Leucine Supplementation Decreases HDAC4 Expression and Nuclear Localization in Skeletal Muscle Fiber of Rats Submitted to Hindlimb Immobilization
Abstract
:1. Introduction
2. Materials and Methods
2.1. Experimental Animals
2.2. Hindlimb Immobilization
2.3. RNA-Seq Analysis
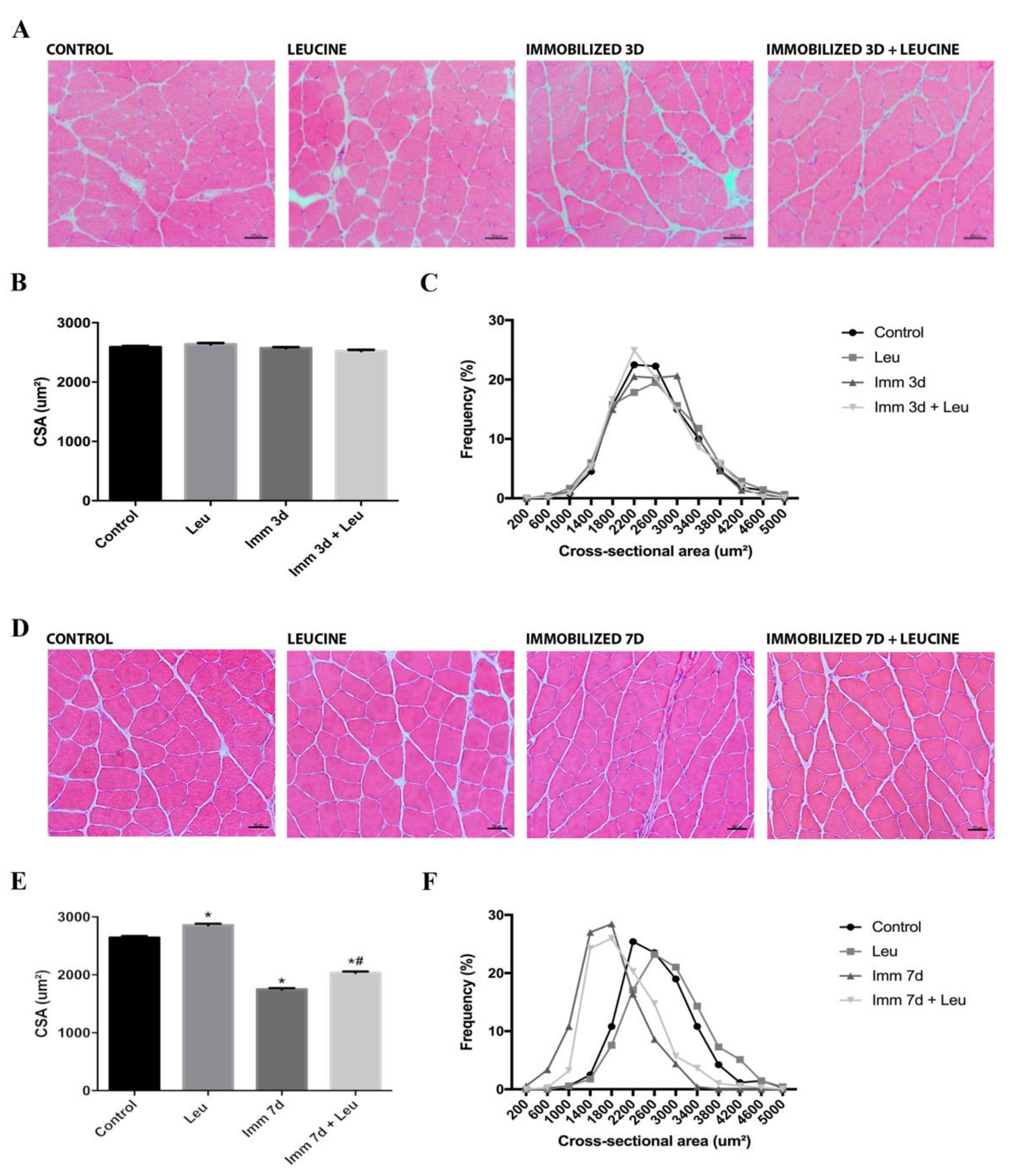
2.4. Cross-Sectional Area (CSA) Analysis
2.5. Dry Weight
2.6. Immunofluorescence
2.7. Western Blotting
2.8. RNA Extraction and Real-Time PCR
2.9. Statistical Analysis
3. Results
3.1. Biometric Measurements
3.2. Hindlimb Immobilization Induced Skeletal Muscle Atrophy, Which Was Attenuated by Leucine Supplementation
3.3. RNA-Seq Analysis and HDAC4 mRNA Expression Validation for 3 and 7 Days of Hindlimb Immobilization
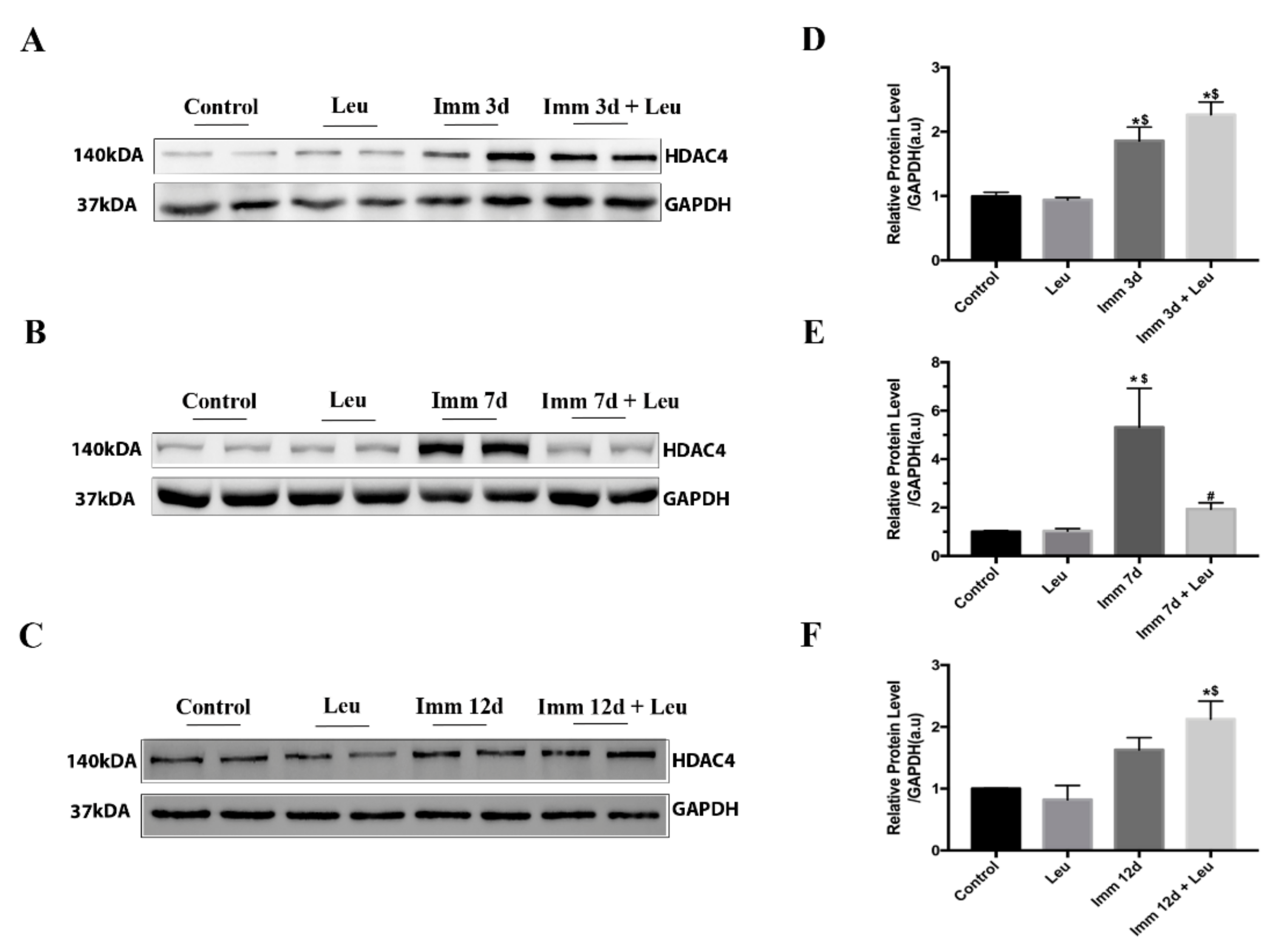
3.4. Leucine Downregulates HDAC4 Protein Expression at 7 Days of Hindlimb Immobilization
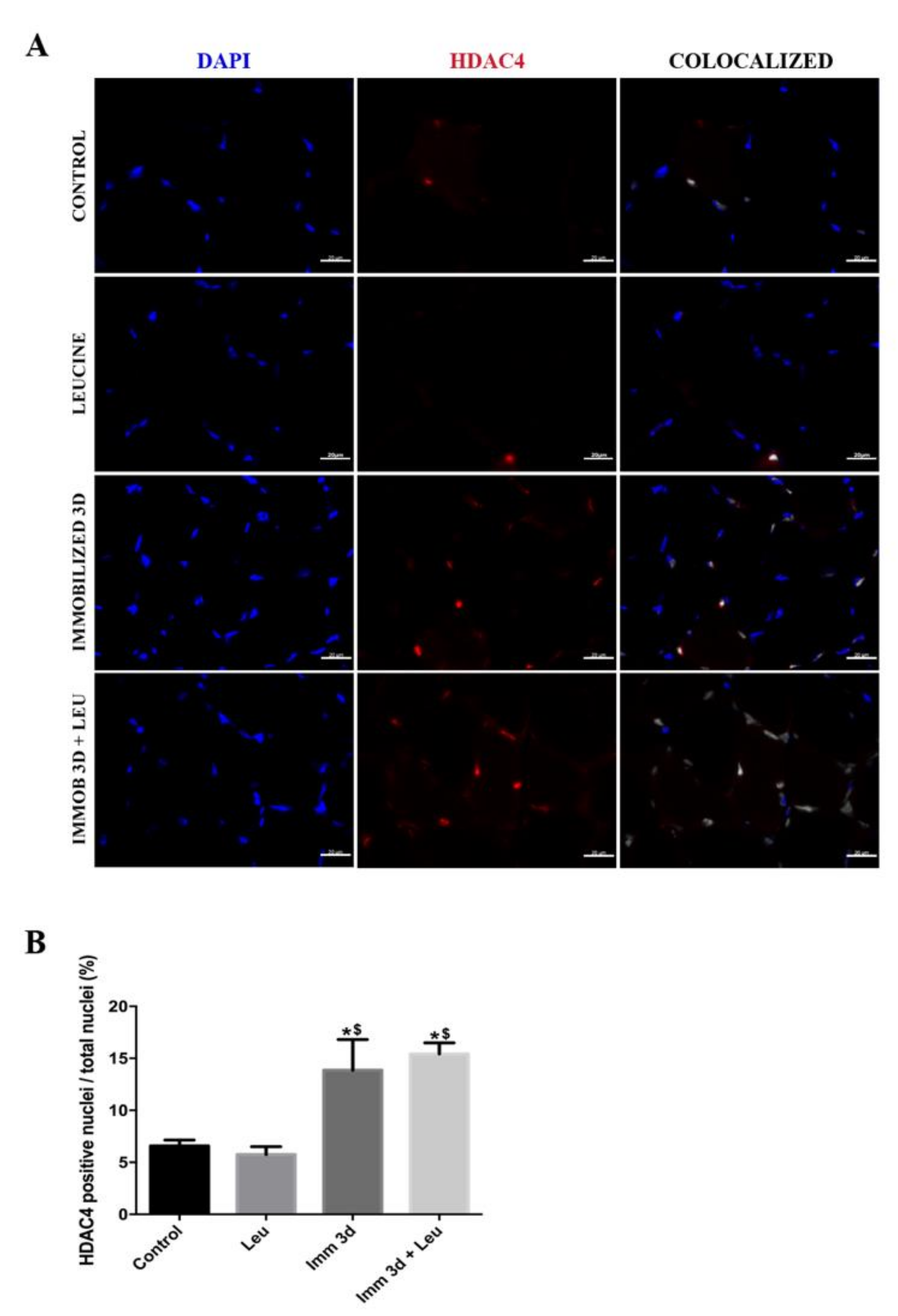
3.5. Leucine Reduces the Percentage of HDAC4 Positive Nuclei Accumulation Induced by Immobilization
3.6. Impact of Leucine on Gene Expression of HDAC4 Pathway-Related Components during 7 Days of Hindlimb Immobilization
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Margolis, L.M.; Rivas, D.A. Implications of Exercise Training and Distribution of Protein Intake on Molecular Processes Regulating Skeletal Muscle Plasticity. Behav. Genet. 2015, 45, 211–221. [Google Scholar] [CrossRef]
- Glass, D.J. Signalling pathways that mediate skeletal muscle hypertrophy and atrophy. Nat. Cell Biol. 2003, 5, 87–90. [Google Scholar] [CrossRef]
- Yoshihara, T.; Machida, S.; Kurosaka, Y.; Kakigi, R.; Sugiura, T.; Naito, H. Immobilization induces nuclear accumulation of HDAC4 in rat skeletal muscle. J. Physiol. Sci. 2016, 66, 337–343. [Google Scholar] [CrossRef]
