Regulatory microRNAs in Brown, Brite and White Adipose Tissue
Abstract
:1. Introduction
1.1. White Adipose Tissue (WAT)
1.2. Brown Adipose Tissue (BAT)
1.3. Brite Adipose Tissue (Beige or Brown-in-White Adipose Tissue)
1.4. Transcription Factors and Browning Agents
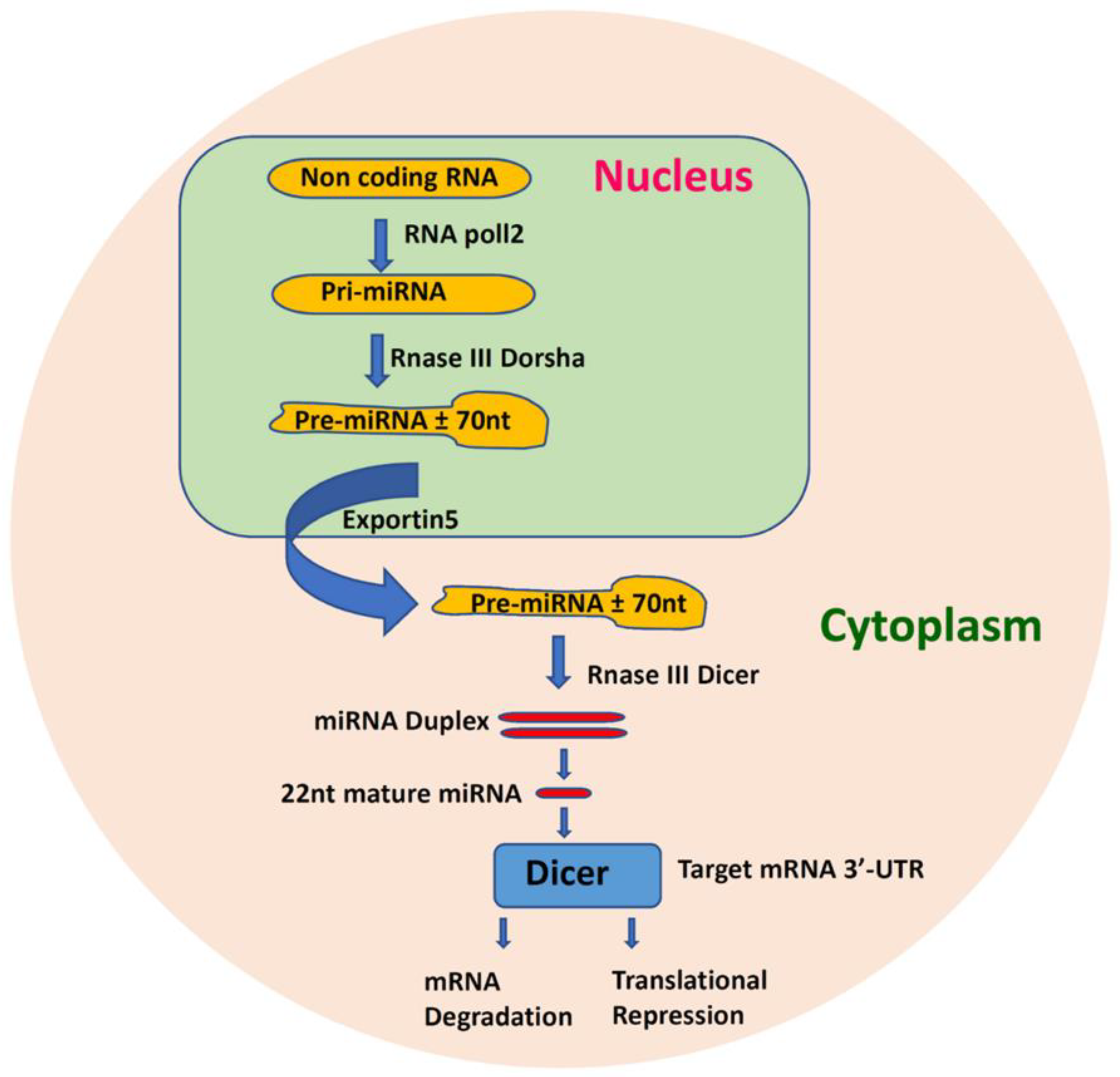
1.5. MicroRNA Function and Regulatory Mechanisms
2. MicroRNAs in BAT
3. MicroRNAs Regulating BAT and Brite
4. Micro RNAs Regulating Brite AT
5. MicroRNA in WAT
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| 3’ UTR | 3’ untranslated regions |
| ABCA1 | Adenosine triphosphate–binding cassette transporter |
| ADAM | A disintegrin and metalloprotease domain |
| ADAM17 | ADAM metallopeptidase domain 17 |
| ADIPOQ | Adiponectin gene |
| AdipoR1 | Adiponectin receptor 1 |
| AKT | Protein-kinase B |
| AMPKα1 | AMP-activated protein kinase alpha1 |
| AP2 | Adipocyte protein 2 |
| APCDD1 | Adenomatosis polyposis coli downregulated 1 |
| ASK1 | Apoptosis signal-regulating kinase 1 |
| AT | Adipose tissue |
| ATF2 | Activating transcription factor 2 |
| ATF6 | Activating transcription factor 6 |
| ATG7 | Autophagy-related gene 7 |
| ATP | Adenosine triphosphate |
| Bace1 | Beta-site amyloid precursor protein cleaving enzyme 1 |
| BAT | Brown adipose tissue |
| BMI | Body mass index |
| BMP7 | Bone morphogenetic protein 7 |
| C/EBP | CCAAT/enhancer-binding protein |
| CAD | Coronary artery disease |
| cAMP | Cyclic adenosine 3′,5′-monophosphate |
| Cav-1 | Caveolin-1 |
| Cdc34 | Cell division cycle 34 |
| CDK6 | Cell division protein kinase 6 |
| CIDEA | Cell death-inducing DNA fragmentation factor-like effector A |
| Creb | cAMP-response element binding protein |
| CVD | Cardiovascular disease |
| DGCR8 | DiGeorge syndrome critical region 8 |
| DLL4 | Delta-like protein 4 |
| E2f6 | E2F transcription factor 6 |
| EBF2 | Early B-cell factor transcription factor 2 |
| Egr1 | Early growth factor response 1 |
| EHMT1 | Euchromatic Histone Lysine Methyltransferase 1 |
| ER | Endoplasmic reticulum |
| ERK | Extracellular-signal-regulated kinase |
| ETS1 | E26 transformation–specific-1 |
| FABP4 | Fatty acid-binding protein 4 |
| Fbxl19 | F-box and leucine rich repeat protein 19 |
| FFA | Free fatty acids |
| FFAR4 | Free fatty acid receptor 4 |
| FGF7 | Fibroblast growth factor 7 |
| FGFR1 | Fibroblast growth factor receptor 1 |
| FNDC5 | Fibronectin type III domain containing 5 |
| GLUT4 | Glucose Transporter Type 4 |
| HDAC3 | Histone deacetylase 3 |
| HFD | High-fat diet |
| HIF1an | Hypoxia inducible factor 1 subunit alpha inhibitor |
| hMADS | Human multipotent adipose-derived stem |
| HMGA2 | High-mobility group AT-hook 2 |
| HOX | Homeobox |
| HOXC8 | Homeobox C8 |
| IgF2 | Insulin like growth factor 2 |
| Igf2bp1 | Insulin-like growth factor 2 mRNA-binding protein 1 |
| IGF2R | Insulin-like growth factor 2 receptor |
| Insig1 | Insulin-induced gene 1 |
| IR | Insulin resistance |
| IRS-1 | Insulin receptor substrate 1 |
| MAP3K7 | Mitogen-activated protein kinase kinase kinase 7 |
| MAPK | Mitogen-activated protein kinase |
| Mef2 | Myocyte enhancer factor-2 |
| MMP11 | Matrix metalloproteinase 11 |
| MiR- | MicroRNA |
| miRNA | MicroRNA |
| mRNA | Messenger RNA |
| MYF5+ | Myogenic factor 5 positive cells |
| NAD+ | Nicotinamide adenine dinucleotide |
| NAFLD | Non-alcoholic fatty liver disease |
| NFKB | Nuclear factor kappa B |
| OPR8 | Oxysterol-binding protein-related protein 8 |
| Pde1b | Phosphodiesterase 1b |
| Pdgfr2 | Platelet-derived growth factor receptor 2 |
| PGC1A | Peroxisome proliferator-activated receptor gamma coactivator 1-alpha |
| PGC1B | Peroxisome proliferator-activated receptor gamma, coactivator 1-beta |
| PI3K | Phosphatidylinositol 3-kinase |
| PKA | Protein kinase cAMP dependent |
| PNPLA2 | Patatin-like phospholipase domain-containing protein 2 |
| PPAR | Peroxisome proliferator-activated receptor |
| PRDM16 | PR domain-containing protein 16 |
| pri-miRNA | Primary miRNA |
| PTEN | Phosphatase and tensin homolog deleted on chromosome 10 |
| RAI14 | Retinoic acid-induced protein 14 |
| RIP140 | Receptor-interacting protein 140 |
| RISC | RNA-induced silencing complex |
| RUNX1t1 | Runt-related transcription factor 1; translocated to, 1 |
| SCF | Skp–Cullin–F-box-containing |
| Sc | Subcutaneous |
| ScAT | Subcutaneous adipose tissue |
| SIRT1 | Sirtuin-1 |
| SNS | Sympathetic nervous system |
| SREBPs | Sterol regulatory element-binding proteins |
| STAT1 | Signal transducer and activator of transcription 1 |
| SVF | Stromal vascular fraction |
| T2DM | Type 2 diabetes mellitus |
| TBX1 | T-Box Transcription Factor 1 |
| TGFB | Transforming growth factor-beta |
| TgfBr3 | Type III transforming growth factor-β receptor |
| Th1 | T-helper 1 |
| TMEM26 | Transmembrane protein 26 |
| TNFRSF9 | Tumour necrosis factor receptor superfamily member 9 |
| TNFA | Tumour necrosis factor alpha |
| TOB1 | Transducer of erbB-2 1 |
| UCP1 | Uncoupling protein 1 |
| WAT | White adipose tissue |
References
- Giralt, M.; Cereijo, R.; Villarroya, F.F. Adipokines and the Endocrine Role of Adipose Tissues. Muscarinic Recept. 2015, 233, 265–282. [Google Scholar] [CrossRef]
- Kwok, K.H.M.; Lam, K.S.L.; Xu, A. Heterogeneity of white adipose tissue: Molecular basis and clinical implications. Exp. Mol. Med. 2016, 48, e215. [Google Scholar] [CrossRef] [Green Version]
- Kyrou, I.; Randeva, H.S.; Tsigos, C.; Kaltsas, G.; Weickert, M.O. Clinical Problems Caused by Obesity. In Endotext; Feingold, K.R., Anawalt, B., Boyce, A., Chrousos, G., Dungan, K., Grossman, A., Hershman, J.M., Kaltsas, G., Koch, C., Kopp, P., et al., Eds.; MDText Inc.: South Dartmouth, MA, USA, 2018. [Google Scholar]
- Herz, C.T.; Kiefer, F.W. Adipose tissue browning in mice and humans. J. Endocrinol. 2019, 241, R97–R109. [Google Scholar] [CrossRef]
- Bargut, T.C.; Souza-Mello, V.; Aguila, M.B.; Mandarim-De-Lacerda, C.A. Browning of white adipose tissue: Lessons from experimental models. Horm. Mol. Biol. Clin. Investig. 2017, 31, 31. [Google Scholar] [CrossRef]
- Abdullahi, A.; Jeschke, M.G. White Adipose Tissue Browning: A Double-edged Sword. Trends Endocrinol. Metab. 2016, 27, 542–552. [Google Scholar] [CrossRef] [Green Version]
- Bartelt, A.; Heeren, J. Adipose tissue browning and metabolic health. Nat. Rev. Endocrinol. 2014, 10, 24–36. [Google Scholar] [CrossRef]
- Symonds, M.E. Brown Adipose Tissue Growth and Development. Science 2013, 2013, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Schulz, T.J.; Tseng, Y.-H. Brown adipose tissue: Development, metabolism and beyond. Biochem. J. 2013, 453, 167–178. [Google Scholar] [CrossRef] [Green Version]
- Kaisanlahti, A.; Glumoff, T. Browning of white fat: Agents and implications for beige adipose tissue to type 2 diabetes. J. Physiol. Biochem. 2019, 75, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Petrovic, N.; Walden, T.B.; Shabalina, I.G.; Timmons, J.A.; Cannon, B.; Nedergaard, J. Chronic Peroxisome Proliferator-activated Receptor γ (PPARγ) Activation of Epididymally Derived White Adipocyte Cultures Reveals a Population of Thermogenically Competent, UCP1-containing Adipocytes Molecularly Distinct from Classic Brown Adipocytes. J. Biol. Chem. 2010, 285, 7153–7164. [Google Scholar] [CrossRef] [Green Version]
- Karpe, F.; Dickmann, J.R.; Frayn, K.N. Fatty Acids, Obesity, and Insulin Resistance: Time for a Reevaluation. Diabetes 2011, 60, 2441–2449. [Google Scholar] [CrossRef] [Green Version]
- Tran, T.T.; Kahn, C.R. Transplantation of adipose tissue and stem cells: Role in metabolism and disease. Nat. Rev. Endocrinol. 2010, 6, 195–213. [Google Scholar] [CrossRef] [Green Version]
- Kyrou, I.; Mattu, H.S.; Chatha, K.; Randeva, H.S. Chapter 7—Fat Hormones, Adipokines. In Endocrinology of the Heart in Health and Disease; Schisler, J.C., Lang, C.H., Willis, M.S., Eds.; Academic Press: Cambridge, MA, USA, 2017; pp. 167–205. [Google Scholar]
- Kalra, S.; Priya, G. Lipocrinology—The relationship between lipids and endocrine function. Drugs Context 2018, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Wronska, A.; Kmieć, Z. Structural and biochemical characteristics of various white adipose tissue depots. Acta Physiol. 2012, 205, 194–208. [Google Scholar] [CrossRef]
- Giordano, A.; Smorlesi, A.; Frontini, A.; Barbatelli, G.; Cinti, S. Mechanisms in Endocrinology: White, brown and pink adipocytes: The extraordinary plasticity of the adipose organ. Eur. J. Endocrinol. 2014, 170, R159–R171. [Google Scholar] [CrossRef] [Green Version]
- Fuster, J.J.; Ouchi, N.; Gokce, N.; Walsh, K. Obesity-Induced Changes in Adipose Tissue Microenvironment and Their Impact on Cardiovascular Disease. Circ. Res. 2016, 118, 1786–1807. [Google Scholar] [CrossRef] [Green Version]
- Ibrahim, M.M. Subcutaneous and visceral adipose tissue: Structural and functional differences. Obes. Rev. 2010, 11, 11–18. [Google Scholar] [CrossRef]
- Mor, E.; Shomron, N. Species-specific microRNA regulation influences phenotypic variability. BioEssays 2013, 35, 881–888. [Google Scholar] [CrossRef]
