Abstract
PRDI-BF1 (positive regulatory domain I-binding factor 1) and RIZ1 (retinoblastoma protein-interacting zinc finger gene 1) (PR) homologous domain containing (PRDM) transcription factors are expressed in neuronal and stem cell systems, and they exert multiple functions in a spatiotemporal manner. Therefore, it is believed that PRDM factors cooperate with a number of protein partners to regulate a critical set of genes required for maintenance of stem cell self-renewal and differentiation through genetic and epigenetic mechanisms. In this review, we summarize recent findings about the expression of PRDM factors and function in stem cell and neuronal systems with a focus on cofactor-dependent regulation of PRDM3/16 and FOG1/2. We put special attention on summarizing the effects of the PRDM proteins interaction with chromatin modulators (NuRD complex and CtBPs) on the stem cell characteristic and neuronal differentiation. Although PRDM factors are known to possess intrinsic enzyme activity, our literature analysis suggests that cofactor-dependent regulation of PRDM3/16 and FOG1/2 is also one of the important mechanisms to orchestrate bidirectional target gene regulation. Therefore, determining stem cell and neuronal-specific cofactors will help better understanding of PRDM3/16 and FOG1/2-controlled stem cell maintenance and neuronal differentiation. Finally, we discuss the clinical aspect of these PRDM factors in different diseases including cancer. Overall, this review will help further sharpen our knowledge of the function of the PRDM3/16 and FOG1/2 with hopes to open new research fields related to these factors in stem cell biology and neuroscience.
1. Introduction
PRDI-BF1 (positive regulatory domain I-binding factor 1) and RIZ1 (retinoblastoma protein-interacting zinc finger gene 1) (PR) homologous-domain-containing (PRDM) transcription factors have received considerable attention recently due to their importance in regulating the development and function of various tissues and organ systems. The PRDM protein family is a group of 19 poorly studied factors that are involved in a wide range of cellular processes [1,2,3,4]. The PR domain is associated with the catalytic SET (suppressor of variegation 3–9, enhancer of zeste and trithorax) domain, which possesses histone lysine methyltransferase (HMT) activity [5]. Although some PRDM proteins have not been shown to have intrinsic HMTase activity [6,7,8], several studies have confirmed that PRDM2, PRDM3, PRDM8, PRDM9, and PRDM16 possess this capability [9,10,11,12,13,14].
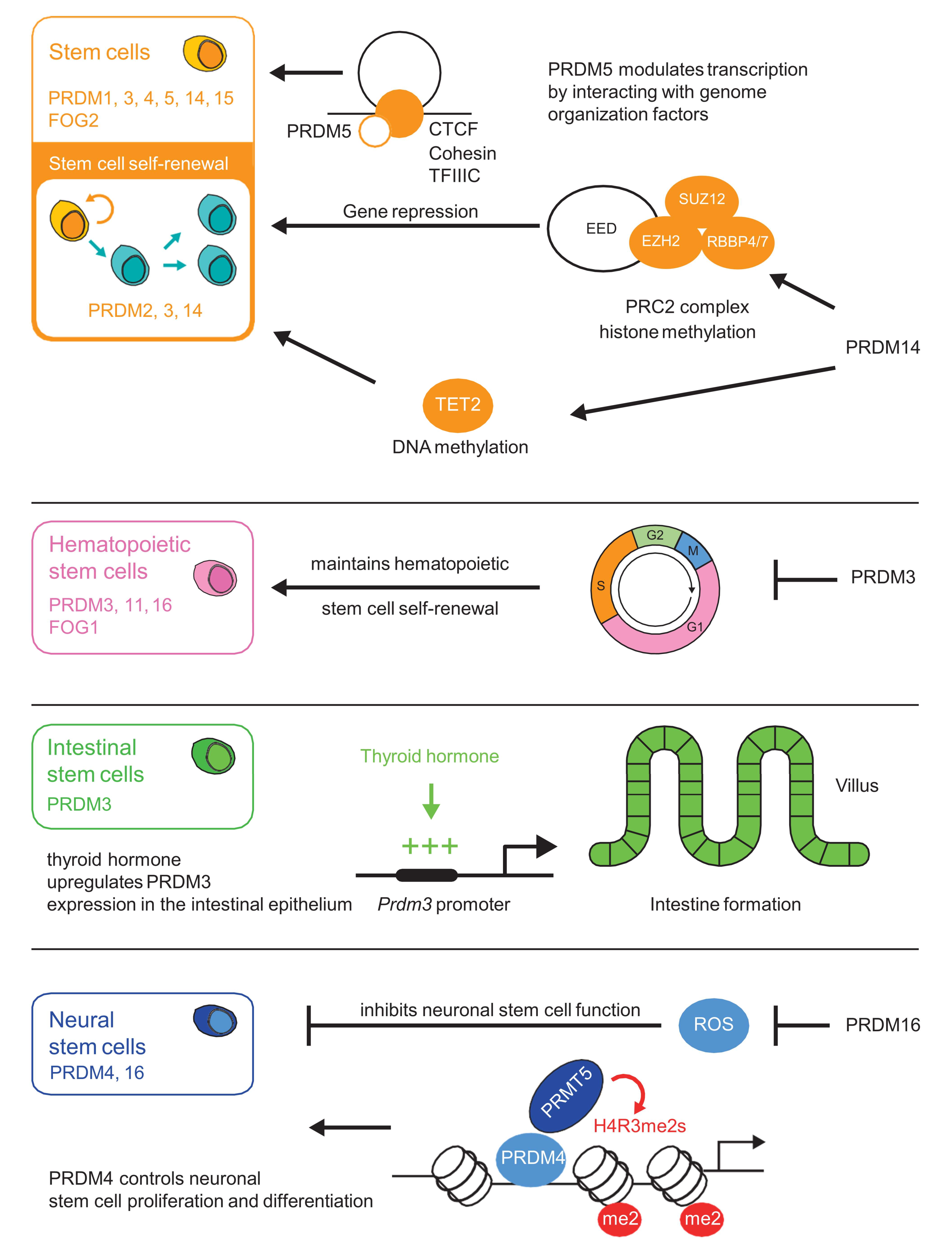
Depending on the cellular or tissue context, PRDM proteins mediate either transcriptional repression or activation. As several PRDM proteins appear to be enzymatically inactive, they achieve transcriptional regulation through interaction with transcription factors and histone-modifying enzymes. Interacting proteins include the Polycomb repressive complex 2 (PRC2), HMTs, histone acetyltransferases (HATs), histone deacetylases (HDACs), protein arginine N-methyltransferase 5 (PRMT5), and lysine-specific demethylase 1 (LSD1) [7,10,15,16,17,18,19,20]. For instance, the interplay between PRDM3 and the Suv39H1 HMT [21] leads to gene repression through H3K9 methylation. PRDM1, PRDM5, PRDM6 and PRDM12 are also known to interact with G9a HMT and repress gene expression through methylation of H3 lysine 9 [6,7,8,12,22]. PRDM proteins are involved in several developmental processes such as stem cell maintenance (Figure 1, Table 1), hematopoiesis, and adipogenesis [12,23]. Recent studies have highlighted the importance of these factors during neuronal development [24,25,26], including brain or spinal cord formation [26,27].
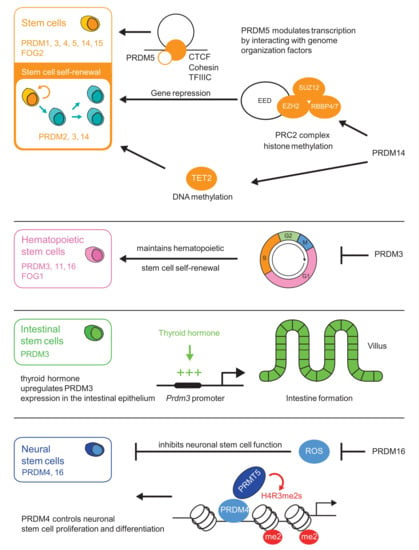
Figure 1.
PRDI-BF1 (positive regulatory domain I-binding factor 1) and RIZ1 (retinoblastoma protein-interacting zinc finger gene 1) (PR) homologous domain containing (PRDM) factors play important roles in stem cell maintenance. PRDM3 and PRDM16 exhibit a crucial regulatory role in hematopoietic stem cell (HSC) and progenitor cell maintenance during fetal development [28,29,30,31,32,33]. PRDM1 determines the fate of embryonic stem cells and their progenitors [34,35]. PRDM14 plays an important role in governing the gene machinery responsible for maintaining the pluripotent state of embryonic stem cells. PRDM14 reprograms somatic cells to induce pluripotent stem cells through epigenetic pathways [36,37]. PRDM15 is also a transcriptional regulator of key genes involved in the maintenance of naive pluripotency of embryonic stem cells [38].

Table 1.
Roles of PRDM factors in stem cell system.
The PR domain is followed by repeated zinc fingers (proline-rich domains) mediating sequence- specific DNA binding and protein-protein interactions with other histone-modifying enzymes, and plays a role in nuclear import [23,64,65,66,67,68,69]. PRDM3 and PRDM16 display 63% nucleotide and 56% amino acid homology [70]. They exhibit intrinsic HMT activity towards histone 3 lysine 9 (H3K9), a mark typically associated with repressed transcription [9]. Other in vitro studies show that PRDM3 and 16 are involved in gene expression activation via methylation of histone 3 lysine 4 (H3K4) [9,24,71,72]. An additional pathway by which PRDM3 and PRDM16 govern gene expression through modification of chromatin structure is via the formation of protein complexes, such as the CtBP and NuRD [73,74,75]. Interestingly, FOG proteins, recently defined as PRDM factors, have also been confirmed to exert their function through the interaction with the NuRD complex and the CtBP protein [76,77,78], and it has been proposed that these factors may control cell fate decisions in stem cells and neuronal cells through the CtBP and NuRD complexes in a similar fashion to PRDM3 and PRDM16 [73,79]. In this review we will discuss the possible mechanism for their function in stem cell and neuronal systems through the interaction with their cofactors.
2. PRDM Factors Are Substantial Players in Stem Cells
In recent years, the number of studies on the role of PRDM proteins in stem cells and cell differentiation has increased significantly. In this section we discuss studies on the role of PRDM proteins in maintaining self-renewal and pluripotency of stem cells, and in the neuronal system with potential molecular mechanisms that regulate the action of PRDM proteins. Highly pluripotent stem cells derived from the inner cell mass (ICM) are generated from precursors with high PRDM1 expression. Moreover, PRDM1-positive cells display a gene expression profile associated with the early specification of embryonic cells toward germ cell identity [35]. Additionally, the silencing of PRDM1 in human embryonic stem cells (hESCs) changes germline potential and directs cell differentiation towards neuronal specification by increasing SOX2 expression [80], suggesting that PRDM1 acts as a switch for neural or germline cell fate by inhibiting SOX2 expression during human development. The pluripotent state in stem cells can also be controlled by epigenetic regulation. PRDM14 is an important player regulating the epigenetic state and the transcription network in stem cells [56,81]. A genome-wide RNAi screen study revealed that PRDM14 colocalizes with stemness transcription factors such as OCT3/4, SOX2 and NANOG to maintain stem cell identity [37]. Moreover, it has been shown that the recruitment of OCT3/4 to the demethylated regulatory regions of pluripotency genes is driven by PRDM14 [36]. PRDM14-dependent pluripotency is mediated by reducing Dnmt3a/b and Dnmt3l expression and globally correlates with the CpG methylation landscape [55,82,83]. PRDM14 also maintains an active DNA demethylation status in the embryonic stem cells via TET (ten-eleven translocation) proteins [54]. These results suggest that the demethylation status observed during the induction of pluripotency is controlled by PRDM14. Besides PRDM14, PRDM15 was shown to be highly expressed in the embryo inner cell mass (ICM) [84] and plays a role as a safeguard of pluripotency in stem cells by regulating MAPK–ERK and WNT signaling [38]. Stimulation of the MAPK–ERK pathway triggers ESC lineage commitment [85], whereas the WNT pathway prevents differentiation of embryonic stem cells [86]. Stem cells with PRDM15 depletion showed a marked rise in nucleosome occupancy along with increased methylation and decreased acetylation at lysine 27 on histone H3 (H3K27ac to H3K27me3) at the promoter region of Rspo1 (R-spondin 1) and Spry1 (protein sprouty homolog 1), which are regulators of WNT signaling and MAPK–ERK pathways [38]. In order to maintain pluripotency and self-renewal of ESC, PRDM15 increases Spry1 and Rspo1 expression by decreasing nucleosome occupancy at the promoter sequence and allows RNA polymerase II recruitment [38].
3. An Overview of the Roles of PRDM Factors in the Neuronal System
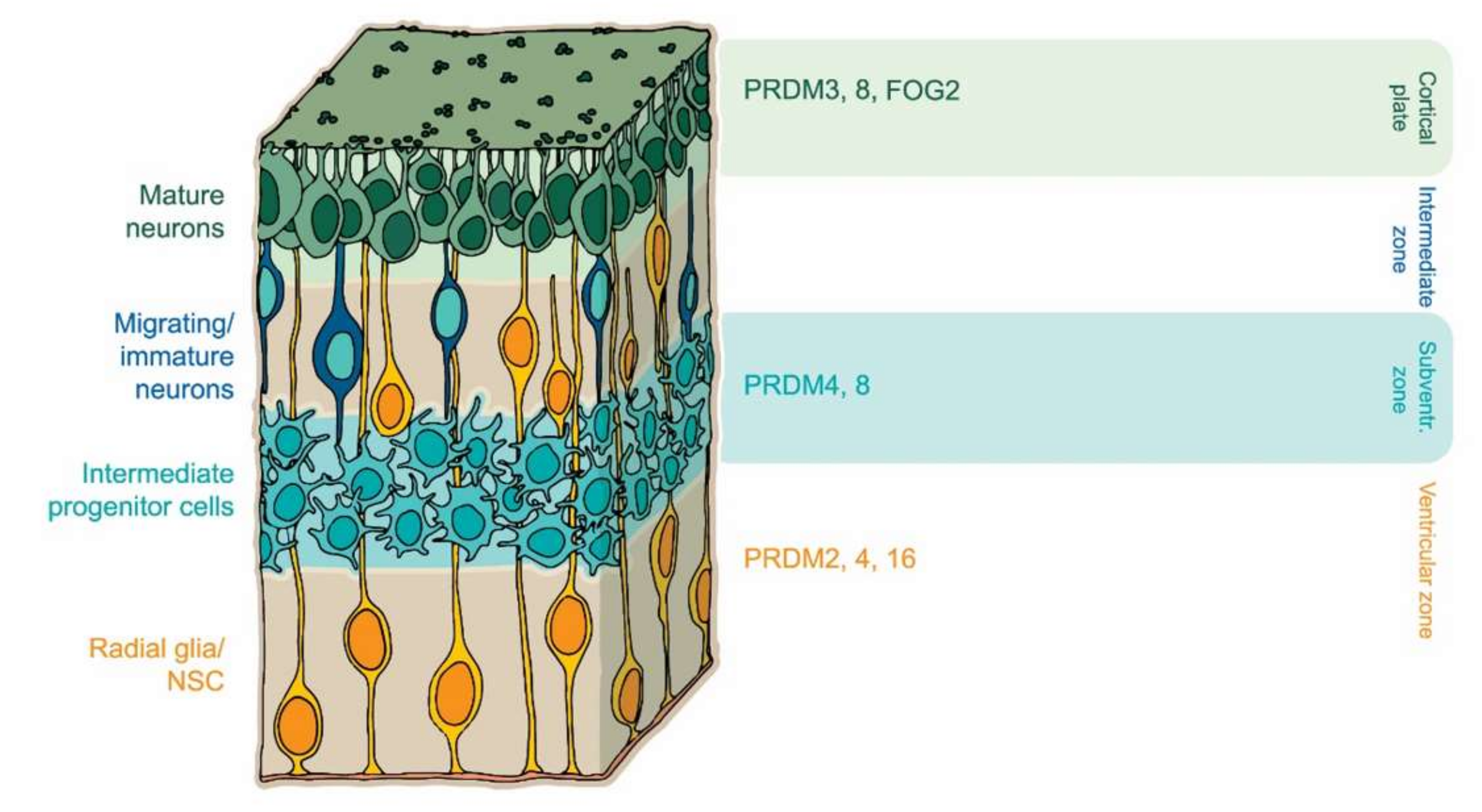
Newly generated cortical neurons are derived from the division of radial glia progenitors in the ventricular zone (VZ). During neuronal maturation, progenitor cells move from VZ to the subventricular zone (SVZ) and then to the intermediate zone (IZ), where they adopt multipolar morphology (MP). In the subplate (SP) zone they acquire a bipolar shape and then settle in the cortical plate (CP) as mature neurons with defined characteristics [87,88]. Role of the PRDM factors and their expression in central nervous system (CNS) is depicted in Figure 2, Table 2. PRDM8 was shown to play a role in the development of brain structures. High expression of PRDM8 is found in the upper part of the IZ where it regulates the transition from multipolar to bipolar morphology of cortical neurons [89]. Moreover, mice carrying a Prdm8 gene deletion displayed a significant reduction of brain growth along with a decreased number of neocortical neurons, indicating its essential role in neocortical development [26]. It has also been shown that basic helix-loop-helix family member E22 (BHLHB5), another cofactor of PRDM8, can significantly influence the formation of neuronal plasticity. PRDM8 and BHLHB5 form a repressor complex that orchestrates the neuronal circuit [90]. Mice with PRDM8 or BHLHB5 deficiency display highly similar axonal mistargeting and behavioral abnormalities [90], indicating the inherent spatiotemporal role of these factors in the formation of the nervous system. Much like their functions in stem cell maintenance, PRDM proteins tune progenitors of neuronal proliferation and differentiation through epigenetic modifications. For instance, PRDM4 is part of an epigenetic complex regulating the proliferative capacity and modulating cell cycle progression in neural stem cells (NSCs). PRDM4 interacts with PRMT5 through the PR/SET domain and the latter modifies chromatin structure by symmetric dimethylation on arginine 3 of histone H4 (H4R3me2s), specifically in the undifferentiated NSCs. Furthermore, a decrease in PRDM4 expression in NSC demonstrated precocious neurogenesis [50].
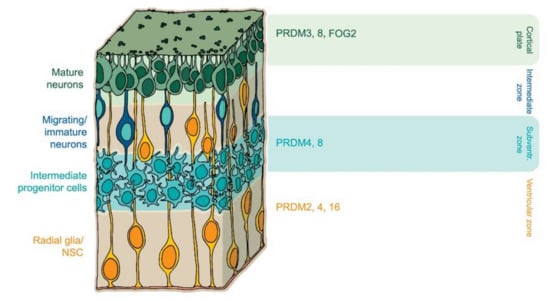
Figure 2.