- Baptista, I.L.; Silva, W.J.; Artioli, G.G.; Guilherme, J.P.L.F.; Leal, M.L.; Aoki, M.S.; Miyabara, E.H.; Moriscot, A.S. Leucine and HMB Differentially Modulate Proteasome System in Skeletal Muscle under Different Sarcopenic Conditions. PLoS ONE 2013, 8, e76752. [Google Scholar] [CrossRef]
- Cadena, S.M.; Zhang, Y.; Fang, J.; Brachat, S.; Kuss, P.; Giorgetti, E.; Stodieck, L.S.; Kneissel, M.; Glass, D.J. Skeletal muscle in MuRF1 null mice is not spared in low-gravity conditions, indicating atrophy proceeds by unique mechanisms in space. Sci. Rep. 2019, 9, 1–11. [Google Scholar] [CrossRef]
- Dirks, M.L.; Wall, B.T.; Van De Valk, B.; Holloway, T.M.; Holloway, G.P.; Chabowski, A.; Goossens, G.H.; Van Loon, L.J. One week of bed rest leads to substantial muscle atrophy and induces whole-body insulin resistance in the absence of skeletal muscle lipid accumulation. Diabetes 2016, 65, 2862–2875. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.-F.; Zhang, Y.; Li, B.; Chen, N. Physical inactivity induces the atrophy of skeletal muscle of rats through activating AMPK/FoxO3 signal pathway. Eur. Rev. Med. Pharmacol. Sci. 2018, 22, 199–209. [Google Scholar]
- Huang, Z.; Fang, Q.; Ma, W.; Zhang, Q.; Qiu, J.; Gu, X.; Yang, H.; Sun, H. Skeletal muscle atrophy was alleviated by salidroside through suppressing oxidative stress and inflammation during denervation. Front. Pharmacol. 2019, 10, 1–15. [Google Scholar] [CrossRef]
- Yamada, T.; Ashida, Y.; Tatebayashi, D.; Abe, M.; Himori, K. Cancer Cachexia Induces Preferential Skeletal Muscle Myosin Loss When Combined With Denervation. Front. Physiol. 2020, 11, 1–9. [Google Scholar] [CrossRef]
- Sato, A.Y.; Richardson, D.; Cregor, M.; Davis, H.M.; Au, E.D.; McAndrews, K.; Zimmers, T.A.; Organ, J.M.; Peacock, M.; Plotkin, L.I.; et al. Glucocorticoids induce bone and muscle atrophy by tissue-specific mechanisms upstream of E3 ubiquitin ligases. Endocrinology 2017, 158, 664–677. [Google Scholar] [CrossRef]
