T.; Chiba, K. Peroxisome Proliferator-activated Receptor Alpha (PPARα) agonists induce Constitutive Androstane Receptor (CAR) and cytochrome P450 2B in rat primary hepatocytes. Drug Metab. Pharmacokinet. 2010, 25, 108–111. [Google Scholar] [CrossRef]
- Aouabdi, S.; Gibson, G.; Plant, N. Transcriptional regulation of the PXR gene: Identification and characterization of a functional peroxisome proliferator-activated receptor α binding site within the proximal promoter of pxr. Drug Metab. Dispos. 2005, 34, 138–144. [Google Scholar] [CrossRef] [Green Version]
- Buler, M.; Aatsinki, S.-M.; Skoumal, R.; Hakkola, J. Energy sensing factors PGC-1α and SIRT1 modulate PXR expression and function. Biochem. Pharmacol. 2011, 82, 2008–2015. [Google Scholar] [CrossRef]
- Bhutia, Y.; Ogura, J.; Sivaprakasam, S.; Ganapathy, V. Gut microbiome and colon cancer: Role of bacterial metabolites and their molecular targets in the host. Curr. Color. Cancer Rep. 2017, 13, 111–118. [Google Scholar] [CrossRef]
- Björkholm, B.; Bok, C.M.; Lundin, A.; Rafter, J.; Hibberd, M.L.; Pettersson, S. Intestinal microbiota regulate xenobiotic metabolism in the liver. PLoS ONE 2009, 4, e6958. [Google Scholar] [CrossRef]
- Selwyn, F.P.; Cheng, S.L.; Bammler, T.K.; Prasad, B.; Vrana, M.; Klaassen, C.; Cui, J.Y. Developmental regulation of drug-processing genes in livers of germ-free mice. Toxicol. Sci. 2015, 147, 84–103. [Google Scholar] [CrossRef] [Green Version]
- Zemanová, N.; Anzenbacher, P.; Zapletalová, I.; Jourová, L.; Hermanová, P.; Hudcovic, T.; Kozáková, H.; Vodička, M.; Pácha, J.; Anzenbacherová, E. The role of the microbiome and psychosocial stress in the expression and activity of drug metabolizing enzymes in mice. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Venu, V.K.P.; Saifeddine, M.; Mihara, K.; Tsai, Y.-C.; Nieves, K.; Alston, L.; Mani, S.; McCoy, K.D.; Hollenberg, M.D.; Hirota, S.A. The pregnane X receptor and its microbiota-derived ligand indole 3-propionic acid regulate endothelium-dependent vasodilation. Am. J. Physiol. Metab. 2019, 317, E350–E361. [Google Scholar] [CrossRef]
- Ranhotra, H.S.; Flannigan, K.L.; Brave, M.; Mukherjee, S.; Lukin, D.J.; Hirota, S.A.; Mani, S. Xenobiotic receptor-mediated regulation of intestinal barrier function and innate immunity. Nucl. Recept. Res. 2016, 3, 3. [Google Scholar] [CrossRef] [Green Version]
- Weger, B.D.; Gobet, C.; Yeung, J.; Martin, E.; Jimenez, S.; Betrisey, B.; Foata, F.; Berger, B.; Balvay, A.; Foussier, A.; et al. The mouse microbiome is required for sex-specific diurnal rhythms of gene expression and metabolism. Cell Metab. 2019, 29, 362–382. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascussi, J.-M.; Gerbal-Chaloin, S.; Duret, C.; Daujat-Chavanieu, M.; Vilarem, M.-J.; Maurel, P. The tangle of nuclear receptors that controls xenobiotic metabolism and transport: Crosstalk and consequences. Annu. Rev. Pharmacol. Toxicol. 2008, 48, 1–32. [Google Scholar] [CrossRef] [Green Version]
- Parviz, F.; Matullo, C.; Garrison, W.D.; Savatski, L.; Adamson, J.W.; Ning, G.; Kaestner, K.H.; Rossi, J.M.; Zaret, K.S.; Duncan, S. Hepatocyte nuclear factor 4α controls the development of a hepatic epithelium and liver morphogenesis. Nat. Genet. 2003, 34, 292–296. [Google Scholar] [CrossRef]
- Hayhurst, G.P.; Lee, Y.-H.; Lambert, G.; Ward, J.M.; Gonzalez, F.J. Hepatocyte nuclear factor 4α (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell. Biol. 2001, 21, 1393–1403. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Ning, G.; Duncan, S.A. Mammalian hepatocyte differentiation requires the transcription factor HNF-4α. Genome Res. 2000, 14, 464–474. [Google Scholar]
- Tirona, R.G.; Lee, W.; Leake, B.F.; Lan, L.-B.; Cline, C.B.; Lamba, V.; Parviz, F.; Duncan, S.A.; Inoue, Y.; Gonzalez, F.J.; et al. The orphan nuclear receptor HNF4α determines PXR- and CAR-mediated xenobiotic induction of CYP3A. Nat. Med. 2003, 9, 220–224. [Google Scholar] [CrossRef]
- Iwazaki, N.; Kobayashi, K.; Morimoto, K.; Hirano, M.; Kawashima, S.; Furihata, T.; Chiba, K. Involvement of hepatocyte nuclear factor 4alpha in transcriptional regulation of the human pregnane X receptor gene in the human liver. Drug Metab. Pharmacokinet. 2008, 23, 59–66. [Google Scholar] [CrossRef]
- Oladimeji, P.; Wright, W.C.; Wu, J.; Chen, T. RNA interference screen identifies NAA10 as a regulator of PXR transcription. Biochem. Pharmacol. 2019, 160, 92–109. [Google Scholar] [CrossRef] [PubMed]
- Babeu, J.-P. Hepatocyte nuclear factor 4-alpha involvement in liver and intestinal inflammatory networks. World J. Gastroenterol. 2014, 20, 22–30. [Google Scholar] [CrossRef] [PubMed]
- Nakhei, H.; Lingott, A.; Lemm, I.; Ryffel, G.U. An alternative splice variant of the tissue specific transcription factor HNF4 predominates in undifferentiated murine cell types. Nucleic Acids Res. 1998, 26, 497–504. [Google Scholar] [CrossRef] [PubMed]
- Pascussi, J.-M.; Robert, A.; Moreau, A.; Ramos, J.; Bioulac-Sage, P.; Navarro, F.; Blanc, P.; Assenat, E.; Maurel, P.; Vilarem, M.J. Differential regulation of constitutive androstane receptor expression by hepatocyte nuclear factor4α isoforms. Hepatology 2007, 45, 1146–1153. [Google Scholar] [CrossRef]
- Duret, C.; Gerbal-Chaloin, S.; Ramos, J.; Fabre, J.-M.; Jacquet, E.; Navarro, F.; Blanc, P.; Cunha, A.S.; Maurel, P.; Daujat-Chavanieu, M.; et al. Isolation, characterization, and differentiation to hepatocyte-like cells of nonparenchymal epithelial cells from adult human liver. STEM CELLS 2007, 25, 1779–1790. [Google Scholar] [CrossRef]
- Pascussi, J.-M.; Gerbal-Chaloin, S.; Fabre, J.-M.; Maurel, P.; Vilarem, M.-J. Dexamethasone enhances constitutive androstane receptor expression in human hepatocytes: Consequences on cytochrome P450 gene regulation. Mol. Pharmacol. 2000, 58, 1441–1450. [Google Scholar] [CrossRef] [Green Version]
- Pascussi, J.; Drocourt, L.; Fabre, J.-M.; Maurel, P.; Vilarem, M.-J. Dexamethasone induces pregnane X receptor and retinoid X receptor-α expression in human hepatocytes: Synergistic increase of CYP3A4 induction by pregnane X receptor activators. Mol. Pharmacol. 2000, 58, 361–372. [Google Scholar] [CrossRef] [Green Version]
- Duret, C.; Daujat-Chavanieu, M.; Pascussi, J.-M.; Pichard-Garcia, L.; Balaguer, P.; Fabre, J.-M.; Vilarem, M.-J.; Maurel, P.; Gerbal-Chaloin, S. Ketoconazole and miconazole are antagonists of the human glucocorticoid receptor: Consequences on the expression and function of the constitutive androstane receptor and the pregnane X receptor. Mol. Pharmacol. 2006, 70, 329–339. [Google Scholar] [CrossRef] [Green Version]
- Cooper, B.W.; Cho, T.M.; Thompson, P.M.; Wallace, A.D. Phthalate induction of CYP3A4 is dependent on glucocorticoid regulation of PXR expression. Toxicol. Sci. 2008, 103, 268–277. [Google Scholar] [CrossRef] [Green Version]
- Pascussi, J.-M.; Coniat, M.B.-L.; Maurel, P.; Vilarem, M.-J. Transcriptional analysis of the orphan Nuclear Receptor Constitutive Androstane Receptor (NR1I3) gene promoter: Identification of a distal glucocorticoid response element. Mol. Endocrinol. 2003, 17, 42–55. [Google Scholar] [CrossRef] [Green Version]
- Smutny, T.; Dusek, J.; Hyrsova, L.; Nekvindova, J.; Horvatova, A.; Micuda, S.; Gerbal-Chaloin, S.; Pavek, P. The 3′-untranslated region contributes to the pregnane X receptor (PXR) expression down-regulation by PXR ligands and up-regulation by glucocorticoids. Acta Pharm. Sin. B 2019, 10, 136–152. [Google Scholar] [CrossRef] [PubMed]
- Huss, J.M.; Kasper, C.B. Two-stage glucocorticoid induction ofCYP3A23through both the glucocorticoid and pregnane X receptors. Mol. Pharmacol. 2000, 58, 48–57. [Google Scholar] [CrossRef] [Green Version]
- Bhadhprasit, W.; Sakuma, T.; Hatakeyama, N.; Fuwa, M.; Kitajima, K.; Nemoto, N. Involvement of glucocorticoid receptor and pregnane X receptor in the regulation of mouse CYP3A44 female-predominant expression by glucocorticoid hormone. Drug Metab. Dispos. 2007, 35, 1880–1885. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, D.; Yang, D.; Yan, B. Dexamethasone transcriptionally increases the expression of the pregnane X receptor and synergistically enhances pyrethroid esfenvalerate in the induction of cytochrome P450 3A23. Biochem. Pharmacol. 2010, 80, 1274–1283. [Google Scholar] [CrossRef] [Green Version]