PRDM factors play multiple roles during central nervous system (CNS) development. PRDM16 controls the migration and differentiation of neuronal progenitors during cortical development [24,25]. FOG2 arranges a neuronal subtype identity [93] whereas PRDM8 controls axonal targeting [90].

Table 2.
The roles of PRDM factors in the neuronal system.
It is interesting to note that PRDM12 is involved in the development of nociceptors. Mutation in human PRDM12 has been found in patients with congenital pain insensitivity (CIP) [91]. This mutation was found to be located in His289 of PRDM12 and it disrupts the interaction between PRDM12 and HMT G9a (EHMT2). PRDM12 has been reported to interact with G9a (EHMT2) and leads to dimethylation of histone H3 on lysine 9 (H3K9me2) in P19 cells [22]. On the other hand, it was shown that PRDM12 ablation negatively affects the Ngn1/2-TrkA pathway which interferes with nociceptor maturation [92]. These observations suggest that PRDM12 functions through epigenetic mechanisms and could serve as a molecular target in the therapeutic treatment of pain.
4. The Function of PRDM3 and 16 in Stem Cells
Among the PRDM factors, emerging roles of PRDM3 and PRDM16 in diverse systems, including stem cells and the neuronal system, have been proposed [25,33,46,103]. Mouse embryos with Prdm3 deletion exhibit various developmental defects and caused lethality at 10.5 days post coitum [100]. This is mainly attributed to impaired hematopoietic stem cell (HSC) proliferation. PRDM3 regulates hematopoietic stem cell proliferation by activating the Gata2 promoter [46], and Gata2 gene expression is greatly reduced in Prdm3 mutant mice [100]. Interestingly, mice lacking GATA2 expression showed a phenotype similar to PRDM3-deficient embryos. This suggests that these factors cooperate in HSC gene regulation [46,116]. Moreover, PRDM3 plays important roles in long-term HSC self-renewal [45]. These findings suggest that PRDM3 is one of the major regulators of GATA2 gene expression and that it is also important for HSC maintenance. In Xenopus tropicalis, Prdm3 is required for adult intestinal formation and for the maintenance of epithelial stem cell proliferation [28]. PRDM3 has been reported as a histone 3 lysine 9 mono-methyltransferase (H3K9me1) in mammals and controls heterochromatin integrity [9]. In zebrafish, Prdm3 controls the chromatin landscape by influencing H3K9me3 and H3K4me3 marks. Furthermore, Prdm3-deficient zebrafish embryos display a 40% reduction in H3K4me3 marks [117]. These findings imply that PRDM3 plays a crucial role in stem cell self-renewal and differentiation through histone methylation. Further investigation to elucidate how PRDM3 impacts global methylation patterns to regulate the stem cell state is warranted.
Much like Prdm3, Prdm16 expression is required for HSC maintenance [32,33]. PRDM16 is also responsible for many developmental processes in brown fat tissue [118,119], heart [120] and craniofacial formation [121,122,123]. Prdm16 deletion in mice induces dysregulation of HSC renewal and increases apoptosis [32]. Moreover, mutation of Prdm16 causes a disturbance in gene expression related to hematopoietic stem cell function [32]. As mentioned previously, studies in mouse embryonic fibroblasts have identified that PRDM3 is an H3K9me1 methyltransferase. PRDM16 displays similar characteristics to PRDM3. Both proteins methylate histone H3 in the cytoplasm. SUV39H1 and SUV39H2 enzymes then convert H3K9me1 to H3K9me3 in the nucleus. These modifications reinforce the heterochromatin to be assembled in pericentric DNA and the nuclear lamina [9]. The integrity of heterochromatin is important for spatial genome organization and gene expression programs [9]. Thus, PRDM16 could be involved in the modification of epigenetic markers, thereby regulating the differentiation of stem cells and progenitor cells. Indeed, this phenomenon is observed in a myoblast differentiation model, suggesting a significant role for PRDM16 in cellular transformation. Ectopic overexpression of Prdm16 reprograms C2C12 myoblasts into brown fat cells [119]. Direct reprogramming induced by PRDM16 is accompanied by hypermethylation of myogenin and MyoD promoters [124]. Taken together, PRDM3 and PRDM16 both play important roles in stem cell maintenance in several systems through epigenetic mechanisms.
5. A Novel PRDM Factors, Friend of GATA (FOG) and its Function in Stem Cells
Recent studies have revealed that FOG1 is a PRDM family member [125]. FOG1/2 and PRDM3/16, carry a CtBP-binding sequence in their protein structure and repress transcription. Both proteins also contain a PR domain [64,125]. Historically, it was known that FOG1 regulates GATA1 transcription factor function and FOG2 governs GATA2 function. The interaction between the FOG family and GATA transcription factors is crucial in various tissues [126,127,128,129] where FOG proteins repress the transcriptional activity of GATA factors. Serious problems, such as failure in heart development, occur as a result of blocking the interaction between GATA4 and FOG2 [130]. Moreover, mutations in FOG2 hinder integration with GATA4 leading to congenital heart disease [131,132]. While FOG2 interacts with GATA4-6, little is known about its role in stem cells and the nervous system. Interestingly, much like AML1-PRDM3 [133], the AML1-FOG2 fusion protein have implications in myelodysplasia [134]. In this regard, the FOG2 protein may also regulate hematopoietic stem cell function. Human bone marrow mesenchymal stem cells (BM-MSC) are a heterogeneous population and only some have cardiomyogenic potential. BM-MSC subpopulations with high cardiomyogenic potential display high FOG2 gene expression [63]. This suggests that FOG2 is involved in cardiomyocyte progenitor cell function. Moreover, since the FOG family’s major role is to inhibit GATA factors, FOGs may be involved in regulatory processes in stem cells and progenitors in which GATA factors play major roles.
It is known that Fog1 deficiency is lethal for mice. Mouse embryos with this deficiency die between days 10.5 and 12.5 of gestation [135]. In these mice, erythropoiesis is highly disrupted and megakaryocytes are absent. Intriguingly, GATA1 acts as a hematopoietic transcription factor that induces erythrocyte and megakaryocyte differentiation. These findings suggest similar phenotypes in FOG1 and GATA1-deficiencies. Another study has clearly shown that GATA1 and FOG1 interaction is essential to promote megakaryocyte/erythrocyte lineage differentiation [135]. Moreover, FOG1 is considered a reprogramming factor that stimulates the stemness state in differentiated cells. In avian eosinophils, FOG1 overexpression leads to the dedifferentiation and generation of multipotent cells [136]. Furthermore, overexpression of Fog1 in mouse hematopoietic lineages resulted in a decreased number of eosinophils [137]. Thus, FOG1 is likely an important factor that controls the stem cell state and its function is tightly associated with GATA1.
6. The Function of PRDM3 and PRDM16 in Neuronal Cells
In Caenorhabditis elegans, it has been suggested that Egl-43 (the PRDM3 and PRDM16 orthologue in C. elegans) has a significant influence on nervous system development. During embryonic growth, two serotonergic hermaphrodite specific neurons migrate from the caudal position to the central part of the body. Disruption of Egl-43 gene expression stops neuronal migration and further development [99]. Follow-up reports in higher organisms highlighted the importance of PRDM3 in the formation of neuronal identity. The cellular specification of olfactory receptor neurons in Drosophila melanogaster is coordinated by a context-dependent response to Notch signaling. Hamlet (the PRDM3 and PRDM16 orthologue in Drosophila) mediates this pathway and contributes to the development of a specific type of neuron [103]. During the initiation of olfactory receptor neuron (ORN) development, Hamlet proteins erase the Notch-active state in differentiating cells. This phenomenon provides new insights into the Notch-dependent signaling pathway. The activity of Hamlet protein influences gene expression by altering the methylation profiles at promoter regions, histone packing density and chromatin organization. Hamlet alters chromatin accessibility by enabling Su(H) (suppressor of hairless protein) binding to Notch-specific enhancers. Mouse embryos with Prdm3 deletion displayed severe defects in nervous system development, but detailed studies on brain structures were not conducted [100]. In mammals, Prdm3 transcription is strongly activated by retinoic acid (RA) in murine embryonal carcinoma P19 cells [101]. Moreover, Prdm3 gene expression is upregulated in NSCs compared to human embryonic stem cells [138]. Additionally, ectopic overexpression of Prdm3 induces neurogenesis in P19 cells without RA stimulation [101]. However, the neuronal-specific role of PRDM3 remains to be addressed in mammals, and PRDM3 could be implicated in the onset of neurogenesis. Recently, our study showed that P19 cells with Prdm3 gene knock-out displayed earlier maturation of neurons along with the rapid proliferation of non-neuronal cells [102]. These findings strongly showed the significant role of PRDM3 in the formation of the mammalian nervous system in vivo. Chromatin structure and epigenetic modifications have been reported to be crucial for regulating gene expression during brain development [139]. In this regard, PRDM3 plays a role in the formation of synaptic plasticity via epigenetic regulation of gene transcription. Investigation of synaptic plasticity using an in vitro model demonstrated that PRDM3 is expressed in the nucleus of hippocampal neurons and may be implicated in neuronal activity associated with α-amino-3-hydroxy-5-methyl-4-isoxazolepropionic acid receptor (AMPAR) regulation [104]. miR-124 expression has been strongly associated with a homeostatic response during neuronal activity [104]. Interestingly, miR-124 transcription is directly dependent on the active complex of PRDM3 and HDAC1. Regulation of miR-124 expression in hippocampal and cortical neurons is partially explained by PRDM3-dependent reduction of mir-124 promoter activity [104]. It is known that gene expression in neurons highly depends on chromatin remodeling factors [140]. Therefore, the PRDM3–HDAC1 complex could be part of a bigger, specific chromatin remodeling complex involved in the fine-tuning of synaptic plasticity. PRDM3 activates genes associated with the self-renewal mechanism in hematopoietic stem cells in mouse myelodysplastic syndrome through an increase in methylation level at the miR-124 promoter [141]. As such, the relationship between PRDM3 and miR-124 is clearly supported in two different cellular models. Therefore, further studies are needed to determine whether PRDM3 directly affects the promoter status of other target genes in the neuronal system.
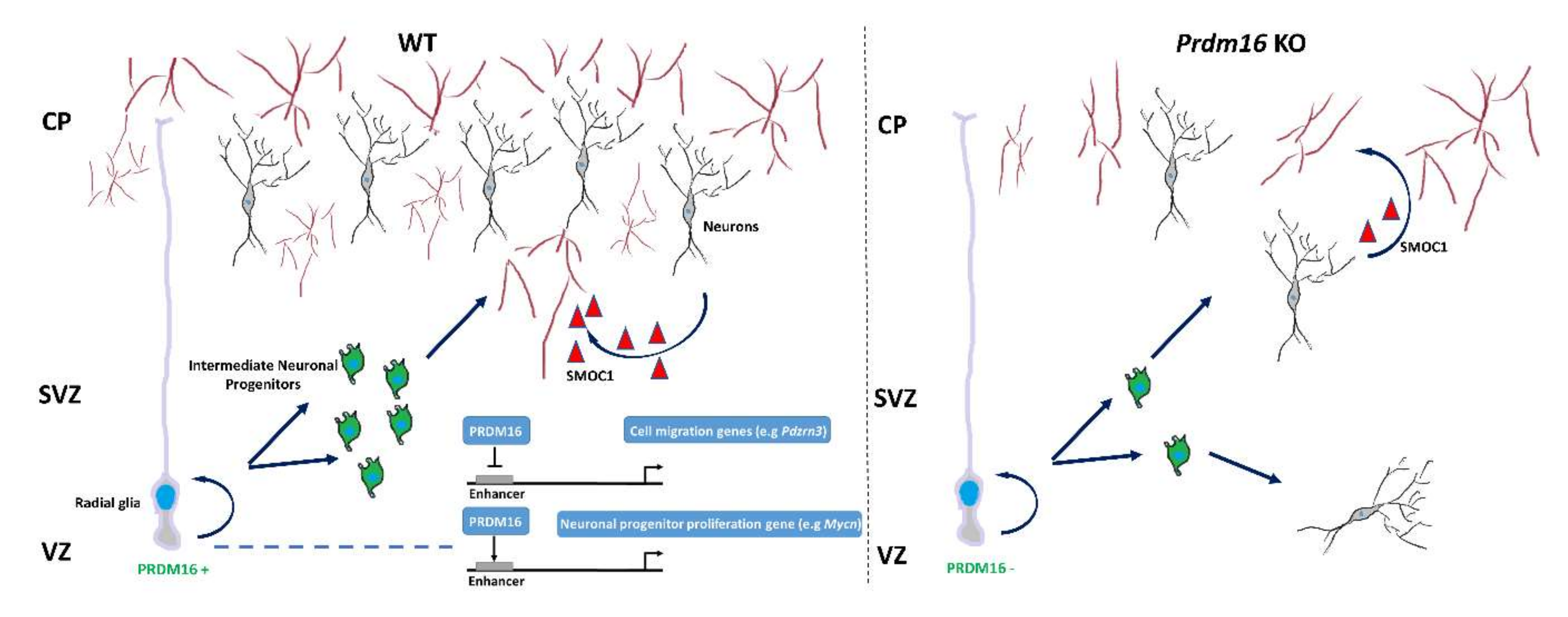
In the developing nervous system, PRDM16 is expressed in the SVZ of the neocortex and is crucial for neural stem cell maintenance. PRDM16 is a key player in maintaining neural stem cell and progenitor cells in the brain by controlling their temporal and spatial gene regulation networks [31]. In the mouse embryo, PRDM16 is mainly expressed in the VZ and SVZ but its expression decreases during brain maturation [114]. A conditional Prdm16 deletion exhibits significant depletion of mouse neural stem cells in the SVZ as well as a limited ability to form self-renewing neurospheres in vitro [31]. Moreover, reduced Prdm16 expression and complementary upregulation of the proneural gene NeuroD1 in embryonic neural stem cells is crucial for the regulation of peroxisome proliferative activated receptor 1 (PGC1)-mediated changes in reactive oxygen species (ROS) levels, and this mechanism has been suggested to be important for neural migration in the developing cortex [25]. On the other hand, PRDM16 is preferentially expressed in adult neural stem cells and is required for their maintenance, partly by suppressing oxidative stress through the promotion of hepatocyte growth factor (HGF) expression [33]. Another study revealed that PRDM16 is involved in the migration and differentiation of neurons during embryonic cerebral cortex development [24]. Here, it was shown that epigenetic status during the early stages of neuronal differentiation is closely related to PRDM16-dependent mechanisms. Interestingly, the association of gene expression level with H3K27ac modification in radial cells (PRDM16 positive) is also found in mature cortical neurons where PRDM16 is not expressed. This phenomenon suggests that the previously acquired epigenetic memory remains during cortical neuronal development [24]. It has been suggested that the determination of neuronal position in the upper layer of the cerebral cortex is controlled by PRDM16-mediated repression of the gene encoding E3 ubiquitin ligase PDZ domain-containing RING finger protein 3 (PDZRN3). PRDM16 significantly reduces PDZRN3 expression in brain progenitor cells. This can be partially explained by the reduction in H3K27ac levels at the enhancer and promoter regions of Pdzrn3. Conversely, a significant increase in H3K27ac levels is observed in cortical cells in the absence of PRDM16. Impaired PRDM16 expression causes a significant increase in PDZRN3 expression in newly formed neurons, but the consequence of this genetic deregulation is the decreased ability of these cells to migrate to the upper brain layers. Moreover, a lack of PRDM16 in neuronal progenitors leads to abnormal dendritic morphology of mature neurons [24]. Prdm16 deletion in neuronal stem cells causes dysregulation of angiogenesis. It was found that neurovascular communication depends in part on SMOC1, which is secreted by certain types of neurons. Neuronal SMOC1 interacts with TGFBR1 and activates the TGF-β-SMAD signaling pathway in endothelial cells. The loss of PRDM16 in neural progenitor disrupts this process and significantly impairs vascular growth in the developing brain [114]. The PRDM16 influence on neuronal cell migration and angiogenesis during central nervous system development is illustrated in Figure 3. Interestingly, RNA sequencing data show that disabling PRDM16 in progenitor cells (radial glia) resulted in a seven-fold increase in Prdm3 expression [24]. Therefore, crosstalk between PRDM16 and PRDM3 during neuronal differentiation merits further investigation. Taken together, although functional studies in the neuronal system have just started, PRDM3 and PRDM16 most likely play paramount roles in neuronal cell fate decision and function.
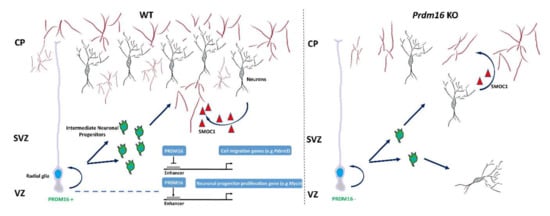
Figure 3.
PRDM16 orchestrates neuronal migration and angiogenesis in the developing brain. PRDM16 organizes the migration of cortical neurons through enhancer-dependent silencing of Pdzrn3 gene expression. In addition, PRDM16 controls the expression of genes associated with the amplification of neuronal progenitors (e.g., Mycn) [24]. PRDM16 positively regulates neuronal angiogenesis by enhancing the TGF-β signaling pathway via neuronally secreted SMOC1 [114]. VZ (ventricular zone), SVZ (subventricular zone), CP (cortical plate).
7. The Function of FOG1 and FOG2 in Neuronal Cells
FOG1 expression was found in the mouse mid-brain [129] as well as in the Danio rerio developing brain [142]. FOG1 affects the heart ventricular wall structure by regulating cardiomyocyte proliferation via the neuregulin (NRG)-ErbB-dependent signaling pathway [143]. NRG is a growth factor involved in the stimulation of ErbB tyrosine kinase receptors and thus affects cell survival, proliferation and differentiation in neuronal and non-neuronal systems. The NRG-ErbB axis is associated with susceptibility to mental illnesses such as bipolar disorder and schizophrenia [144,145]. A tight relationship between disease state and the NRG-ErbB pathway is indicated by the significant dysregulation of synaptic plasticity and neurotransmission (reviewed in [146]). Thus, the FOG1-controlled NRG-ErbB pathway may theoretically be involved in the development of synaptic plasticity and neuronal identity. However, due to the limited number of reports, the role of FOG1 in the nervous system development remains to be clarified.