- Hajer, G.R.; Van Haeften, T.W.; Visseren, F.L.J. Adipose tissue dysfunction in obesity, diabetes, and vascular diseases. Eur. Hear. J. 2008, 29, 2959–2971. [Google Scholar] [CrossRef] [Green Version]
- Fontana, L.; Eagon, J.C.; Trujillo, M.E.; Scherer, P.E.; Klein, S. Visceral Fat Adipokine Secretion Is Associated with Systemic Inflammation in Obese Humans. Diabetes 2007, 56, 1010–1013. [Google Scholar] [CrossRef] [Green Version]
- Veilleux, A.; Caron-Jobin, M.; Noël, S.; Laberge, P.Y.; Tchernof, A. Visceral Adipocyte Hypertrophy is Associated with Dyslipidemia Independent of Body Composition and Fat Distribution in Women. Diabetes 2011, 60, 1504–1511. [Google Scholar] [CrossRef] [Green Version]
- Neeland, I.J.; Ayers, C.R.; Rohatgi, A.K.; Turer, A.T.; Berry, J.D.; Das, S.R.; Vega, G.L.; Khera, A.; McGuire, D.K.; Grundy, S.M.; et al. Associations of visceral and abdominal subcutaneous adipose tissue with markers of cardiac and metabolic risk in obese adults. Obesity 2013, 21, E439–E447. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Verdejo, R.; Marlatt, K.L.; Ravussin, E.; Galgani, J.E. Contribution of brown adipose tissue to human energy metabolism. Mol. Asp. Med. 2019, 68, 82–89. [Google Scholar] [CrossRef]
- Villarroya, J.; Cereijo, R.; Gavaldà-Navarro, A.; Peyrou, M.; Giralt, M.; Villarroya, F. New insights into the secretory functions of brown adipose tissue. J. Endocrinol. 2019, 243, R19–R27. [Google Scholar] [CrossRef] [Green Version]
- Klingenspor, M.; Fromme, T. Brown Adipose Tissue; Springer Science and Business Media LLC.: Berlin, Germany, 2011; pp. 39–69. [Google Scholar]
- Lichtenbelt, W.D.V.M.; Vanhommerig, J.W.; Smulders, N.M.; Drossaerts, J.M.; Kemerink, G.J.; Bouvy, N.D.; Schrauwen, P.; Teule, G.J. Cold-Activated Brown Adipose Tissue in Healthy Men. N. Engl. J. Med. 2009, 360, 1500–1508. [Google Scholar] [CrossRef] [Green Version]
- Reddy, N.L.; Jones, T.A.; Wayte, S.; Adesanya, O.O.; Sankar, S.; Yeo, Y.C.; Tripathi, G.; McTernan, P.G.; Randeva, H.S.; Kumar, S.; et al. Identification of Brown Adipose Tissue Using MR Imaging in a Human Adult with Histological and Immunohistochemical Confirmation. J. Clin. Endocrinol. Metab. 2014, 99, E117–E121. [Google Scholar] [CrossRef] [Green Version]
- Reddy, N.L.; Tan, B.K.; Barber, T.M.; Randeva, H.S. Brown adipose tissue: Endocrine determinants of function and therapeutic manipulation as a novel treatment strategy for obesity. BMC Obes. 2014, 1, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Alcalá, M.; Calderon-Dominguez, M.; Serra, D.; Herrero, L.; Viana, M. Mechanisms of Impaired Brown Adipose Tissue Recruitment in Obesity. Front. Physiol. 2019, 10, 94. [Google Scholar] [CrossRef]
- Bartness, T.J.; Vaughan, C.H.; Song, C.K. Sympathetic and sensory innervation of brown adipose tissue. Int. J. Obes. 2010, 34, S36–S42. [Google Scholar] [CrossRef] [Green Version]
- Epoher, A.-L.; Ealtirriba, J.; Eveyrat-Durebex, C.; Rohner-Jeanrenaud, F. Brown adipose tissue activity as a target for the treatment of obesity/insulin resistance. Front. Physiol. 2015, 6, 4. [Google Scholar] [CrossRef] [Green Version]
- Valente, M.H.; Gomes, F.M.D.S.; Benseñor, I.J.M.; Brentani, A.; Escobar, A.M.D.U.; Grisi, S. Relation between Birth Weight, Growth, and Subclinical Atherosclerosis in Adulthood. BioMed. Res. Int. 2015, 2015, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Wu, L.; Van Kaer, L. Contribution of lipid-reactive natural killer T cells to obesity-associated inflammation and insulin resistance. Adipocyte 2013, 2, 12–16. [Google Scholar] [CrossRef] [Green Version]
- Wu, J.; Boström, P.; Sparks, L.M.; Ye, L.; Choi, J.H.; Giang, A.-H.; Khandekar, M.; Virtanen, K.A.; Nuutila, P.; Schaart, G.; et al. Beige Adipocytes Are a Distinct Type of Thermogenic Fat Cell in Mouse and Human. Cell 2012, 150, 366–376. [Google Scholar] [CrossRef] [Green Version]
- Vijgen, G.H.E.J.; Bouvy, N.D.; Teule, G.J.J.; Brans, B.; Schrauwen, P.; Lichtenbelt, W.D.V.M. Brown Adipose Tissue in Morbidly Obese Subjects. PLoS ONE 2011, 6, e17247. [Google Scholar] [CrossRef] [Green Version]
- Wang, Y.; Cai, L.; Wu, Y.; Wilson, R.F.; Weston, C.; Fawole, O.; Bleich, S.N.; Cheskin, L.J.; Showell, N.N.; Lau, B.D.; et al. What childhood obesity prevention programmes work? A systematic review and meta-analysis. Obes. Rev. 2015, 16, 547–565. [Google Scholar] [CrossRef]
- Sanchez-Gurmaches, J.; Hung, C.-M.; Sparks, C.A.; Tang, Y.; Li, H.; Guertin, D.A. PTEN Loss in the Myf5 Lineage Redistributes Body Fat and Reveals Subsets of White Adipocytes that Arise from Myf5 Precursors. Cell Metab. 2012, 16, 348–362. [Google Scholar] [CrossRef] [Green Version]
- Shan, T.; Liang, X.; Bi, P.; Zhang, P.; Liu, W.; Kuang, S.; Chitraju, C.; Trötzmüller, M.; Hartler, J.; Wolinski, H.; et al. Distinct populations of adipogenic and myogenic Myf5-lineage progenitors in white adipose tissues. J. Lipid Res. 2013, 54, 2214–2224. [Google Scholar] [CrossRef] [Green Version]
- Celi, F.S. Human Brown Adipose Tissue Plasticity: Hormonal and Environmental Manipulation. In Research and Perspectives in Endocrine Interactions; Springer Science and Business Media LLC.: Berlin, Germany, 2017; pp. 1–11. [Google Scholar]
- Rosenwald, M.; Wolfrum, C. The origin and definition of brite versus white and classical brown adipocytes. Adipocyte 2014, 3, 4–9. [Google Scholar] [CrossRef]
- Garcia, R.A.; Roemmich, J.N.; Claycombe, K.J. Evaluation of markers of beige adipocytes in white adipose tissue of the mouse. Nutr. Metab. 2016, 13, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Chi, J.; Cohen, P. The Multifaceted Roles of PRDM16: Adipose Biology and Beyond. Trends Endocrinol. Metab. 2016, 27, 11–23. [Google Scholar] [CrossRef]
- Rosen, E.D.; Sarraf, P.; E Troy, A.; Bradwin, G.; Moore, K.; Milstone, D.S.; Spiegelman, B.M.; Mortensen, R.M. PPARγ Is Required for the Differentiation of Adipose Tissue In Vivo and In Vitro. Mol. Cell 1999, 4, 611–617. [Google Scholar] [CrossRef]
- Linhart, H.G.; Ishimura-Oka, K.; DeMayo, F.; Kibe, T.; Repka, D.; Poindexter, B.; Bick, R.J.; Darlington, G.J. C/EBP is required for differentiation of white, but not brown, adipose tissue. Proc. Natl. Acad. Sci. USA 2001, 98, 12532–12537. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seale, P. Transcriptional Regulatory Circuits Controlling Brown Fat Development and Activation. Diabetes 2015, 64, 2369–2375. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giralt, M.; Villarroya, F. White, Brown, Beige/Brite: Different Adipose Cells for Different Functions? Endocrinology 2013, 154, 2992–3000. [Google Scholar] [CrossRef] [Green Version]
- Chu, D.-T.; Gawronska-Kozak, B. Brown and brite adipocytes: Same function, but different origin and response. Biochimie 2017, 138, 102–105. [Google Scholar] [CrossRef] [Green Version]
- Van Harmelen, V.; Röhrig, K.; Hauner, H. Comparison of proliferation and differentiation capacity of human adipocyte precursor cells from the omental and subcutaneous adipose tissue depot of obese subjects. Metabolism 2004, 53, 632–637. [Google Scholar] [CrossRef]
- Price, N.L.; Fernández-Hernando, C. miRNA regulation of white and brown adipose tissue differentiation and function. Biochim. Biophys. Acta 2016, 1861, 2104–2110. [Google Scholar] [CrossRef] [Green Version]
- Landrier, J.-F.; Derghal, A.; Mounien, L. MicroRNAs in Obesity and Related Metabolic Disorders. Cells 2019, 8, 859. [Google Scholar] [CrossRef] [Green Version]
- Chen, L.; Yang, G. PPARs Integrate the Mammalian Clock and Energy Metabolism. PPAR Res. 2014, 2014, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Hondares, E.; Iglesias, R.; Giralt, A.; Gonzalez, F.J.; Giralt, M.; Mampel, T.; Villarroya, F. Thermogenic Activation Induces FGF21 Expression and Release in Brown Adipose Tissue. J. Biol. Chem. 2011, 286, 12983–12990. [Google Scholar] [CrossRef] [Green Version]
- Seale, P.; Bjork, B.C.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scimè, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nat. Cell Biol. 2008, 454, 961–967. [Google Scholar] [CrossRef] [Green Version]
- Harms, M.J.; Ishibashi, J.; Wang, W.; Lim, H.-W.; Goyama, S.; Sato, T.; Kurokawa, M.; Won, K.-J.; Seale, P. Prdm16 Is Required for the Maintenance of Brown Adipocyte Identity and Function in Adult Mice. Cell Metab. 2014, 19, 593–604. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nerlov, C. The C/EBP family of transcription factors: A paradigm for interaction between gene expression and proliferation control. Trends Cell Biol. 2007, 17, 318–324. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.-E.; Ge, K. Transcriptional and epigenetic regulation of PPARγ expression during adipogenesis. Cell Biosci. 2014, 4, 29. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ringholm, S.; Olesen, J.; Pedersen, J.T.; Brandt, C.T.; Halling, J.F.; Hellsten, Y.; Prats, C.; Pilegaard, H. Effect of lifelong resveratrol supplementation and exercise training on skeletal muscle oxidative capacity in aging mice; impact of PGC-1α. Exp. Gerontol. 2013, 48, 1311–1318. [Google Scholar] [CrossRef] [PubMed]