- Londhe, P.; Guttridge, D.C. Inflammation induced loss of skeletal muscle. Bone 2015, 80, 131–142. [Google Scholar] [CrossRef] [Green Version]
- Solomon, V.; Lecker, S.H.; Goldberg, A.L. The n-end rule pathway catalyzes a major fraction of the protein degradation in skeletal muscle. J. Biol. Chem. 1998, 273, 25216–25222. [Google Scholar] [CrossRef] [Green Version]
- Myung, J.; Kim, K.B.; Crews, C.M. The ubiquitin-proteasome pathway and proteasome inhibitors. Med. Res. Rev. 2001, 21, 245–273. [Google Scholar] [CrossRef]
- Rock, K.L.; Gramm, C.; Rothstein, L.; Clark, K.; Stein, R.; Dick, L.; Hwang, D.; Goldberg, A.L. Inhibitors of the proteasome block the degradation of most cell proteins and the generation of peptides presented on MHC class I molecules. Cell 1994, 78, 761–771. [Google Scholar] [CrossRef]
- Gomes, M.D.; Lecker, S.H.; Jagoe, R.T.; Navon, A.; Goldberg, A.L. Atrogin-1, a muscle-specific F-box protein highly expressed during muscle atrophy. Proc. Natl. Acad. Sci. USA 2001, 98, 14440–14445. [Google Scholar] [CrossRef] [Green Version]
- Bodine, S.C.; Latres, E.; Baumhueter, S.; Lai, V.K.M.; Nunez, L.; Clarke, B.A.; Poueymirou, W.T.; Panaro, F.J.; Erqian, N.; Dharmarajan, K.; et al. Identification of ubiquitin ligases required for skeletal Muscle Atrophy. Science 2001, 294, 1704–1708. [Google Scholar] [CrossRef]
- Bodine, S.C.; Baehr, L.M. Skeletal muscle atrophy and the E3 ubiquitin ligases MuRF1 and MAFbx/atrogin-1. Am. J. Physiol. Endocrinol. Metab. 2014, 307, E469–E484. [Google Scholar] [CrossRef] [Green Version]
- Bolster, D.R.; Vary, T.C.; Kimball, S.R.; Jefferson, L.S. Leucine regulates translation initiation in rat skeletal muscle via enhanced eIF4G phosphorylation. J. Nutr. 2004, 134, 1704–1710. [Google Scholar] [CrossRef] [Green Version]