- Probert, P.M.E.; Chung, G.W.; Cockell, S.J.; Agius, L.; Mosesso, P.; White, S.A.; Oakley, F.; Brown, C.D.A.; Wright, M.C. Utility of B-13 progenitor-derived hepatocytes in hepatotoxicity and genotoxicity studies. Toxicol. Sci. 2013, 137, 350–370. [Google Scholar] [CrossRef] [Green Version]
- Chen, Q.; Jia, A.; Snyder, S.A.; Gong, Z.; Lam, S.H. Glucocorticoid activity detected by in vivo zebrafish assay and in vitro glucocorticoid receptor bioassay at environmental relevant concentrations. Chemosphere 2016, 144, 1162–1169. [Google Scholar] [CrossRef]
- Haag, M.; Fautrel, A.; Guillouzo, A.; Frossard, N.; Pons, F. Expression of cytochromes P450 3A in mouse lung: Effects of dexamethasone and pregnenolone 16α-carbonitrile. Arch. Toxicol. 2002, 77, 145–149. [Google Scholar] [CrossRef]
- Khan, A.A.; Chow, E.C.; Van Loenen-Weemaes, A.-M.M.; Porte, R.J.; Pang, K.S.; Groothuis, G.M. Comparison of effects of VDR versus PXR, FXR and GR ligands on the regulation of CYP3A isozymes in rat and human intestine and liver. Eur. J. Pharm. Sci. 2009, 37, 115–125. [Google Scholar] [CrossRef] [PubMed]
- Narang, V.S.; Fraga, C.; Kumar, N.; Shen, J.; Throm, S.; Stewart, C.F.; Waters, C.M. Dexamethasone increases expression and activity of multidrug resistance transporters at the rat blood-brain barrier. Am. J. Physiol. Physiol. 2008, 295, C440–C450. [Google Scholar] [CrossRef] [Green Version]
- Larigot, L.; Juricek, L.; Dairou, J.; Coumoul, X. AhR signaling pathways and regulatory functions. Biochim. Open 2018, 7, 1–9. [Google Scholar] [CrossRef]
- Schraplau, A.; Schewe, B.; Neuschäfer-Rube, F.; Ringel, S.; Neuber, C.; Kleuser, B.; Püschel, G.P. Enhanced thyroid hormone breakdown in hepatocytes by mutual induction of the constitutive androstane receptor (CAR, NR1I3) and arylhydrocarbon receptor by benzo[a]pyrene and phenobarbital. Toxicology 2015, 328, 21–28. [Google Scholar] [CrossRef]
- Rasmussen, M.K.; Daujat-Chavanieu, M.; Gerbal-Chaloin, S. Activation of the aryl hydrocarbon receptor decreases rifampicin-induced CYP3A4 expression in primary human hepatocytes and HepaRG. Toxicol. Lett. 2017, 277, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Modica, S.; Gadaleta, R.M.; Moschetta, A. Deciphering the nuclear bile acid receptor FXR paradigm. Nucl. Recept. Signal. 2010, 8, e005. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, T.; Chiang, J.Y.L. Nuclear receptors in bile acid metabolism. Drug Metab. Rev. 2013, 45, 145–155. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jung, D.; Mangelsdorf, D.J.; Meyer, U.A. Pregnane X receptor is a target of farnesoid X receptor. J. Biol. Chem. 2006, 281, 19081–19091. [Google Scholar] [CrossRef] [Green Version]
- Wieneke, N.; Neuschafer-Rube, F.; Bode, L.M.; Kuna, M.; Andres, J.; Carnevali, L.C., Jr.; Hirsch-Ernst, K.I.; Puschel, G.P. Synergistic acceleration of thyroid hormone degradation by phenobarbital and the PPAR alpha agonist WY14643 in rat hepatocytes. Toxicol. Appl. Pharmacol. 2009, 240, 99–107. [Google Scholar] [CrossRef]
- Bing, Y.; Zhu, S.; Jiang, K.; Dong, G.; Li, J.; Yang, Z.; Yang, J.; Yue, J. Reduction of thyroid hormones triggers down-regulation of hepatic CYP2B through nuclear receptors CAR and TR in a rat model of acute stroke. Biochem. Pharmacol. 2014, 87, 636–649. [Google Scholar] [CrossRef]
- Ooe, H.; Kon, J.; Oshima, H.; Mitaka, T. Thyroid hormone is necessary for expression of constitutive androstane receptor in rat hepatocytes. Drug Metab. Dispos. 2009, 37, 1963–1969. [Google Scholar] [CrossRef] [Green Version]
- Wahlang, B.; Prough, R.A.; Falkner, K.C.; Hardesty, J.E.; Song, M.; Clair, H.B.; Clark, B.J.; States, J.C.; Arteel, G.E.; Cave, M.C. Polychlorinated biphenyl-xenobiotic nuclear receptor interactions regulate energy metabolism, behavior, and inflammation in non-alcoholic-steatohepatitis. Toxicol. Sci. 2015, 149, 396–410. [Google Scholar] [CrossRef] [Green Version]
- Maglich, J.M.; Stoltz, C.M.; Goodwin, B.; Hawkins-Brown, D.; Moore, J.T.; Kliewer, S.A. Nuclear pregnane x receptor and constitutive androstane receptor regulate overlapping but distinct sets of genes involved in xenobiotic detoxification. Mol. Pharmacol. 2002, 62, 638–646. [Google Scholar] [CrossRef] [Green Version]
- Kumari, S.; Saradhi, M.; Rana, M.; Chatterjee, S.; Aumercier, M.; Mukhopadhyay, G.; Tyagi, R.K. Pregnane and Xenobiotic Receptor gene expression in liver cells is modulated by Ets-1 in synchrony with transcription factors Pax5, LEF-1 and c-jun. Exp. Cell Res. 2015, 330, 398–411. [Google Scholar] [CrossRef] [PubMed]
- Saradhi, M.; Kumari, S.; Rana, M.; Mukhopadhyay, G.; Tyagi, R.K. Identification and interplay of sequence specific DNA binding proteins involved in regulation of human Pregnane and Xenobiotic Receptor gene. Exp. Cell Res. 2015, 339, 187–196. [Google Scholar] [CrossRef]
- Assenat, E.; Gerbal-Chaloin, S.; Larrey, D.; Saric, J.; Fabre, J.M.; Maurel, P.; Vilarem, M.J.; Pascussi, J.M. Interleukin 1beta inhibits CAR-induced expression of hepatic genes involved in drug and bilirubin clearance. Hepatology 2004, 40, 951–960. [Google Scholar] [CrossRef]
- Meng, Z.-X.; Wang, L.; Chang, L.; Sun, J.; Bao, J.; Li, Y.; Chen, Y.E.; Lin, J.D. A diet-sensitive BAF60a-mediated pathway links hepatic bile acid metabolism to cholesterol absorption and atherosclerosis. Cell Rep. 2015, 13, 1658–1669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ishihara, H.; Yamashita, S.; Amano, R.; Kimura, K.; Hirakawa, K.; Ueda, T.; Murakami, Y.; Tamori, A.; Tanabe, K.; Kawada, N.; et al. Pancreatic cancer cell fraction estimation in a DNA sample. Oncology 2018, 95, 370–379. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Miao, X.-J.; Wang, X.; Pan, H.-H.; Li, P.; Ren, H.; Jia, Y.-R.; Lu, C.; Wang, H.-B.; Yuan, L.; et al. Antiproliferation of berberine is mediated by epigenetic modification of constitutive androstane receptor (CAR) metabolic pathway in hepatoma cells. Sci. Rep. 2016, 6, 28116. [Google Scholar] [CrossRef]
- Tian, Y. Epigenetic regulation of pregnane X receptor activity. Drug Metab. Rev. 2013, 45, 166–172. [Google Scholar] [CrossRef] [PubMed]
- Misawa, A.; Inoue, J.; Sugino, Y.; Hosoi, H.; Sugimoto, T.; Hosoda, F.; Ohki, M.; Imoto, I.; Inazawa, J. Methylation-associated silencing of the nuclear receptor 1I2 gene in advanced-type neuroblastomas, identified by bacterial artificial chromosome array-based methylated CpG island amplification. Cancer Res. 2005, 65, 10233–10242. [Google Scholar] [CrossRef] [Green Version]
- Habano, W.; Gamo, T.; Terashima, J.; Sugai, T.; Otsuka, K.; Takahara, T.; Ozawa, S. Involvement of promoter methylation in the regulation of Pregnane X receptor in colon cancer cells. BMC Cancer 2011, 11, 81. [Google Scholar] [CrossRef] [Green Version]
- Cabrerizo, R.; Castaño, G.O.; Burgueño, A.L.; Gianotti, T.F.; Ledesma, M.M.G.L.; Flichman, D.; Pirola, C.J.; Sookoian, S. Promoter DNA methylation of farnesoid X receptor and pregnane X receptor modulates the intrahepatic cholestasis of pregnancy phenotype. PLoS ONE 2014, 9, e87697. [Google Scholar] [CrossRef]
- Takagi, S.; Nakajima, M.; Mohri, T.; Yokoi, T. Post-transcriptional regulation of human pregnane X receptor by micro-RNA affects the expression of cytochrome P450 3A4. J. Biol. Chem. 2008, 283, 9674–9680. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lamba, V.; Ghodke-Puranik, Y.; Guan, W.; Tracy, T. microRNA-34a is associated with expression of key hepatic transcription factors and cytochromes P450. Biochem. Biophys. Res. Commun. 2014, 445, 404–411. [Google Scholar] [CrossRef] [Green Version]
- Rieger, J.K.; Klein, K.; Winter, S.; Zanger, U.M. Expression variability of absorption, distribution, metabolism, excretion–related MicroRNAs in human liver: Influence of nongenetic factors and association with gene expression. Drug Metab. Dispos. 2013, 41, 1752–1762. [Google Scholar] [CrossRef]
- Wei, Z.; Chen, M.; Zhang, Y.; Wang, X.; Jiang, S.; Wang, Y.; Wu, X.; Qin, S.; He, L.; Zhang, L.; et al. No Correlation of hsa-miR-148a with expression of PXR or CYP3A4 in human livers from chinese han population. PLoS ONE 2013, 8, e59141. [Google Scholar] [CrossRef] [Green Version]
- Ramamoorthy, A.; Li, L.; Gaedigk, A.; Bradford, L.D.; Benson, E.A.; Flockhart, D.A.; Skaar, T.C. In silico and in vitro identification of microRNAs that regulate hepatic nuclear factor 4α expression. Drug Metab. Dispos. 2012, 40, 726–733. [Google Scholar] [CrossRef] [Green Version]