In the developing brain, FOG-2 expression is detectable from around day 10 of embryonic development (E10.5) [147]. Recent studies have reported that FOG2 may be involved in shaping neuronal identity [93]. FOG2 was found exclusively in postmitotic neurons in the cortex as well as in the thalamic reticular nucleus, the hippocampus, the amygdala and the hypothalamus. These findings strongly suggest that FOG2 plays a role as a transcriptional regulator during the final stage of neuronal maturation [93]. In addition, high expression of GATA transcription factors (e.g., GATA4 and GATA6) was found within the central nervous system in mice and humans, and this expression was mainly observed in post-mitotic neurons of the cerebral cortex and the hippocampus [148,149]. Nevertheless, the function of GATA proteins in shaping the identity of neurons has not been sufficiently investigated. GATA-dependent mechanisms that form neuronal identity could be partly explained through interaction with FOG1 and FOG2 proteins. FOG2, in cooperation with GATA6, significantly increases promoter activity of the Kv4.2 (KCND2) gene (voltage-dependent potassium channels) in PC12 neuron-like cells. This study highlighted the importance of FOG2 in neuronal function and plasticity [115]. Tying FOG2-GATA6 to the regulation of voltage-gated ion channels could reveal new avenues in investigating the regulation of brain neuroplasticity and thus the formation of memory. Accurate tuning of gene expression during cortical development requires precise regulation of molecular machinery. In this context, FOG2 appears to be a mediator in the process of generating final cellular identities in the brain. FOG2 is involved in the control of corticothalamic projection neuron (CThPN) identity and positioning [93]. CThPNs are a diverse set of neurons that are important for the function of cortical circuitry. They are responsible for the access of sensory information to the cerebral cortex by modulating thalamic activity [150,151]. Interestingly, regulation of CThPN projection is controlled by the FOG2 and GATA4 complex via Ctip2 (Coup-Tf interacting protein 1) promoter activity [93]. CTIP2 belongs to a group of factors crucial in postnatal brain development [152]. CTIP2 is also involved in the differentiation of postmitotic neurons and thus in memory and learning [152]. In humans, FOG2 mutations have been recognized primarily in congenital heart disease, but neurological and behavioral abnormalities have also been observed. Patients with a FOG2 deletion exhibit delayed or impaired speech ability, intellectual disability and seizures [153,154]. Based on the above information, it is postulated that FOG2 could be a crucial factor during the assembly of neural circuits and the acquisition of identities in postmitotic neurons. Further research is needed to determine the FOG2 role in brain development and neuronal plasticity.
8. NuRD Interacts with PRDM3, PRDM16, FOG1, and FOG2
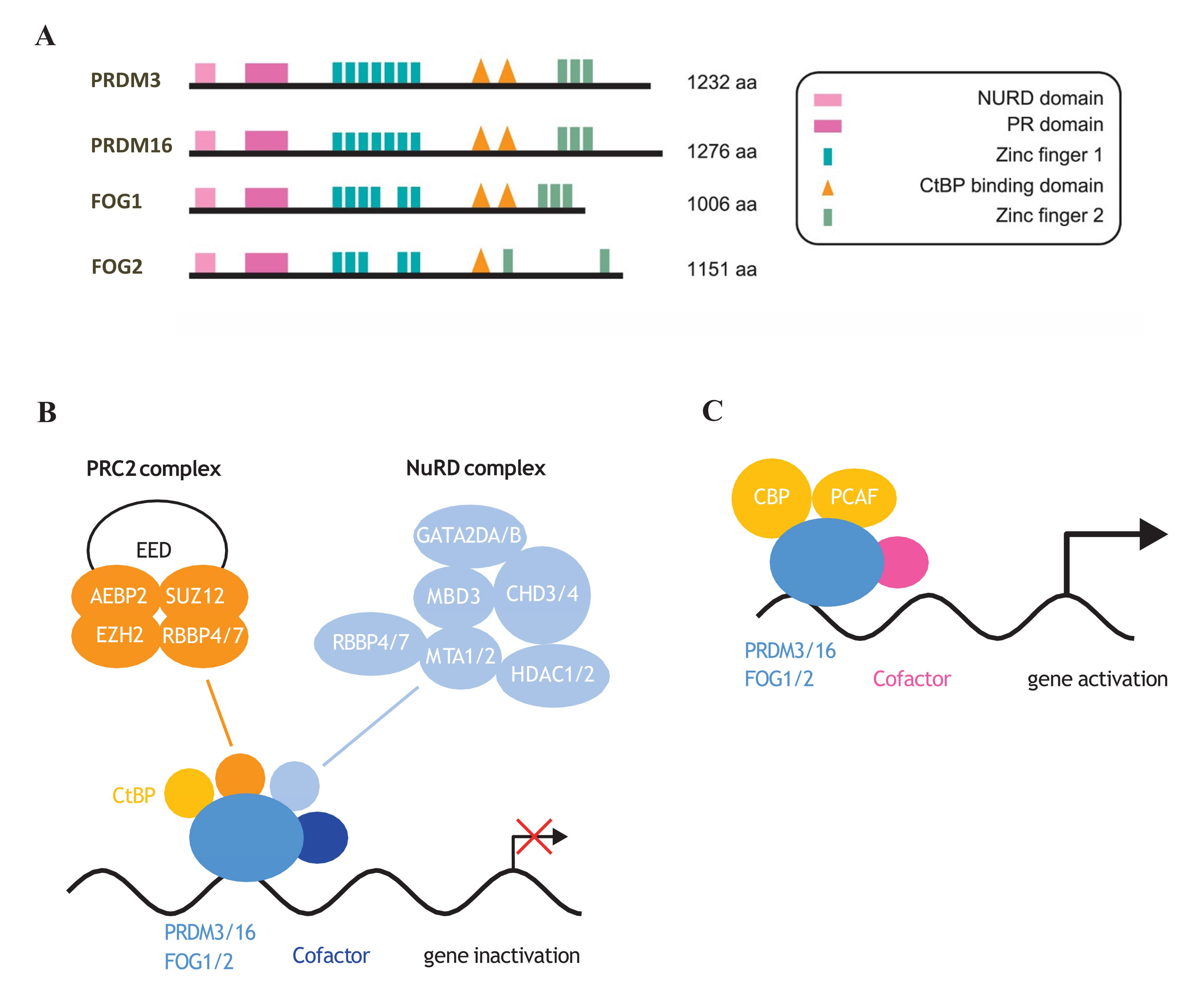
Chromatin remodeling complex NuRD (nucleosome remodeling and deacetylase) primarily exhibits histone deacetylase activity and, therefore, exerts a repressor function [155,156]. The base composition of NuRD includes the metastasis-associated proteins MTA1/2/3, the histone deacetylases HDAC1 and HDAC2, the methyl-CpG-binding domain proteins (MBD2 or MBD3), CHD4, and the histone binding proteins RBBP4/7 [157,158,159]. NuRD regulates gene transcription associated with pluripotency and mediates the cellular response to differentiation signals in mouse embryonic stem cells (ESCs) [160,161]. NuRD activity has been reported to mediate the reduction of H3K27 acetylation facilitating recruitment of PRC2 and subsequent trimethylation of H3K27 in NuRD-dependent promoters [160,161]. The NuRD/MBD3 complex significantly shapes the final developmental stages of the brain. Despite high Mbd3 expression in neuroepithelial cells (NECs) of the embryonic cortex [162], NuRD/MBD3 has been shown to be particularly important in regulating cell differentiation during neuronal specification [163]. Moreover, the NuRD complex affects synaptic plasticity in the mammalian brain and controls cortical neuron identity [140,164]. A good example of how NuRD influences the shape of terminal neuronal differentiation is by the regulation of the Satb2-Ctip2 axis. CTIP2 and SATB2 are key transcription factors that define the development of two classes of projection neurons. Ctip2 expression is terminated by the cooperation of SATB2 and NuRD. This, in turn, induces NuRD recruitment of HDAC1 and finally deacetylation of the Ctip2 locus. Decreased levels of CTIP2 lead to the formation of a different subclass of projection neuron [165]. Recent studies have demonstrated that both PRDM3 and PRDM16 proteins interact with the chromatin remodeling NuRD complex through the RBBP4 (RB binding protein 4, chromatin remodeling factor; also known as NURF55) protein [73]. RBBP4 is recognized as a mediator that facilitates the association of chromatin with the NuRD complex by binding to histone H3 tails [166]. Amino acids from the N-terminus of PRDM3 and PRDM16 are responsible for binding to RBBP4 [73]. Interestingly, it has been reported that FOG1 and FOG2 also interact with the NuRD complex through their N-terminal amino acid sequence (Figure 4A,B) [76,78]. The N-terminal amino acid residues that interact with NuRD are conserved between PRDM3 and PRDM16 (Figure 4A). The first 12 residues of both proteins show high sequence similarity with histone 3 N-terminal residues [73]. It is known that RBBP4 interacts with LHX2 and regulates the expression of the Sox11 and Fezf2 genes; the most important factors determining the identity of neuronal subtype in the mouse cortex [164]. Since PRDM3 and PRDM16 interact with RBBP4 and the NuRD complex, these proteins may play a regulatory role in shaping the identity of neurons and their position in various brain structures. Moreover, since CtBP is also a major PRDM3 and PRDM16 modulator, and controls cell fate decisions (Figure 4B) [74], investigation is needed into how PRDM3 and PRDM16 select their binding to CtBP and NuRD complex.
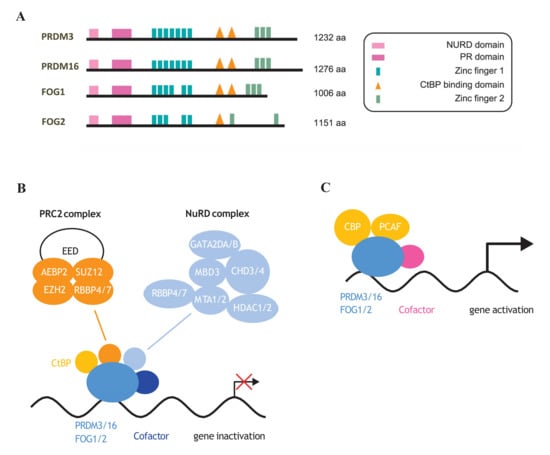
Figure 4.
Protein structure of NuRD complex-dependent PRDM factors. (A) PR domain, CtBP binding sites, zinc fingers as well as NuRD complex binding elements are indicated. (B) PRDM3/16 and FOG1/2 by interaction with CtBP or NuRD complex negatively regulate gene expression during cellular differentiation [15,61,74,167,168]. (C) CBP- and PCAF-mediated acetylation of PRDM3 [169,170] increases its transcriptional activity [168].
FOG1/GATA1-dependent transcriptional repression is mediated by the NuRD complex. FOG-1 binds to NuRD via a 12-amino acid N-terminal motif [171]. FOG1 forms an active complex with NuRD to promote hematopoiesis. Depending on the composition of the protein complex, it also regulates cell lineage specification [61,171]. The FOG1/NuRD complex acts as a repressor of GATA1 and GATA2. This repression induces hematopoiesis by inhibiting GATA factors and subsequently halts mast cell differentiation [61]. Similarly, FOG2 modulates cardiomyocyte proliferation and heart morphogenesis by interacting with GATA4 to reduce GATA4-dependent gene expression [167,172]. However, the specific role of FOG2/NuRD interaction remains unknown.
PRDM genes can generate alternative forms of transcripts, which mainly include an isoform without a PR domain and a long product with a PR domain at the N-terminus. These two forms of transcripts can be generated using different promoter sites or by alternative splicing [64,65,67]. It is striking that the PRDM short product almost always acts as an oncogene and the long-form acts as a tumor suppressor [64,65]. PRDM3 is generated by combining two distinct genes- MDS1 and EVI1. The construct without the PR domain is transcribed from one locus and is called EVI1 or sPRDM3 (short PRDM3) [173]. Mutations in the PRDM3 gene are common in acute myeloid leukemia and are related to reduced overall survival [174,175,176]. High expression of short PRDM3 is relatively often observed in myeloid or solid tumors. Intriguingly, the PR domain in PRDM3 appears to play a tumor suppressor function [177,178,179]. In addition, sPRDM3 appears to be frequently mutated in skin melanomas, colon, lung, bladder and endometrial cancers with simultaneous decreased expression of the long PRDM3 form [180]. Moreover, deficiency of the long version of PRDM3 also leads to a decrease in the number of hemopoietic stem cells and a loss of long-term repopulation capacity through deregulation of the p57-kip2 pathway [45]. Apparently, the PR domain of the PRDM16 protein plays a significant role in regulating gene expression through modification of epigenetic signatures. Loss of the PR domain of PRDM16 leads to a reduction in histone H3 acetylation (H3ac), H3K4me3 and H3K27me3 modifications at the Pparγ promoter. These changes attenuate the potential of adipogenic transdifferentiation in C2C12 myoblasts [124]. Furthermore, the isoforms of PRDM16 show distinct impact on leukemia hematopoiesis. While full-length PRDM16 suppresses the inflammatory pathway, its short-isoform exhibits the opposite effect in HSC [30]. On the other hand, PRDM16 lacking a PR domain triggers leukemic transformation in mice progenitor cells that carry a p53 deletion. Hence, it is presumed that the long isoform of PRDM16 conceivably acts as a tumor suppressor [181]. The function of the PR domain in PRDM proteins during neuronal differentiation is unclear due to the small number of related studies, but the PR domain of PRDM16 is known to control epigenetic silencing determining the migration and position of neurons in the brain cortex [24]. In the same study, it was demonstrated that only a long version of PRDM16 is able to reverse the neuronal migration defects in PRDM16-deficient mouse brains [24]. Interestingly, the removal of a PR domain in the PRDM3/16 and FOG1/2 should also eliminate the NuRD domain, which is located at the N-terminus of the protein (Figure 4A). NuRD has shown paramount importance in the maintenance of hematopoietic stem cells [182] and neuronal development [183]. Therefore, it would be interesting to determine whether the effect of loss of PR domain results from the lack of interaction with cofactors such as the NuRD complex.
9. CtBP Controls PRDM3, PRDM16, and FOG Function
CtBP controls the function of PRDM3, 16, and the FOG family through its specific binding sites [74,75,77,126]. CtBP1 and 2 are known as major transcriptional corepressors [184]. Ablation of CtBP proteins in early development is known to cause embryonic lethality [184], and information on the function of CtBP in the neural system is, therefore, limited. However, expression of CtBP1 has been reported in cultured hippocampal neurons, suggesting a potential function of CtBP1 in learning and memory [185]. Moreover, functional roles for CtBP in the neural system have been described in a number of studies in D. melanogaster. Drosophila CtBP (dCtBP) can control cell fate decisions in the sensory organ system as its loss of function leads to the formation of extra bristles, whereas flies with mutated dCtBP show a loss of bristles. Hamlet, fly PRDM3/16, loses its repressive activity when both CtBP domain and Zn fingers are deleted [103]. These results suggest that the function of PRDM3/16 and Hamlet is largely dependent on the CtBP protein, and their physical interaction is one of the key mechanisms by which PRDM3/16 orchestrates target gene expressions in stem cells and neuronal systems.
Crosstalk between CtBP and sumoylation has been reported to be important for repressing transcriptional activity of PRDM16 protein [186]. Sumoylation is characterized by the reversible attachment of small ubiquitin-related modifier (SUMO) family members to lysine residue(s) located on target proteins via SUMO-activating enzyme subunit 1/2 (SAE1/2) and the SUMO-1-conjugating enzyme UBC9. The role of sumoylation has been studied in neurodegenerative diseases including Alzheimer’s disease [187,188]. Sumo1, 2, and 3 are expressed in the developing mouse brain [189] and sumoylation acts as a modulator of several neural activities and of neuronal stem cell maintenance [190,191,192]. Among PRDM factors, PRDM3 and PRDM16 are known to be modulated by sumoylation. Sumoylation inhibits the DNA binding activity of PRDM3 to the Bcl-xL promoter, which eventually might be involved in mediating apoptosis [193]. Sumoylation of PRDM16 regulates its interaction with CtBP and activates PRDM16’s repressor activity. Moreover, the role of sumoylation in the regulation of PRDM16 has been well characterized in adipose tissue thermogenesis [194] and acute myeloid leukemia progression [195]. Thus, it has been proposed that sumoylation plays an important role in modulating PRDM3 and PRDM16 activity. Still, the role of sumoylation-dependent regulation of PRDM3 and PRDM16 in the nervous system remains to be elucidated. The new PRDM family member FOG1 is also regulated by sumoylation and specifically its interaction with CtBP. Interestingly, the sumoylation of FOG1 is modulated by the presence of phosphorylated residues in its sequence. Yang et al. [196] have proposed that several transcription factors possess a ψKxExxS/T (K = sumoylation target, S/T = phosphorylation target) motif which could be responsible for the interplay between sumoylation and phosphorylation marks on FOG1 activity. Thus, the interplay between post-translational modifications may orchestrate transcriptional repression via CtBP-dependent PRDM factors including PRDM3, 16, FOG1, and FOG2.
10. PRDM3/16 Function as Activators in Gene Regulation
Interestingly, PRDM3 and 16 interact not only with transcriptional repressors but also activators. Therefore, they are considered to be able to bidirectionally control gene expression (gene upregulation and downregulation) to exert their function. Acetylation of proteins acts as a positive regulator of transcription factors. PCAF and CBP acetylate PRDM3 [169,170], which enhances its transcriptional activity at the GATA2 promoter (Figure 4C) [168]. Accordingly, the active PRDM3-GATA2 axis maintains a pool of hematopoietic stem cells. An interesting example of activation of gene expression by PRDM3 can be depicted by the interaction of PRDM3 with BRG1 in the embryonic fibroblast cell line (NIH 3T3). BRG1 significantly reduces the E2F1 promoter activity, thus reducing the level of cell proliferation. By binding to BRG1, PRDM3 blocks this repression thereby accelerating the cell cycle [197]. BRG1 is a component of the SWI/SNF complex. SWI/SNF predominantly enhances gene expression by remodeling chromatin structure and making it more accessible to transcription factors but, depending on its protein partners, it may exhibit a repressive effect [198,199]. Interestingly, it has been shown that BRG1 can be also associate with the NuRD complex [200]. Therefore, the functional relationships between PRDM3, BRG1 and NuRD complex [73,197] remain to be clarified. Overexpression of C/EBPβ and PRDM16 reprograms murine and human fibroblasts into fat cells. It is suggested that C/EBP-β acts as a PRDM16 coactivator to enable the interaction of PGC-1α and PPARγ and activates cell differentiation [201]. It is known that C/EBP-β mediates the tuning of the transcription program to induce neurogenesis and to inhibit glial growth [202]. In mice, C/EBP-β has been shown to be involved in the survival and proliferation of neural stem cells in the hippocampus [203]. A similar effect was described by Shimada et al. where PRDM16 was indispensable for the maintenance of neural stem cells in the postnatal brain [31]. These similarities imply that C/EBP-β may also interact with PRDM16 during brain development and thus tune the genetic program in order to generate neuronal precursors.