- Pereira, M.G.; Silva, M.T.; Carlassara, E.O.C.; Gonçalves, D.A.; Abrahamsohn, P.A.; Kettelhut, I.C.; Moriscot, A.S.; Aoki, M.S.; Miyabara, E.H. Leucine supplementation accelerates connective tissue repair of injured tibialis anterior muscle. Nutrients 2014, 6, 3981–4001. [Google Scholar] [CrossRef] [Green Version]
- Nagasawa, T.; Kido, T.; Yoshizawa, F.; Ito, Y.; Nishizawa, N. Rapid suppression of protein degradation in skeletal muscle after oral feeding of leucine in rats. J. Nutr. Biochem. 2002, 13, 121–127. [Google Scholar] [CrossRef]
- Sugawara, T.; Ito, Y.; Nishizawa, N.; Nagasawa, T. Supplementation with dietary leucine to a protein-deficient diet suppresses myofibrillar protein degradation in rats. J. Nutr. Sci. Vitaminol. 2007, 53, 552–555. [Google Scholar] [CrossRef] [Green Version]
- Cohen, T.J.; Waddell, D.S.; Barrientos, T.; Lu, Z.; Feng, G.; Cox, G.A.; Bodine, S.C.; Yao, T.P. The histone deacetylase HDAC4 connects neural activity to muscle transcriptional reprogramming. J. Biol. Chem. 2007, 282, 33752–33759. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Macpherson, P.; Marvin, M.; Meadows, E.; Klein, W.H.; Yang, X.-J.; Goldman, D. A Histone Deacetylase 4/Myogenin Positive Feedback Loop Coordinates Denervation-dependent Gene Induction and Suppression. Mol. Biol. Cell 2009, 20, 1120–1131. [Google Scholar] [CrossRef] [Green Version]
- Tang, H.; Goldman, D. Activity-dependent gene regulation in skeletal muscle is mediated by a histone deacetylase (HDAC)-Dach2-myogenin signal transduction cascade. Proc. Natl. Acad. Sci. USA 2006, 103, 16977–16982. [Google Scholar] [CrossRef] [Green Version]
- Moresi, V.; Williams, A.H.; Meadows, E.; Flynn, J.M.; Potthoff, M.J.; McAnally, J.; Shelton, J.M.; Backs, J.; Klein, W.H.; Richardson, J.A.; et al. Myogenin and class II HDACs control neurogenic muscle atrophy by inducing E3 ubiquitin ligases. Cell 2010, 143, 35–45. [Google Scholar] [CrossRef] [Green Version]