- Sharma, D.; Turkistani, A.A.; Chang, T.K.; Hu, C.; Xu, Z.; Chang, T.K.H. Negative regulation of human pregnane X receptor by MicroRNA-18a-5p: Evidence for suppression of MicroRNA-18a-5p expression by rifampin and rilpivirine. Mol. Pharmacol. 2017, 92, 48–56. [Google Scholar] [CrossRef] [Green Version]
- Rao, Z.-Z.; Zhang, X.-W.; Ding, Y.-L.; Yang, M.-Y. miR-148a-mediated estrogen-induced cholestasis in intrahepatic cholestasis of pregnancy: Role of PXR/MRP. PLoS ONE 2017, 12, e0178702. [Google Scholar] [CrossRef] [Green Version]
- Reuter, T.; Herold-Mende, C.; Dyckhoff, G.; Rigalli, J.P.; Weiss, J. Functional role of miR-148a in oropharyngeal cancer: Influence on pregnane X receptor and P-glycoprotein expression. J. Recept. Signal Transduct. 2019, 39, 451–459. [Google Scholar] [CrossRef] [PubMed]
- Li, M.; Yang, Y.; He, Z.-X.; Zhou, Z.-W.; Yang, T.; Guo, P.; Zhang, X.; Zhou, S.-F. MicroRNA-561 promotes acetaminophen-induced hepatotoxicity in HepG2 cells and primary human hepatocytes through downregulation of the nuclear receptor corepressor dosage-sensitive sex-reversal adrenal hypoplasia congenital critical region on the X chromosome, gene 1 (DAX-1). Drug Metab. Dispos. 2013, 42, 44–61. [Google Scholar] [CrossRef] [Green Version]
- Takwi, A.; Wang, Y.-M.; Wu, J.; Michaelis, M.; Cinatl, J.; Chen, T. miR-137 regulates the constitutive androstane receptor and modulates doxorubicin sensitivity in parental and doxorubicin-resistant neuroblastoma cells. Oncogene 2014, 33, 3717–3729. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vachirayonstien, T.; Yan, B. MicroRNA-30c-1-3p is a silencer of the pregnane X receptor by targeting the 3′-untranslated region and alters the expression of its target gene cytochrome P450 3A4. Biochim. Biophys. Acta (BBA) Bioenerg. 2016, 1859, 1238–1244. [Google Scholar] [CrossRef] [Green Version]
- Li, J.; Zhao, J.; Wang, H.; Li, X.; Liu, A.; Qin, Q.; Li, B. MicroRNA-140-3p enhances the sensitivity of hepatocellular carcinoma cells to sorafenib by targeting pregnenolone X receptor. OncoTargets Ther. 2018, 11, 5885–5894. [Google Scholar] [CrossRef] [Green Version]
- Deng, Y.; Wei, Z.; Huang, M.; Xu, G.; Wei, W.; Peng, B.; Nong, S.; Qin, H. Long non-coding RNA F11-AS1 inhibits HBV-related hepatocellular carcinoma progression by regulating NR1I3 via binding to microRNA-211-5p. J. Cell. Mol. Med. 2019, 24, 1848–1865. [Google Scholar] [CrossRef]
- Li, D.; Wu, L.; Knox, B.; Chen, S.; Tolleson, W.H.; Liu, F.; Yu, D.; Guo, L.; Tong, W.; Ning, B. Long noncoding RNA LINC00844-mediated molecular network regulates expression of drug metabolizing enzymes and nuclear receptors in human liver cells. Arch. Toxicol. 2020, 94, 1637–1653. [Google Scholar] [CrossRef]
- Qi, X.; Zhang, D.-H.; Wu, N.; Xiao, J.-H.; Wang, X.; Ma, W. ceRNA in cancer: Possible functions and clinical implications. J. Med. Genet. 2015, 52, 710–718. [Google Scholar] [CrossRef]
- Wang, Y.; Yan, L.; Liu, J.; Chen, S.; Liu, G.; Nie, Y.; Wang, P.; Yang, W.; Chen, L.; Zhong, X.; et al. The HNF1α-regulated LncRNA HNF1α-AS1 Is involved in the regulation of cytochrome P450 expression in human liver tissues and Huh7 cells. J. Pharmacol. Exp. Ther. 2019, 368, 353–362. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, M.; Fukami, T.; Nakajima, M. Adenosine deaminases acting on RNA downregulate the expression of constitutive androstane receptor in the human liver-derived cells by attenuating splicing. J. Pharmacol. Exp. Ther. 2019, 370, 408–415. [Google Scholar] [CrossRef]
- Masuyama, H.; Inoshita, H.; Hiramatsu, Y.; Kudo, T. Ligands have various potential effects on the degradation of pregnane X receptor by proteasome. Endocrinology 2002, 143, 55–61. [Google Scholar] [CrossRef]
- Rana, R.; Coulter, S.; Kinyamu, H.; Goldstein, J.A. RBCK1, an E3 ubiquitin ligase, interacts with and ubiquinates the human pregnane X receptor. Drug Metab. Dispos. 2012, 41, 398–405. [Google Scholar] [CrossRef]
- Ong, S.S.; Goktug, A.N.; Elias, A.; Wu, J.; Saunders, D.N.; Chen, T. Stability of the human pregnane X receptor is regulated by E3 ligase UBR5 and serine/threonine kinase DYRK. Biochem. J. 2014, 459, 193–203. [Google Scholar] [CrossRef]
- Pettersson, F.; Hanna, N.; Lagodich, M.; Dupéré-Richer, D.; Couture, M.-C.; Choi, C.; Miller, W.H. Rexinoids modulate steroid and xenobiotic receptor activity by increasing its protein turnover in a calpain-dependent manner. J. Biol. Chem. 2008, 283, 21945–21952. [Google Scholar] [CrossRef] [Green Version]
- Chen, T.; Laurenzana, E.M.; Coslo, D.M.; Chen, F.; Omiecinski, C.J. Proteasomal interaction as a critical activity modulator of the human constitutive androstane receptor. Biochem. J. 2014, 458, 95–107. [Google Scholar] [CrossRef] [Green Version]
- Gerbal-Chaloin, S.; Iankova, I.; Maurel, P.; Daujat-Chavanieu, M. Nuclear receptors in the cross-talk of drug metabolism and inflammation. Drug Metab. Rev. 2013, 45, 122–144. [Google Scholar] [CrossRef]
- Beigneux, A.P.; Moser, A.H.; Shigenaga, J.K.; Grunfeld, C.; Feingold, K.R. Reduction in cytochrome P-450 enzyme expression is associated with repression of CAR (constitutive androstane receptor) and PXR (pregnane X receptor) in mouse liver during the acute phase response. Biochem. Biophys. Res. Commun. 2002, 293, 145–149. [Google Scholar] [CrossRef]
- Abualsunun, W.A.; Piquette-Miller, M. Involvement of nuclear factor κB, not pregnane X receptor, in inflammation-mediated regulation of hepatic transporters. Drug Metab. Dispos. 2017, 45, 1077–1083. [Google Scholar] [CrossRef] [Green Version]
- Xu, D.-X.; Wei, W.; Sun, M.-F.; Wu, C.-Y.; Wang, J.-P.; Wei, L.-Z.; Zhou, C.-F. Kupffer cells and reactive oxygen species partially mediate lipopolysaccharide-induced downregulation of nuclear receptor pregnane x receptor and its target gene CYP3a in mouse liver. Free. Radic. Biol. Med. 2004, 37, 10–22. [Google Scholar] [CrossRef]
- Xu, D.-X.; Chen, Y.-H.; Wang, J.-P.; Sun, M.-F.; Wang, H.; Wei, L.-Z.; Wei, W. Perinatal lipopolysaccharide exposure downregulates pregnane X receptor and Cyp3a11 expression in fetal mouse liver. Toxicol. Sci. 2005, 87, 38–45. [Google Scholar] [CrossRef]
- Xu, D.-X.; Wei, W.; Sun, M.-F.; Wei, L.-Z.; Wang, J.-P. Melatonin attenuates lipopolysaccharide-induced down-regulation of pregnane X receptor and its target gene CYP3A in mouse liver. J. Pineal Res. 2005, 38, 27–34. [Google Scholar] [CrossRef]
- Ghose, R.; Guo, T.; Vallejo, J.G.; Gandhi, A. Differential role of Toll-interleukin 1 receptor domain-containing adaptor protein in Toll-like receptor 2-mediated regulation of gene expression of hepatic cytokines and drug-metabolizing enzymes. Drug Metab. Dispos. 2011, 39, 874–881. [Google Scholar] [CrossRef] [Green Version]
- Pascussi, J.-M.; Gerbal-Chaloin, S.; Pichard-Garcia, L.; Daujat, M.; Fabre, J.-M.; Maurel, P.; Vilarem, M.-J. Interleukin-6 negatively regulates the expression of pregnane x receptor and constitutively activated receptor in primary human hepatocytes. Biochem. Biophys. Res. Commun. 2000, 274, 707–713. [Google Scholar] [CrossRef]
- Ogura, J.; Terada, Y.; Tsujimoto, T.; Koizumi, T.; Kuwayama, K.; Maruyama, H.; Fujikawa, A.; Takaya, A.; Kobayashi, M.; Itagaki, S.; et al. The decrease in farnesoid X receptor, pregnane X receptor and constitutive androstane receptor in the liver after intestinal ischemia-reperfusion. J. Pharm. Pharm. Sci. 2012, 15, 616–631. [Google Scholar] [CrossRef] [Green Version]
- Tanner, N.; Kubik, L.; Luckert, C.; Thomas, M.; Hofmann, U.; Zanger, U.M.; Bohmert, L.; Lampen, A.; Braeuning, A. Regulation of drug metabolism by the interplay of inflammatory signaling, steatosis, and xeno-sensing receptors in HepaRG cells. Drug Metab. Dispos. 2018, 46, 326–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ning, R.; Zhan, Y.; He, S.; Hu, J.; Zhu, Z.; Hu, G.; Yan, B.; Yang, J.; Liu, W. Interleukin-6 induces DEC1, promotes DEC1 interaction with RXRalpha and suppresses the expression of PXR, CAR and their target genes. Front. Pharmacol. 2017, 8, 866. [Google Scholar] [CrossRef] [Green Version]
- Teng, S.; Piquette-Miller, M. The involvement of the pregnane X receptor in hepatic gene regulation during inflammation in mice. J. Pharmacol. Exp. Ther. 2004, 312, 841–848. [Google Scholar] [CrossRef] [Green Version]
- Rasmussen, M.K.; Bertholdt, L.; Gudiksen, A.; Pilegaard, H.; Knudsen, J.G. Impact of fasting followed by short-term exposure to interleukin-6 on cytochrome P450 mRNA in mice. Toxicol. Lett. 2018, 282, 93–99. [Google Scholar] [CrossRef]