- Christian, M.; White, R.; Parker, M.G. Metabolic regulation by the nuclear receptor corepressor RIP140. Trends Endocrinol. Metab. 2006, 17, 243–250. [Google Scholar] [CrossRef]
- Rajakumari, S.; Wu, J.; Ishibashi, J.; Lim, H.-W.; Giang, A.-H.; Won, K.-J.; Reed, R.R.; Seale, P. EBF2 Determines and Maintains Brown Adipocyte Identity. Cell Metab. 2013, 17, 562–574. [Google Scholar] [CrossRef] [Green Version]
- Bartel, D.P. MicroRNAs. Cell 2004, 116, 281–297. [Google Scholar] [CrossRef] [Green Version]
- Lee, R.C.; Feinbaum, R.L.; Ambros, V. The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 1993, 75, 843–854. [Google Scholar] [CrossRef]
- Gebert, L.F.R.; Macrae, I.J. Regulation of microRNA function in animals. Nat. Rev. Mol. Cell Biol. 2019, 20, 21–37. [Google Scholar] [CrossRef]
- Vienberg, S.; Geiger, J.; Madsen, S.; Dalgaard, L.T. MicroRNAs in metabolism. Acta Physiol. 2016, 219, 346–361. [Google Scholar] [CrossRef] [PubMed]
- Barraclough, J.Y.; Joan, M.; Joglekar, M.V.; Hardikar, A.A.; Patel, S. MicroRNAs as Prognostic Markers in Acute Coronary Syndrome Patients—A Systematic Review. Cells 2019, 8, 1572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Joglekar, M.V.; Januszewski, A.S.; Jenkins, A.J.; Hardikar, A.A. Circulating microRNA Biomarkers of Diabetic Retinopathy. Diabetes 2015, 65, 22–24. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, Y. RNA-induced Silencing Complex (RISC). In Encyclopedia of Systems Biology; Springer Science and Business Media LLC.: Berlin, Germany, 2013. [Google Scholar]
- Pratt, A.J.; Macrae, I.J. The RNA-induced Silencing Complex: A Versatile Gene-silencing Machine. J. Biol. Chem. 2009, 284, 17897–17901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Roush, S.; Slack, F.J. The let-7 family of microRNAs. Trends Cell Biol. 2008, 18, 505–516. [Google Scholar] [CrossRef] [PubMed]
- Arias, N.; Aguirre, L.; Fernández-Quintela, A.; Gonzalez, M.; Lasa, A.; Miranda, J.; Macarulla, M.T.; Portillo, M.P. MicroRNAs involved in the browning process of adipocytes. J. Physiol. Biochem. 2015, 72, 509–521. [Google Scholar] [CrossRef]
- Kim, Y.-K.; Kim, B.; Kim, V.N. Re-evaluation of the roles of DROSHA, Exportin 5, and DICER in microRNA biogenesis. Proc. Natl. Acad. Sci. USA 2016, 113, E1881–E1889. [Google Scholar] [CrossRef] [Green Version]
- Choo, K.-B.; Soon, Y.L.; Nguyen, P.N.N.; Hiew, M.S.Y.; Huang, C.-J. MicroRNA-5p and -3p co-expression and cross-targeting in colon cancer cells. J. Biomed. Sci. 2014, 21, 95. [Google Scholar] [CrossRef] [Green Version]
- Mori, M.A.; Ludwig, R.G.; Garcia-Martin, R.; Brandão, B.B.; Kahn, C.R. Extracellular miRNAs: From Biomarkers to Mediators of Physiology and Disease. Cell Metab. 2019, 30, 656–673. [Google Scholar] [CrossRef]
- Thomou, T.; Mori, M.A.; Dreyfuss, J.M.; Konishi, M.; Sakaguchi, M.; Wolfrum, C.; Rao, T.N.; Winnay, J.N.; Garcia-Martin, R.; Grinspoon, S.K.; et al. Adipose-Derived Circulating miRNAs Regulate Gene Expression in Other Tissues. Nature 2017, 542, 450–455. [Google Scholar] [CrossRef]
- Sohel, M.H. Extracellular/Circulating MicroRNAs: Release Mechanisms, Functions and Challenges. Achiev. Life Sci. 2016, 10, 175–186. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.-J.; Cho, H.; Alexander, R.; Patterson, H.C.; Gu, M.; Lo, K.A.; Xu, D.; Goh, V.J.; Nguyen, L.N.; Chai, X.; et al. MicroRNAs Are Required for the Feature Maintenance and Differentiation of Brown Adipocytes. Diabetes 2014, 63, 4045–4056. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.; Nakagami, H.; Rodriguez-Araujo, G.; Nimura, K.; Kaneda, Y. Essential Role for miR-196a in Brown Adipogenesis of White Fat Progenitor Cells. PLoS Biol. 2012, 10, e1001314. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mori, M.A.; Thomou, T.; Boucher, J.; Lee, K.Y.; Lallukka, S.; Kim, J.K.; Torriani, M.; Yki-Jaervinen, H.; Grinspoon, S.K.; Cypess, A.M.; et al. Altered miRNA processing disrupts brown/white adipocyte determination and associates with lipodystrophy. J. Clin. Investig. 2014, 124, 3339–3351. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Icli, B.; Feinberg, M.W. MicroRNAs in dysfunctional adipose tissue: Cardiovascular implications. Cardiovasc. Res. 2017, 113, 1024–1034. [Google Scholar] [CrossRef]
- Goody, D.; Pfeifer, A. MicroRNAs in brown and beige fat. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 29–36. [Google Scholar] [CrossRef]
- Shamsi, F.; Zhang, H.; Tseng, Y.-H. MicroRNA Regulation of Brown Adipogenesis and Thermogenic Energy Expenditure. Front. Endocrinol. 2017, 8, 205. [Google Scholar] [CrossRef] [Green Version]
- Ahonen, M.A.; Haridas, P.N.; Mysore, R.; Wabitsch, M.; Fischer-Posovszky, P.; Olkkonen, V.M. miR-107 inhibits CDK6 expression, differentiation, and lipid storage in human adipocytes. Mol. Cell. Endocrinol. 2019, 479, 110–116. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Pan, R.; Pfeifer, A. Regulation of brown and beige fat by microRNAs. Pharmacol. Ther. 2017, 170, 1–7. [Google Scholar] [CrossRef]
- Sun, L.; Xie, H.; Mori, M.A.; Alexander, R.; Yuan, B.B.; Hattangadi, S.M.; Liu, Q.; Kahn, C.R.; Lodish, H.F. Mir193b–365 is essential for brown fat differentiation. Nat. Cell Biol. 2011, 13, 958–965. [Google Scholar] [CrossRef] [Green Version]
- Oliverio, M.; Schmidt, E.; Mauer, J.; Baitzel, C.; Hansmeier, N.; Khani, S.; Konieczka, S.; Pradas-Juni, M.; Brodesser, S.; Van, T.-M.; et al. Dicer1–miR-328–Bace1 signalling controls brown adipose tissue differentiation and function. Nat. Cell Biol. 2016, 18, 328–336. [Google Scholar] [CrossRef] [PubMed]
- Cioffi, M.; Vallespinos-Serrano, M.; Trabulo, S.M.; Fernandez-Marcos, P.J.; Firment, A.N.; Vazquez, B.N.; Vieira, C.R.; Mulero, F.; Camara, J.A.; Cronin, U.P.; et al. MiR-93 Controls Adiposity via Inhibition of Sirt7 and Tbx3. Cell Rep. 2015, 12, 1594–1605. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meakin, P.J.; Harper, A.J.; Hamilton, D.L.; Gallagher, J.; McNeilly, A.D.; Burgess, L.A.; Vaanholt, L.M.; Bannon, K.A.; Latcham, J.; Hussain, I.; et al. Reduction in BACE1 decreases body weight, protects against diet-induced obesity and enhances insulin sensitivity in mice. Biochem. J. 2011, 441, 285–296. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fu, X.; Jin, L.; Han, L.; Yuan, Y.; Mu, Q.; Wang, H.; Yang, J.; Ning, G.; Zhou, D.; Zhang, Z. miR-129-5p Inhibits Adipogenesis through Autophagy and May Be a Potential Biomarker for Obesity. Int. J. Endocrinol. 2019, 2019, 5069578. [Google Scholar] [CrossRef]
- Zhang, H.; Guan, M.; Townsend, K.L.; Huang, T.L.; An, D.; Yan, X.; Xue, R.; Schulz, T.J.; Winnay, J.; Mori, M.; et al. Micro RNA -455 regulates brown adipogenesis via a novel HIF 1an-AMPK-PGC 1α signaling network. EMBO Rep. 2015, 16, 1378–1393. [Google Scholar] [CrossRef] [Green Version]
- Cai, Z.; Liu, J.; Bian, H.; Cai, J.; Guo, X. MiR-455 enhances adipogenic differentiation of 3T3-L1 cells through targeting uncoupling protein-1. Die Pharm. 2016, 71, 625–628. [Google Scholar]
- Pahlavani, M.; Wijayatunga, N.N.; Kalupahana, N.S.; Ramalingam, L.; Gunaratne, P.H.; Coarfa, C.; Rajapakshe, K.; Kottapalli, P.; Moustaid-Moussa, N. Transcriptomic and microRNA analyses of gene networks regulated by eicosapentaenoic acid in brown adipose tissue of diet-induced obese mice. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2018, 1863, 1523–1531. [Google Scholar] [CrossRef]
- Hu, F.; Wang, M.; Xiao, T.; Yin, B.; He, L.; Meng, W.; Dong, M.; Liu, F. miR-30 Promotes Thermogenesis and the Development of Beige Fat by Targeting RIP140. Diabetes 2015, 64, 2056–2068. [Google Scholar] [CrossRef] [Green Version]
- Zaragosi, L.-E.; Wdziekonski, B.; Lebrigand, K.; Villageois, P.; Mari, B.; Waldmann, R.; Dani, C.; Barbry, P. Small RNA sequencing reveals miR-642a-3p as a novel adipocyte-specific microRNA and miR-30 as a key regulator of human adipogenesis. Genome Biol. 2011, 12, R64. [Google Scholar] [CrossRef] [Green Version]
- Ge, X.; Sathiakumar, D.; Lua, B.J.G.; Kukreti, H.; Lee, M.; McFarlane, C. Myostatin signals through miR-34a to regulate Fndc5 expression and browning of white adipocytes. Int. J. Obes. 2016, 41, 137–148. [Google Scholar] [CrossRef] [Green Version]
- Karbiener, M.; Fischer, C.; Nowitsch, S.; Opriessnig, P.; Papak, C.; Ailhaud, G.; Dani, C.; Amri, E.-Z.; Scheideler, M. microRNA miR-27b impairs human adipocyte differentiation and targets PPARγ. Biochem. Biophys. Res. Commun. 2009, 390, 247–251. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.; Trajkovski, M. MiR-27 orchestrates the transcriptional regulation of brown adipogenesis. Metabolism 2014, 63, 272–282. [Google Scholar] [CrossRef] [PubMed]