Table 3 displays the most recognized cofactors interacting with PRDM3/16 and FOG1/2, and their roles.

Table 3.
Summary of PRDM3/16 and FOG1/2 interaction with their known cofactors. The PRDM3/16 and FOG1/2 proteins affect the genetic program and cell functions by interaction with their cofactors. This influence may have a positive or negative effect.
11. Role of PRDM Proteins in Cancer Development and their Gene Mutations Found in Neuronal Diseases
Although PRDM proteins are recognized as regulators involved in cell differentiation [23], their commitment (suppressive or oncogenic) in the pathogenesis of human diseases such as carcinogenesis is also under investigation. PRDM1 is recognized as a tumor suppressor that inhibits the development of cancer cells, including lymphomas [232,233,234,235]. A chromosomal deletion or epigenetic silencing of PRDM1 expression is common in diffuse large B cell lymphoma subtypes [234,236,237]. Abnormal PRDM1 expression is also associated with other nonhematopoietic cancer cells, such as glioblastoma malignancies [238]. It has been identified that the downregulation of PRDM1 correlates with increased malignancy of lung tumors, where PRDM1 disruption promoted neoplastic invasiveness [235]. The PRDM3 is a fused complex of two different transcripts, MDS1 and EVI1. It is a frequent site of viral insertion and is associated with the development of myeloid leukemia [239,240]. PRDM14 overexpression, or retroviral integration in the gene locus, is often found in various types of cancer, and the molecular mechanism is set up to promote pluripotent traits [241]. PRDM14 overexpression is detected in approximately 25% of human lymphoid tumors. Mice bone marrow cells transduced with the PRDM14 expression vector often develop leukemia. The analysis of the gene expression profile indicated that PRDM14 overexpressing cells showed significant enrichment of pluripotent genes and enhancement of the tumor-initiating pathway (WNT and RAS signaling) [242]. Although PRDM14 seems to be associated with the acquisition of an immortal phenotype, the neoplastic process driven by PRDM14 might depend on the tissue. Assessment of gene methylation levels from cervical scratches positive for human papillomavirus (HPV) at high risk of malignant disease showed an increased methylation level of the PRDM14 gene [243]. It can, therefore, be assumed that PRDM14 acts as a tumor suppressor in cervical cancer. Hence, the role of PRDM14 in tumor development is enigmatic and requires further research. In lung cancer, the high methylation signature of the PRDM16 gene caused a significant reduction of its expression [244,245]. Another study indicated that relatively high expression of PRDM16 in patients with nonsmall cell lung cancer was associated with a preferable survival score [246,247,248]. At least in part, the PRDM16-mediated tumor inhibition could be explained by hammering the epithelial-to-mesenchymal transition in lung adenocarcinomas [246]. The role of FOG1 in the development of neoplasms is still ambiguous, albeit mutations within FOG1 locus are extremely common (approximately 50% of cases) in patients with adrenocortical carcinoma [180]. Moreover, preliminary analysis indicates that FOG1 is also frequently mutated in colorectal cancer [180]. Despite the small number of studies conducted on the relationship between FOG1 and initiation or tumor progression, the abovementioned findings indicate an unexplored phenomenon of the high frequency of FOG1 mutations in tumors. FOG2, along with GATA4 and GATA6, is relatively highly expressed in sex cord-derived ovarian tumors [249]. Deregulation of the expression of FOG2 and its cofactors has also been observed in ovarian granular cell tumors and ovarian stromal tumors in children [250,251]. Moreover, recent studies have shown that FOG2 is one of the most common mutated genes in the PRDM protein family. A high frequency of FOG2 mutations is found in skin melanomas, uterine cancer, rectal cancer, esophageal cancer, gastric adenocarcinoma and lung tumors [180].
Human PRDM gene mutations in the nervous system are poorly recognized. Nevertheless, genetic abnormalities found within PRDM genes are significantly associated with neurological disabilities (Table 4). Clinical effects of PRDM12 mutations in patients within the congenital insensitivity to pain are caused by defects in the development of nociceptors [91]. FOG2 mutations were found to have a deleterious effect on brain structure development and patients exhibited a motor, linguistic and cognitive delay with seizure events [154,252].

Table 4.
Clinical effect of mutation in PRDM genes within the neurological system.
12. Conclusions
Emerging evidence has suggested that PRDM factors cooperate with a number of protein partners to regulate a critical set of genes required for the maintenance of stem cell self-renewal and differentiation through multiple mechanisms. In this review, we proposed a NuRD and CtBP-dependent function of PRDM3/16 and FOG1/2 with respect to stem cell maintenance and neuronal differentiation. Moreover, we listed possible mechanisms of how these factors can regulate their target gene expression in a spatiotemporal and bidirectional manner. Although the PR domain that is contained in PRDM factors exerts methylation enzyme activity, our study suggests that cofactor-dependent regulation of PRDM3/16 and FOG1/2 is also one of the most important mechanisms to regulate PRDM factors function. Stem cell and neuronal cell fate are orchestrated by fine-tuned molecular mechanisms in which several transcription factors are encountered and dissociated. Furthermore, dysfunction of these factors causes abnormality in several tissues, and even leads to increased cancer risk. Therefore, identifying their stem cell and neuronal-specific cofactors will help to improve understanding of how they function in healthy and diseased conditions.
Author Contributions
Conceptualization, P.L. and H.T.; writing—original draft preparation, P.L., M.Ś., and H.T.; writing—review and editing, M.Ś., H.T., E.P., C.W., and K.-i.M.; figure design; P.L., M.Ś., and H.T.; funding acquisition, P.L. All authors have read and agreed to the published version of the manuscript.
Funding
This review was financially supported by National Science Centre, Poland (grant no. 2017/25/N/NZ3/01886).
Acknowledgments
The authors thank Tobias Hohenauer for help with editing the manuscript and illustrations.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Buyse, I.M.; Shao, G.; Huang, S. The retinoblastoma protein binds to RIZ, a zinc-finger protein that shares an epitope with the adenovirus E1A protein. Proc. Natl. Acad. Sci. USA 1995, 92, 4467–4471. [Google Scholar] [CrossRef] [PubMed]
- Huang, S. Blimp-1 is the murine homolog of the human transcriptional repressor PRDI-BF1. Cell 1994, 78, 9–10. [Google Scholar] [CrossRef]
- Keller, A.D.; Maniatis, T. Identification and characterization of a novel repressor of beta-interferon gene expression. Genes Dev. 1991, 5, 868–879. [Google Scholar] [CrossRef] [PubMed]
- Turner, C.A., Jr.; Mack, D.H.; Davis, M.M. Blimp-1, a novel zinc finger-containing protein that can drive the maturation of B lymphocytes into immunoglobulin-secreting cells. Cell 1994, 77, 297–306. [Google Scholar] [CrossRef]
- Xiao, B.; Wilson, J.R.; Gamblin, S.J. SET domains and histone methylation. Curr. Opin. Struct. Biol. 2003, 13, 699–705. [Google Scholar] [CrossRef]
- Duan, Z.; Person, R.E.; Lee, H.H.; Huang, S.; Donadieu, J.; Badolato, R.; Grimes, H.L.; Papayannopoulou, T.; Horwitz, M.S. Epigenetic regulation of protein-coding and microRNA genes by the Gfi1-interacting tumor suppressor PRDM5. Mol. Cell. Biol. 2007, 27, 6889–6902. [Google Scholar] [CrossRef]
- Davis, C.A.; Haberland, M.; Arnold, M.A.; Sutherland, L.B.; McDonald, O.G.; Richardson, J.A.; Childs, G.; Harris, S.; Owens, G.K.; Olson, E.N. PRISM/PRDM6, a transcriptional repressor that promotes the proliferative gene program in smooth muscle cells. Mol. Cell Biol. 2006, 26, 2626–2636. [Google Scholar] [CrossRef]
- Gyory, I.; Wu, J.; Fejer, G.; Seto, E.; Wright, K.L. PRDI-BF1 recruits the histone H3 methyltransferase G9a in transcriptional silencing. Nat. Immunol. 2004, 5, 299–308. [Google Scholar] [CrossRef]
- Pinheiro, I.; Margueron, R.; Shukeir, N.; Eisold, M.; Fritzsch, C.; Richter, F.M.; Mittler, G.; Genoud, C.; Goyama, S.; Kurokawa, M.; et al. Prdm3 and Prdm16 are H3K9me1 methyltransferases required for mammalian heterochromatin integrity. Cell 2012, 150, 948–960. [Google Scholar] [CrossRef]
- Kim, K.C.; Geng, L.; Huang, S. Inactivation of a histone methyltransferase by mutations in human cancers. Cancer Res 2003, 63, 7619–7623. [Google Scholar]
- Hayashi, K.; Yoshida, K.; Matsui, Y. A histone H3 methyltransferase controls epigenetic events required for meiotic prophase. Nature 2005, 438, 374–378. [Google Scholar] [CrossRef] [PubMed]
- Fog, C.K.; Galli, G.G.; Lund, A.H. PRDM proteins: Important players in differentiation and disease. Bioessays 2012, 34, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Eom, G.H.; Kim, K.; Kim, S.M.; Kee, H.J.; Kim, J.Y.; Jin, H.M.; Kim, J.R.; Kim, J.H.; Choe, N.; Kim, K.B.; et al. Histone methyltransferase PRDM8 regulates mouse testis steroidogenesis. Biochem. Biophys. Res. Commun. 2009, 388, 131–136. [Google Scholar] [CrossRef] [PubMed]
- Derunes, C.; Briknarova, K.; Geng, L.; Li, S.; Gessner, C.R.; Hewitt, K.; Wu, S.; Huang, S.; Woods, V.I., Jr.; Ely, K.R. Characterization of the PR domain of RIZ1 histone methyltransferase. Biochem. Biophys. Res. Commun. 2005, 333, 925–934. [Google Scholar] [CrossRef]
- Yoshimi, A.; Goyama, S.; Watanabe-Okochi, N.; Yoshiki, Y.; Nannya, Y.; Nitta, E.; Arai, S.; Sato, T.; Shimabe, M.; Nakagawa, M.; et al. Evi1 represses PTEN expression and activates PI3K/AKT/mTOR via interactions with polycomb proteins. Blood 2011, 117, 3617–3628. [Google Scholar] [CrossRef]
- Su, S.T.; Ying, H.Y.; Chiu, Y.K.; Lin, F.R.; Chen, M.Y.; Lin, K.I. Involvement of histone demethylase LSD1 in Blimp-1-mediated gene repression during plasma cell differentiation. Mol. Cell Biol. 2009, 29, 1421–1431. [Google Scholar] [CrossRef]
- Ancelin, K.; Lange, U.C.; Hajkova, P.; Schneider, R.; Bannister, A.J.; Kouzarides, T.; Surani, M.A. Blimp1 associates with Prmt5 and directs histone arginine methylation in mouse germ cells. Nat. Cell Biol. 2006, 8, 623–630. [Google Scholar] [CrossRef]
- Alliston, T.; Ko, T.C.; Cao, Y.; Liang, Y.Y.; Feng, X.H.; Chang, C.; Derynck, R. Repression of bone morphogenetic protein and activin-inducible transcription by Evi-1. J. Biol. Chem. 2005, 280, 24227–24237. [Google Scholar] [CrossRef]
- Chittka, A.; Arevalo, J.C.; Rodriguez-Guzman, M.; Perez, P.; Chao, M.V.; Sendtner, M. The p75NTR-interacting protein SC1 inhibits cell cycle progression by transcriptional repression of cyclin E. J. Cell Biol. 2004, 164, 985–996. [Google Scholar] [CrossRef]
- Yu, J.; Angelin-Duclos, C.; Greenwood, J.; Liao, J.; Calame, K. Transcriptional repression by blimp-1 (PRDI-BF1) involves recruitment of histone deacetylase. Mol. Cell Biol. 2000, 20, 2592–2603. [Google Scholar] [CrossRef]
- Cattaneo, F.; Nucifora, G. EVI1 recruits the histone methyltransferase SUV39H1 for transcription repression. J. Cell Biochem. 2008, 105, 344–352. [Google Scholar] [CrossRef] [PubMed]
- Yang, C.M.; Shinkai, Y. Prdm12 is induced by retinoic acid and exhibits anti-proliferative properties through the cell cycle modulation of P19 embryonic carcinoma cells. Cell Struct. Funct. 2013, 38, 197–206. [Google Scholar] [CrossRef] [PubMed]
- Hohenauer, T.; Moore, A.W. The Prdm family: Expanding roles in stem cells and development. Development 2012, 139, 2267–2282. [Google Scholar] [CrossRef] [PubMed]
- Baizabal, J.M.; Mistry, M.; Garcia, M.T.; Gomez, N.; Olukoya, O.; Tran, D.; Johnson, M.B.; Walsh, C.A.; Harwell, C.C. The Epigenetic State of PRDM16-Regulated Enhancers in Radial Glia Controls Cortical Neuron Position. Neuron 2018, 98, 945–962.e8. [Google Scholar] [CrossRef] [PubMed]
- Inoue, M.; Iwai, R.; Tabata, H.; Konno, D.; Komabayashi-Suzuki, M.; Watanabe, C.; Iwanari, H.; Mochizuki, Y.; Hamakubo, T.; Matsuzaki, F.; et al. Prdm16 is crucial for progression of the multipolar phase during neural differentiation of the developing neocortex. Development 2017, 144, 385–399. [Google Scholar] [CrossRef]
- Inoue, M.; Iwai, R.; Yamanishi, E.; Yamagata, K.; Komabayashi-Suzuki, M.; Honda, A.; Komai, T.; Miyachi, H.; Kitano, S.; Watanabe, C.; et al. Deletion of Prdm8 impairs development of upper-layer neocortical neurons. Genes Cells 2015, 20, 758–770. [Google Scholar] [CrossRef]
- Eguchi, R.; Yoshigai, E.; Koga, T.; Kuhara, S.; Tashiro, K. Spatiotemporal expression of Prdm genes during Xenopus development. Cytotechnology 2015, 67, 711–719. [Google Scholar] [CrossRef]
- Okada, M.; Shi, Y.B. EVI and MDS/EVI are required for adult intestinal stem cell formation during postembryonic vertebrate development. FASEB J. 2018, 32, 431–439. [Google Scholar] [CrossRef]
- Miller, T.C.; Sun, G.; Hasebe, T.; Fu, L.; Heimeier, R.A.; Das, B.; Ishizuya-Oka, A.; Shi, Y.B. Tissue-specific upregulation of MDS/EVI gene transcripts in the intestine by thyroid hormone during Xenopus metamorphosis. PLoS ONE 2013, 8, e55585. [Google Scholar] [CrossRef]
- Corrigan, D.J.; Luchsinger, L.L.; Justino de Almeida, M.; Williams, L.J.; Strikoudis, A.; Snoeck, H.W. PRDM16 isoforms differentially regulate normal and leukemic hematopoiesis and inflammatory gene signature. J. Clin. Investig. 2018, 128, 3250–3264. [Google Scholar] [CrossRef]
- Shimada, I.S.; Acar, M.; Burgess, R.J.; Zhao, Z.; Morrison, S.J. Prdm16 is required for the maintenance of neural stem cells in the postnatal forebrain and their differentiation into ependymal cells. Genes Dev. 2017, 31, 1134–1146. [Google Scholar] [CrossRef] [PubMed]
- Aguilo, F.; Avagyan, S.; Labar, A.; Sevilla, A.; Lee, D.F.; Kumar, P.; Lemischka, I.R.; Zhou, B.Y.; Snoeck, H.W. Prdm16 is a physiologic regulator of hematopoietic stem cells. Blood 2011, 117, 5057–5066. [Google Scholar] [CrossRef] [PubMed]
- Chuikov, S.; Levi, B.P.; Smith, M.L.; Morrison, S.J. Prdm16 promotes stem cell maintenance in multiple tissues, partly by regulating oxidative stress. Nat. Cell Biol. 2010, 12, 999–1006. [Google Scholar] [CrossRef] [PubMed]