- Bricceno, K.V.; Sampognaro, P.J.; Van meerbeke, J.P.; Sumner, C.J.; Fischbeck, K.H.; Burnett, B.G. Histone deacetylase inhibition suppresses myogenin-dependent atrogene activation in spinal muscular atrophy mice. Hum. Mol. Genet. 2012, 21, 4448–4459. [Google Scholar] [CrossRef] [Green Version]
- Walsh, M.E.; Bhattacharya, A.; Sataranatarajan, K.; Qaisar, R.; Sloane, L.; Rahman, M.M.; Kinter, M.; Van Remmen, H. The histone deacetylase inhibitor butyrate improves metabolism and reduces muscle atrophy during aging. Aging Cell 2015, 14, 957–970. [Google Scholar] [CrossRef]
- Kim, D.; Paggi, J.M.; Park, C.; Bennett, C.; Salzberg, S.L. Graph-based genome alignment and genotyping with HISAT2 and HISAT-genotype. Nat. Biotechnol. 2019, 37, 907–915. [Google Scholar] [CrossRef]
- Kim, D.; Langmead, B.; Salzberg, S.L. HISAT: A fast spliced aligner with low memory requirements. Nat. Methods 2015, 12, 357–360. [Google Scholar] [CrossRef] [Green Version]
- Pertea, M.; Kim, D.; Pertea, G.M.; Leek, J.T.; Salzberg, S.L. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat. Protoc. 2016, 11, 1650–1667. [Google Scholar] [CrossRef]
- Liao, Y.; Smyth, G.K.; Shi, W. FeatureCounts: An efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014, 30, 923–930. [Google Scholar] [CrossRef] [Green Version]
- Robinson, M.D.; McCarthy, D.J.; Smyth, G.K. edgeR: A Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 2010, 26, 139–140. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, D.J.; Chen, Y.; Smyth, G.K. Differential expression analysis of multifactor RNA-Seq experiments with respect to biological variation. Nucleic Acids Res. 2012, 40, 4288–4297. [Google Scholar] [CrossRef] [Green Version]
- Gissel, H. Effects of varying pulse parameters on ion homeostasis, cellular integrity, and force following electroporation of rat muscle in vivo. Am. J. Physiol. Regul. Integr. Comp. Physiol. 2010, 298, 918–929. [Google Scholar] [CrossRef] [Green Version]