- Kang, T.; Lu, W.; Xu, W.; Anderson, L.; Bacanamwo, M.; Thompson, W.; Chen, Y.E.; Liu, D. MicroRNA-27 (miR-27) Targets Prohibitin and Impairs Adipocyte Differentiation and Mitochondrial Function in Human Adipose-derived Stem Cells. J. Biol. Chem. 2013, 288, 34394–34402. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, J.; Okla, M.; Erickson, A.; Carr, T.; Natarajan, S.K.; Chung, S. Eicosapentaenoic Acid Potentiates Brown Thermogenesis through FFAR4-dependent Up-regulation of miR-30b and miR-378. J. Biol. Chem. 2016, 291, 20551–20562. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liu, W.; Bi, P.; Shan, T.; Yang, X.; Yin, H.; Wang, Y.-X.; Liu, N.; Rudnicki, M.A.; Kuang, S. miR-133a Regulates Adipocyte Browning In Vivo. PLoS Genet. 2013, 9, e1003626. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Pasut, A.; Soleimani, V.D.; Bentzinger, C.F.; Antoun, G.; Thorn, S.; Seale, P.; Fernando, P.; Van Ijcken, W.; Grosveld, F.; et al. MicroRNA-133 Controls Brown Adipose Determination in Skeletal Muscle Satellite Cells by Targeting Prdm16. Cell Metab. 2013, 17, 210–224. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Trajkovski, M.; Ahmed, K.; Esau, C.C.; Stoffel, M. MyomiR-133 regulates brown fat differentiation through Prdm16. Nat. Cell Biol. 2012, 14, 1330–1335. [Google Scholar] [CrossRef] [PubMed]
- Chen, Y.; Siegel, F.; Kipschull, S.; Haas, B.; Fröhlich, H.; Meister, G.; Pfeifer, A. miR-155 regulates differentiation of brown and beige adipocytes via a bistable circuit. Nat. Commun. 2013, 4, 1769. [Google Scholar] [CrossRef] [Green Version]
- Gaudet, A.D.; Fonken, L.K.; Gushchina, L.V.; Aubrecht, T.G.; Maurya, S.K.; Periasamy, M.; Nelson, R.J.; Popovich, P.G. miR-155 Deletion in Female Mice Prevents Diet-Induced Obesity. Sci. Rep. 2016, 6, 22862. [Google Scholar] [CrossRef] [Green Version]
- Karkeni, E.; Astier, J.; Tourniaire, F.; El Abed, M.; Romier, B.; Gouranton, E.; Wan, L.; Borel, P.; Salles, J.; Walrand, S.; et al. Obesity-associated Inflammation Induces microRNA-155 Expression in Adipocytes and Adipose Tissue: Outcome on Adipocyte Function. J. Clin. Endocrinol. Metab. 2016, 101, 1615–1626. [Google Scholar] [CrossRef]
- Ng, R.; Hussain, N.A.; Zhang, Q.; Chang, C.; Li, H.; Fu, Y.; Cao, L.; Han, W.; Stunkel, W.; Xu, F. miRNA-32 Drives Brown Fat Thermogenesis and Trans-activates Subcutaneous White Fat Browning in Mice. Cell Rep. 2017, 19, 1229–1246. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Feuermann, Y.; Kang, K.; Gavrilova, O.; Haetscher, N.; Jang, S.J.; Yoo, K.H.; Jiang, C.; Gonzalez, F.J.; Robinson, G.W.; Hennighausen, L. MiR-193b and miR-365-1 are not required for the development and function of brown fat in the mouse. RNA Biol. 2013, 10, 1807–1814. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guller, I.; A McNaughton, S.; Crowley, T.M.; Gilsanz, V.; Kajimura, S.; Watt, M.J.; Russell, A.P. Comparative analysis of microRNA expression in mouse and human brown adipose tissue. BMC Genom. 2015, 16, 820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wu, Y.; Zuo, J.; Zhang, Y.; Xie, Y.; Hu, F.; Chen, L.; Liu, B.; Liu, F. Identification of miR-106b-93 as a negative regulator of brown adipocyte differentiation. Biochem. Biophys. Res. Commun. 2013, 438, 575–580. [Google Scholar] [CrossRef]
- Murri, M.; El Azzouzi, H. MicroRNAs as regulators of mitochondrial dysfunction and obesity. Am. J. Physiol. Circ. Physiol. 2018, 315, H291–H302. [Google Scholar] [CrossRef]
- Kajimura, S.; Seale, P.; Tomaru, T.; Erdjument-Bromage, H.; Cooper, M.P.; Ruas, J.L.; Chin, S.; Tempst, P.; Lazar, M.A.; Spiegelman, B.M. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008, 22, 1397–1409. [Google Scholar] [CrossRef]
- Døssing, K.B.V.; Binderup, T.; Kaczkowski, B.; Skanderup, A.J.; Rossing, M.; Winther, O.; Federspiel, B.; Knigge, U.; Kjaer, A.; Friis-Hansen, L. Down-Regulation of miR-129-5p and the let-7 Family in Neuroendocrine Tumors and Metastases Leads to Up-Regulation of Their Targets Egr1, G3bp1, Hmga2 and Bach1. Genes 2014, 6, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Koh, E.-H.; Chernis, N.; Saha, P.K.; Xiao, L.; Bader, D.A.; Zhu, B.; Rajapakshe, K.; Hamilton, M.P.; Liu, X.; Perera, D.; et al. miR-30a Remodels Subcutaneous Adipose Tissue Inflammation to Improve Insulin Sensitivity in Obesity. Diabetes 2018, 67, 2541–2553. [Google Scholar] [CrossRef] [Green Version]
- Fu, T.; Seok, S.; Choi, S.; Huang, Z.; Suino-Powell, K.; Xu, H.E.; Kemper, B.; Kemper, J.K. MicroRNA 34a Inhibits Beige and Brown Fat Formation in Obesity in Part by Suppressing Adipocyte Fibroblast Growth Factor 21 Signaling and SIRT1 Function. Mol. Cell. Biol. 2014, 34, 4130–4142. [Google Scholar] [CrossRef] [Green Version]
- Bostroem, P.; Wu, J.; Jedrychowski, M.P.; Korde, A.; Ye, L.; Lo, J.C.; Rasbach, K.A.; Bostroem, E.A.; Choi, J.H.; Long, J.Z.; et al. A PGC1-α-dependent myokine that drives brown-fat-like development of white fat and thermogenesis. Nat. Cell Biol. 2012, 481, 463–468. [Google Scholar] [CrossRef]
- Yu, J.; Lv, Y.; Wang, F.; Kong, X.; Di, W.; Liu, J.; Sheng, Y.; Lv, S.; Ding, G. MiR-27b-3p Inhibition Enhances Browning of Epididymal Fat in High-Fat Diet Induced Obese Mice. Front. Endocrinol. 2019, 10, 38. [Google Scholar] [CrossRef] [PubMed]
- Pan, D.; Mao, C.; Quattrochi, B.; Friedline, R.H.; Zhu, L.J.; Jung, D.Y.; Kim, J.K.; Lewis, B.C.; Wang, Y.-X. MicroRNA-378 controls classical brown fat expansion to counteract obesity. Nat. Commun. 2014, 5, 1–12. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Francis, S.H.; Blount, M.A.; Corbin, J.D. Mammalian Cyclic Nucleotide Phosphodiesterases: Molecular Mechanisms and Physiological Functions. Physiol. Rev. 2011, 91, 651–690. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Li, C.; Li, H.; Song, Y.; Zhao, Y.; Zhai, L.; Wang, H.; Zhong, R.; Tang, H.; Zhu, D. miR-378 Activates the Pyruvate-PEP Futile Cycle and Enhances Lipolysis to Ameliorate Obesity in Mice. EBioMedicine 2016, 5, 93–104. [Google Scholar] [CrossRef] [Green Version]
- Machado, I.F.; Teodoro, J.S.; Palmeira, C.M.; Rolo, A.P. miR-378a: A new emerging microRNA in metabolism. Cell. Mol. Life Sci. 2019, 77, 1947–1958. [Google Scholar] [CrossRef] [PubMed]
- Xie, M.; Dart, D.A.; Owen, S.; Wen, X.; Ji, J.-F.; Jiang, W.G. Insights into roles of the miR-1, -133 and -206 family in gastric cancer (Review). Oncol. Rep. 2016, 36, 1191–1198. [Google Scholar] [CrossRef]
- Carmona, M.C.; Hondares, E.; De La Concepción, M.L.R.; Rodríguez-Sureda, V.; Peinado-Onsurbe, J.; Poli, V.; Iglesias, R.; Villarroya, F.F.; Giralt, M.M. Defective thermoregulation, impaired lipid metabolism, but preserved adrenergic induction of gene expression in brown fat of mice lacking C/EBPβ. Biochem. J. 2005, 389, 47–56. [Google Scholar] [CrossRef] [Green Version]
- Karbiener, M.; Pisani, D.F.; Frontini, A.; Oberreiter, L.M.; Lang, E.; Vegiopoulos, A.; Mössenböck, K.; Bernhardt, G.A.; Mayr, T.; Hildner, F.; et al. MicroRNA-26 Family Is Required for Human Adipogenesis and Drives Characteristics of Brown Adipocytes. Stem Cells 2014, 32, 1578–1590. [Google Scholar] [CrossRef]
- Acharya, A.; Berry, D.C.; Zhang, H.; Jiang, Y.; Jones, B.T.; Hammer, R.E.; Graff, J.M.; Mendell, J.T. miR-26 suppresses adipocyte progenitor differentiation and fat production by targeting Fbxl19. Genes Dev. 2019, 33, 1367–1380. [Google Scholar] [CrossRef] [Green Version]
- Rockstroh, D.; Löffler, D.; Kiess, W.; Landgraf, K.; Körner, A. Regulation of human adipogenesis by miR125b-5p. Adipocyte 2016, 5, 283–297. [Google Scholar] [CrossRef] [Green Version]
- Giroud, M.; Karbiener, M.; Pisani, D.F.; Ghandour, R.A.; Beranger, G.E.; Niemi, T.; Taittonen, M.; Nuutila, P.; Virtanen, K.A.; Langin, D.; et al. Let-7i-5p represses brite adipocyte function in mice and humans. Sci. Rep. 2016, 6, 28613. [Google Scholar] [CrossRef] [Green Version]
- Divoux, A.; Xie, H.; Adeline, D.; Karastergiou, K.; Perera, R.J.; Chang, R.J.; Fried, S.K.; Smith, S.R. MicroRNA-196 Regulates HOX Gene Expression in Human Gluteal Adipose Tissue. Obesity 2017, 25, 1375–1383. [Google Scholar] [CrossRef]
- Hilton, C.; Neville, M.J.; Wittemans, L.B.; Todorcevic, M.; Pinnick, K.E.; Pulit, S.L.; Luan, J.; Kulyté, A.; Dahlman, I.; Wareham, N.J.; et al. MicroRNA-196a links human body fat distribution to adipose tissue extracellular matrix composition. EBioMedicine 2019, 44, 467–475. [Google Scholar] [CrossRef] [Green Version]
- Moss, M.L.; Minond, D. Recent Advances in ADAM17 Research: A Promising Target for Cancer and Inflammation. Mediat. Inflamm. 2017, 2017, 1–21. [Google Scholar] [CrossRef] [Green Version]
- Jin, J.; Cardozo, T.; Lovering, R.C.; Elledge, S.J.; Pagano, M.; Harper, J.W. Systematic analysis and nomenclature of mammalian F-box proteins. Genes Dev. 2004, 18, 2573–2580. [Google Scholar] [CrossRef] [Green Version]
- Yin, H.; Sun, Y.; Wang, X.; Park, J.; Zhang, Y.; Li, M.; Yin, J.; Liu, Q.; Wei, M. Progress on the relationship between miR-125 family and tumorigenesis. Exp. Cell Res. 2015, 339, 252–260. [Google Scholar] [CrossRef]
- Giroud, M.; Pisani, D.F.; Karbiener, M.; Barquissau, V.; Ghandour, R.A.; Tews, D.; Fischer-Posovszky, P.; Chambard, J.C.; Knippschild, U.; Niemi, T.; et al. miR-125b affects mitochondrial biogenesis and impairs brite adipocyte formation and function. Mol. Metab. 2016, 5, 615–625. [Google Scholar] [CrossRef]
- Sun, T.; Fu, M.; Bookout, A.L.; Kliewer, S.A.; Mangelsdorf, D.J. MicroRNA let-7 Regulates 3T3-L1 Adipogenesis. Mol. Endocrinol. 2009, 23, 925–931. [Google Scholar] [CrossRef] [Green Version]