- Mould, A.; Morgan, M.A.; Li, L.; Bikoff, E.K.; Robertson, E.J. Blimp1/Prdm1 governs terminal differentiation of endovascular trophoblast giant cells and defines multipotent progenitors in the developing placenta. Genes Dev. 2012, 26, 2063–2074. [Google Scholar] [CrossRef]
- Chu, L.F.; Surani, M.A.; Jaenisch, R.; Zwaka, T.P. Blimp1 expression predicts embryonic stem cell development in vitro. Curr. Biol. 2011, 21, 1759–1765. [Google Scholar] [CrossRef]
- Okashita, N.; Suwa, Y.; Nishimura, O.; Sakashita, N.; Kadota, M.; Nagamatsu, G.; Kawaguchi, M.; Kashida, H.; Nakajima, A.; Tachibana, M.; et al. PRDM14 Drives OCT3/4 Recruitment via Active Demethylation in the Transition from Primed to Naive Pluripotency. Stem. Cell Reports 2016, 7, 1072–1086. [Google Scholar] [CrossRef]
- Chia, N.Y.; Chan, Y.S.; Feng, B.; Lu, X.; Orlov, Y.L.; Moreau, D.; Kumar, P.; Yang, L.; Jiang, J.; Lau, M.S.; et al. A genome-wide RNAi screen reveals determinants of human embryonic stem cell identity. Nature 2010, 468, 316–320. [Google Scholar] [CrossRef]
- Mzoughi, S.; Zhang, J.; Hequet, D.; Teo, S.X.; Fang, H.; Xing, Q.R.; Bezzi, M.; Seah, M.K.Y.; Ong, S.L.M.; Shin, E.M.; et al. PRDM15 safeguards naive pluripotency by transcriptionally regulating WNT and MAPK-ERK signaling. Nat. Genet 2017, 49, 1354–1363. [Google Scholar] [CrossRef]
- Elias, S.; Morgan, M.A.; Bikoff, E.K.; Robertson, E.J. Long-lived unipotent Blimp1-positive luminal stem cells drive mammary gland organogenesis throughout adult life. Nat. Commun. 2017, 8, 1–12. [Google Scholar] [CrossRef]
- Cheedipudi, S.; Gala, H.P.; Puri, D.; Dhawan, J. Identification of PRDM2 regulated genes in quiescent C2C12 myoblasts. Genom. Data 2015, 6, 264–266. [Google Scholar] [CrossRef][Green Version]
- Cheedipudi, S.; Puri, D.; Saleh, A.; Gala, H.P.; Rumman, M.; Pillai, M.S.; Sreenivas, P.; Arora, R.; Sellathurai, J.; Schroder, H.D.; et al. A fine balance: Epigenetic control of cellular quiescence by the tumor suppressor PRDM2/RIZ at a bivalent domain in the cyclin a gene. Nucleic. Acids Res. 2015, 43, 6236–6256. [Google Scholar] [CrossRef] [PubMed]
- Kustikova, O.S.; Schwarzer, A.; Stahlhut, M.; Brugman, M.H.; Neumann, T.; Yang, M.; Li, Z.; Schambach, A.; Heinz, N.; Gerdes, S.; et al. Activation of Evi1 inhibits cell cycle progression and differentiation of hematopoietic progenitor cells. Leukemia 2013, 27, 1127–1138. [Google Scholar] [CrossRef] [PubMed]
- Konantz, M.; Alghisi, E.; Muller, J.S.; Lenard, A.; Esain, V.; Carroll, K.J.; Kanz, L.; North, T.E.; Lengerke, C. Evi1 regulates Notch activation to induce zebrafish hematopoietic stem cell emergence. EMBO J. 2016, 35, 2315–2331. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, S.; Hoggatt, J.; Singh, P.; Abe, M.; Speth, J.M.; Hu, P.; Conway, E.M.; Nucifora, G.; Yamaguchi, S.; Pelus, L.M. Survivin modulates genes with divergent molecular functions and regulates proliferation of hematopoietic stem cells through Evi-1. Leukemia 2015, 29, 433–440. [Google Scholar] [CrossRef]
- Zhang, Y.; Stehling-Sun, S.; Lezon-Geyda, K.; Juneja, S.C.; Coillard, L.; Chatterjee, G.; Wuertzer, C.A.; Camargo, F.; Perkins, A.S. PR-domain-containing Mds1-Evi1 is critical for long-term hematopoietic stem cell function. Blood 2011, 118, 3853–3861. [Google Scholar] [CrossRef]
- Yuasa, H.; Oike, Y.; Iwama, A.; Nishikata, I.; Sugiyama, D.; Perkins, A.; Mucenski, M.L.; Suda, T.; Morishita, K. Oncogenic transcription factor Evi1 regulates hematopoietic stem cell proliferation through GATA-2 expression. EMBO J. 2005, 24, 1976–1987. [Google Scholar] [CrossRef]
- Goyama, S.; Yamamoto, G.; Shimabe, M.; Sato, T.; Ichikawa, M.; Ogawa, S.; Chiba, S.; Kurokawa, M. Evi-1 is a critical regulator for hematopoietic stem cells and transformed leukemic cells. Cell Stem. Cell 2008, 3, 207–220. [Google Scholar] [CrossRef]
- Arai, S.; Yoshimi, A.; Shimabe, M.; Ichikawa, M.; Nakagawa, M.; Imai, Y.; Goyama, S.; Kurokawa, M. Evi-1 is a transcriptional target of mixed-lineage leukemia oncoproteins in hematopoietic stem cells. Blood 2011, 117, 6304–6314. [Google Scholar] [CrossRef]
- Bogani, D.; Morgan, M.A.; Nelson, A.C.; Costello, I.; McGouran, J.F.; Kessler, B.M.; Robertson, E.J.; Bikoff, E.K. The PR/SET domain zinc finger protein Prdm4 regulates gene expression in embryonic stem cells but plays a nonessential role in the developing mouse embryo. Mol. Cell Biol. 2013, 33, 3936–3950. [Google Scholar] [CrossRef][Green Version]
- Chittka, A.; Nitarska, J.; Grazini, U.; Richardson, W.D. Transcription factor positive regulatory domain 4 (PRDM4) recruits protein arginine methyltransferase 5 (PRMT5) to mediate histone arginine methylation and control neural stem cell proliferation and differentiation. J. Biol. Chem. 2012, 287, 42995–43006. [Google Scholar] [CrossRef]
- Galli, G.G.; Carrara, M.; Francavilla, C.; De Lichtenberg, K.H.; Olsen, J.V.; Calogero, R.A.; Lund, A.H. Genomic and proteomic analyses of Prdm5 reveal interactions with insulator binding proteins in embryonic stem cells. Mol Cell Biol 2013, 33, 4504–4516. [Google Scholar] [CrossRef] [PubMed]
- Riddell, J.; Gazit, R.; Garrison, B.S.; Guo, G.; Saadatpour, A.; Mandal, P.K.; Ebina, W.; Volchkov, P.; Yuan, G.C.; Orkin, S.H.; et al. Reprogramming committed murine blood cells to induced hematopoietic stem cells with defined factors. Cell 2014, 157, 549–564. [Google Scholar] [CrossRef] [PubMed]
- Thoren, L.A.; Fog, C.K.; Jensen, K.T.; Buza-Vidas, N.; Come, C.; Lund, A.H.; Porse, B.T. PRDM11 is dispensable for the maintenance and function of hematopoietic stem and progenitor cells. Stem. Cell Res. 2013, 11, 1129–1136. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Okashita, N.; Sakashita, N.; Ito, K.; Mitsuya, A.; Suwa, Y.; Seki, Y. PRDM14 maintains pluripotency of embryonic stem cells through TET-mediated active DNA demethylation. Biochem. Biophys. Res. Commun. 2015, 466, 138–145. [Google Scholar] [CrossRef] [PubMed]
- Yamaji, M.; Ueda, J.; Hayashi, K.; Ohta, H.; Yabuta, Y.; Kurimoto, K.; Nakato, R.; Yamada, Y.; Shirahige, K.; Saitou, M. PRDM14 ensures naive pluripotency through dual regulation of signaling and epigenetic pathways in mouse embryonic stem cells. Cell Stem. Cell 2013, 12, 368–382. [Google Scholar] [CrossRef]
- Ma, Z.; Swigut, T.; Valouev, A.; Rada-Iglesias, A.; Wysocka, J. Sequence-specific regulator Prdm14 safeguards mouse ESCs from entering extraembryonic endoderm fates. Nat. Struct. Mol. Biol. 2011, 18, 120–127. [Google Scholar] [CrossRef] [PubMed]
- Tsuneyoshi, N.; Sumi, T.; Onda, H.; Nojima, H.; Nakatsuji, N.; Suemori, H. PRDM14 suppresses expression of differentiation marker genes in human embryonic stem cells. Biochem. Biophys. Res. Commun. 2008, 367, 899–905. [Google Scholar] [CrossRef]
- Payer, B.; Rosenberg, M.; Yamaji, M.; Yabuta, Y.; Koyanagi-Aoi, M.; Hayashi, K.; Yamanaka, S.; Saitou, M.; Lee, J.T. Tsix RNA and the germline factor, PRDM14, link X reactivation and stem cell reprogramming. Mol Cell 2013, 52, 805–818. [Google Scholar] [CrossRef]
- Yang, H.Y.; Kim, S.H.; Kim, S.H.; Kim, D.J.; Kim, S.U.; Yu, D.Y.; Yeom, Y.I.; Lee, D.S.; Kim, Y.J.; Park, B.J.; et al. The suppression of zfpm-1 accelerates the erythropoietic differentiation of human CD34+ cells. Biochem. Biophys. Res. Commun. 2007, 353, 978–984. [Google Scholar] [CrossRef]
- Amigo, J.D.; Ackermann, G.E.; Cope, J.J.; Yu, M.; Cooney, J.D.; Ma, D.; Langer, N.B.; Shafizadeh, E.; Shaw, G.C.; Horsely, W.; et al. The role and regulation of friend of GATA-1 (FOG-1) during blood development in the zebrafish. Blood 2009, 114, 4654–4663. [Google Scholar] [CrossRef]
- Gregory, G.D.; Miccio, A.; Bersenev, A.; Wang, Y.; Hong, W.; Zhang, Z.; Poncz, M.; Tong, W.; Blobel, G.A. FOG1 requires NuRD to promote hematopoiesis and maintain lineage fidelity within the megakaryocytic-erythroid compartment. Blood 2010, 115, 2156–2166. [Google Scholar] [CrossRef] [PubMed]
- Mancini, E.; Sanjuan-Pla, A.; Luciani, L.; Moore, S.; Grover, A.; Zay, A.; Rasmussen, K.D.; Luc, S.; Bilbao, D.; O’Carroll, D.; et al. FOG-1 and GATA-1 act sequentially to specify definitive megakaryocytic and erythroid progenitors. EMBO J. 2012, 31, 351–365. [Google Scholar] [CrossRef] [PubMed]
- Tripathy, N.K.; Rizvi, S.H.M.; Singh, S.P.; Garikpati, V.N.S.; Nityanand, S. Cardiomyogenic Heterogeneity of Clonal Subpopulations of Human Bone Marrow Mesenchymal Stem Cells. J. Stem. Cells Regen. Med. 2018, 14, 27–33. [Google Scholar] [PubMed]
- Mzoughi, S.; Tan, Y.X.; Low, D.; Guccione, E. The role of PRDMs in cancer: One family, two sides. Curr. Opin. Genet Dev. 2016, 36, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Sorrentino, A.; Rienzo, M.; Ciccodicola, A.; Casamassimi, A.; Abbondanza, C. Human PRDM2: Structure, function and pathophysiology. Biochim. Biophys. Acta Gene. Regul. Mech. 2018, 1861, 657–671. [Google Scholar] [CrossRef]
- Ren, B.; Chee, K.J.; Kim, T.H.; Maniatis, T. PRDI-BF1/Blimp-1 repression is mediated by corepressors of the Groucho family of proteins. Genes Dev. 1999, 13, 125–137. [Google Scholar] [CrossRef]
- Di Zazzo, E.; De Rosa, C.; Abbondanza, C.; Moncharmont, B. PRDM Proteins: Molecular Mechanisms in Signal Transduction and Transcriptional Regulation. Biology 2013, 2, 107–141. [Google Scholar] [CrossRef]
- Huang, S.; Shao, G.; Liu, L. The PR domain of the Rb-binding zinc finger protein RIZ1 is a protein binding interface and is related to the SET domain functioning in chromatin-mediated gene expression. J. Biol. Chem. 1998, 273, 15933–15939. [Google Scholar] [CrossRef]
- Bartholomew, C.; Kilbey, A.; Clark, A.M.; Walker, M. The Evi-1 proto-oncogene encodes a transcriptional repressor activity associated with transformation. Oncogene 1997, 14, 569–577. [Google Scholar] [CrossRef]
- Mochizuki, N.; Shimizu, S.; Nagasawa, T.; Tanaka, H.; Taniwaki, M.; Yokota, J.; Morishita, K. A novel gene, MEL1, mapped to 1p36.3 is highly homologous to the MDS1/EVI1 gene and is transcriptionally activated in t(1;3)(p36;q21)-positive leukemia cells. Blood 2000, 96, 3209–3214. [Google Scholar] [CrossRef]
- Hanotel, J.; Bessodes, N.; Thelie, A.; Hedderich, M.; Parain, K.; Van Driessche, B.; Brandao Kde, O.; Kricha, S.; Jorgensen, M.C.; Grapin-Botton, A.; et al. The Prdm13 histone methyltransferase encoding gene is a Ptf1a-Rbpj downstream target that suppresses glutamatergic and promotes GABAergic neuronal fate in the dorsal neural tube. Dev. Biol. 2014, 386, 340–357. [Google Scholar] [CrossRef] [PubMed]
- Zhou, B.; Wang, J.; Lee, S.Y.; Xiong, J.; Bhanu, N.; Guo, Q.; Ma, P.; Sun, Y.; Rao, R.C.; Garcia, B.A.; et al. PRDM16 Suppresses MLL1r Leukemia via Intrinsic Histone Methyltransferase Activity. Mol. Cell 2016, 62, 222–236. [Google Scholar] [CrossRef] [PubMed]
- Ivanochko, D.; Halabelian, L.; Henderson, E.; Savitsky, P.; Jain, H.; Marcon, E.; Duan, S.; Hutchinson, A.; Seitova, A.; Barsyte-Lovejoy, D.; et al. Direct interaction between the PRDM3 and PRDM16 tumor suppressors and the NuRD chromatin remodeling complex. Nucleic Acids Res. 2019, 47, 1225–1238. [Google Scholar] [CrossRef]
- Kajimura, S.; Seale, P.; Tomaru, T.; Erdjument-Bromage, H.; Cooper, M.P.; Ruas, J.L.; Chin, S.; Tempst, P.; Lazar, M.A.; Spiegelman, B.M. Regulation of the brown and white fat gene programs through a PRDM16/CtBP transcriptional complex. Genes Dev. 2008, 22, 1397–1409. [Google Scholar] [CrossRef] [PubMed]
- Izutsu, K.; Kurokawa, M.; Imai, Y.; Maki, K.; Mitani, K.; Hirai, H. The corepressor CtBP interacts with Evi-1 to repress transforming growth factor beta signaling. Blood 2001, 97, 2815–2822. [Google Scholar] [CrossRef]
- Garnatz, A.S.; Gao, Z.; Broman, M.; Martens, S.; Earley, J.U.; Svensson, E.C. FOG-2 mediated recruitment of the NuRD complex regulates cardiomyocyte proliferation during heart development. Dev. Biol. 2014, 395, 50–61. [Google Scholar] [CrossRef]
- Snow, J.W.; Kim, J.; Currie, C.R.; Xu, J.; Orkin, S.H. Sumoylation regulates interaction of FOG1 with C-terminal-binding protein (CTBP). J. Biol. Chem. 2010, 285, 28064–28075. [Google Scholar] [CrossRef]
- Wang, Y.; Meng, R.; Hayes, V.; Fuentes, R.; Yu, X.; Abrams, C.S.; Heijnen, H.F.; Blobel, G.A.; Marks, M.S.; Poncz, M. Pleiotropic platelet defects in mice with disrupted FOG1-NuRD interaction. Blood 2011, 118, 6183–6191. [Google Scholar] [CrossRef]
- Nitta, E.; Izutsu, K.; Yamaguchi, Y.; Imai, Y.; Ogawa, S.; Chiba, S.; Kurokawa, M.; Hirai, H. Oligomerization of Evi-1 regulated by the PR domain contributes to recruitment of corepressor CtBP. Oncogene 2005, 24, 6165–6173. [Google Scholar] [CrossRef]
- Lin, I.Y.; Chiu, F.L.; Yeang, C.H.; Chen, H.F.; Chuang, C.Y.; Yang, S.Y.; Hou, P.S.; Sintupisut, N.; Ho, H.N.; Kuo, H.C.; et al. Suppression of the SOX2 neural effector gene by PRDM1 promotes human germ cell fate in embryonic stem cells. Stem. Cell Reports 2014, 2, 189–204. [Google Scholar] [CrossRef]
- Seki, Y. PRDM14 Is a Unique Epigenetic Regulator Stabilizing Transcriptional Networks for Pluripotency. Front. Cell Dev. Biol. 2018, 6, 12. [Google Scholar] [CrossRef] [PubMed]
- Ficz, G.; Hore, T.A.; Santos, F.; Lee, H.J.; Dean, W.; Arand, J.; Krueger, F.; Oxley, D.; Paul, Y.L.; Walter, J.; et al. FGF signaling inhibition in ESCs drives rapid genome-wide demethylation to the epigenetic ground state of pluripotency. Cell Stem. Cell 2013, 13, 351–359. [Google Scholar] [CrossRef] [PubMed]
- Habibi, E.; Brinkman, A.B.; Arand, J.; Kroeze, L.I.; Kerstens, H.H.; Matarese, F.; Lepikhov, K.; Gut, M.; Brun-Heath, I.; Hubner, N.C.; et al. Whole-genome bisulfite sequencing of two distinct interconvertible DNA methylomes of mouse embryonic stem cells. Cell Stem. Cell 2013, 13, 360–369. [Google Scholar] [CrossRef]
- Burton, A.; Muller, J.; Tu, S.; Padilla-Longoria, P.; Guccione, E.; Torres-Padilla, M.E. Single-cell profiling of epigenetic modifiers identifies PRDM14 as an inducer of cell fate in the mammalian embryo. Cell Rep. 2013, 5, 687–701. [Google Scholar] [CrossRef] [PubMed]
- Kunath, T.; Saba-El-Leil, M.K.; Almousailleakh, M.; Wray, J.; Meloche, S.; Smith, A. FGF stimulation of the Erk1/2 signalling cascade triggers transition of pluripotent embryonic stem cells from self-renewal to lineage commitment. Development 2007, 134, 2895–2902. [Google Scholar] [CrossRef]
- Hao, J.; Li, T.G.; Qi, X.; Zhao, D.F.; Zhao, G.Q. WNT/beta-catenin pathway up-regulates Stat3 and converges on LIF to prevent differentiation of mouse embryonic stem cells. Dev. Biol. 2006, 290, 81–91. [Google Scholar] [CrossRef]
- Hippenmeyer, S. Molecular pathways controlling the sequential steps of cortical projection neuron migration. Adv. Exp. Med. Biol. 2014, 800, 1–24. [Google Scholar]