- Weaver, K.F.; Morales, V.C.; Dunn, S.L.; Godde, K.; Weaver, P.F. Kruskal—Wallis. In An Introduction to Statistical Analysis in Research; Wiley Online Books: Hoboken, NJ, USA, 2017; pp. 353–391. ISBN 9781119454205. [Google Scholar]
- Baptista, I.L.; Silvestre, J.G.; Silva, W.J.; Labeit, S.; Moriscot, A.S. FoxO3a suppression and VPS34 activity are essential to anti-atrophic effects of leucine in skeletal muscle. Cell Tissue Res. 2017, 369, 381–394. [Google Scholar] [CrossRef]
- Baptista, I.L.; Leal, M.L.; Artioli, G.G.; Aoki, M.S.; Fiamoncini, J.; Turri, A.O.; Curi, R.; Miyabara, E.H.; Moriscot, A.S. Leucine attenuates skeletal muscle wasting via inhibition of ubiquitin ligases. Muscle Nerve 2010, 41, 800–808. [Google Scholar] [CrossRef]
- He, L.; Zhang, J.; Zhao, J.; Ma, N.; Kim, S.W.; Qiao, S.; Ma, X. Autophagy: The last defense against cellular nutritional stress. Adv. Nutr. 2018, 9, 493–504. [Google Scholar] [CrossRef] [Green Version]
- Segalés, J.; Perdiguero, E.; Serrano, A.L.; Sousa-Victor, P.; Ortet, L.; Jardí, M.; Budanov, A.V.; Garcia-Prat, L.; Sandri, M.; Thomson, D.M.; et al. Sestrin prevents atrophy of disused and aging muscles by integrating anabolic and catabolic signals. Nat. Commun. 2020, 11, 1–13. [Google Scholar] [CrossRef]
- Castets, P.; Rion, N.; Théodore, M.; Falcetta, D.; Lin, S.; Reischl, M.; Wild, F.; Guérard, L.; Eickhorst, C.; Brockhoff, M.; et al. mTORC1 and PKB/Akt control the muscle response to denervation by regulating autophagy and HDAC4. Nat. Commun. 2019, 10, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Yoon, M.S.; Son, K.; Arauz, E.; Han, J.M.; Kim, S.; Chen, J. Leucyl-tRNA Synthetase Activates Vps34 in Amino Acid-Sensing mTORC1 Signaling. Cell Rep. 2016, 16, 1510–1517. [Google Scholar] [CrossRef] [Green Version]