- Zhu, H.; Shyh-Chang, N.; Segrè, A.V.; Shinoda, G.; Shah, S.P.; Einhorn, W.S.; Takeuchi, A.; Engreitz, J.M.; Hagan, J.P.; Kharas, M.G.; et al. The Lin28/let-7 Axis Regulates Glucose Metabolism. Cell 2011, 147, 81–94. [Google Scholar] [CrossRef] [Green Version]
- Youssef, E.M.; ElFiky, A.M.; Abu-Shahba, N.; Elhefnawi, M.; Soliman, B. Expression profiling and analysis of some miRNAs in subcutaneous white adipose tissue during development of obesity. Genes Nutr. 2020, 15, 8–14. [Google Scholar] [CrossRef]
- Johnson, C.D.; Esquela-Kerscher, A.; Stefani, G.; Byrom, M.; Kelnar, K.; Ovcharenko, D.; Wilson, M.; Wang, X.; Shelton, J.; Shingara, J.; et al. The let-7 MicroRNA Represses Cell Proliferation Pathways in Human Cells. Cancer Res. 2007, 67, 7713–7722. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, C.-T.; Risom, T.; Strauss, W.M. Evolutionary Conservation of MicroRNA Regulatory Circuits: An Examination of MicroRNA Gene Complexity and Conserved MicroRNA-Target Interactions through Metazoan Phylogeny. DNA Cell Biol. 2007, 26, 209–218. [Google Scholar] [CrossRef] [PubMed]
- Mayr, C.; Hemann, M.T.; Bartel, D.P. Disrupting the Pairing Between let-7 and Hmga2 Enhances Oncogenic Transformation. Science 2007, 315, 1576–1579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boyerinas, B.; Park, S.-M.; Shomron, N.; Hedegaard, M.M.; Vinther, J.; Andersen, J.S.; Feig, C.; Xu, J.; Burge, C.B.; Peter, M.E. Identification of Let-7-Regulated Oncofetal Genes. Cancer Res. 2008, 68, 2587–2591. [Google Scholar] [CrossRef] [Green Version]
- Arner, P.; Kulyté, A. MicroRNA regulatory networks in human adipose tissue and obesity. Nat. Rev. Endocrinol. 2015, 11, 276–288. [Google Scholar] [CrossRef]
- Choe, S.S.; Huh, J.Y.; Hwang, I.J.; Kim, J.I.; Kim, J.B. Adipose Tissue Remodeling: Its Role in Energy Metabolism and Metabolic Disorders. Front. Endocrinol. 2016, 7, 30. [Google Scholar] [CrossRef] [Green Version]
- Kajimoto, K. MicroRNA and 3T3-L1 pre-adipocyte differentiation. RNA 2006, 12, 1626–1632. [Google Scholar] [CrossRef] [Green Version]
- Xie, H.; Lim, B.; Lodish, H.F. MicroRNAs Induced During Adipogenesis that Accelerate Fat Cell Development Are Downregulated in Obesity. Diabetes 2009, 58, 1050–1057. [Google Scholar] [CrossRef] [Green Version]
- Ling, H.-Y.; Wen, G.-B.; Feng, S.-D.; Tuo, Q.-H.; Ou, H.-S.; Yao, C.H.; Zhu, B.-Y.; Gao, Z.-P.; Zhang, L.; Liao, D.-F. MicroRNA-375 promotes 3T3-L1 adipocyte differentiation through modulation of extracellular signal-regulated kinase signalling. Clin. Exp. Pharmacol. Physiol. 2011, 38, 239–246. [Google Scholar] [CrossRef] [Green Version]
- Kang, M.; Yan, L.-M.; Zhang, W.-Y.; Li, Y.-M.; Tang, A.-Z.; Ou, H.-S. Role of microRNA-21 in regulating 3T3-L1 adipocyte differentiation and adiponectin expression. Mol. Biol. Rep. 2013, 40, 5027–5034. [Google Scholar] [CrossRef]
- Song, G.; Xu, G.; Ji, C.; Shi, C.; Shen, Y.; Chen, L.; Zhu, L.; Yang, L.; Zhao, Y.; Guo, X. The role of microRNA-26b in human adipocyte differentiation and proliferation. Gene 2014, 533, 481–487. [Google Scholar] [CrossRef] [PubMed]
- Shi, C.; Zhang, M.; Tong, M.; Yang, L.; Pang, L.; Chen, L.; Xu, G.; Chi, X.; Hong, Q.; Ni, Y.; et al. miR-148a is Associated with Obesity and Modulates Adipocyte Differentiation of Mesenchymal Stem Cells through Wnt Signaling. Sci. Rep. 2015, 5, srep09930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ortega, F.J.; Moreno-Navarrete, J.M.; Pardo, G.; Sabater, M.; Hummel, M.; Ferrer, A.; Rodriguez-Hermosa, J.I.; Ruiz, B.; Ricart, W.; Peral, B.; et al. MiRNA Expression Profile of Human Subcutaneous Adipose and during Adipocyte Differentiation. PLoS ONE 2010, 5, e9022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, C.; Huang, F.; Gu, X.; Zhang, M.; Wen, J.; Wang, X.; You, L.; Cui, X.; Ji, C.; Guo, X. Adipogenic miRNA and meta-signature miRNAs involved in human adipocyte differentiation and obesity. Oncotarget 2016, 7, 40830–40845. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, H.; Chen, X.; Guan, L.; Qi, Q.; Shu, G.; Jiang, Q.; Yuan, L.; Xi, Q.; Zhang, Y.-L. MiRNA-181a Regulates Adipogenesis by Targeting Tumor Necrosis Factor-α (TNF-α) in the Porcine Model. PLoS ONE 2013, 8, e71568. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Williams, A.; Henao-Mejia, J.; Harman, C.C.D.; Flavell, R.A. miR-181 and Metabolic Regulation in the Immune System. Cold Spring Harb. Symp. Quant. Biol. 2013, 78, 223–230. [Google Scholar] [CrossRef] [Green Version]
- Virtue, A.T.; McCright, S.J.; Wright, J.M.; Jimenez, M.T.; Mowel, W.K.; Kotzin, J.; Joannas, L.; Basavappa, M.G.; Spencer, S.P.; Clark, M.L.; et al. The gut microbiota regulates white adipose tissue inflammation and obesity via a family of microRNAs. Sci. Transl. Med. 2019, 11, eaav1892. [Google Scholar] [CrossRef]
- Miranda, K.; Yang, X.; Bam, M.; Murphy, E.A.; Nagarkatti, P.S.; Nagarkatti, M. MicroRNA-30 modulates metabolic inflammation by regulating Notch signaling in adipose tissue macrophages. Int. J. Obes. 2018, 42, 1140–1150. [Google Scholar] [CrossRef]
- Chen, L.; Hou, J.; Ye, L.; Chen, Y.; Cui, J.; Tian, W.; Li, C.; Liu, L. MicroRNA-143 Regulates Adipogenesis by Modulating the MAP2K5–ERK5 Signaling. Sci. Rep. 2014, 4, 3819. [Google Scholar] [CrossRef] [Green Version]
- Esau, C.; Kang, X.; Peralta, E.; Hanson, E.; Marcusson, E.G.; Ravichandran, L.V.; Sun, Y.; Koo, S.; Perera, R.J.; Jain, R.; et al. MicroRNA-143 Regulates Adipocyte Differentiation. J. Biol. Chem. 2004, 279, 52361–52365. [Google Scholar] [CrossRef] [Green Version]
- Jordan, S.D.; Krüger, M.; Willmes, D.M.; Redemann, N.; Wunderlich, F.T.; Brönneke, H.S.; Merkwirth, C.; Kashkar, H.; Olkkonen, V.M.; Böttger, T.; et al. Obesity-induced overexpression of miRNA-143 inhibits insulin-stimulated AKT activation and impairs glucose metabolism. Nat. Cell Biol. 2011, 13, 434–446. [Google Scholar] [CrossRef] [PubMed]
- Xihua, L.; Shengjie, T.; Weiwei, G.; Matro, E.; Tingting, T.; Lin, L.; Fang, W.; Jiaqiang, Z.; Fenping, Z.; Hong, L. Circulating miR-143-3p inhibition protects against insulin resistance in Metabolic Syndrome via targeting of the insulin-like growth factor 2 receptor. Transl. Res. 2019, 205, 33–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Kong, X.; Liu, J.; Lv, Y.; Sheng, Y.; Lv, S.; Di, W.; Wang, C.; Zhang, F.; Ding, G.X. Expression Profiling of PPARγ-Regulated MicroRNAs in Human Subcutaneous and Visceral Adipogenesis in both Genders. Endocrinology 2014, 155, 2155–2165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, G.; Wu, Z.; Li, X.; Ning, X.; Li, Y.; Yang, G. Biological role of MicroRNA-103 based on expression profile and target genes analysis in pigs. Mol. Biol. Rep. 2010, 38, 4777–4786. [Google Scholar] [CrossRef]
- Trajkovski, M.; Hausser, J.; Soutschek, J.; Bhat, B.; Akin, A.; Zavolan, M.; Heim, M.H.; Stoffel, M. MicroRNAs 103 and 107 regulate insulin sensitivity. Nat. Cell Biol. 2011, 474, 649–653. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Z.; Wu, S.; Muhammad, S.; Ren, Q.; Chao, S. miR-103/107 promote ER stress-mediated apoptosis via targeting the Wnt3a/β-catenin/ATF6 pathway in preadipocytes. J. Lipid Res. 2018, 59, 843–853. [Google Scholar] [CrossRef] [Green Version]
- Meerson, A.; Traurig, M.; Ossowski, V.; Fleming, J.M.; Mullins, M.; Baier, L.J. Human adipose microRNA-221 is upregulated in obesity and affects fat metabolism downstream of leptin and TNF-α. Diabetologia 2013, 56, 1971–1979. [Google Scholar] [CrossRef] [Green Version]
- Lustig, Y.; Barhod, E.; Ashwal-Fluss, R.; Gordin, R.; Shomron, N.; Baruch-Umansky, K.; Hemi, R.; Karasik, A.; Kanety, H. RNA-Binding Protein PTB and MicroRNA-221 Coregulate AdipoR1 Translation and Adiponectin Signaling. Diabetes 2013, 63, 433–445. [Google Scholar] [CrossRef] [Green Version]
- Zhang, C.; Qian, D.; Zhao, H.; Lv, N.; Yu, P.; Sun, Z. MiR17 improves insulin sensitivity through inhibiting expression of ASK1 and anti-inflammation of macrophages. Biomed. Pharmacother. 2018, 100, 448–454. [Google Scholar] [CrossRef]
- Zhang, M.; Liu, Q.; Mi, S.; Liang, X.; Zhang, Z.; Su, X.; Liu, J.; Chen, Y.; Wang, M.; Zhang, Y.; et al. Both miR-17-5p and miR-20a Alleviate Suppressive Potential of Myeloid-Derived Suppressor Cells by Modulating STAT3 Expression. J. Immunol. 2011, 186, 4716–4724. [Google Scholar] [CrossRef] [Green Version]
- Lin, Q.; Gao, Z.; Alarcon, R.M.; Ye, J.; Yun, Z. A role ofmiR-27in the regulation of adipogenesis. FEBS J. 2009, 276, 2348–2358. [Google Scholar] [CrossRef]
- Herrera, B.M.; Lockstone, H.E.; Taylor, J.M.; Ria, M.; Barrett, A.; Collins, S.; Kaisaki, P.; Argoud, K.; Fernandez, C.; Travers, M.E.; et al. Global microRNA expression profiles in insulin target tissues in a spontaneous rat model of type 2 diabetes. Diabetologia 2010, 53, 1099–1109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, C.; Lee, H.; Cho, Y.M.; Kwon, O.-J.; Kim, W.; Lee, E.K. TNFα-induced miR-130 resulted in adipocyte dysfunction during obesity-related inflammation. FEBS Lett. 2013, 587, 3853–3858. [Google Scholar] [CrossRef] [PubMed]