- Noctor, S.C.; Martinez-Cerdeno, V.; Ivic, L.; Kriegstein, A.R. Cortical neurons arise in symmetric and asymmetric division zones and migrate through specific phases. Nat. Neurosci. 2004, 7, 136–144. [Google Scholar] [CrossRef]
- Inoue, M.; Kuroda, T.; Honda, A.; Komabayashi-Suzuki, M.; Komai, T.; Shinkai, Y.; Mizutani, K. Prdm8 regulates the morphological transition at multipolar phase during neocortical development. PLoS ONE 2014, 9, e86356. [Google Scholar] [CrossRef]
- Ross, S.E.; McCord, A.E.; Jung, C.; Atan, D.; Mok, S.I.; Hemberg, M.; Kim, T.K.; Salogiannis, J.; Hu, L.; Cohen, S.; et al. Bhlhb5 and Prdm8 form a repressor complex involved in neuronal circuit assembly. Neuron 2012, 73, 292–303. [Google Scholar] [CrossRef]
- Chen, Y.C.; Auer-Grumbach, M.; Matsukawa, S.; Zitzelsberger, M.; Themistocleous, A.C.; Strom, T.M.; Samara, C.; Moore, A.W.; Cho, L.T.; Young, G.T.; et al. Transcriptional regulator PRDM12 is essential for human pain perception. Nat. Genet. 2015, 47, 803–808. [Google Scholar] [CrossRef] [PubMed]
- Desiderio, S.; Vermeiren, S.; Van Campenhout, C.; Kricha, S.; Malki, E.; Richts, S.; Fletcher, E.V.; Vanwelden, T.; Schmidt, B.Z.; Henningfeld, K.A.; et al. Prdm12 Directs Nociceptive Sensory Neuron Development by Regulating the Expression of the NGF Receptor TrkA. Cell Rep. 2019, 26, 3522–3536.e5. [Google Scholar] [CrossRef] [PubMed]
- Galazo, M.J.; Emsley, J.G.; Macklis, J.D. Corticothalamic Projection Neuron Development beyond Subtype Specification: Fog2 and Intersectional Controls Regulate Intraclass Neuronal Diversity. Neuron 2016, 91, 90–106. [Google Scholar] [CrossRef] [PubMed]
- Brzezinski, J.A.; Lamba, D.A.; Reh, T.A. Blimp1 controls photoreceptor versus bipolar cell fate choice during retinal development. Development 2010, 137, 619–629. [Google Scholar] [CrossRef] [PubMed]
- Chang, D.H.; Cattoretti, G.; Calame, K.L. The dynamic expression pattern of B lymphocyte induced maturation protein-1 (Blimp-1) during mouse embryonic development. Mech. Dev. 2002, 117, 305–309. [Google Scholar] [CrossRef]
- Katoh, K.; Omori, Y.; Onishi, A.; Sato, S.; Kondo, M.; Furukawa, T. Blimp1 suppresses Chx10 expression in differentiating retinal photoreceptor precursors to ensure proper photoreceptor development. J. Neurosci. 2010, 30, 6515–6526. [Google Scholar] [CrossRef]
- Roy, S.; Ng, T. Blimp-1 specifies neural crest and sensory neuron progenitors in the zebrafish embryo. Curr. Biol. 2004, 14, 1772–1777. [Google Scholar] [CrossRef]
- Barbier, E.; Johnstone, A.L.; Khomtchouk, B.B.; Tapocik, J.D.; Pitcairn, C.; Rehman, F.; Augier, E.; Borich, A.; Schank, J.R.; Rienas, C.A.; et al. Dependence-induced increase of alcohol self-administration and compulsive drinking mediated by the histone methyltransferase PRDM2. Mol. Psychiatry 2017, 22, 1746–1758. [Google Scholar] [CrossRef]
- Garriga, G.; Guenther, C.; Horvitz, H.R. Migrations of the Caenorhabditis elegans HSNs are regulated by egl-43, a gene encoding two zinc finger proteins. Genes Dev. 1993, 7, 2097–2109. [Google Scholar] [CrossRef][Green Version]
- Hoyt, P.R.; Bartholomew, C.; Davis, A.J.; Yutzey, K.; Gamer, L.W.; Potter, S.S.; Ihle, J.N.; Mucenski, M.L. The Evi1 proto-oncogene is required at midgestation for neural, heart, and paraxial mesenchyme development. Mech. Dev. 1997, 65, 55–70. [Google Scholar] [CrossRef]
- Kazama, H.; Kodera, T.; Shimizu, S.; Mizoguchi, H.; Morishita, K. Ecotropic viral integration site-1 is activated during, and is sufficient for, neuroectodermal P19 cell differentiation. Cell Growth Differ. 1999, 10, 565–573. [Google Scholar] [PubMed]
- Leszczynski, P.; Smiech, M.; Salam Teeli, A.; Haque, E.; Viger, R.; Ogawa, H.; Pierzchala, M.; Taniguchi, H. Deletion of the Prdm3 Gene Causes a Neuronal Differentiation Deficiency in P19 Cells. Int. J. Mol. Sci. 2020, 21, 7192. [Google Scholar] [CrossRef] [PubMed]
- Endo, K.; Karim, M.R.; Taniguchi, H.; Krejci, A.; Kinameri, E.; Siebert, M.; Ito, K.; Bray, S.J.; Moore, A.W. Chromatin modification of Notch targets in olfactory receptor neuron diversification. Nat. Neurosci. 2011, 15, 224–233. [Google Scholar] [CrossRef] [PubMed]
- Hou, Q.; Ruan, H.; Gilbert, J.; Wang, G.; Ma, Q.; Yao, W.D.; Man, H.Y. MicroRNA miR124 is required for the expression of homeostatic synaptic plasticity. Nat. Commun. 2015, 6, 10045. [Google Scholar] [CrossRef]
- Zhang, Y.; Liu, X.; Xue, H.; Liu, X.; Dai, A.; Song, Y.; Ke, K.; Cao, M. Upregulation of PRDM5 Is Associated with Astrocyte Proliferation and Neuronal Apoptosis Caused by Lipopolysaccharide. J. Mol. Neurosci. 2016, 59, 146–157. [Google Scholar] [CrossRef]
- Ling, W.; Xu, X.; Liu, J. A causal relationship between the neurotherapeutic effects of miR182/7a and decreased expression of PRDM5. Biochem. Biophys. Res. Commun. 2017, 490, 1–7. [Google Scholar] [CrossRef]
- Meani, N.; Pezzimenti, F.; Deflorian, G.; Mione, M.; Alcalay, M. The tumor suppressor PRDM5 regulates Wnt signaling at early stages of zebrafish development. PLoS ONE 2009, 4, e4273. [Google Scholar] [CrossRef]
- Jung, C.C.; Atan, D.; Ng, D.; Ploder, L.; Ross, S.E.; Klein, M.; Birch, D.G.; Diez, E.; McInnes, R.R. Transcription factor PRDM8 is required for rod bipolar and type 2 OFF-cone bipolar cell survival and amacrine subtype identity. Proc. Natl. Acad. Sci. USA 2015, 112, E3010–E3019. [Google Scholar] [CrossRef]
- Nagy, V.; Cole, T.; Van Campenhout, C.; Khoung, T.M.; Leung, C.; Vermeiren, S.; Novatchkova, M.; Wenzel, D.; Cikes, D.; Polyansky, A.A.; et al. The evolutionarily conserved transcription factor PRDM12 controls sensory neuron development and pain perception. Cell Cycle 2015, 14, 1799–1808. [Google Scholar] [CrossRef]
- Mona, B.; Uruena, A.; Kollipara, R.K.; Ma, Z.; Borromeo, M.D.; Chang, J.C.; Johnson, J.E. Repression by PRDM13 is critical for generating precision in neuronal identity. Elife 2017, 6. [Google Scholar] [CrossRef]
- Chang, J.C.; Meredith, D.M.; Mayer, P.R.; Borromeo, M.D.; Lai, H.C.; Ou, Y.H.; Johnson, J.E. Prdm13 mediates the balance of inhibitory and excitatory neurons in somatosensory circuits. Dev. Cell 2013, 25, 182–195. [Google Scholar] [CrossRef] [PubMed]
- Liu, C.; Ma, W.; Su, W.; Zhang, J. Prdm14 acts upstream of islet2 transcription to regulate axon growth of primary motoneurons in zebrafish. Development 2012, 139, 4591–4600. [Google Scholar] [CrossRef] [PubMed]
- Mzoughi, S.; Di Tullio, F.; Low, D.H.P.; Motofeanu, C.M.; Ong, S.L.M.; Wollmann, H.; Wun, C.M.; Kruszka, P.; Muenke, M.; Hildebrandt, F.; et al. PRDM15 loss of function links NOTCH and WNT/PCP signaling to patterning defects in holoprosencephaly. Sci. Adv. 2020, 6, eaax9852. [Google Scholar] [CrossRef] [PubMed]
- Su, L.; Lei, X.; Ma, H.; Feng, C.; Jiang, J.; Jiao, J. PRDM16 orchestrates angiogenesis via neural differentiation in the developing brain. Cell Death Differ. 2020, 27, 2313–2329. [Google Scholar] [CrossRef] [PubMed]
- Jia, Y.; Takimoto, K. GATA and FOG2 transcription factors differentially regulate the promoter for Kv4.2 K(+) channel gene in cardiac myocytes and PC12 cells. Cardiovasc. Res. 2003, 60, 278–287. [Google Scholar] [CrossRef]
- Tsai, F.Y.; Keller, G.; Kuo, F.C.; Weiss, M.; Chen, J.; Rosenblatt, M.; Alt, F.W.; Orkin, S.H. An early haematopoietic defect in mice lacking the transcription factor GATA-2. Nature 1994, 371, 221–226. [Google Scholar] [CrossRef]
- Shull, L.C.; Sen, R.; Menzel, J.; Goyama, S.; Kurokawa, M.; Artinger, K.B. The conserved and divergent roles of Prdm3 and Prdm16 in zebrafish and mouse craniofacial development. Dev. Biol. 2020, 461, 132–144. [Google Scholar] [CrossRef]
- Seale, P.; Bjork, B.; Yang, W.; Kajimura, S.; Chin, S.; Kuang, S.; Scime, A.; Devarakonda, S.; Conroe, H.M.; Erdjument-Bromage, H.; et al. PRDM16 controls a brown fat/skeletal muscle switch. Nature 2008, 454, 961–967. [Google Scholar] [CrossRef]
- Seale, P.; Kajimura, S.; Yang, W.; Chin, S.; Rohas, L.M.; Uldry, M.; Tavernier, G.; Langin, D.; Spiegelman, B.M. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007, 6, 38–54. [Google Scholar] [CrossRef]
- Arndt, A.K.; Schafer, S.; Drenckhahn, J.D.; Sabeh, M.K.; Plovie, E.R.; Caliebe, A.; Klopocki, E.; Musso, G.; Werdich, A.A.; Kalwa, H.; et al. Fine mapping of the 1p36 deletion syndrome identifies mutation of PRDM16 as a cause of cardiomyopathy. Am. J. Hum. Genet. 2013, 93, 67–77. [Google Scholar] [CrossRef]
- Shaffer, J.R.; Orlova, E.; Lee, M.K.; Leslie, E.J.; Raffensperger, Z.D.; Heike, C.L.; Cunningham, M.L.; Hecht, J.T.; Kau, C.H.; Nidey, N.L.; et al. Genome-Wide Association Study Reveals Multiple Loci Influencing Normal Human Facial Morphology. PLoS Genet 2016, 12, e1006149. [Google Scholar] [CrossRef]
- Ding, H.L.; Clouthier, D.E.; Artinger, K.B. Redundant roles of PRDM family members in zebrafish craniofacial development. Dev. Dyn. 2013, 242, 67–79. [Google Scholar] [CrossRef] [PubMed]
- Bjork, B.C.; Turbe-Doan, A.; Prysak, M.; Herron, B.J.; Beier, D.R. Prdm16 is required for normal palatogenesis in mice. Hum. Mol. Genet. 2010, 19, 774–789. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Wang, J.; Jiang, Z.; Guo, F.; Soloway, P.D.; Zhao, R. Role of PRDM16 and its PR domain in the epigenetic regulation of myogenic and adipogenic genes during transdifferentiation of C2C12 cells. Gene 2015, 570, 191–198. [Google Scholar] [CrossRef] [PubMed]
- Clifton, M.K.; Westman, B.J.; Thong, S.Y.; O’Connell, M.R.; Webster, M.W.; Shepherd, N.E.; Quinlan, K.G.; Crossley, M.; Blobel, G.A.; Mackay, J.P. The identification and structure of an N-terminal PR domain show that FOG1 is a member of the PRDM family of proteins. PLoS ONE 2014, 9, e106011. [Google Scholar] [CrossRef] [PubMed]
- Jack, B.H.; Crossley, M. GATA proteins work together with friend of GATA (FOG) and C-terminal binding protein (CTBP) co-regulators to control adipogenesis. J. Biol. Chem. 2010, 285, 32405–32414. [Google Scholar] [CrossRef]
- Rodriguez, P.; Bonte, E.; Krijgsveld, J.; Kolodziej, K.E.; Guyot, B.; Heck, A.J.; Vyas, P.; De Boer, E.; Grosveld, F.; Strouboulis, J. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J. 2005, 24, 2354–2366. [Google Scholar] [CrossRef]
- Hong, W.; Nakazawa, M.; Chen, Y.Y.; Kori, R.; Vakoc, C.R.; Rakowski, C.; Blobel, G.A. FOG-1 recruits the NuRD repressor complex to mediate transcriptional repression by GATA-1. EMBO J. 2005, 24, 2367–2378. [Google Scholar] [CrossRef]
- Tsang, A.P.; Visvader, J.E.; Turner, C.A.; Fujiwara, Y.; Yu, C.; Weiss, M.J.; Crossley, M.; Orkin, S.H. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 1997, 90, 109–119. [Google Scholar] [CrossRef]
- Zhou, B.; Ma, Q.; Kong, S.W.; Hu, Y.; Campbell, P.H.; McGowan, F.X.; Ackerman, K.G.; Wu, B.; Zhou, B.; Tevosian, S.G.; et al. Fog2 is critical for cardiac function and maintenance of coronary vasculature in the adult mouse heart. J. Clin. Investig. 2009, 119, 1462–1476. [Google Scholar] [CrossRef]
- Tan, Z.P.; Huang, C.; Xu, Z.B.; Yang, J.F.; Yang, Y.F. Novel ZFPM2/FOG2 variants in patients with double outlet right ventricle. Clin. Genet. 2012, 82, 466–471. [Google Scholar] [CrossRef] [PubMed]
- Pizzuti, A.; Sarkozy, A.; Newton, A.L.; Conti, E.; Flex, E.; Digilio, M.C.; Amati, F.; Gianni, D.; Tandoi, C.; Marino, B.; et al. Mutations of ZFPM2/FOG2 gene in sporadic cases of tetralogy of Fallot. Hum. Mutat. 2003, 22, 372–377. [Google Scholar] [CrossRef] [PubMed]
- Cuenco, G.M.; Nucifora, G.; Ren, R. Human AML1/MDS1/EVI1 fusion protein induces an acute myelogenous leukemia (AML) in mice: A model for human AML. Proc. Natl. Acad. Sci. USA 2000, 97, 1760–1765. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.M.; Comer, E.M.; Brown, F.C.; Richkind, K.E.; Holmes, M.L.; Chong, B.H.; Shiffman, R.; Zhang, D.E.; Slovak, M.L.; Willman, C.L.; et al. AML1-FOG2 fusion protein in myelodysplasia. Blood 2005, 105, 4523–4526. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Tsang, A.P.; Fujiwara, Y.; Hom, D.B.; Orkin, S.H. Failure of megakaryopoiesis and arrested erythropoiesis in mice lacking the GATA-1 transcriptional cofactor FOG. Genes Dev. 1998, 12, 1176–1188. [Google Scholar] [CrossRef] [PubMed]
- Querfurth, E.; Schuster, M.; Kulessa, H.; Crispino, J.D.; Doderlein, G.; Orkin, S.H.; Graf, T.; Nerlov, C. Antagonism between C/EBPbeta and FOG in eosinophil lineage commitment of multipotent hematopoietic progenitors. Genes Dev. 2000, 14, 2515–2525. [Google Scholar] [CrossRef]
- Du Roure, C.; Versavel, A.; Doll, T.; Cao, C.; Pillonel, V.; Matthias, G.; Kaller, M.; Spetz, J.F.; Kopp, P.; Kohler, H.; et al. Hematopoietic overexpression of FOG1 does not affect B-cells but reduces the number of circulating eosinophils. PLoS ONE 2014, 9, e92836. [Google Scholar] [CrossRef]
- Elkabetz, Y.; Panagiotakos, G.; Al Shamy, G.; Socci, N.D.; Tabar, V.; Studer, L. Human ES cell-derived neural rosettes reveal a functionally distinct early neural stem cell stage. Genes Dev. 2008, 22, 152–165. [Google Scholar] [CrossRef]
- Hirabayashi, Y.; Gotoh, Y. Epigenetic control of neural precursor cell fate during development. Nat. Rev. Neurosci. 2010, 11, 377–388. [Google Scholar] [CrossRef]
- Yamada, T.; Yang, Y.; Hemberg, M.; Yoshida, T.; Cho, H.Y.; Murphy, J.P.; Fioravante, D.; Regehr, W.G.; Gygi, S.P.; Georgopoulos, K.; et al. Promoter decommissioning by the NuRD chromatin remodeling complex triggers synaptic connectivity in the mammalian brain. Neuron 2014, 83, 122–134. [Google Scholar] [CrossRef]
- Dickstein, J.; Senyuk, V.; Premanand, K.; Laricchia-Robbio, L.; Xu, P.; Cattaneo, F.; Fazzina, R.; Nucifora, G. Methylation and silencing of miRNA-124 by EVI1 and self-renewal exhaustion of hematopoietic stem cells in murine myelodysplastic syndrome. Proc. Natl. Acad. Sci. USA 2010, 107, 9783–9788. [Google Scholar] [CrossRef] [PubMed]
- Walton, R.Z.; Bruce, A.E.; Olivey, H.E.; Najib, K.; Johnson, V.; Earley, J.U.; Ho, R.K.; Svensson, E.C. Fog1 is required for cardiac looping in zebrafish. Dev. Biol. 2006, 289, 482–493. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Yang, Y.; Li, B.; Zhang, X.; Zhao, Q.; Lou, X. The zinc finger protein Zfpm1 modulates ventricular trabeculation through Neuregulin-ErbB signalling. Dev. Biol. 2019, 446, 142–150. [Google Scholar] [CrossRef] [PubMed]
- Meier, S.; Strohmaier, J.; Breuer, R.; Mattheisen, M.; Degenhardt, F.; Muhleisen, T.W.; Schulze, T.G.; Nothen, M.M.; Cichon, S.; Rietschel, M.; et al. Neuregulin 3 is associated with attention deficits in schizophrenia and bipolar disorder. Int. J. Neuropsychopharmacol. 2013, 16, 549–556. [Google Scholar] [CrossRef] [PubMed]