- Xie, L.; He, Y.; Zhou, X.; Li, X.; Jin, X.; Wang, X.; Shi, D. Porcine interleukin-6 enhances the expression of CYP2C33 through a constitutive androstane receptor/retinoid X receptor-mediated pathway. Xenobiotica 2018, 49, 257–264. [Google Scholar] [CrossRef]
- Geier, A.; Dietrich, C.G.; Voigt, S.; Ananthanarayanan, M.; Lammert, F.; Schmitz, A.; Trauner, M.; Wasmuth, H.E.; Boraschi, D.; Balasubramaniyan, N.; et al. Cytokine-dependent regulation of hepatic organic anion transporter gene transactivators in mouse liver. Am. J. Physiol. Liver Physiol. 2005, 289, G831–G841. [Google Scholar] [CrossRef]
- Langmann, T.; Moehle, C.; Mauerer, R.; Scharl, M.; Liebisch, G.; Zahn, A.; Stremmel, W.; Schmitz, G. Loss of detoxification in inflammatory bowel disease: Dysregulation of pregnane X receptor target genes. Gastroenterology 2004, 127, 26–40. [Google Scholar] [CrossRef]
- Bossche, L.V.D.; Borsboom, D.; Devriese, S.; Van Welden, S.; Holvoet, T.; Devisscher, L.; Hindryckx, P.; De Vos, M.; Laukens, D. Tauroursodeoxycholic acid protects bile acid homeostasis under inflammatory conditions and dampens Crohn’s disease-like ileitis. Lab. Investig. 2017, 97, 519–529. [Google Scholar] [CrossRef]
- Deuring, J.J.; Li, M.; Cao, W.; Chen, S.; Wang, W.; de Haar, C.; van der Woude, C.J.; Peppelenbosch, M. Pregnane X receptor activation constrains mucosal NF-kappaB activity in active inflammatory bowel disease. PLoS ONE 2019, 14, e0221924. [Google Scholar] [CrossRef] [Green Version]
- Kusunoki, Y.; Ikarashi, N.; Hayakawa, Y.; Ishii, M.; Kon, R.; Ochiai, W.; Machida, Y.; Sugiyama, K. Hepatic early inflammation induces downregulation of hepatic cytochrome P450 expression and metabolic activity in the dextran sulfate sodium-induced murine colitis. Eur. J. Pharm. Sci. 2014, 54, 17–27. [Google Scholar] [CrossRef]
- Kusunoki, Y.; Ikarashi, N.; Matsuda, S.; Matsukawa, Y.; Kitaoka, S.; Kon, R.; Tajima, M.; Wakui, N.; Ochiai, W.; Machida, Y.; et al. Expression of hepatic cytochrome P450 in a mouse model of ulcerative colitis changes with pathological conditions. J. Gastroenterol. Hepatol. 2015, 30, 1618–1626. [Google Scholar] [CrossRef] [PubMed]
- Xu, D.-X.; Wang, J.-P.; Sun, M.-F.; Chen, Y.-H.; Wei, W. Lipopolysaccharide downregulates the expressions of intestinal pregnane X receptor and cytochrome P450 3a11. Eur. J. Pharmacol. 2006, 536, 162–170. [Google Scholar] [CrossRef]
- Chen, Y.-H.; Wang, J.-P.; Wang, H.; Sun, M.-F.; Wei, L.-Z.; Wei, W.; Xu, D.-X. Lipopolysaccharide treatment downregulates the expression of the pregnane X receptor, cyp3a11 and mdr1a genes in mouse placenta. Toxicology 2005, 211, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Yoshinari, K.; Takagi, S.; Sugatani, J.; Miwa, M. Changes in the expression of cytochromes P450 and nuclear receptors in the liver of genetically diabetic db/db mice. Biol. Pharm. Bull. 2006, 29, 1634–1638. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.; Zhao, T.; Li, Z.; Yan, M.; Zhao, H.; Zhu, B.; Li, P. Transcriptional profile of kidney from type 2 diabetic db/db mice. J. Diabetes Res. 2017, 2017, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Lam, J.L.; Jiang, Y.; Zhang, T.; Zhang, E.Y.; Smith, B.J. Expression and functional analysis of hepatic cytochromes P450, nuclear receptors, and membrane transporters in 10- and 25-week-old db/db mice. Drug Metab. Dispos. 2010, 38, 2252–2258. [Google Scholar] [CrossRef]
- Xiong, H.; Yoshinari, K.; Brouwer, K.L.; Negishi, M. Role of constitutive androstane receptor in the in vivo induction of Mrp3 and CYP2B1/2 by phenobarbital. Drug Metab. Dispos. 2002, 30, 918–923. [Google Scholar] [CrossRef] [Green Version]
- Zhang, L.; Xu, P.; Cheng, Y.; Wang, P.; Ma, X.; Liu, M.; Wang, X.; Xu, F. Diet-induced obese alters the expression and function of hepatic drug-metabolizing enzymes and transporters in rats. Biochem. Pharmacol. 2019, 164, 368–376. [Google Scholar] [CrossRef]
- Ghoneim, R.H.; Sock, E.T.N.; Lavoie, J.-M.; Piquette-Miller, M. Effect of a high-fat diet on the hepatic expression of nuclear receptors and their target genes: Relevance to drug disposition. Br. J. Nutr. 2015, 113, 507–516. [Google Scholar] [CrossRef] [Green Version]
- Wang, P.; Shao, X.; Bao, Y.; Zhu, J.; Chen, L.; Zhang, L.; Ma, X.; Zhong, X.-B. Impact of obese levels on the hepatic expression of nuclear receptors and drug-metabolizing enzymes in adult and offspring mice. Acta Pharm. Sin. B 2020, 10, 171–185. [Google Scholar] [CrossRef] [PubMed]
- Zhang, F.; Duan, Y.; Wei, Y.; Zhang, J.; Ma, X.; Tian, H.; Wang, X.; Saad, A.A.A.; Li, B.; Wu, X. The inhibition of hepatic Pxr-Oatp2 pathway mediating decreased hepatic uptake of rosuvastatin in rats with high-fat diet-induced obesity. Life Sci. 2020, 257, 118079. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, Z.; Klaunig, J.E. Modulation of xenobiotic nuclear receptors in high-fat diet induced non-alcoholic fatty liver disease. Toxicology 2018, 410, 199–213. [Google Scholar] [CrossRef]
- Ghose, R.; Omoluabi, O.; Gandhi, A.; Shah, P.; Strohacker, K.; Carpenter, K.C.; McFarlin, B.; Guo, T. Role of high-fat diet in regulation of gene expression of drug metabolizing enzymes and transporters. Life Sci. 2011, 89, 57–64. [Google Scholar] [CrossRef] [Green Version]
- Kudo, T.; Shimada, T.; Toda, T.; Igeta, S.; Suzuki, W.; Ikarashi, N.; Ochiai, W.; Ito, K.; Aburada, M.; Sugiyama, K. Altered expression of CYP in TSOD mice: A model of type 2 diabetes and obesity. Xenobiotica 2009, 39, 889–902. [Google Scholar] [CrossRef]
- Bushkofsky, J.R.; Maguire, M.; Larsen, M.C.; Fong, Y.H.; Jefcoate, C.R. Cyp1b1 affects external control of mouse hepatocytes, fatty acid homeostasis and signaling involving HNF4alpha and PPARalpha. Arch. Biochem. Biophys. 2016, 597, 30–47. [Google Scholar] [CrossRef] [Green Version]
- Yao, H.; Gu, J.; Shan, Y.; Wang, Y.; Chen, X.; Sun, D.; Guo, Y. Type 2 diabetes mellitus decreases systemic exposure of clopidogrel active metabolite through upregulation of P-glycoprotein in rats. Biochem. Pharmacol. 2020, 180, 114142. [Google Scholar] [CrossRef]
- Watanabe, A.; Marumo, T.; Kawarazaki, W.; Nishimoto, M.; Ayuzawa, N.; Ueda, K.; Hirohama, D.; Tanaka, T.; Yagi, S.; Ota, S.; et al. Aberrant DNA methylation of pregnane X receptor underlies metabolic gene alterations in the diabetic kidney. Am. J. Physiol. Physiol. 2018, 314, F551–F560. [Google Scholar] [CrossRef] [Green Version]
- Cusi, K. Nonalcoholic fatty liver disease in type 2 diabetes mellitus. Curr. Opin. Endocrinol. Diabetes Obes. 2009, 16, 141–149. [Google Scholar] [CrossRef]
- Oladimeji, P.; Lin, W.; Brewer, C.T.; Chen, T. Glucose-dependent regulation of pregnane X receptor is modulated by AMP-activated protein kinase. Sci. Rep. 2017, 7, 46751. [Google Scholar] [CrossRef] [Green Version]
- Xiong, J.; Shang, W.; Wu, L.; Chen, R.; Liu, W.; Ning, R.; Hu, G.; Yang, J. Glucose dominates the regulation of carboxylesterases induced by lipopolysaccharide or interleukin-6 in primary mouse hepatocytes. Life Sci. 2014, 112, 41–48. [Google Scholar] [CrossRef]
- Davidson, M.D.; Ballinger, K.R.; Khetani, S.R. Long-term exposure to abnormal glucose levels alters drug metabolism pathways and insulin sensitivity in primary human hepatocytes. Sci. Rep. 2016, 6, 28178. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.; Zhang, X.; Liu, Y.; Xi, T.; Xiong, J. Insulin transcriptionally down-regulates carboxylesterases through pregnane X receptor in an Akt-dependent manner. Toxicology 2019, 422, 60–68. [Google Scholar] [CrossRef]
- Shan, E.; Zhu, Z.; He, S.; Chu, D.; Ge, D.; Zhan, Y.; Liu, W.; Yang, J.; Xiong, J. Involvement of pregnane X receptor in the suppression of carboxylesterases by metformin in vivo and in vitro, mediated by the activation of AMPK and JNK signaling pathway. Eur. J. Pharm. Sci. 2017, 102, 14–23. [Google Scholar] [CrossRef] [PubMed]
- Luo, W.; Xin, Y.; Zhao, X.; Zhang, F.; Liu, C.; Fan, H.; Xi, T.; Xiong, J. Suppression of carboxylesterases by imatinib mediated by the down-regulation of pregnane X receptor. Br. J. Pharmacol. 2017, 174, 700–717. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y.; Li, C.; Sun, X.; Kuang, X.; Ruan, X.-C. High glucose decreases expression and activity of p-glycoprotein in cultured human retinal pigment epithelium possibly through iNOS induction. PLoS ONE 2012, 7, e31631. [Google Scholar] [CrossRef] [PubMed]