- Lee, E.K.; Lee, M.J.; Abdelmohsen, K.; Kim, W.; Kim, M.M.; Srikantan, S.; Martindale, J.L.; Hutchison, E.R.; Kim, H.H.; Marasa, B.S.; et al. miR-130 Suppresses Adipogenesis by Inhibiting Peroxisome Proliferator-Activated Receptor Expression. Mol. Cell. Biol. 2010, 31, 626–638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yiew, N.K.H.; Chatterjee, T.K.; Tang, Y.; Pellenberg, R.; Stansfield, B.K.; Bagi, Z.; Fulton, D.J.; Stepp, D.W.; Chen, W.; Patel, V.; et al. A novel role for the Wnt inhibitor APCDD1 in adipocyte differentiation: Implications for diet-induced obesity. J. Biol. Chem. 2017, 292, 6312–6324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayner, K.J.; Suárez, Y.; Dávalos, A.; Parathath, S.; Fitzgerald, M.L.; Tamehiro, N.; Fisher, E.A.; Moore, K.J.; Fernández-Hernando, C. MiR-33 Contributes to the Regulation of Cholesterol Homeostasis. Science 2010, 328, 1570–1573. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rayner, K.J.; Esau, C.C.; Hussain, F.N.; McDaniel, A.L.; Marshall, S.M.; Van Gils, J.M.; Ray, T.D.; Sheedy, F.J.; Goedeke, L.; Liu, X.; et al. Inhibition of miR-33a/b in non-human primates raises plasma HDL and lowers VLDL triglycerides. Nat. Cell Biol. 2011, 478, 404–407. [Google Scholar] [CrossRef] [Green Version]
- Price, N.L.; Holtrup, B.; Kwei, S.L.; Wabitsch, M.; Rodeheffer, M.; Bianchini, L.; Suárez, Y.; Fernández-Hernando, C. SREBP-1c/MicroRNA 33b Genomic Loci Control Adipocyte Differentiation. Mol. Cell. Biol. 2016, 36, 1180–1193. [Google Scholar] [CrossRef] [Green Version]
- Bork, S.; Horn, P.; Castoldi, M.; Hellwig, I.; Ho, A.D.; Wagner, W. Adipogenic differentiation of human mesenchymal stromal cells is down-regulated by microRNA-369-5p and up-regulated by microRNA-371. J. Cell. Physiol. 2011, 226, 2226–2234. [Google Scholar] [CrossRef]
- Ji, J.; Yamashita, T.; Budhu, A.; Forgues, M.; Jia, H.-L.; Li, C.; Deng, C.; Wauthier, E.; Reid, L.M.; Ye, Q.-H.; et al. Identification of microRNA-181 by genome-wide screening as a critical player in EpCAM-positive hepatic cancer stem cells. Hepatology 2009, 50, 472–480. [Google Scholar] [CrossRef] [Green Version]
- Fukuda, D.; Aikawa, E.; Swirski, F.K.; Novobrantseva, T.I.; Kotelianski, V.; Gorgun, C.Z.; Chudnovskiy, A.; Yamazaki, H.; Croce, K.; Weissleder, R.; et al. Notch ligand Delta-like 4 blockade attenuates atherosclerosis and metabolic disorders. Proc. Natl. Acad. Sci. USA 2012, 109, E1868–E1877. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bridge, G.; Monteiro, R.; Henderson, S.; Emuss, V.; Lagos, D.; Georgopoulou, D.; Patient, R.; Boshoff, C. The microRNA-30 family targets DLL4 to modulate endothelial cell behavior during angiogenesis. Blood 2012, 120, 5063–5072. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, D.; Aikawa, M. Expanding Role of Delta-Like 4 Mediated Notch Signaling in Cardiovascular and Metabolic Diseases. Circ. J. 2013, 77, 2462–2468. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, B.; Fan, J.; Chen, N. A Novel Regulator of Type II Diabetes: MicroRNA-143. Trends Endocrinol. Metab. 2018, 29, 380–388. [Google Scholar] [CrossRef] [PubMed]
- Kilic, I.D.; Dodurga, Y.; Uludag, B.; Alihanoglu, Y.I.; Yıldız, B.S.; Enli, Y.; Secme, M.; Bostancı, H.E. microRNA -143 and -223 in obesity. Gene 2015, 560, 140–142. [Google Scholar] [CrossRef]
- Bost, F.; Aouadi, M.; Caron, L.; Binétruy, B. The role of MAPKs in adipocyte differentiation and obesity. Biochimie 2005, 87, 51–56. [Google Scholar] [CrossRef] [Green Version]
- Choy, L.; Skillington, J.; Derynck, R. Roles of Autocrine TGF-β Receptor and Smad Signaling in Adipocyte Differentiation. J. Cell Biol. 2000, 149, 667–682. [Google Scholar] [CrossRef]
- Tsurutani, Y.; Fujimoto, M.; Takemoto, M.; Irisuna, H.; Koshizaka, M.; Onishi, S.; Ishikawa, T.; Mezawa, M.; He, P.; Honjo, S.; et al. The roles of transforming growth factor-β and Smad3 signaling in adipocyte differentiation and obesity. Biochem. Biophys. Res. Commun. 2011, 407, 68–73. [Google Scholar] [CrossRef]
- Zhu, L.; Shi, C.; Ji, C.; Xu, G.; Chen, L.; Yang, L.; Fu, Z.; Ji, C.; Lu, Y.; Guo, X. FFAs and adipokine-mediated regulation of hsa-miR-143 expression in human adipocytes. Mol. Biol. Rep. 2013, 40, 5669–5675. [Google Scholar] [CrossRef]
- Ambati, S.; Kim, H.-K.; Yang, J.-Y.; Lin, J.; A Della-Fera, M.; Baile, C.A. Effects of leptin on apoptosis and adipogenesis in 3T3-L1 adipocytes. Biochem. Pharmacol. 2007, 73, 378–384. [Google Scholar] [CrossRef]
- Blumensatt, M.; Wronkowitz, N.; Wiza, C.; Cramer, A.; Mueller, H.; Rabelink, M.J.; Hoeben, R.C.; Eckel, J.; Sell, H.; Ouwens, D.M. Adipocyte-derived factors impair insulin signaling in differentiated human vascular smooth muscle cells via the upregulation of miR-143. Biochim. Biophys. Acta Mol. Basis Dis. 2014, 1842, 275–283. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wilfred, B.R.; Wang, W.-X.; Nelson, P.T. Energizing miRNA research: A review of the role of miRNAs in lipid metabolism, with a prediction that miR-103/107 regulates human metabolic pathways. Mol. Genet. Metab. 2007, 91, 209–217. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- John, E.; Wienecke-Baldacchino, A.; Liivrand, M.; Heinäniemi, M.; Carlberg, C.; Sinkkonen, L. Dataset integration identifies transcriptional regulation of microRNA genes by PPARγ in differentiating mouse 3T3-L1 adipocytes. Nucleic Acids Res. 2012, 40, 4446–4460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, M.; Liu, Z.; Zhang, Z.; Liu, G.; Sun, S.; Sun, C. miR-103 promotes 3T3-L1 cell adipogenesis through AKT/mTOR signal pathway with its target being MEF2D. Biol. Chem. 2015, 396, 235–244. [Google Scholar] [CrossRef] [PubMed]
- Brandebourg, T.D.; Hu, C.Y. Regulation of differentiating pig preadipocytes by retinoic acid. J. Anim. Sci. 2005, 83, 98–107. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.E.; Chen, J. Regulation of Peroxisome Proliferator-Activated Receptor- Activity by Mammalian Target of Rapamycin and Amino Acids in Adipogenesis. Diabetes 2004, 53, 2748–2756. [Google Scholar] [CrossRef] [Green Version]
- Baudry, A.; Yang, Z.; Hemmings, B.A. PKB is required for adipose differentiation of mouse embryonic fibroblasts. J. Cell Sci. 2006, 119, 889–897. [Google Scholar] [CrossRef] [Green Version]
- Zhang, H.H.; Huang, J.; Düvel, K.; Boback, B.; Wu, S.; Squillace, R.M.; Wu, C.-L.; Manning, B.D. Insulin Stimulates Adipogenesis through the Akt-TSC2-mTORC1 Pathway. PLoS ONE 2009, 4, e6189. [Google Scholar] [CrossRef]
- Zhu, H.; Leung, S.W. Identification of microRNA biomarkers in type 2 diabetes: A meta-analysis of controlled profiling studies. Diabetologia 2015, 58, 900–911. [Google Scholar] [CrossRef]
- Alkhouri, N.; Gornicka, A.; Berk, M.P.; Thapaliya, S.; Dixon, L.J.; Kashyap, S.; Schauer, P.R.; Feldstein, A.E. Adipocyte Apoptosis, a Link between Obesity, Insulin Resistance, and Hepatic Steatosis. J. Biol. Chem. 2010, 285, 3428–3438. [Google Scholar] [CrossRef] [Green Version]
- Skårn, M.; Namløs, H.M.; Noordhuis, P.; Wang, M.; Meza-Zepeda, L.A.; Myklebost, O. Adipocyte Differentiation of Human Bone Marrow-Derived Stromal Cells Is Modulated by MicroRNA-155, MicroRNA-221, and MicroRNA-222. Stem Cells Dev. 2012, 21, 873–883. [Google Scholar] [CrossRef] [PubMed]
- Ouchi, N.; Parker, J.L.; Lugus, J.J.; Walsh, K. Adipokines in inflammation and metabolic disease. Nat. Rev. Immunol. 2011, 11, 85–97. [Google Scholar] [CrossRef] [PubMed]
- E Brown, J. Dysregulated adipokines in the pathogenesis of type 2 diabetes and vascular disease. Br. J. Diabetes Vasc. Dis. 2012, 12, 249–254. [Google Scholar] [CrossRef] [Green Version]
- Zhu, N.; Zhang, D.; Chen, S.; Liu, X.; Lin, L.; Huang, X.; Guo, Z.; Liu, J.; Wang, Y.; Yuan, W.; et al. Endothelial enriched microRNAs regulate angiotensin II-induced endothelial inflammation and migration. Atherosclerosis 2011, 215, 286–293. [Google Scholar] [CrossRef] [PubMed]
- Grenningloh, R.; Kang, B.Y.; Ho, I.-C.; Vermi, W.; Riboldi, E.; Wittamer, V.; Gentili, F.; Luini, W.; Marrelli, S.; Vecchi, A.; et al. Ets-1, a functional cofactor of T-bet, is essential for Th1 inflammatory responses. J. Exp. Med. 2005, 201, 615–626. [Google Scholar] [CrossRef]
- Zhan, Y.; Brown, C.; Maynard, E.; Anshelevich, A.; Ni, W.; Ho, I.-C.; Oettgen, P. Ets-1 is a critical regulator of Ang II-mediated vascular inflammation and remodeling. J. Clin. Investig. 2005, 115, 2508–2516. [Google Scholar] [CrossRef] [Green Version]
- Peng, J.; Zhou, Y.; Deng, Z.; Zhang, H.; Wu, Y.; Song, T.; Yang, Y.; Wei, H.; Peng, J. miR-221 negatively regulates inflammation and insulin sensitivity in white adipose tissue by repression of sirtuin-1 (SIRT1). J. Cell. Biochem. 2018, 119, 6418–6428. [Google Scholar] [CrossRef]
- Yoshizaki, T.; Milne, J.C.; Imamura, T.; Schenk, S.; Sonoda, N.; Babendure, J.L.; Lu, J.-C.; Smith, J.J.; Jirousek, M.R.; Olefsky, J.M. SIRT1 Exerts Anti-Inflammatory Effects and Improves Insulin Sensitivity in Adipocytes. Mol. Cell. Biol. 2008, 29, 1363–1374. [Google Scholar] [CrossRef] [Green Version]
- Yamauchi, T.; Kamon, J.; Waki, H.; Terauchi, Y.; Kubota, N.; Hara, K.; Mori, Y.; Ide, T.; Murakami, K.; Tsuboyamakasaoka, N.; et al. The fat-derived hormone adiponectin reverses insulin resistance associated with both lipoatrophy and obesity. Nat. Med. 2001, 7, 941–946. [Google Scholar] [CrossRef]