- Benzel, I.; Bansal, A.; Browning, B.L.; Galwey, N.W.; Maycox, P.R.; McGinnis, R.; Smart, D.; St Clair, D.; Yates, P.; Purvis, I. Interactions among genes in the ErbB-Neuregulin signalling network are associated with increased susceptibility to schizophrenia. Behav. Brain Funct. 2007, 3, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Mei, L.; Nave, K.A. Neuregulin-ERBB signaling in the nervous system and neuropsychiatric diseases. Neuron 2014, 83, 27–49. [Google Scholar] [CrossRef] [PubMed]
- Lu, J.R.; McKinsey, T.A.; Xu, H.; Wang, D.Z.; Richardson, J.A.; Olson, E.N. FOG-2, a heart- and brain-enriched cofactor for GATA transcription factors. Mol. Cell Biol. 1999, 19, 4495–4502. [Google Scholar] [CrossRef]
- Agnihotri, S.; Wolf, A.; Picard, D.; Hawkins, C.; Guha, A. GATA4 is a regulator of astrocyte cell proliferation and apoptosis in the human and murine central nervous system. Oncogene 2009, 28, 3033–3046. [Google Scholar] [CrossRef]
- Kamnasaran, D.; Guha, A. Expression of GATA6 in the human and mouse central nervous system. Brain Res. Dev. Brain Res. 2005, 160, 90–95. [Google Scholar] [CrossRef]
- Bortone, D.S.; Olsen, S.R.; Scanziani, M. Translaminar inhibitory cells recruited by layer 6 corticothalamic neurons suppress visual cortex. Neuron 2014, 82, 474–485. [Google Scholar] [CrossRef]
- Olsen, S.R.; Bortone, D.S.; Adesnik, H.; Scanziani, M. Gain control by layer six in cortical circuits of vision. Nature 2012, 483, 47–52. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Brylka, H.; Schwegler, H.; Venkataramanappa, S.; Andratschke, J.; Wiegreffe, C.; Liu, P.; Fuchs, E.; Jenkins, N.A.; Copeland, N.G.; et al. A dual function of Bcl11b/Ctip2 in hippocampal neurogenesis. EMBO J. 2012, 31, 2922–2936. [Google Scholar] [CrossRef] [PubMed]
- Thierry, G.; Pichon, O.; Briand, A.; Poulain, D.; Sznajer, Y.; David, A.; Le Caignec, C. Autosomal insertional translocation mimicking an X-linked mode of inheritance. Eur. J. Med. Genet. 2013, 56, 46–49. [Google Scholar] [CrossRef] [PubMed]
- Wat, M.J.; Veenma, D.; Hogue, J.; Holder, A.M.; Yu, Z.; Wat, J.J.; Hanchard, N.; Shchelochkov, O.A.; Fernandes, C.J.; Johnson, A.; et al. Genomic alterations that contribute to the development of isolated and non-isolated congenital diaphragmatic hernia. J. Med. Genet. 2011, 48, 299–307. [Google Scholar] [CrossRef]
- Ee, L.S.; McCannell, K.N.; Tang, Y.; Fernandes, N.; Hardy, W.R.; Green, M.R.; Chu, F.; Fazzio, T.G. An Embryonic Stem Cell-Specific NuRD Complex Functions through Interaction with WDR5. Stem. Cell Rep. 2017, 8, 1488–1496. [Google Scholar] [CrossRef]
- Xue, Y.; Wong, J.; Moreno, G.T.; Young, M.K.; Cote, J.; Wang, W. NURD, a novel complex with both ATP-dependent chromatin-remodeling and histone deacetylase activities. Mol. Cell 1998, 2, 851–861. [Google Scholar] [CrossRef]
- Allen, H.F.; Wade, P.A.; Kutateladze, T.G. The NuRD architecture. Cell Mol. Life Sci. 2013, 70, 3513–3524. [Google Scholar] [CrossRef]
- Zhang, Y.; LeRoy, G.; Seelig, H.P.; Lane, W.S.; Reinberg, D. The dermatomyositis-specific autoantigen Mi2 is a component of a complex containing histone deacetylase and nucleosome remodeling activities. Cell 1998, 95, 279–289. [Google Scholar] [CrossRef]
- Zhang, Y.; Ng, H.H.; Erdjument-Bromage, H.; Tempst, P.; Bird, A.; Reinberg, D. Analysis of the NuRD subunits reveals a histone deacetylase core complex and a connection with DNA methylation. Genes Dev. 1999, 13, 1924–1935. [Google Scholar] [CrossRef]
- Bornelov, S.; Reynolds, N.; Xenophontos, M.; Gharbi, S.; Johnstone, E.; Floyd, R.; Ralser, M.; Signolet, J.; Loos, R.; Dietmann, S.; et al. The Nucleosome Remodeling and Deacetylation Complex Modulates Chromatin Structure at Sites of Active Transcription to Fine-Tune Gene Expression. Mol. Cell 2018, 71, 56–72.e4. [Google Scholar] [CrossRef]
- Reynolds, N.; Latos, P.; Hynes-Allen, A.; Loos, R.; Leaford, D.; O’Shaughnessy, A.; Mosaku, O.; Signolet, J.; Brennecke, P.; Kalkan, T.; et al. NuRD suppresses pluripotency gene expression to promote transcriptional heterogeneity and lineage commitment. Cell Stem. Cell 2012, 10, 583–594. [Google Scholar] [CrossRef] [PubMed]
- Jung, B.P.; Zhang, G.; Nitsch, R.; Trogadis, J.; Nag, S.; Eubanks, J.H. Differential expression of methyl CpG-binding domain containing factor MBD3 in the developing and adult rat brain. J. Neurobiol. 2003, 55, 220–232. [Google Scholar] [CrossRef] [PubMed]
- Knock, E.; Pereira, J.; Lombard, P.D.; Dimond, A.; Leaford, D.; Livesey, F.J.; Hendrich, B. The methyl binding domain 3/nucleosome remodelling and deacetylase complex regulates neural cell fate determination and terminal differentiation in the cerebral cortex. Neural Dev. 2015, 10, 13. [Google Scholar] [CrossRef] [PubMed]
- Muralidharan, B.; Khatri, Z.; Maheshwari, U.; Gupta, R.; Roy, B.; Pradhan, S.J.; Karmodiya, K.; Padmanabhan, H.; Shetty, A.S.; Balaji, C.; et al. LHX2 Interacts with the NuRD Complex and Regulates Cortical Neuron Subtype Determinants Fezf2 and Sox11. J. Neurosci. 2017, 37, 194–203. [Google Scholar] [CrossRef] [PubMed]
- Harb, K.; Magrinelli, E.; Nicolas, C.S.; Lukianets, N.; Frangeul, L.; Pietri, M.; Sun, T.; Sandoz, G.; Grammont, F.; Jabaudon, D.; et al. Area-specific development of distinct projection neuron subclasses is regulated by postnatal epigenetic modifications. Elife 2016, 5, e09531. [Google Scholar] [CrossRef] [PubMed]
- Schmitges, F.W.; Prusty, A.B.; Faty, M.; Stutzer, A.; Lingaraju, G.M.; Aiwazian, J.; Sack, R.; Hess, D.; Li, L.; Zhou, S.; et al. Histone methylation by PRC2 is inhibited by active chromatin marks. Mol. Cell 2011, 42, 330–341. [Google Scholar] [CrossRef] [PubMed]
- Roche, A.E.; Bassett, B.J.; Samant, S.A.; Hong, W.; Blobel, G.A.; Svensson, E.C. The zinc finger and C-terminal domains of MTA proteins are required for FOG-2-mediated transcriptional repression via the NuRD complex. J. Mol. Cell Cardiol. 2008, 44, 352–360. [Google Scholar] [CrossRef][Green Version]
- Shimahara, A.; Yamakawa, N.; Nishikata, I.; Morishita, K. Acetylation of lysine 564 adjacent to the C-terminal binding protein-binding motif in EVI1 is crucial for transcriptional activation of GATA2. J. Biol. Chem. 2010, 285, 16967–16977. [Google Scholar] [CrossRef]
- Senyuk, V.; Sinha, K.K.; Chakraborty, S.; Buonamici, S.; Nucifora, G. P/CAF and GCN5 acetylate the AML1/MDS1/EVI1 fusion oncoprotein. Biochem. Biophys. Res. Commun. 2003, 307, 980–986. [Google Scholar] [CrossRef]
- Chakraborty, S.; Senyuk, V.; Sitailo, S.; Chi, Y.; Nucifora, G. Interaction of EVI1 with cAMP-responsive element-binding protein-binding protein (CBP) and p300/CBP-associated factor (P/CAF) results in reversible acetylation of EVI1 and in co-localization in nuclear speckles. J. Biol. Chem. 2001, 276, 44936–44943. [Google Scholar] [CrossRef]
- Miccio, A.; Wang, Y.; Hong, W.; Gregory, G.D.; Wang, H.; Yu, X.; Choi, J.K.; Shelat, S.; Tong, W.; Poncz, M.; et al. NuRD mediates activating and repressive functions of GATA-1 and FOG-1 during blood development. EMBO J. 2010, 29, 442–456. [Google Scholar] [CrossRef] [PubMed]
- Svensson, E.C.; Tufts, R.L.; Polk, C.E.; Leiden, J.M. Molecular cloning of FOG-2: A modulator of transcription factor GATA-4 in cardiomyocytes. Proc. Natl. Acad. Sci. USA 1999, 96, 956–961. [Google Scholar] [CrossRef] [PubMed]
- Fears, S.; Mathieu, C.; Zeleznik-Le, N.; Huang, S.; Rowley, J.D.; Nucifora, G. Intergenic splicing of MDS1 and EVI1 occurs in normal tissues as well as in myeloid leukemia and produces a new member of the PR domain family. Proc. Natl. Acad. Sci. USA 1996, 93, 1642–1647. [Google Scholar] [CrossRef] [PubMed]
- Groschel, S.; Lugthart, S.; Schlenk, R.F.; Valk, P.J.; Eiwen, K.; Goudswaard, C.; Van Putten, W.J.; Kayser, S.; Verdonck, L.F.; Lubbert, M.; et al. High EVI1 expression predicts outcome in younger adult patients with acute myeloid leukemia and is associated with distinct cytogenetic abnormalities. J. Clin. Oncol. 2010, 28, 2101–2107. [Google Scholar] [CrossRef] [PubMed]
- Lugthart, S.; Van Drunen, E.; Van Norden, Y.; Van Hoven, A.; Erpelinck, C.A.; Valk, P.J.; Beverloo, H.B.; Lowenberg, B.; Delwel, R. High EVI1 levels predict adverse outcome in acute myeloid leukemia: Prevalence of EVI1 overexpression and chromosome 3q26 abnormalities underestimated. Blood 2008, 111, 4329–4337. [Google Scholar] [CrossRef] [PubMed]
- Suzukawa, K.; Parganas, E.; Gajjar, A.; Abe, T.; Takahashi, S.; Tani, K.; Asano, S.; Asou, H.; Kamada, N.; Yokota, J. Identification of a breakpoint cluster region 3’ of the ribophorin I gene at 3q21 associated with the transcriptional activation of the EVI1 gene in acute myelogenous leukemias with inv(3)(q21q26). Blood 1994, 84, 2681–2688. [Google Scholar] [CrossRef] [PubMed]
- Brooks, D.J.; Woodward, S.; Thompson, F.H.; Dos Santos, B.; Russell, M.; Yang, J.M.; Guan, X.Y.; Trent, J.; Alberts, D.S.; Taetle, R. Expression of the zinc finger gene EVI-1 in ovarian and other cancers. Br. J. Cancer 1996, 74, 1518–1525. [Google Scholar] [CrossRef]
- Sattler, H.P.; Lensch, R.; Rohde, V.; Zimmer, E.; Meese, E.; Bonkhoff, H.; Retz, M.; Zwergel, T.; Bex, A.; Stoeckle, M.; et al. Novel amplification unit at chromosome 3q25-q27 in human prostate cancer. Prostate 2000, 45, 207–215. [Google Scholar] [CrossRef]
- Yasui, K.; Konishi, C.; Gen, Y.; Endo, M.; Dohi, O.; Tomie, A.; Kitaichi, T.; Yamada, N.; Iwai, N.; Nishikawa, T.; et al. EVI1, a target gene for amplification at 3q26, antagonizes transforming growth factor-beta-mediated growth inhibition in hepatocellular carcinoma. Cancer Sci. 2015, 106, 929–937. [Google Scholar] [CrossRef][Green Version]
- Sorrentino, A.; Federico, A.; Rienzo, M.; Gazzerro, P.; Bifulco, M.; Ciccodicola, A.; Casamassimi, A.; Abbondanza, C. PR/SET Domain Family and Cancer: Novel Insights from the Cancer Genome Atlas. Int. J. Mol. Sci. 2018, 19, 3250. [Google Scholar] [CrossRef]
- Shing, D.C.; Trubia, M.; Marchesi, F.; Radaelli, E.; Belloni, E.; Tapinassi, C.; Scanziani, E.; Mecucci, C.; Crescenzi, B.; Lahortiga, I.; et al. Overexpression of sPRDM16 coupled with loss of p53 induces myeloid leukemias in mice. J. Clin. Investig. 2007, 117, 3696–3707. [Google Scholar] [CrossRef] [PubMed]
- Ramirez, J.; Hagman, J. The Mi-2/NuRD complex: A critical epigenetic regulator of hematopoietic development, differentiation and cancer. Epigenetics 2009, 4, 532–536. [Google Scholar] [CrossRef] [PubMed]
- Hoffmann, A.; Spengler, D. Chromatin Remodeling Complex NuRD in Neurodevelopment and Neurodevelopmental Disorders. Front. Genet. 2019, 10, 682. [Google Scholar] [CrossRef]
- Chinnadurai, G. CtBP family proteins: More than transcriptional corepressors. Bioessays 2003, 25, 9–12. [Google Scholar] [CrossRef] [PubMed]
- Ivanova, D.; Dirks, A.; Montenegro-Venegas, C.; Schone, C.; Altrock, W.D.; Marini, C.; Frischknecht, R.; Schanze, D.; Zenker, M.; Gundelfinger, E.D.; et al. Synaptic activity controls localization and function of CtBP1 via binding to Bassoon and Piccolo. EMBO J. 2015, 34, 1056–1077. [Google Scholar] [CrossRef]
- Nishikata, I.; Nakahata, S.; Saito, Y.; Kaneda, K.; Ichihara, E.; Yamakawa, N.; Morishita, K. Sumoylation of MEL1S at lysine 568 and its interaction with CtBP facilitates its repressor activity and the blockade of G-CSF-induced myeloid differentiation. Oncogene 2011, 30, 4194–4207. [Google Scholar] [CrossRef]
- Lee, L.; Dale, E.; Staniszewski, A.; Zhang, H.; Saeed, F.; Sakurai, M.; Fa’, M.; Orozco, I.; Michelassi, F.; Akpan, N.; et al. Regulation of synaptic plasticity and cognition by SUMO in normal physiology and Alzheimer’s disease. Sci. Rep. 2014, 4, 7190. [Google Scholar] [CrossRef]
- Henley, J.M.; Craig, T.J.; Wilkinson, K.A. Neuronal SUMOylation: Mechanisms, physiology, and roles in neuronal dysfunction. Physiol. Rev. 2014, 94, 1249–1285. [Google Scholar] [CrossRef]
- Hasegawa, Y.; Yoshida, D.; Nakamura, Y.; Sakakibara, S. Spatiotemporal distribution of SUMOylation components during mouse brain development. J. Comp. Neurol. 2014, 522, 3020–3036. [Google Scholar] [CrossRef]
- Maruyama, T.; Wada, H.; Abe, Y.; Niikura, T. Alteration of global protein SUMOylation in neurons and astrocytes in response to Alzheimer’s disease-associated insults. Biochem. Biophys. Res. Commun. 2018, 500, 470–475. [Google Scholar] [CrossRef]
- Henley, J.M.; Carmichael, R.E.; Wilkinson, K.A. Extranuclear SUMOylation in Neurons. Trends Neurosci. 2018, 41, 198–210. [Google Scholar] [CrossRef] [PubMed]
- Thiruvalluvan, M.; Barghouth, P.G.; Tsur, A.; Broday, L.; Oviedo, N.J. SUMOylation controls stem cell proliferation and regional cell death through Hedgehog signaling in planarians. Cell Mol. Life Sci. 2018, 75, 1285–1301. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.; Pradhan, A.K.; Chakraborty, S. SUMO1 negatively regulates the transcriptional activity of EVI1 and significantly increases its co-localization with EVI1 after treatment with arsenic trioxide. Biochim. Biophys. Acta 2013, 1833, 2357–2368. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Chen, Q.; Huang, L.; Pan, D.; Zhu, L.J.; Wang, Y.X. Cbx4 Sumoylates Prdm16 to Regulate Adipose Tissue Thermogenesis. Cell Rep. 2018, 22, 2860–2872. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.; Chen, J. SUMOylation of sPRDM16 promotes the progression of acute myeloid leukemia. BMC Cancer 2015, 15, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Yang, X.J.; Gregoire, S. A recurrent phospho-sumoyl switch in transcriptional repression and beyond. Mol. Cell 2006, 23, 779–786. [Google Scholar] [CrossRef]
- Chi, Y.; Senyuk, V.; Chakraborty, S.; Nucifora, G. EVI1 promotes cell proliferation by interacting with BRG1 and blocking the repression of BRG1 on E2F1 activity. J. Biol. Chem. 2003, 278, 49806–49811. [Google Scholar] [CrossRef]
- Trotter, K.W.; Archer, T.K. The BRG1 transcriptional coregulator. Nucl. Recept. Signal. 2008, 6, e004. [Google Scholar] [CrossRef]
- Kadam, S.; Emerson, B.M. Transcriptional specificity of human SWI/SNF BRG1 and BRM chromatin remodeling complexes. Mol. Cell 2003, 11, 377–389. [Google Scholar] [CrossRef]
- Hoffmeister, H.; Fuchs, A.; Erdel, F.; Pinz, S.; Grobner-Ferreira, R.; Bruckmann, A.; Deutzmann, R.; Schwartz, U.; Maldonado, R.; Huber, C.; et al. CHD3 and CHD4 form distinct NuRD complexes with different yet overlapping functionality. Nucleic Acids Res. 2017, 45, 10534–10554. [Google Scholar] [CrossRef]
- Kajimura, S.; Seale, P.; Kubota, K.; Lunsford, E.; Frangioni, J.V.; Gygi, S.P.; Spiegelman, B.M. Initiation of myoblast to brown fat switch by a PRDM16-C/EBP-beta transcriptional complex. Nature 2009, 460, 1154–1158. [Google Scholar] [CrossRef] [PubMed]