- Rowe, I.A. Lessons from epidemiology: The burden of liver disease. Dig. Dis. 2017, 35, 304–309. [Google Scholar] [CrossRef]
- Jungst, C.; Lammert, F. Cholestatic liver disease. Dig Dis. 2013, 31, 152–154. [Google Scholar] [CrossRef]
- Kurzawski, M.; Dziedziejko, V.; Post, M.; Wójcicki, M.; Urasińska, E.; Miętkiewski, J.; Drozdzik, M. Expression of genes involved in xenobiotic metabolism and transport in end-stage liver disease: Up-regulation of ABCC4 and CYP1B. Pharmacol. Rep. 2012, 64, 927–939. [Google Scholar] [CrossRef]
- Congiu, M.; Mashford, M.L.; Slavin, J.L.; Desmond, P.V. Coordinate regulation of metabolic enzymes and transporters by nuclear transcription factors in human liver disease. J. Gastroenterol. Hepatol. 2009, 24, 1038–1044. [Google Scholar] [CrossRef]
- Hanada, K.; Nakai, K.; Tanaka, H.; Suzuki, F.; Kumada, H.; Ohno, Y.; Ozawa, S.; Ogata, H. Effect of nuclear receptor downregulation on hepatic expression of cytochrome P450 and transporters in chronic hepatitis C in association with fibrosis development. Drug Metab. Pharmacokinet. 2011, 27, 301–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- More, V.R.; Cheng, Q.; Donepudi, A.C.; Buckley, D.B.; Lu, Z.J.; Cherrington, N.J.; Slitt, A.L. Alcohol cirrhosis alters nuclear receptor and drug transporter expression in human liver. Drug Metab. Dispos. 2013, 41, 1148–1155. [Google Scholar] [CrossRef] [Green Version]
- Elbel, E.E.; LaVine, J.E.; Downes, M.; Van Natta, M.; Yu, R.; Schwimmer, J.B.; Behling, C.; Brunt, E.M.; Tonascia, J.; Evans, R. Hepatic nuclear receptor expression associates with features of histology in pediatric nonalcoholic fatty liver disease. Hepatol. Commun. 2018, 2, 1213–1226. [Google Scholar] [CrossRef] [Green Version]
- Angulo, P.; Kleiner, D.E.; Dam-Larsen, S.; Adams, L.A.; Björnsson, E.S.; Charatcharoenwitthaya, P.; Mills, P.R.; Keach, J.C.; Lafferty, H.D.; Stahler, A.; et al. Liver fibrosis, but no other histologic features, is associated with long-term outcomes of patients with nonalcoholic fatty liver disease. Gastroenterology 2015, 149, 389–397. [Google Scholar] [CrossRef] [Green Version]
- Tutau, F.; Rodríguez, C.; Puche, J.E.; Juanarena, N.; Monreal, I.; Garcia-Fernandez, M.I.; Clavijo, E.; Castilla, A.; Castilla-Cortázar, I. Enhanced actions of insulin-like growth factor-I and interferon-α co-administration in experimental cirrhosis. Liver Int. 2009, 29, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Mohandas, S.; Vairappan, B. Ginkgolide-A attenuates bacterial translocation through activating PXR and improving antimicrobial peptide Reg 3A in experimental cirrhosis. Life Sci. 2020, 257, 118111. [Google Scholar] [CrossRef]
- Mohandas, S.; Vairappan, B. Pregnane X receptor activation by its natural ligand Ginkgolide-A improves tight junction proteins expression and attenuates bacterial translocation in cirrhosis. Chem. Interact. 2020, 315, 108891. [Google Scholar] [CrossRef]
- Xie, Y.; Hao, H.-P.; Wang, H.; Wang, Z.-X.; Wang, G.-J. Reversing effects of silybin on TAA-induced hepatic CYP3A dysfunction through PXR regulation. Chin. J. Nat. Med. 2013, 11, 645–652. [Google Scholar] [CrossRef]
- Naito, H.; Jia, X.; Yetti, H.; Yanagiba, Y.; Tamada, H.; Kitamori, K.; Hayashi, Y.; Wang, D.; Kato, M.; Ishii, A.; et al. Importance of detoxifying enzymes in differentiating fibrotic development between SHRSP5/Dmcr and SHRSP rats. Environ. Health Prev. Med. 2016, 21, 368–381. [Google Scholar] [CrossRef]
- Yetti, H.; Naito, H.; Yuan, Y.; Jia, X.; Hayashi, Y.; Tamada, H.; Kitamori, K.; Ikeda, K.; Yamori, Y.; Nakajima, T. Bile acid detoxifying enzymes limit susceptibility to liver fibrosis in female SHRSP5/Dmcr rats fed with a high-fat-cholesterol diet. PLoS ONE 2018, 13, e0192863. [Google Scholar] [CrossRef] [Green Version]
- Avouac, J. Mouse model of experimental dermal fibrosis: The bleomycin-induced dermal fibrosis. Adv. Struct. Saf. Stud. 2014, 1142, 91–98. [Google Scholar] [CrossRef]
- Wunsch, E.; Klak, M.; Wasik, U.; Milkiewicz, M.; Blatkiewicz, M.; Urasinska, E.; Barbier, O.; Bielicki, D.; Bogdanos, D.P.; Elias, E.; et al. Liver expression of sulphotransferase 2A1 enzyme is impaired in patients with primary sclerosing cholangitis: Lack of the response to enhanced expression of PXR. J. Immunol. Res. 2015, 2015, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Zollner, G.; Wagner, M.; Fickert, P.; Silbert, D.; Gumhold, J.; Zatloukal, K.; Denk, H.; Trauner, M. Expression of bile acid synthesis and detoxification enzymes and the alternative bile acid efflux pump MRP4 in patients with primary biliary cirrhosis. Liver Int. 2007, 27, 920–929. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Luo, D.; Wu, X.; Wang, H.; He, Y.; Li, Q.; Zhang, Y.; Chen, L.; Peng, Z.-H.; Xiao, T.; et al. Changes of organic anion transporter MRP4 and related nuclear receptors in human obstructive cholestasis. J. Gastrointest. Surg. 2011, 15, 996–1004. [Google Scholar] [CrossRef] [PubMed]
- Chai, J.; Feng, X.; Zhang, L.; Chen, S.; Cheng, Y.; He, X.; Yang, Y.; He, Y.; Wang, H.; Wang, R.; et al. Hepatic expression of detoxification enzymes is decreased in human obstructive cholestasis due to gallstone biliary obstruction. PLoS ONE 2015, 10, e0120055. [Google Scholar] [CrossRef] [PubMed]
- Chen, H.-L.; Liu, Y.-J.; Chen, H.-L.; Wu, S.-H.; Ni, Y.-H.; Ho, M.-C.; Lai, H.-S.; Hsu, W.-M.; Chang, M.-H.; Tseng, H.-C.; et al. Expression of hepatocyte transporters and nuclear receptors in children with early and late-stage biliary atresia. Pediatr. Res. 2008, 63, 667–673. [Google Scholar] [CrossRef] [Green Version]
- Honda, A.; Ikegami, T.; Nakamuta, M.; Miyazaki, T.; Iwamoto, J.; Hirayama, T.; Saito, Y.; Takikawa, H.; Imawari, M.; Matsuzaki, Y. Anticholestatic effects of bezafibrate in patients with primary biliary cirrhosis treated with ursodeoxycholic acid. Hepatology 2013, 57, 1931–1941. [Google Scholar] [CrossRef]
- Gabbia, D.; Pozzo, L.; Zigiotto, G.; Roverso, M.; Sacchi, D.; Pozza, A.D.; Carrara, M.; Bogialli, S.; Floreani, A.; Guido, M.; et al. Dexamethasone counteracts hepatic inflammation and oxidative stress in cholestatic rats via CAR activation. PLoS ONE 2018, 13, e0204336. [Google Scholar] [CrossRef]
- Li, T.; Xu, L.; Zheng, R.; Wang, X.; Li, L.; Ji, H.; Hu, Q. Picroside II protects against cholestatic liver injury possibly through activation of farnesoid X receptor. Phytomedicine 2020, 68, 153153. [Google Scholar] [CrossRef]
- Teng, S.; Piquette-Miller, M. Hepatoprotective role of PXR activation and MRP3 in cholic acid-induced cholestasis. Br. J. Pharmacol. 2007, 151, 367–376. [Google Scholar] [CrossRef] [Green Version]
- Gabbia, D.; Pozza, A.D.; Albertoni, L.; Lazzari, R.; Zigiotto, G.; Carrara, M.; Baldo, V.; Baldovin, T.; Floreani, A.; De Martin, S. Pregnane X receptor and constitutive androstane receptor modulate differently CYP3A-mediated metabolism in early- and late-stage cholestasis. World J. Gastroenterol. 2017, 23, 7519–7530. [Google Scholar] [CrossRef]
- Gupta, D.; Venkatesh, M.; Wang, H.; Kim, S.; Sinz, M.; Goldberg, G.L.; Whitney, K.; Longley, C.; Mani, S. Expanding the roles for pregnane X receptor in cancer: Proliferation and drug resistance in ovarian cancer. Clin. Cancer Res. 2008, 14, 5332–5340. [Google Scholar] [CrossRef] [Green Version]
- Dong, Y.; Wang, Z.; Xie, G.; Li, C.; Zuo, W.-W.; Meng, G.; Xu, C.-P.; Li, J. Pregnane X receptor is associated with unfavorable survival and induces chemotherapeutic resistance by transcriptional activating multidrug resistance-related protein 3 in colorectal cancer. Mol. Cancer 2017, 16, 71. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van de Winkel, A.; Menke, V.; Capello, A.; Moons, L.M.; Pot, R.G.; van Dekken, H.; Siersema, P.D.; Kusters, J.G.; van der Laan, L.J.; Kuipers, E.J. Expression, localization and polymorphisms of the nuclear receptor PXR in Barrett’s esophagus and esophageal adenocarcinoma. BMC Gastroenterol. 2011, 11, 108. [Google Scholar] [CrossRef] [Green Version]
- Dotzlaw, H.; Leygue, E.; Watson, P.; Murphy, L.C. The human orphan receptor PXR messenger RNA is expressed in both normal and neoplastic breast tissue. Clin. Cancer Res. 1999, 5, 2103–2107. [Google Scholar] [PubMed]