- Yamauchi, T.; Kamon, J.; Minokoshi, Y.; Ito, Y.; Waki, H.; Uchida, S.; Yamashita, S.; Noda, M.; Kita, S.; Ueki, K.; et al. Adiponectin stimulates glucose utilization and fatty-acid oxidation by activating AMP-activated protein kinase. Nat. Med. 2002, 8, 1288–1295. [Google Scholar] [CrossRef]
- Berg, A.H.; Combs, T.P.; Du, X.; Brownlee, M.; Scherer, P.E. The adipocyte-secreted protein Acrp30 enhances hepatic insulin action. Nat. Med. 2001, 7, 947–953. [Google Scholar] [CrossRef] [PubMed]
- Lihn, A.S.; Pedersen, S.B.; Richelsen, B. Adiponectin: Action, regulation and association to insulin sensitivity. Obes. Rev. 2005, 6, 13–21. [Google Scholar] [CrossRef] [PubMed]
- Heneghan, H.M.; Miller, N.; McAnena, O.J.; O’Brien, T.; Kerin, M.J. Differential miRNA Expression in Omental Adipose Tissue and in the Circulation of Obese Patients Identifies Novel Metabolic Biomarkers. J. Clin. Endocrinol. Metab. 2011, 96, E846–E850. [Google Scholar] [CrossRef] [PubMed]
- A Deiuliis, J. MicroRNAs as regulators of metabolic disease: Pathophysiologic significance and emerging role as biomarkers and therapeutics. Int. J. Obes. 2016, 40, 88–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Osaka, N.; Takahashi, T.; Murakami, S.; Matsuzawa, A.; Noguchi, T.; Fujiwara, T.; Aburatani, H.; Moriyama, K.; Takeda, K.; Ichijo, H. ASK1-dependent recruitment and activation of macrophages induce hair growth in skin wounds. J. Cell Biol. 2007, 176, 903–909. [Google Scholar] [CrossRef] [Green Version]
- Philippe, L.; Alsaleh, G.; Bahram, S.; Pfeffer, S.; Georgel, P. The miR-17 ∼ 92 Cluster: A Key Player in the Control of Inflammation during Rheumatoid Arthritis. Front. Immunol. 2013, 4, 70. [Google Scholar] [CrossRef] [Green Version]
- O’Connell, R.M.; Rao, D.S.; Chaudhuri, A.A.; Baltimore, D. Physiological and pathological roles for microRNAs in the immune system. Nat. Rev. Immunol. 2010, 10, 111–122. [Google Scholar] [CrossRef]
- Chen, T.-C.; Sung, M.-L.; Kuo, H.-C.; Chien, S.-J.; Yen, C.-K.; Chen, C.-N. Differential Regulation of Human Aortic Smooth Muscle Cell Proliferation by Monocyte-Derived Macrophages from Diabetic Patients. PLoS ONE 2014, 9, e113752. [Google Scholar] [CrossRef]
- Hilton, C.; Neville, M.J.; Karpe, F. MicroRNAs in adipose tissue: Their role in adipogenesis and obesity. Int. J. Obes. 2012, 37, 325–332. [Google Scholar] [CrossRef] [Green Version]
- Gu, C.; Xu, Y.; Zhang, S.; Guan, H.; Song, S.; Wang, X.; Wang, Y.; Li, Y.; Zhao, G. miR-27a attenuates adipogenesis and promotes osteogenesis in steroid-induced rat BMSCs by targeting PPARγ and GREM1. Sci. Rep. 2016, 6, 38491. [Google Scholar] [CrossRef]
- Gan, C.-C.; Ni, T.-W.; Yu, Y.; Qin, N.; Chen, Y.; Jin, M.-N.; Duan, H.-Q. Flavonoid derivative (Fla-CN) inhibited adipocyte differentiation via activating AMPK and up-regulating microRNA-27 in 3T3-L1 cells. Eur. J. Pharmacol. 2017, 797, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Villard, A.; Marchand, L. Diagnostic Value of Cell-free Circulating Micrornas for Obesity and Type 2 Diabetes: A Meta-analysis. J. Mol. Biomark. Diagn. 2015, 6, 251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Butler, M.G.; Wang, K.; Naggert, J.K.; A Rethmeyer, J.; Gunewardena, S.S.; Manzardo, A.M.; Marshall, J.D. Coding and noncoding expression patterns associated with rare obesity-related disorders: Prader–Willi and Alström syndromes. Adv. Genom. Genet. 2015, 5, 53–75. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xie, N.; Cui, H.; Banerjee, S.; Tan, Z.; Salomao, R.; Fu, M.; Abraham, E.; Thannickal, V.J.; Liu, G. miR-27a Regulates Inflammatory Response of Macrophages by Targeting IL-10. J. Immunol. 2014, 193, 327–334. [Google Scholar] [CrossRef] [Green Version]
- McGregor, R.A.; Choi, M.-S. microRNAs in the Regulation of Adipogenesis and Obesity. Curr. Mol. Med. 2011, 11, 304–316. [Google Scholar] [CrossRef]
- Zou, B.; Ge, Z.; Zhu, W.; Xu, Z.; Li, C.-M. Persimmon tannin represses 3T3-L1 preadipocyte differentiation via up-regulating expression of miR-27 and down-regulating expression of peroxisome proliferator-activated receptor-γ in the early phase of adipogenesis. Eur. J. Nutr. 2014, 54, 1333–1343. [Google Scholar] [CrossRef]
- Kim, S.Y.; Kim, A.Y.; Lee, H.W.; Son, Y.H.; Lee, G.Y.; Lee, J.-W.; Lee, Y.S.; Kim, J.B. miR-27a is a negative regulator of adipocyte differentiation via suppressing PPARγ expression. Biochem. Biophys. Res. Commun. 2010, 392, 323–328. [Google Scholar] [CrossRef]
- Yao, F.; Yu, Y.; Feng, L.; Li, J.; Zhang, M.; Lan, X.; Yan, X.; Liu, Y.; Guan, F.; Zhang, M.; et al. Adipogenic miR-27a in adipose tissue upregulates macrophage activation via inhibiting PPARγ of insulin resistance induced by high-fat diet-associated obesity. Exp. Cell Res. 2017, 355, 105–112. [Google Scholar] [CrossRef]
- Zhang, B. Negative regulation of peroxisome proliferator-activated receptor-gamma gene expression contributes to the antiadipogenic effects of tumor necrosis factor-alpha. Mol. Endocrinol. 1996, 10, 1457–1466. [Google Scholar] [CrossRef]
- Gustafson, B.; Gogg, S.; Hedjazifar, S.; Jenndahl, L.; Hammarstedt, A.; Smith, U. Inflammation and impaired adipogenesis in hypertrophic obesity in man. Am. J. Physiol. Metab. 2009, 297, E999–E1003. [Google Scholar] [CrossRef] [Green Version]
- Almuraikhy, S.; Kafienah, W.; Bashah, M.; Diboun, I.; Jaganjac, M.; Al-Khelaifi, F.; Abdesselem, H.; Mazloum, N.A.; Alsayrafi, M.; Mohamed-Ali, V.; et al. Interleukin-6 induces impairment in human subcutaneous adipogenesis in obesity-associated insulin resistance. Diabetologia 2016, 59, 2406–2416. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Wang, H.; Hegde, V.; Dubuisson, O.; Gao, Z.; Dhurandhar, N.V.; Ye, J. Interplay of pro- and anti-inflammatory cytokines to determine lipid accretion in adipocytes. Int. J. Obes. 2013, 37, 1490–1498. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, M.; Zhou, Z.; Wang, J.; Li, S. MiR-130b promotes obesity associated adipose tissue inflammation and insulin resistance in diabetes mice through alleviating M2 macrophage polarization via repression of PPAR-γ. Immunol. Lett. 2016, 180, 1–8. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.-C.; Li, Y.; Huang, X.-C.; Zhang, D.; Zhang, H.; Wu, Q.; He, Y.-Q.; Wang, J.-Y.; Zhang, L.; Xia, H.; et al. Circulating miR-130b mediates metabolic crosstalk between fat and muscle in overweight/obesity. Diabetologia 2013, 56, 2275–2285. [Google Scholar] [CrossRef]
- Prats-Puig, A.; Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Moreno, M.; Bonet, N.; Ricart, W.; López-Bermejo, A.; Fernández-Real, J.M. Changes in Circulating MicroRNAs Are Associated With Childhood Obesity. J. Clin. Endocrinol. Metab. 2013, 98, E1655–E1660. [Google Scholar] [CrossRef]
- Al-Rawaf, H.A. Circulating microRNAs and adipokines as markers of metabolic syndrome in adolescents with obesity. Clin. Nutr. 2019, 38, 2231–2238. [Google Scholar] [CrossRef]
- Samad, F.; Yamamoto, K.; Pandey, M.; Loskutoff, D.J. Elevated Expression of Transforming Growth Factor-β in Adipose Tissue from Obese Mice. Mol. Med. 1997, 3, 37–48. [Google Scholar] [CrossRef] [Green Version]
- Lee, M.-J. Transforming growth factor beta superfamily regulation of adipose tissue biology in obesity. Biochim. Biophys. Acta Mol. Basis Dis. 2018, 1864, 1160–1171. [Google Scholar] [CrossRef]
- Ortega, F.J.; Mercader, J.M.; Moreno-Navarrete, J.M.; Rovira, O.; Guerra, E.; Esteve, E.; Xifra, G.; Martínez, C.; Ricart, W.; Rieusset, J.; et al. Profiling of Circulating MicroRNAs Reveals Common MicroRNAs Linked to Type 2 Diabetes That Change with Insulin Sensitization. Diabetes Care 2014, 37, 1375–1383. [Google Scholar] [CrossRef] [Green Version]
- Yuan, Y.; Peng, W.; Liu, Y.; Xu, Z.-S. Circulating miR-130 and its target PPAR-γ may be potential biomarkers in patients of coronary artery disease with type 2 diabetes mellitus. Mol. Genet. Genom. Med. 2019, 7, e909. [Google Scholar] [CrossRef] [Green Version]
- Peng, Y.; Yu, S.; Li, H.; Xiang, H.; Peng, J.; Jiang, S. MicroRNAs: Emerging roles in adipogenesis and obesity. Cell Signal. 2014, 26, 1888–1896. [Google Scholar] [CrossRef] [PubMed]
- Taniguchi, M.; Nakajima, I.; Chikuni, K.; Kojima, M.; Awata, T.; Mikawa, S. MicroRNA-33b downregulates the differentiation and development of porcine preadipocytes. Mol. Biol. Rep. 2014, 41, 1081–1090. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xi, Y.; Shen, W.; Ma, L.; Zhao, M.; Zheng, J.; Bu, S.; Hino, S.; Nakao, M. HMGA2 promotes adipogenesis by activating C/EBPβ-mediated expression of PPARγ. Biochem. Biophys. Res. Commun. 2016, 472, 617–623. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Y.; Xi, Y.; Chen, J.; Zhu, P.; Kang, J.; Tan, R.-H.; Wang, F.; Bu, S. STAT3 stimulates adipogenic stem cell proliferation and cooperates with HMGA2 during the early stage of differentiation to promote adipogenesis. Biochem. Biophys. Res. Commun. 2017, 482, 1360–1366. [Google Scholar] [CrossRef] [PubMed]
- Horie, T.; Nishino, T.; Baba, O.; Kuwabara, Y.; Nakao, T.; Nishiga, M.; Usami, S.; Izuhara, M.; Sowa, N.; Yahagi, N.; et al. MicroRNA-33 regulates sterol regulatory element-binding protein 1 expression in mice. Nat. Commun. 2013, 4, 2883. [Google Scholar] [CrossRef] [Green Version]