- McKinsey, T.A.; Zhang, C.; Lu, J.; Olson, E.N. Signal-dependent nuclear export of a histone deacetylase regulates muscle differentiation. Nature 2000, 408, 106–111. [Google Scholar] [CrossRef]
- Backs, J.; Backs, T.; Bezprozvannaya, S.; McKinsey, T.A.; Olson, E.N. Histone Deacetylase 5 Acquires Calcium/Calmodulin-Dependent Kinase II Responsiveness by Oligomerization with Histone Deacetylase 4. Mol. Cell. Biol. 2008, 28, 3437–3445. [Google Scholar] [CrossRef] [Green Version]
- Al-Shanti, N.; Stewart, C.E. Ca2+/calmodulin-dependent transcriptional pathways: Potential mediators of skeletal muscle growth and development. Biol. Rev. 2009, 84, 637–652. [Google Scholar] [CrossRef]
- Ispoglou, T.; White, H.; Preston, T.; McElhone, S.; McKenna, J.; Hind, K. Double-blind, placebo-controlled pilot trial of L-Leucine-enriched amino-acid mixtures on body composition and physical performance in men and women aged 65-75 years. Eur. J. Clin. Nutr. 2016, 70, 182–188. [Google Scholar] [CrossRef] [PubMed]
- English, K.L.; Mettler, J.A.; Ellison, J.B.; Mamerow, M.M.; Arentson-Lantz, E.; Pattarini, J.M.; Ploutz-Snyder, R.; Sheffield-Moore, M.; Paddon-Jones, D. Leucine partially protects muscle mass and function during bed rest in middle-aged adults. Am. J. Clin. Nutr. 2016, 103, 465–473. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolfe, R.R. Branched-chain amino acids and muscle protein synthesis in humans: Myth or reality? J. Int. Soc. Sports Nutr. 2017, 14, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Mitchell, W.K.; Phillips, B.E.; Hill, I.; Greenhaff, P.; Lund, J.N.; Williams, J.P.; Rankin, D.; Wilkinson, D.J.; Smith, K.; Atherton, P.J. Human skeletal muscle is refractory to the anabolic effects of leucine during the postprandial muscle-full period in older men. Clin. Sci. 2017, 131, 2643–2653. [Google Scholar] [CrossRef] [Green Version]