- Chen, Y.; Tang, Y.; Wang, M.-T.; Zeng, S.; Nie, D. Human pregnane X receptor and resistance to chemotherapy in prostate cancer. Cancer Res. 2007, 67, 10361–10367. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Masuyama, H.; Hiramatsu, Y.; Kodama, J.-I.; Kudo, T. Expression and potential roles of pregnane X receptor in endometrial cancer. J. Clin. Endocrinol. Metab. 2003, 88, 4446–4454. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Planque, C.; Rajabi, F.; Grillet, F.; Finetti, P.; Bertucci, F.; Gironella, M.; Lozano, J.J.; Beucher, B.; Giraud, J.; Garambois, V.; et al. Pregnane X-receptor promotes stem cell-mediated colon cancer relapse. Oncotarget 2016, 7, 56558–56573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiao, E.; Ji, M.; Wu, J.; Ma, R.; Zhang, X.; He, Y.; Zha, Q.; Song, X.; Zhu, L.-W.; Tang, J. Expression of the PXR gene in various types of cancer and drug resistance. Oncol. Lett. 2013, 5, 1093–1100. [Google Scholar] [CrossRef] [Green Version]
- Bhagyaraj, E.; Ahuja, N.; Kumar, S.; Tiwari, D.; Gupta, S.; Nanduri, R.; Gupta, P. TGF-beta induced chemoresistance in liver cancer is modulated by xenobiotic nuclear receptor PXR. Cell Cycle 2019, 18, 3589–3602. [Google Scholar] [CrossRef]
- Kotiya, D.; Jaiswal, B.; Ghose, S.; Kaul, R.; Datta, K.; Tyagi, R.K. Role of PXR in hepatic cancer: Its influences on liver detoxification capacity and cancer progression. PLoS ONE 2016, 11, e0164087. [Google Scholar] [CrossRef]
- Haines, C.; Elcombe, B.M.; Chatham, L.R.; Vardy, A.; Higgins, L.G.; Elcombe, C.R.; Lake, B.G. Comparison of the effects of sodium phenobarbital in wild type and humanized constitutive androstane receptor (CAR)/pregnane X receptor (PXR) mice and in cultured mouse, rat and human hepatocytes. Toxicology 2018, 396–397, 23–32. [Google Scholar] [CrossRef]
- Chen, Y.; Tang, Y.; Guo, C.; Wang, J.; Boral, D.; Nie, D. Nuclear receptors in the multidrug resistance through the regulation of drug-metabolizing enzymes and drug transporters. Biochem. Pharmacol. 2012, 83, 1112–1126. [Google Scholar] [CrossRef] [Green Version]
- Robbins, D.; Chen, T. Tissue-specific regulation of pregnane X receptor in cancer development and therapy. Cell Biosci. 2014, 4, 17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chakraborty, S.; Kanakasabai, S.; Bright, J.J. Constitutive androstane receptor agonist CITCO inhibits growth and expansion of brain tumour stem cells. Br. J. Cancer 2011, 104, 448–459. [Google Scholar] [CrossRef] [PubMed]
- Fukumasu, H.; Cordeiro, Y.; Rochetti, A.; Barra, C.; Sámora, T.; Strefezzi, R.; Dagli, M.L.Z. Expression of NR1I3 in mouse lung tumors induced by the tobacco-specific nitrosamine 4-(methylnitrosamino)-4-(3-pyridyl)-1-butanone. Braz. J. Med. Biol. Res. 2015, 48, 240–244. [Google Scholar] [CrossRef] [Green Version]
- Nagata, T.; Takahashi, Y.; Ishii, Y.; Asai, S.; Nishida, Y.; Murata, A.; Koshinaga, T.; Fukuzawa, M.; Hamazaki, M.; Asami, K.; et al. Transcriptional profiling in hepatoblastomas using high-density oligonucleotide DNA array. Cancer Genet. Cytogenet. 2003, 145, 152–160. [Google Scholar] [CrossRef]
- Tang, H.Q.; Xu, M.; Rong, Q.; Jin, R.W.; Liu, Q.J.; Li, Y.L. The effect of ZnO nanoparticles on liver function in rats. Int. J. Nanomed. 2016, 11, 4275–4285. [Google Scholar]
- Tang, H.; Xu, M.; Shi, F.; Ye, G.; Lv, C.; Luo, J.; Zhao, L.; Li, Y. Effects and mechanism of nano-copper exposure on hepatic cytochrome P450 enzymes in rats. Int. J. Mol. Sci. 2018, 19, 2140. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Tang, H.; Xu, M.; Luo, J.; Zhao, L.; Shi, F.; Ye, G.; Lv, C.; Li, Y. Effect of copper nanoparticles on brain cytochrome P450 enzymes in rats. Mol. Med. Rep. 2019, 20, 771–778. [Google Scholar] [CrossRef] [PubMed]
- Tian, J.; Hu, J.; Liu, G.; Yin, H.; Chen, M.; Miao, P.; Bai, P.; Yin, J. Corrigendum to “Altered gene expression of ABC transporters, nuclear receptors and oxidative stress signaling in zebrafish embryos exposed to CdTe quantum dots”. Environ. Pollut. 2019, 245, 1136. [Google Scholar] [CrossRef]
- Zenata, O.; Vrzalova, A.; Bachleda, P.; Janečková, J.; Panáček, A.; Kvítek, L.; Vrzal, R. The effect of graphene oxide on signalling of xenobiotic receptors involved in biotransformation. Chemosphere 2020, 253, 126753. [Google Scholar] [CrossRef]
- Kermanizadeh, A.; Gaiser, B.K.; Johnston, H.; Brown, D.M.; Stone, V. Toxicological effect of engineered nanomaterials on the liver. Br. J. Pharmacol. 2014, 171, 3980–3987. [Google Scholar] [CrossRef] [PubMed]
- Yilmaz, B.; Terekeci, H.; Sandal, S.; Kelestimur, F. Endocrine disrupting chemicals: Exposure, effects on human health, mechanism of action, models for testing and strategies for prevention. Rev. Endocr. Metab. Disord. 2019, 21, 127–147. [Google Scholar] [CrossRef]
- Hernandez, J.P.; Huang, W.; Chapman, L.M.; Chua, S.; Moore, D.D.; Baldwin, W.S. The environmental estrogen, nonylphenol, activates the constitutive androstane receptor. Toxicol. Sci. 2007, 98, 416–426. [Google Scholar] [CrossRef] [Green Version]
- Toporova, L.; Balaguer, P. Nuclear receptors are the major targets of endocrine disrupting chemicals. Mol. Cell. Endocrinol. 2020, 502, 110665. [Google Scholar] [CrossRef]
- Meucci, V.; Arukwe, A. The xenoestrogen 4-nonylphenol modulates hepatic gene expression of pregnane X receptor, aryl hydrocarbon receptor, CYP3A and CYP1A1 in juvenile Atlantic salmon (Salmo salar). Comp. Biochem. Physiol. Part C: Toxicol. Pharmacol. 2006, 142, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Litwa, E.; Kajta, M.; Wnuk, A.; Lasoń, W.; Krzeptowski, W.; Kajta, M. RXRα, PXR and CAR xenobiotic receptors mediate the apoptotic and neurotoxic actions of nonylphenol in mouse hippocampal cells. J. Steroid Biochem. Mol. Biol. 2016, 156, 43–52. [Google Scholar] [CrossRef] [PubMed]
- Fu, H.; Qiu, S.; Yao, X.; Gao, F.; Tan, P.; Teng, T.; Shi, B. Toxicity of glyphosate in feed for weanling piglets and the mechanism of glyphosate detoxification by the liver nuclear receptor CAR/PXR pathway. J. Hazard. Mater. 2020, 387, 121707. [Google Scholar] [CrossRef] [PubMed]
- Xiang, D.; Chu, T.; Li, M.; Wang, Q.; Zhu, G. Effects of pyrethroid pesticide cis-bifenthrin on lipogenesis in hepatic cell line. Chemosphere 2018, 201, 840–849. [Google Scholar] [CrossRef]
- Medina-Díaz, I.; Estrada-Muñiz, E.; Reyes-Hernández, O.; Ramírez, P.; Vega, L.; Elizondo, G. Arsenite and its metabolites, MMAIII and DMAIII, modify CYP3A4, PXR and RXR alpha expression in the small intestine of CYP3A4 transgenic mice. Toxicol. Appl. Pharmacol. 2009, 239, 162–168. [Google Scholar] [CrossRef] [PubMed]
- Noreault, T.L.; Kostrubsky, V.E.; Wood, S.G.; Nichols, R.C.; Strom, S.C.; Trask, H.W.; Wrighton, S.A.; Evans, R.M.; Jacobs, J.M.; Sinclair, P.R.; et al. Arsenite decreases CYP3A4 and RXRalpha in primary human hepatocytes. Drug Metab. Dispos. 2005, 33, 993–1003. [Google Scholar] [CrossRef]
- Liu, G.; Tian, J.; Yin, H.; Yin, J.; Tang, Y. Self-protective transcriptional alterations in ZF4 cells exposed to Pb(NO3)2 and AgNO3. J. Biochem. Mol. Toxicol. 2019, 33, e22408. [Google Scholar] [CrossRef] [PubMed]
- Li, X.-Y.; Ma, J.; Jing, C.; Wang, J. Expression alterations of cytochromes P4501A1, 2E1, and 3A, and their receptors AhR and PXR caused by 1-octyl-3-methylimidazolium chloride in mouse mammary carcinoma cells. Chemosphere 2013, 93, 2488–2492. [Google Scholar] [CrossRef]
- Souidi, M.; Gueguen, Y.; Linard, C.; Dudoignon, N.; Grison, S.; Baudelin, C.; Marquette, C.; Gourmelon, P.; Aigueperse, J.; Dublineau, I. In vivo effects of chronic contamination with depleted uranium on CYP3A and associated nuclear receptors PXR and CAR in the rat. Toxicology 2005, 214, 113–122. [Google Scholar] [CrossRef]
- Ayed-Boussema, I.; Pascussi, J.-M.; Maurel, P.; Bacha, H.; Hassen, W. Effect of aflatoxin B1 on nuclear receptors PXR, CAR, and AhR and their target cytochromes P450 mRNA expression in primary cultures of human hepatocytes. Int. J. Toxicol. 2011, 31, 86–93. [Google Scholar] [CrossRef]
- Ayed-Boussema, I.; Pascussi, J.-M.; Rjiba, K.; Maurel, P.; Bacha, H.; Hassen, W. The mycotoxin, patulin, increases the expression of PXR and AhR and their target cytochrome P450s in primary cultured human hepatocytes. Drug Chem. Toxicol. 2011, 35, 241–250. [Google Scholar] [CrossRef]
- Dořičáková, A.; Vrzal, R. A food contaminant ochratoxin A suppresses pregnane X receptor (PXR)-mediated CYP3A4 induction in primary cultures of human hepatocytes. Toxicology 2015, 337, 72–78. [Google Scholar] [CrossRef]