- Price, N.L.; Singh, A.K.; Rotllan, N.; Goedeke, L.; Wing, A.; Canfrán-Duque, A.; Diaz-Ruiz, A.; Araldi, E.; Baldán, Á.; Camporez, J.-P.; et al. Genetic Ablation of miR-33 Increases Food Intake, Enhances Adipose Tissue Expansion, and Promotes Obesity and Insulin Resistance. Cell Rep. 2018, 22, 2133–2145. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Karunakaran, D.; Richards, L.; Geoffrion, M.; Barrette, D.; Gotfrit, R.J.; Harper, M.-E.; Rayner, K.J. Therapeutic Inhibition of miR-33 Promotes Fatty Acid Oxidation but Does Not Ameliorate Metabolic Dysfunction in Diet-Induced Obesity. Arter. Thromb. Vasc. Biol. 2015, 35, 2536–2543. [Google Scholar] [CrossRef] [Green Version]
- Furuhashi, M.; Saitoh, S.; Shimamoto, K.; Miura, T. Fatty Acid-Binding Protein 4 (FABP4): Pathophysiological Insights and Potent Clinical Biomarker of Metabolic and Cardiovascular Diseases. Clin. Med. Insights Cardiol. 2014, 8 (Suppl. 3), 23–33. [Google Scholar] [CrossRef]
- Drenth, J.P.H.; Schattenberg, J.M. The nonalcoholic steatohepatitis (NASH) drug development graveyard: Established hurdles and planning for future success. Expert Opin. Investig. Drugs 2020, 1–11. [Google Scholar] [CrossRef]
- Rech, M.; Kuhn, A.R.; Lumens, J.; Carai, P.; Van Leeuwen, R.; Verhesen, W.; Verjans, R.; LeComte, J.; Liu, Y.; Luiken, J.J.; et al. AntagomiR-103 and -107 Treatment Affects Cardiac Function and Metabolism. Mol. Ther. Nucleic Acids 2019, 14, 424–437. [Google Scholar] [CrossRef] [Green Version]

| miRNA | Function(s) | Target(s) * | Reference(s) and Corresponding Study Model(s) |
|---|---|---|---|
| MiR-193b-365 | Involved in the regulation of BAT differentiation | Runx1t1 | [85]: Mouse; in vitro [86]: Mouse; in vitro |
| MiR-182 | Positive regulator of BAT adipogenesis | Insig1, Pdgfr2 | [77]: Mouse; in vitro |
| MiR-203 | Positive regulator of BAT adipogenesis | Insig1, Pdgfr2 | [77]: Mouse; in vitro |
| MiR-106b-93 | Negative regulator of BAT differentiation, involved in energy homeostasis | Ppara | [87]: Mouse |
| MiR-328 | Positive regulator of BAT differentiation, controls brown adipogenesis by regulating the switch between muscle-specific (myogenic) and brown adipogenic lineages | Bace1 | [86]: Mouse; in vitro [88]: Mouse; in vitro |
| MiR-129 | Positive regulator of BAT function, involved in thermogenesis and energy expenditure, potential obesity biomarker | Igf2, Egr1 | [89]: Human; mouse; in vitro |
| MiR-455 | Positive regulator of brown/beige AT, promotes cells differentiation | UCP1, Runx1t1, Necdin, Hif1an, Tgfbr3 | [90]: Human; mouse; in vitro [91]: In vitro [92]: Mouse |
| MiR-30b/c | Positive regulator of brown and brite adipogenesis | Rip140 | [93]: Mouse; in vitro [94]: In vitro |
| MiR-34a | Negative regulator of brown and brite adipogenesis, exhibiting increased expression in obesity | Fgfr1 | [95]: Mouse; in vitro |
| MiR-27b | Inhibitor of brown and beige adipogenesis, exhibiting decreased expression in response to cold exposure and β-adrenergic activation | Prdm16, Ppar, Pparg, Creb, Pgc1b, Prohibitin | [96]: In vitro [97]: Mouse; in vitro [98]: In vitro |
| MiR-378 | Positive regulator of BAT that promotes Ucp1 expression, BAT mass and BAT oxygen consumption, showing increased expression during brown adipocyte differentiation | Pde1b | [99]: Mouse; in vitro |
| MiR-133 | Negative regulator of brown adipogenesis, acting as an inhibitor of BAT differentiation | Prdm16 | [100]: Mouse; in vitro [101]: Mouse; in vitro [102]: In vitro |
| MiR-155 | Negative regulator of brown adipogenesis, enriched in BAT and highly expressed in proliferating brown pre-adipocytes, exhibiting declining expression with cell differentiation | C/ebpb | [103]: Mouse; in vitro [104]: Mouse; in vitro [105]: Human; mouse; in vitro |
| MiR-32 | Positive regulator of brown adipogenesis with significantly increased expression in BAT during cold exposure | Tob1 | [106]: Mouse; in vitro |
| miRNA | Function(s) | Target(s) * | Reference(s) and Corresponding Study Model(s) |
|---|---|---|---|
| MiR-196a | Plays a role in browning of white progenitor cells, inducing WAT browning and involved in regulating human body fat distribution | Hoxc8 | [78]: Mouse; in vitro |
| MiR-26 | Key regulator of human white and beige adipocyte differentiation and positive regulator of brown adipogenesis | ADAM17, Fbxl19 | [123]: Human; in vitro [124]: Mouse; in vitro |
| MiR-125 | Negative regulator of browning and the formation of functional brite adipocytes | MMP11 | [125]: Human; in vitro |
| MiR-Let7i-5p | Thermogenesis inhibitor, strongly inhibiting mitochondrial and browning markers in mice scWAT and human and murine brite adipocytes | Not specified | [126]: Human; mouse; in vitro |
| MiR-455 | Positive regulator of brown/beige AT | Ucp1, Runx1t1, Necdin, Hif1an, Tgfbr3 | [90]: Human; mouse; in vitro [91]: In vitro [92]: Mouse |
| MiR-30b/c | Positive regulator of brown and brite adipogenesis | Rip140 | [93]: Mouse; in vitro [94]: In vitro |
| MiR-34a | Negative regulator of brown and brite adipogenesis, exhibiting increased expression in obesity, disrupts FGF21 signaling in AT and prevents PGC1A activation and browning of WAT | Fgfr1 | [95]: Mouse; in vitro |
| MiR-27b | Inhibitor of brown and beige adipogenesis, downregulated during WAT and brite differentiation | Prmd16, Ppara, Pparg, Creb, Pgc1b, Prohibitin | [96]: In vitro [97]: Mouse; in vitro [98]: In vitro |
| MiR-378 | Negative regulator of brite adipogenesis | Pde1b | [99]: Mouse; in vitro |
| MiR-133 | Negative regulator of brown adipogenesis | Prmd16 | [100]: Mouse; in vitro [101]: Mouse; in vitro [102]: In vitro |
| MiR-155 | Negative regulator of brown adipogenesis | C/ebpb | [103]: Mouse; in vitro [104]: Mouse; in vitro [105]: Human, Mouse; in vitro |
| MiR-32 | Positive regulator of brown adipogenesis, promoting thermogenesis and WAT browning | Tob1 | [106]: Mouse; in vitro |
| miRNA | Function(s) | Target(s) * | Reference(s) and Corresponding Study Model(s) |
|---|---|---|---|
| MiR-181 | Elevated expression in obese WAT, promotes insulin resistance and inflammation in WAT | PTEN, TNFA | [150]: Piglet; in vitro [151]: Review [152]: Human; mouse |
| MiR-30a | Stimulates adipogenesis, protects adipocytes against inflammation, prevents polarization of macrophages to the M1 phenotype | STAT1, DLL4 | [94]: In vitro [153]: Mouse; in vitro [113]: Human; mouse |
| MiR-143 | Promotes adipocyte differentiation and insulin resistance | ERK5, FGF7, MAP3K7, IGF2R, Opr8 | [154]: Rat; in vitro [155]: In vitro [156]: Mouse; in vitro [157]: Human; mouse, in vitro [158]: Human; in vitro |
| MiR-103 | Pro-adipogenic, increases lipid accumulation, attenuates insulin signaling and promotes apoptosis in pre-adipocytes | RAI14, Caveolin-1, Wnt3a | [159]: Piglet; in vitro [160]: Human; mouse; in vitro [161]: In vitro |
| MiR-107 | Anti-adipogenic, attenuates insulin signaling and promotes apoptosis in pre-adipocytes | CDK6, Caveolin-1, Wnt3a | [83]: In vitro [160]: Human; mouse; in vitro [161]: In vitro |
| MiR-221 | Negative regulator of adipogenesis, pro-inflammatory effects | ETS1, Sirt1, AdipoR1 | [162]: Human; in vitro [163]: Mouse; in vitro |
| MiR-17 | Prevents macrophage-mediated AT inflammation and improves insulin resistance | Ask1, STAT3 | [164]: Mouse; in vitro [165]: Mouse; in vitro |
| MiR-27a | Anti-adipogenic, upregulated expression in 3T3-L1 adipocytes and ob/ob mice, increased in the circulation of patients with T2DM or obesity | Pparg | [166]: In vitro [167]: Rat; in vitro |
| MiR-130 | Anti-adipogenic, mediates the inhibitory effects of TNFA on PPARG, pro-inflammatory effects | PPARG, Apcdd1 | [168]: Mouse; in vitro [169]: Human; in vitro [170]: Human; mouse; in vitro |
| MiR-33 | Attenuates adipogenesis and lipid accumulation, regulates cholesterol efflux and HDL synthesis | HMGA2, ABCA1 | [171]: Mouse; in vitro [172]: Monkey; in vitro [173]: Human; in vitro |
| MiR-369-5p | Anti-adipogenic | FABP4 | [174]: Human; in vitro |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gharanei, S.; Shabir, K.; Brown, J.E.; Weickert, M.O.; Barber, T.M.; Kyrou, I.; Randeva, H.S. Regulatory microRNAs in Brown, Brite and White Adipose Tissue. Cells 2020, 9, 2489. https://doi.org/10.3390/cells9112489
Gharanei S, Shabir K, Brown JE, Weickert MO, Barber TM, Kyrou I, Randeva HS. Regulatory microRNAs in Brown, Brite and White Adipose Tissue. Cells. 2020; 9(11):2489. https://doi.org/10.3390/cells9112489
Chicago/Turabian StyleGharanei, Seley, Kiran Shabir, James E. Brown, Martin O. Weickert, Thomas M. Barber, Ioannis Kyrou, and Harpal S. Randeva. 2020. "Regulatory microRNAs in Brown, Brite and White Adipose Tissue" Cells 9, no. 11: 2489. https://doi.org/10.3390/cells9112489
APA StyleGharanei, S., Shabir, K., Brown, J. E., Weickert, M. O., Barber, T. M., Kyrou, I., & Randeva, H. S. (2020). Regulatory microRNAs in Brown, Brite and White Adipose Tissue. Cells, 9(11), 2489. https://doi.org/10.3390/cells9112489