- Backx, E.M.P.; Horstman, A.M.H.; Marzuca-Nassr, G.N.; van Kranenburg, J.; Smeets, J.S.; Fuchs, C.J.; Janssen, A.A.W.; de Groot, L.C.P.G.M.; Snijders, T.; Verdijk, L.B.; et al. Leucine supplementation does not attenuate skeletal muscle loss during leg immobilization in healthy, young men. Nutrients 2018, 10, 635. [Google Scholar] [CrossRef] [Green Version]



| Biometric Feature | Control | Leucine | Immobilized | Immobilized + Leucine | ||||
|---|---|---|---|---|---|---|---|---|
| 3 Days | 7 Days | 3 Days | 7 Days | 3 Days | 7 Days | 3 Days | 7 Days | |
| Body Weight (g) | 297.4 ± 34.7 | 361.2 ± 44.2 | 280.0 ± 20.8 | 342.7 ± 45.8 | 282.8 ± 23.3 | 302.9 ± 48.6 | 280.3 ± 31.6 | 312.1 ± 20.0 |
| Soleus Dry Weight/Tibia (mg/cm) | 7.69 ± 0.69 | 6.11 ± 0.1 | 6.58 ± 0.91 | 7.05 ± 0.7 | 5.91 ± 0.68 * | 3.42 ± 0.1 * | 6.22 ± 0.53 | 4.38 ± 0.2 *,# |
| Whole Area of Soleus Muscle (mm2) | 8.50 ± 1.49 | 9.57 ± 0.28 | 8.53 ± 1.09 | 8.45 ± 0.98 | 7.20 ± 1.68 | 5.33 ± 0.52 * | 7.83 ± 0.87 | 7.75 ± 1.30 # |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, P.K.N.; Cruz, A.; Silva, W.J.; Labeit, S.; Moriscot, A.S. Leucine Supplementation Decreases HDAC4 Expression and Nuclear Localization in Skeletal Muscle Fiber of Rats Submitted to Hindlimb Immobilization. Cells 2020, 9, 2582. https://doi.org/10.3390/cells9122582
Alves PKN, Cruz A, Silva WJ, Labeit S, Moriscot AS. Leucine Supplementation Decreases HDAC4 Expression and Nuclear Localization in Skeletal Muscle Fiber of Rats Submitted to Hindlimb Immobilization. Cells. 2020; 9(12):2582. https://doi.org/10.3390/cells9122582
Chicago/Turabian StyleAlves, Paula K. N., André Cruz, William J. Silva, Siegfried Labeit, and Anselmo S. Moriscot. 2020. "Leucine Supplementation Decreases HDAC4 Expression and Nuclear Localization in Skeletal Muscle Fiber of Rats Submitted to Hindlimb Immobilization" Cells 9, no. 12: 2582. https://doi.org/10.3390/cells9122582