- Shen, Y.; Shi, Z.; Fan, J.T.; Yan, B. Dechlorination and demethylation of ochratoxin A enhance blocking activity of PXR activation, suppress PXR expression and reduce cytotoxicity. Toxicol. Lett. 2020, 332, 171–180. [Google Scholar] [CrossRef] [PubMed]
- Renu, K.; Gopalakrishnan, A.V. Deciphering the molecular mechanism during doxorubicin-mediated oxidative stress, apoptosis through Nrf2 and PGC-1α in a rat testicular milieu. Reprod. Biol. 2019, 19, 22–37. [Google Scholar] [CrossRef]
- Fukumasu, H.; Rochetti, A.L.; Pires, P.R.L.; Da Silva, E.R.; Mesquita, L.G.; Strefezzi, R.; De Carvalho, D.D.; Dagli, M.L.Z. Constitutive androstane receptor ligands modulate the anti-tumor efficacy of paclitaxel in non-small cell lung cancer cells. PLoS ONE 2014, 9, e99484. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swart, M.; Dandara, C. MicroRNA mediated changes in drug metabolism and target gene expression by efavirenz and rifampicin in vitro: Clinical implications. OMICS A J. Integr. Biol. 2019, 23, 496–507. [Google Scholar] [CrossRef] [Green Version]
- Ku, P.; Wang, C.; Nie, X.; Ou, R.; Li, K. Regulation of pregnane-X-receptor and microRNAs on detoxification-related genes expressions in Mugilogobius abei under the exposure to diclofenac. Environ. Pollut. 2018, 233, 395–406. [Google Scholar] [CrossRef]
- Liu, Y.-H.; Mo, S.-L.; Bi, H.; Hu, B.-F.; Li, C.G.; Wang, Y.-T.; Huang, L.; Huang, M.; Duan, W.; Liu, J.-P.; et al. Regulation of human pregnane X receptor and its target gene cytochrome P450 3A4 by Chinese herbal compounds and a molecular docking study. Xenobiotica 2010, 41, 259–280. [Google Scholar] [CrossRef] [PubMed]
- Hu, D.; Wang, Y.; Chen, Z.; Ma, Z.; You, Q.; Zhang, X.; Liang, Q.; Tan, H.; Xiao, C.; Tang, X.; et al. The protective effect of piperine on dextran sulfate sodium induced inflammatory bowel disease and its relation with pregnane X receptor activation. J. Ethnopharmacol. 2015, 169, 109–123. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Gao, Y.; Wang, Y.; Ma, Z.; Liang, Q.; Tang, X.; Hu, D.; Tan, H.; Xiao, C. Tanshinone IIA ameliorates dextran sulfate sodium-induced inflammatory bowel disease via the pregnane X receptor. Drug Des. Dev. Ther. 2015, 9, 6343–6362. [Google Scholar] [CrossRef] [PubMed]
- Dou, W.; Zhang, J.; Li, H.; Kortagere, S.; Sun, K.; Ding, L.; Ren, G.; Hu, F.; Mani, S. Plant flavonol isorhamnetin attenuates chemically induced inflammatory bowel disease via a PXR-dependent pathway. J. Nutr. Biochem. 2014, 25, 923–933. [Google Scholar] [CrossRef] [Green Version]
- Juan, Y.-C.; Chang, C.-C.; Tsai, W.-J.; Lin, Y.-L.; Hsu, Y.-S.; Liu, H.-K. Pharmacological evaluation of insulin mimetic novel suppressors of PEPCK gene transcription from Paeoniae Rubra Radix. J. Ethnopharmacol. 2011, 137, 592–600. [Google Scholar] [CrossRef]
- Murali, G.; Milne, G.L.; Webb, C.D.; Stewart, A.B.; McMillan, R.P.; Lyle, B.C.; Hulver, M.W.; Saraswathi, V. Fish oil and indomethacin in combination potently reduce dyslipidemia and hepatic steatosis in LDLR(-/-) mice. J. Lipid Res. 2012, 53, 2186–2197. [Google Scholar] [CrossRef] [Green Version]
- Saraswathi, V.; Perriotte-Olson, C.; Ganesan, M.; DeSouza, C.V.; Alnouti, Y.; Duryee, M.J.; Thiele, G.M.; Nordgren, T.M.; Clemens, D.L. A combination of dietary N-3 fatty acids and a cyclooxygenase-1 inhibitor attenuates nonalcoholic fatty liver disease in mice. J. Nutr. Biochem. 2017, 42, 149–159. [Google Scholar] [CrossRef]
- He, L.; Li, H.; Huang, N.; Zhou, X.; Tian, J.; Li, T.; Wu, J.; Tian, Y.; Yin, Y.; Yao, K. Alpha-ketoglutarate suppresses the NF-κB-mediated inflammatory pathway and enhances the PXR-regulated detoxification pathway. Oncotarget 2017, 8, 102974–102988. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, L.; Zhou, L.; Geng, S.; Li, M.; Lu, W.; Lu, Y.; Feng, Z.; Zhou, X. Catalpol coordinately regulates phase I and II detoxification enzymes of Triptolide through CAR and NRF2 pathways to reduce Triptolide-induced hepatotoxicity. Biomed. Pharmacother. 2020, 129, 110379. [Google Scholar] [CrossRef] [PubMed]
- Radović, B.; Hussong, R.; Gerhäuser, C.; Meinl, W.; Frank, N.; Becker, H.; Köhrle, J. Xanthohumol, a prenylated chalcone from hops, modulates hepatic expression of genes involved in thyroid hormone distribution and metabolism. Mol. Nutr. Food Res. 2010, 54, S225–S235. [Google Scholar] [CrossRef]
- Xu, H.-B.; Tang, Z.-Q.; Wang, J.; Kong, P.-S. Z-guggulsterone regulates MDR1 expression mainly through the pregnane X receptor-dependent manner in human brain microvessel endothelial cells. Eur. J. Pharmacol. 2020, 874, 173023. [Google Scholar] [CrossRef] [PubMed]
- Fukumasu, H.; Rochetti, A.L.; Latorre, A.O.; Pires, P.R.L.; Silva, T.C.; Dagli, M.L.Z. Caffeine increases Nr1i3 expression and potentiates the effects of its ligand, TCPOBOP, in mice liver. Braz. J. Pharm. Sci. 2015, 51, 295–303. [Google Scholar] [CrossRef] [Green Version]
- Noreault, T.L.; Jacobs, J.M.; Nichols, R.C.; Trask, H.W.; Wrighton, S.A.; Sinclair, P.R.; Sinclair, J.F. Mechanism of arsenite-mediated decreases in CYP3A23 in rat hepatocytes. Biochem. Biophys. Res. Commun. 2005, 333, 1211–1217. [Google Scholar] [CrossRef]
- Chai, S.C.; Cherian, M.T.; Wang, Y.-M.; Chen, T. Small-molecule modulators of PXR and CAR. Biochim. Biophys. Acta (BBA) Bioenerg. 2016, 1859, 1141–1154. [Google Scholar] [CrossRef] [Green Version]
| Transcription Factor | Species | Tissue | Effect | Mechanism | Reference |
|---|---|---|---|---|---|
| PXR | |||||
| HNF4α | Mouse | Liver | + | [118,119] | |
| Human | HepG2 cells | + | DR1-proximal promoter | [120] | |
| NAA10 | Human | AsPC-1 pancreatic cancer cell line | + | HNF4 co-factor | [121] |
| GR | Human | PHHs | + | [127] | |
| Huh7 cells | + | 2.2kb promoter | [106] | ||
| Rat | Primary hepatocytes | + | [129,132,133,134] | ||
| Pancreatic acinar cell line | + | ||||
| Zebrafish | Larvae, liver | + | [136] | ||
| Mouse | Lung | + | [137] | ||
| Rat | Jejunum, ileum, colon | + | [138] | ||
| Rat | Blood–brain barrier, brain endothelial cells | + | [68,69] | ||
| + | [139] | ||||
| AHR | Human | Hepatocytes | - | AHR dependent | [142] |
| FXR | Mouse | Liver | + | IR1 | [145] |
| KLF4 | Mouse | Calvaria osteoblasts | + | [41] | |
| PPARα | Human | Huh7 cells | + | 2.2 kb promoter, PPRE in silico | [106] |
| Rat | Hepatocytes | + | [23] | ||
| PPARα PGC-1α | Mouse | Liver | + | [107] | |
| CAR-PXR | Human | Huh7 cells | − | 2.2 kb promoter | [106] |
| ETS1 | Mouse | Hepa1-6 cells | + | 1 kb promoter | [85] |
| LyF-VI–NF1 | Mouse | Hepa1-6 cells | − | 1 kb promoter | [85] |
| ETS1, PAX5, LEF1, c-Jun | Human | HepG2 cells | + | [151] | |
| Sp1, hnRNPK | Human | HepG2, LS180 cells | + | Composite element | [152] |
| CAR | |||||
| HNF4α1 | Human | Hepatocytes | + | [124] | |
| HNF4α7 | Human | Hepatocytes | + | Poor activator–repressor of HNF4α1 | [124] |
| HNF4α | Mouse | Mouse and human promoters in CV1 | + | Co-activation by PGC-1α | [102] |
| Human | |||||
| GR | Human | PHH | + | Distal GRE | [126,129,130] |
| Rat | Pancreatic acinar cell line | + | [135] | ||
| AhR | Mouse | Liver and extrahepatic tissue | + | [72,73] | |
| Human | PHH | + | [72,73] | ||
| Rat | Hepatocytes | + | [141] | ||
| TR | Rat | Hepatocyte progenitors | + | Indirect | [148] |
| TCF/LEF | Mouse | Liver | + | WRE | [85] |
| PGC-1α | Mouse | Liver, hepatocytes | + | Indirect through HNF4-RE | [102] |
| PPARα | Rat | Hepatocytes | + | DR1 motif | [103,105] |
| Mouse | Liver | ||||
| PXR | Mouse | Liver | −/+ | [149,150] | |
| miRNA | Cell Type/Species | Target | Effects | Reference |
|---|---|---|---|---|
| miRNA-148a | HepG2, LS180 cells | PXR 3′-UTR | Inhibition | [161] |
| Human liver | PXR | No correlation in the Chinese Han population | [164] | |
| Human liver | PXR CAR | No correlation | [163] | |
| Human liver | PXR CAR | Positive correlation | [162] | |
| LO2 liver cells | PXR | Inhibition | [167] | |
| Oropharyngeal cancer cell lines | PXR | Inhibition | [168] | |
| miR-34a | Hela, HepG2 cells Human hepatocytes | PXR | Inhibition through HNF4-α | [165] |
| Human liver | PXR CAR | Negative correlation | [162] | |
| Human liver | CAR | Negative correlation | [163] | |
| miR449-a | Hela, HepG2 cells Human hepatocytes | PXR | Inhibition through HNF4-α | [165] |
| miR-204 | Human liver | CAR | Positive correlation | [163] |