- Menard, C.; Hein, P.; Paquin, A.; Savelson, A.; Yang, X.M.; Lederfein, D.; Barnabe-Heider, F.; Mir, A.A.; Sterneck, E.; Peterson, A.C.; et al. An essential role for a MEK-C/EBP pathway during growth factor-regulated cortical neurogenesis. Neuron 2002, 36, 597–610. [Google Scholar] [CrossRef]
- Cortes-Canteli, M.; Aguilar-Morante, D.; Sanz-Sancristobal, M.; Megias, D.; Santos, A.; Perez-Castillo, A. Role of C/EBPbeta transcription factor in adult hippocampal neurogenesis. PLoS ONE 2011, 6, e24842. [Google Scholar] [CrossRef] [PubMed]
- Bard-Chapeau, E.A.; Gunaratne, J.; Kumar, P.; Chua, B.Q.; Muller, J.; Bard, F.A.; Blackstock, W.; Copeland, N.G.; Jenkins, N.A. EVI1 oncoprotein interacts with a large and complex network of proteins and integrates signals through protein phosphorylation. Proc. Natl. Acad. Sci. USA 2013, 110, E2885–E2894. [Google Scholar] [CrossRef] [PubMed]
- Tokita, K.; Maki, K.; Mitani, K. RUNX1/EVI1, which blocks myeloid differentiation, inhibits CCAAT-enhancer binding protein alpha function. Cancer Sci. 2007, 98, 1752–1757. [Google Scholar] [CrossRef] [PubMed]
- Senyuk, V.; Chakraborty, S.; Mikhail, F.M.; Zhao, R.; Chi, Y.; Nucifora, G. The leukemia-associated transcription repressor AML1/MDS1/EVI1 requires CtBP to induce abnormal growth and differentiation of murine hematopoietic cells. Oncogene 2002, 21, 3232–3240. [Google Scholar] [CrossRef] [PubMed]
- Turner, J.; Crossley, M. Cloning and characterization of mCtBP2, a co-repressor that associates with basic Kruppel-like factor and other mammalian transcriptional regulators. EMBO J. 1998, 17, 5129–5140. [Google Scholar] [CrossRef] [PubMed]
- Senyuk, V.; Sinha, K.K.; Li, D.; Rinaldi, C.R.; Yanamandra, S.; Nucifora, G. Repression of RUNX1 activity by EVI1: A new role of EVI1 in leukemogenesis. Cancer Res. 2007, 67, 5658–5666. [Google Scholar] [CrossRef]
- Laricchia-Robbio, L.; Fazzina, R.; Li, D.; Rinaldi, C.R.; Sinha, K.K.; Chakraborty, S.; Nucifora, G. Point mutations in two EVI1 Zn fingers abolish EVI1-GATA1 interaction and allow erythroid differentiation of murine bone marrow cells. Mol. Cell Biol. 2006, 26, 7658–7666. [Google Scholar] [CrossRef][Green Version]
- Laricchia-Robbio, L.; Premanand, K.; Rinaldi, C.R.; Nucifora, G. EVI1 Impairs myelopoiesis by deregulation of PU.1 function. Cancer Res. 2009, 69, 1633–1642. [Google Scholar] [CrossRef][Green Version]
- Kurokawa, M.; Mitani, K.; Imai, Y.; Ogawa, S.; Yazaki, Y.; Hirai, H. The t(3;21) fusion product, AML1/Evi-1, interacts with Smad3 and blocks transforming growth factor-beta-mediated growth inhibition of myeloid cells. Blood 1998, 92, 4003–4012. [Google Scholar] [CrossRef] [PubMed]
- Kurokawa, M.; Mitani, K.; Yamagata, T.; Takahashi, T.; Izutsu, K.; Ogawa, S.; Moriguchi, T.; Nishida, E.; Yazaki, Y.; Hirai, H. The evi-1 oncoprotein inhibits c-Jun N-terminal kinase and prevents stress-induced cell death. EMBO J. 2000, 19, 2958–2968. [Google Scholar] [CrossRef] [PubMed]
- Lu, Y.; Liang, Y.; Zheng, X.; Deng, X.; Huang, W.; Zhang, G. EVI1 promotes epithelial-to-mesenchymal transition, cancer stem cell features and chemo-/radioresistance in nasopharyngeal carcinoma. J. Exp. Clin. Cancer Res. 2019, 38, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Senyuk, V.; Premanand, K.; Xu, P.; Qian, Z.; Nucifora, G. The oncoprotein EVI1 and the DNA methyltransferase Dnmt3 co-operate in binding and de novo methylation of target DNA. PLoS ONE 2011, 6, e20793. [Google Scholar] [CrossRef] [PubMed]
- Goyama, S.; Nitta, E.; Yoshino, T.; Kako, S.; Watanabe-Okochi, N.; Shimabe, M.; Imai, Y.; Takahashi, K.; Kurokawa, M. EVI-1 interacts with histone methyltransferases SUV39H1 and G9a for transcriptional repression and bone marrow immortalization. Leukemia 2010, 24, 81–88. [Google Scholar] [CrossRef] [PubMed]
- Spensberger, D.; Delwel, R. A novel interaction between the proto-oncogene Evi1 and histone methyltransferases, SUV39H1 and G9a. FEBS Lett. 2008, 582, 2761–2767. [Google Scholar] [CrossRef]
- Pradhan, A.K.; Halder, A.; Chakraborty, S. Physical and functional interaction of the proto-oncogene EVI1 and tumor suppressor gene HIC1 deregulates Bcl-xL mediated block in apoptosis. Int. J. Biochem. Cell Biol. 2014, 53, 320–328. [Google Scholar] [CrossRef]
- Xu, X.; Woo, C.H.; Steere, R.R.; Lee, B.C.; Huang, Y.; Wu, J.; Pang, J.; Lim, J.H.; Xu, H.; Zhang, W.; et al. EVI1 acts as an inducible negative-feedback regulator of NF-kappaB by inhibiting p65 acetylation. J. Immunol. 2012, 188, 6371–6380. [Google Scholar] [CrossRef]
- Spensberger, D.; Vermeulen, M.; Le Guezennec, X.; Beekman, R.; Van Hoven, A.; Bindels, E.; Stunnenberg, H.; Delwel, R. Myeloid transforming protein Evi1 interacts with methyl-CpG binding domain protein 3 and inhibits in vitro histone deacetylation by Mbd3/Mi-2/NuRD. Biochemistry 2008, 47, 6418–6426. [Google Scholar] [CrossRef]
- Ohno, H.; Shinoda, K.; Ohyama, K.; Sharp, L.Z.; Kajimura, S. EHMT1 controls brown adipose cell fate and thermogenesis through the PRDM16 complex. Nature 2013, 504, 163–167. [Google Scholar] [CrossRef]
- Takahata, M.; Inoue, Y.; Tsuda, H.; Imoto, I.; Koinuma, D.; Hayashi, M.; Ichikura, T.; Yamori, T.; Nagasaki, K.; Yoshida, M.; et al. SKI and MEL1 cooperate to inhibit transforming growth factor-beta signal in gastric cancer cells. J. Biol. Chem. 2009, 284, 3334–3344. [Google Scholar] [CrossRef] [PubMed]
- Zeng, X.; Jedrychowski, M.P.; Chen, Y.; Serag, S.; Lavery, G.G.; Gygi, S.P.; Spiegelman, B.M. Lysine-specific demethylase 1 promotes brown adipose tissue thermogenesis via repressing glucocorticoid activation. Genes Dev. 2016, 30, 1822–1836. [Google Scholar] [CrossRef] [PubMed]
- Harms, M.J.; Lim, H.W.; Ho, Y.; Shapira, S.N.; Ishibashi, J.; Rajakumari, S.; Steger, D.J.; Lazar, M.A.; Won, K.J.; Seale, P. PRDM16 binds MED1 and controls chromatin architecture to determine a brown fat transcriptional program. Genes Dev. 2015, 29, 298–307. [Google Scholar] [CrossRef] [PubMed]
- Dempersmier, J.; Sambeat, A.; Gulyaeva, O.; Paul, S.M.; Hudak, C.S.; Raposo, H.F.; Kwan, H.Y.; Kang, C.; Wong, R.H.; Sul, H.S. Cold-inducible Zfp516 activates UCP1 transcription to promote browning of white fat and development of brown fat. Mol. Cell 2015, 57, 235–246. [Google Scholar] [CrossRef]
- Fox, A.H.; Liew, C.; Holmes, M.; Kowalski, K.; Mackay, J.; Crossley, M. Transcriptional cofactors of the FOG family interact with GATA proteins by means of multiple zinc fingers. EMBO J. 1999, 18, 2812–2822. [Google Scholar] [CrossRef]
- Liew, C.K.; Simpson, R.J.; Kwan, A.H.; Crofts, L.A.; Loughlin, F.E.; Matthews, J.M.; Crossley, M.; Mackay, J.P. Zinc fingers as protein recognition motifs: Structural basis for the GATA-1/friend of GATA interaction. Proc. Natl. Acad. Sci. USA 2005, 102, 583–588. [Google Scholar] [CrossRef]
- Deconinck, A.E.; Mead, P.E.; Tevosian, S.G.; Crispino, J.D.; Katz, S.G.; Zon, L.I.; Orkin, S.H. FOG acts as a repressor of red blood cell development in Xenopus. Development 2000, 127, 2031–2040. [Google Scholar]
- Snow, J.W.; Orkin, S.H. Translational isoforms of FOG1 regulate GATA1-interacting complexes. J. Biol. Chem. 2009, 284, 29310–29319. [Google Scholar] [CrossRef]
- Huggins, G.S.; Bacani, C.J.; Boltax, J.; Aikawa, R.; Leiden, J.M. Friend of GATA 2 physically interacts with chicken ovalbumin upstream promoter-TF2 (COUP-TF2) and COUP-TF3 and represses COUP-TF2-dependent activation of the atrial natriuretic factor promoter. J. Biol. Chem. 2001, 276, 28029–28036. [Google Scholar] [CrossRef]
- Carter, D.R.; Buckle, A.D.; Tanaka, K.; Perdomo, J.; Chong, B.H. Art27 interacts with GATA4, FOG2 and NKX2.5 and is a novel co-repressor of cardiac genes. PLoS ONE 2014, 9, e95253. [Google Scholar] [CrossRef]
- Clabby, M.L.; Robison, T.A.; Quigley, H.F.; Wilson, D.B.; Kelly, D.P. Retinoid X receptor alpha represses GATA-4-mediated transcription via a retinoid-dependent interaction with the cardiac-enriched repressor FOG-2. J. Biol. Chem. 2003, 278, 5760–5767. [Google Scholar] [CrossRef] [PubMed]
- Boi, M.; Zucca, E.; Inghirami, G.; Bertoni, F. PRDM1/BLIMP1: A tumor suppressor gene in B and T cell lymphomas. Leuk. Lymphoma 2015, 56, 1223–1228. [Google Scholar] [CrossRef] [PubMed]
- Kucuk, C.; Iqbal, J.; Hu, X.; Gaulard, P.; De Leval, L.; Srivastava, G.; Au, W.Y.; McKeithan, T.W.; Chan, W.C. PRDM1 is a tumor suppressor gene in natural killer cell malignancies. Proc. Natl. Acad. Sci. USA 2011, 108, 20119–20124. [Google Scholar] [CrossRef] [PubMed]
- Nie, K.; Gomez, M.; Landgraf, P.; Garcia, J.F.; Liu, Y.; Tan, L.H.; Chadburn, A.; Tuschl, T.; Knowles, D.M.; Tam, W. MicroRNA-mediated down-regulation of PRDM1/Blimp-1 in Hodgkin/Reed-Sternberg cells: A potential pathogenetic lesion in Hodgkin lymphomas. Am. J. Pathol. 2008, 173, 242–252. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Z.; Wang, H.; Wei, Y.; Meng, F.; Liu, Z.; Zhang, Z. Downregulation of PRDM1 promotes cellular invasion and lung cancer metastasis. Tumour. Biol. 2017, 39, 1010428317695929. [Google Scholar] [CrossRef]
- Nie, K.; Zhang, T.; Allawi, H.; Gomez, M.; Liu, Y.; Chadburn, A.; Wang, Y.L.; Knowles, D.M.; Tam, W. Epigenetic down-regulation of the tumor suppressor gene PRDM1/Blimp-1 in diffuse large B cell lymphomas: A potential role of the microRNA let-7. Am. J. Pathol. 2010, 177, 1470–1479. [Google Scholar] [CrossRef]
- Tam, W.; Gomez, M.; Chadburn, A.; Lee, J.W.; Chan, W.C.; Knowles, D.M. Mutational analysis of PRDM1 indicates a tumor-suppressor role in diffuse large B-cell lymphomas. Blood 2006, 107, 4090–4100. [Google Scholar] [CrossRef]
- Wang, X.; Wang, K.; Han, L.; Zhang, A.; Shi, Z.; Zhang, K.; Zhang, H.; Yang, S.; Pu, P.; Shen, C.; et al. PRDM1 is directly targeted by miR-30a-5p and modulates the Wnt/beta-catenin pathway in a Dkk1-dependent manner during glioma growth. Cancer Lett. 2013, 331, 211–219. [Google Scholar] [CrossRef]
- Morishita, K.; Parker, D.S.; Mucenski, M.L.; Jenkins, N.A.; Copeland, N.G.; Ihle, J.N. Retroviral activation of a novel gene encoding a zinc finger protein in IL-3-dependent myeloid leukemia cell lines. Cell 1988, 54, 831–840. [Google Scholar] [CrossRef]
- Nucifora, G.; Laricchia-Robbio, L.; Senyuk, V. EVI1 and hematopoietic disorders: History and perspectives. Gene 2006, 368, 1–11. [Google Scholar] [CrossRef]
- Dettman, E.J.; Justice, M.J. The zinc finger SET domain gene Prdm14 is overexpressed in lymphoblastic lymphomas with retroviral insertions at Evi32. PLoS ONE 2008, 3, e3823. [Google Scholar] [CrossRef] [PubMed]
- Dettman, E.J.; Simko, S.J.; Ayanga, B.; Carofino, B.L.; Margolin, J.F.; Morse, H.C., 3rd; Justice, M.J. Prdm14 initiates lymphoblastic leukemia after expanding a population of cells resembling common lymphoid progenitors. Oncogene 2011, 30, 2859–2873. [Google Scholar] [CrossRef] [PubMed]
- Steenbergen, R.D.; Ongenaert, M.; Snellenberg, S.; Trooskens, G.; Van der Meide, W.F.; Pandey, D.; Bloushtain-Qimron, N.; Polyak, K.; Meijer, C.J.; Snijders, P.J.; et al. Methylation-specific digital karyotyping of HPV16E6E7-expressing human keratinocytes identifies novel methylation events in cervical carcinogenesis. J. Pathol. 2013, 231, 53–62. [Google Scholar] [CrossRef] [PubMed]
- Tan, S.X.; Hu, R.C.; Liu, J.J.; Tan, Y.L.; Liu, W.E. Methylation of PRDM2, PRDM5 and PRDM16 genes in lung cancer cells. Int. J. Clin. Exp. Pathol. 2014, 7, 2305–2311. [Google Scholar]
- Tan, S.X.; Hu, R.C.; Xia, Q.; Tan, Y.L.; Liu, J.J.; Gan, G.X.; Wang, L.L. The methylation profiles of PRDM promoters in non-small cell lung cancer. Onco Targets Ther. 2018, 11, 2991–3002. [Google Scholar] [CrossRef]
- Fei, L.R.; Huang, W.J.; Wang, Y.; Lei, L.; Li, Z.H.; Zheng, Y.W.; Wang, Z.; Yang, M.Q.; Liu, C.C.; Xu, H.T. PRDM16 functions as a suppressor of lung adenocarcinoma metastasis. J. Exp. Clin. Cancer Res. 2019, 38, 35. [Google Scholar] [CrossRef]
- Lv, W.; Yu, X.; Li, W.; Feng, N.; Feng, T.; Wang, Y.; Lin, H.; Qian, B. Low expression of LINC00982 and PRDM16 is associated with altered gene expression, damaged pathways and poor survival in lung adenocarcinoma. Oncol. Rep. 2018, 40, 2698–2709. [Google Scholar] [CrossRef]
- Zhang, D.L.; Qu, L.W.; Ma, L.; Zhou, Y.C.; Wang, G.Z.; Zhao, X.C.; Zhang, C.; Zhang, Y.F.; Wang, M.; Zhang, M.Y.; et al. Genome-wide identification of transcription factors that are critical to non-small cell lung cancer. Cancer Lett. 2018, 434, 132–143. [Google Scholar] [CrossRef]
- Laitinen, M.P.; Anttonen, M.; Ketola, I.; Wilson, D.B.; Ritvos, O.; Butzow, R.; Heikinheimo, M. Transcription factors GATA-4 and GATA-6 and a GATA family cofactor, FOG-2, are expressed in human ovary and sex cord-derived ovarian tumors. J. Clin. Endocrinol. Metab. 2000, 85, 3476–3483. [Google Scholar]
- Anttonen, M.; Unkila-Kallio, L.; Leminen, A.; Butzow, R.; Heikinheimo, M. High GATA-4 expression associates with aggressive behavior, whereas low anti-Mullerian hormone expression associates with growth potential of ovarian granulosa cell tumors. J. Clin. Endocrinol. Metab. 2005, 90, 6529–6535. [Google Scholar] [CrossRef]
- Efimenko, E.; Padua, M.B.; Manuylov, N.L.; Fox, S.C.; Morse, D.A.; Tevosian, S.G. The transcription factor GATA4 is required for follicular development and normal ovarian function. Dev. Biol. 2013, 381, 144–158. [Google Scholar] [CrossRef] [PubMed]
- Bashamboo, A.; Brauner, R.; Bignon-Topalovic, J.; Lortat-Jacob, S.; Karageorgou, V.; Lourenco, D.; Guffanti, A.; McElreavey, K. Mutations in the FOG2/ZFPM2 gene are associated with anomalies of human testis determination. Hum. Mol. Genet. 2014, 23, 3657–3665. [Google Scholar] [CrossRef] [PubMed]
- Turnbull, J.; Girard, J.M.; Lohi, H.; Chan, E.M.; Wang, P.; Tiberia, E.; Omer, S.; Ahmed, M.; Bennett, C.; Chakrabarty, A.; et al. Early-onset Lafora body disease. Brain 2012, 135, 2684–2698. [Google Scholar] [CrossRef] [PubMed]
- Bowne, S.J.; Sullivan, L.S.; Wheaton, D.K.; Locke, K.G.; Jones, K.D.; Koboldt, D.C.; Fulton, R.S.; Wilson, R.K.; Blanton, S.H.; Birch, D.G.; et al. North Carolina macular dystrophy (MCDR1) caused by a novel tandem duplication of the PRDM13 gene. Mol. Vis. 2016, 22, 1239–1247. [Google Scholar]
- Shapira, S.K.; McCaskill, C.; Northrup, H.; Spikes, A.S.; Elder, F.F.; Sutton, V.R.; Korenberg, J.R.; Greenberg, F.; Shaffer, L.G. Chromosome 1p36 deletions: The clinical phenotype and molecular characterization of a common newly delineated syndrome. Am. J. Hum. Genet. 1997, 61, 642–650. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).