| miR-21 | Human liver | CAR | Negative correlation | [163] |
| mir-150 | Human liver | PXR CAR | Negative correlation | [162] |
| miR27a | Human liver | PXR | Negative correlation | [162] |
| miR-561 | HepG2 | PXR CAR | Induction through DAX-1 inhibition | [169] |
| miR-137 | Mouse primary hepatocytes | PXR | Inhibition | [9] |
| Neuroblastoma, HCC, and CRC | CAR | Inhibition | [170] | |
| miRNA-30c-1-3p | 293T, HepG2, LS180 cells | PXR 3′-UTR | Inhibition | [171] |
| miR-140-3p | HepG2 cells MHCC97-H cells | PXR 3′-UTR | Inhibition | [172] |
| miRNA-18a-5p | LS180 cells | PXR | Inhibition | [166] |
| Human hepatocytes Mouse liver HepG2 cells | PXR 3′-UTR | Inhibition | [131] | |
| miR-211-5p | HepG2.2.15 cells | CAR | Inhibition through sequestration by the lncRNA F11-AS1 | [173] |
| miR-486-5p | HepaRG human hepatocytes | PXR | Inhibition through sequestration by the lncRNA LINC00844 | [174] |
| Species | Model | S | Age/ Treatment | Organ | Nuclear Receptors and CYP Target Genes | REF |
|---|---|---|---|---|---|---|
| Diabetes/Obesity | ||||||
| Mouse | db/db | M | >10 w | liver | (+) Car, Rxrα, Hnf4α, Cyp2b10, Cyp2c29, Cyp4a10, CAR, PXR, RXRα, HNF4α α, CYP2B10, CYP2C29, CYP4A10 | [205] |
| db/db | M | 22 w | kidney | (−) Pxr | [206] | |
| 8 w | (−) Pxr | [218] | ||||
| db/db | M | 10 w | liver | (+) Cyp2b10, Cyp4a10, Cyp2c29 (0) Car, Pxr, Cyp3a11 | [207] | |
| 25 w | (+) Cyp4a10 (−) Cyp2c29 (0) Car, Pxr, Cyp3a11, Cyp2b10 | |||||
| TSOD | M | 12 w 28 w | liver | (+) Cyp3a11 (+) Pxr, Hnf4α, Pgc-1α, Cyp3a11, CYP3A, CYP2C (0) Car | [215] | |
| Rat | Zucker | 7–9 w | liver | (−) Car, CAR (0) Rxra, Ppara, Fxr, Lxr | [208] | |
| HFD/streptozotocin | 8 w/5 w | intestine | (+) Pxr, PXR, Cyp3a2 (−) Cyp2c | [217] | ||
| HFD | M | 4 w/4 w | liver | (+) Pxr | [212] | |
| HFD | M | 5 w/12 w | liver | (+) Car, Pparγ (0) PXR, RXRα, PPARα | [209] | |
| HFD | F | 6 w/13 w | liver | (+) Pxr, Fxr, Lxrs, Cyp3a2, CYP3A2 (0) CAR | [210] | |
| Mouse | HFD | M | 5 w/4 w 5 w/8, 18 w | liver | (+) Car, Pparα, Pxr, Hnf4α, Cyp2b10 (+) Car, Pparα, Cyp2b10 (0) Pxr, Hnf4a | [211] |
| F | 5 w/4, 18 w 5 w/8 w | (0) Car, Pparα, Pxr, Hnf4α, Cyp2b10 (+) Hnf4a (0) Car, Pparα, Pxr, Cyp2b10 | ||||
| HFD | M F | 4 w/11 w 4 w/36 w | liver | (+) CAR (+) CAR | [216] | |
| HFD | M | 6 w/4 w | liver | (+) Fxr, Cyp3a11 (0) Car, Pxr, Cyp2b10 | [213] | |
| 6 w/12 w | (+) Car, Pxr, Cyp2b10, Cyp3a11 | |||||
| 6 w/16, 24, 32 w | (+) Pxr, Cyp3a11 (0) Car, Cyp2b10 | |||||
| CD1/HFD | M | 6 w/14 w | liver | (−) Car, Pxr, Cyp2b10, Cyp2a4, Cyp3a11 (0) Rxra | [214] | |
| Human | Chronic kidney disease, acute renal dysfunction | adult | kidney | (−) PXR | [218] | |
| Fibrosis/Cirrhosis | ||||||
| Human | End-stage liver disease (ALD, HCV, PSC) | adult | liver | (−) CAR, RXR, AHR, PXR | [229] | |
| HCV F3 fibrosis | adult | liver | (−) CAR, PXR | [231] | ||
| Steatosis, diabetes | adult | liver | (0) CAR, PXR, FXR, SHP | [232] | ||
| Alcohol cirrhosis | (0) CAR, PXR, SHP (−) FXR | |||||
| Diabetic cirrhosis | (−) PXR, FXR (0) CAR, SHP | |||||
| NAFLD—fibrosis | Pediatric | liver | (+) CAR, PXR, GCNF, COUP-TF1, NURR1, PPARs, Ers, TR, RAR/RXR | [233] | ||
| Rat | CCl4-induced cirrhosis | M | 3 w/12 w | liver | (−) PXR | [235] |
| Thioacetamide-induced cirrhosis | 200 g/7 w | liver | (−) Pxr, Cyp3a (−) PXR, CYP3A | [238] | ||
| SHRSP5/Dmcr HFC | M F | 10 w/8 w | liver | (−) Car, Pxr (−) CAR, PXR (−) Cyp8b1 (−) CYP8B1 (+) Cyp7a1, Cyp7b1 (+) CYP7A1, CYP7B1 | [239,240] | |
| Mouse | CCl4-induced cirrhosis | M | 25 g/12 w | liver intestine | (−) Pxr, Cyp3a | [236,237] |
| Cholestasis | ||||||
| Human | Chronic cholestasis PBC PSC | adult | liver | (+) PXR (+) PXR | [242] | |
| PBC grade III, IV | adult | liver | (0) PXR, CAR, FXR, RXR, SHP, HNF1α, HNF4α (−) CYP7A1 (0) CYP8B1, CYP3A4 | [243] | ||
| Obstructive cholestasis | adult | liver | (+) PXR, RXR, VDR, HNF4α, RARα (+) PXR, RXR, CAR, VDR, HNF4α, RARα (+) CYP7B1 (+) CYP7B1, CYP8B1 | [244,245] | ||
| Obstructive cholestasis | children | liver | (0) PXR (0) CAR (−) FXR, CYP7A1 | [246] | ||
| Early stage | ||||||
| Late stage | (−) PXR (0) CAR, CYP3A4 | |||||
| Rat | Bile duct ligation | M | 3 w after surgery | liver | (−) PXR, CAR, Cyp3a1/2, CYP3Aa1/2 | [248] |
| Bile duct ligation Mild cholestasis Severe cholestasis | M | 2 w after surgery 4 w after surgery | liver | (+) Pxr, Car, PXR, CAR, Cyp3A1/2, CYP3A1/2 (−) Pxr, Car, PXR, CAR, Cyp3A1/2, CYP3A1/2 | [251] | |
| Mouse | ANIT-induced cholestasis | M | liver | (+) Pxr, Pparα (−) Fxr, Ahr, Shp, Cyp7a1, Cyp8b1, CYP8B1 | [249] | |
| CA fed mice | M | 10 w/5 d | (+) Pxr, Fxr, Cyp3a11, Cyp7a1 (−) Car | [250] | ||
| Compound | Target | Model | Effect/Mechanism | Reference |
|---|---|---|---|---|
| Nanoparticles | ||||
| ZnO | CAR | Rat liver in vivo | Inhibition through inflammatory signaling | [268] |
| Nano-copper | CAR PXR | Rat liver in vivo | Inflammatory signaling and oxidative stress | [269] |
| Rat brain | Inhibition through oxidative stress | [270] | ||
| Quantum dots | PXR | Zebrafish embryos | Induction | [271] |
| Graphene oxide | PXR | PHH LS174 cells | Proteasome-independent inhibition | [272] |
| Environmental pollutants | ||||
| Nonylphenol | PXR | Juvenile salmon | Induction | [277] |
| CAR PXR | Rat hippocampal cells | Induction | [278] | |
| Glyphosate | CAR | Piglet liver | Induction | [279] |
| cis-BF pyrethroid pesticide | PXR | Human HepG2 cells | Induction | [280] |
| Benzo[a]pyrene | CAR | Rat hepatocytes | Induction | [141] |
| Arsenic | PXR | Mouse small intestine | Induction | [281] |
| Arsenite | PXR | Rat and human hepatocytes | No effect | [306] |
| Heavy metal | PXR | Zebrafish fibroblasts | Induction | [283] |
| Uranium | PXR | Rat brain, liver, and kidney | Induction | [285] |
| CAR | Rat lung | Induction | ||
| 1-octyl-3-methylimidazolium | PXR | Mouse mammary carcinoma cell line | Induction | [284] |
| Food contaminants | ||||
| Aflatoxin B1 | CAR PXR | PHH | Induction | [286] |
| Patulin | PXR | PHH | Induction | [287] |
| Ochratoxin A | PXR | PHH | Proteasome-independent inhibition through miR-148a | [288] |
| Ochratoxin A derivative | PXR | PXR-overexpressing HepG2 cells | Inhibition | [289] |
| Drugs | ||||
| Diclofenac | PXR | Benthic small fish | Induction | [293] |
| Doxorubicin | PXR | Rat gonad epithelium | Induction | [290] |
| Paclitaxel | CAR | Mouse lung cancer cell line E9 | Induction | [266] |
| Efavirenz | CAR | Human HepaRG cells | Inhibition | [292] |
| Rifampicin | ||||
| Food components | ||||
| Artemisinin | PXR | Human LS174 cells | Induction | [25] |
| Piperidine | PXR | Human LS174 cells | Induction | [295] |
| Tanshione IIA | PXR | Human LS174 cells | Induction | [296] |
| Isorhamnetin | PXR | Human LS174 cells | Induction | [297] |
| Probiotics VSL#3 | PXR | Colon and adipose/induced colitis mouse model | Reverse colitis-induced PXR inhibition | [40] |
| A-ketoglutarate | PXR | Induced colitis | Induction | [301] |
| Paeoniae radix rubra extracts | PXR | Rat hepatic cell line H4IIE | Downregulation by disruption of GR signaling | [298] |
| n−3 fatty acids (n−3) + SC-560 | PXR | HFD mice liver Human Huh7 cells | Induction through FXR signaling? | [300] |
| Fish oil + indomethacin | PXR | Liver | Induction | [299] |
| Tripolide | CAR | Rat liver Human L-02 and HepG2 cells | Inhibition | [302] |
| Z-guggulsterone | CAR | Human brain micro-vessel endothelial cells | Inhibition | [304] |
| Xanthohumol | CAR | Rat liver | Inhibition | [303] |
| Catalpol | CAR | Rat liver Human L-02 and HepG2 cells | Reverse tripolide-mediated inhibition | [302] |
| Caffeine | CAR | Mouse liver | Induction | [305] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Daujat-Chavanieu, M.; Gerbal-Chaloin, S. Regulation of CAR and PXR Expression in Health and Disease. Cells 2020, 9, 2395. https://doi.org/10.3390/cells9112395
Daujat-Chavanieu M, Gerbal-Chaloin S. Regulation of CAR and PXR Expression in Health and Disease. Cells. 2020; 9(11):2395. https://doi.org/10.3390/cells9112395
Chicago/Turabian StyleDaujat-Chavanieu, Martine, and Sabine Gerbal-Chaloin. 2020. "Regulation of CAR and PXR Expression in Health and Disease" Cells 9, no. 11: 2395. https://doi.org/10.3390/cells9112395
APA StyleDaujat-Chavanieu, M., & Gerbal-Chaloin, S. (2020). Regulation of CAR and PXR Expression in Health and Disease. Cells, 9(11), 2395. https://doi.org/10.3390/cells9112395