HIGD-Driven Regulation of Cytochrome c Oxidase Biogenesis and Function
Abstract
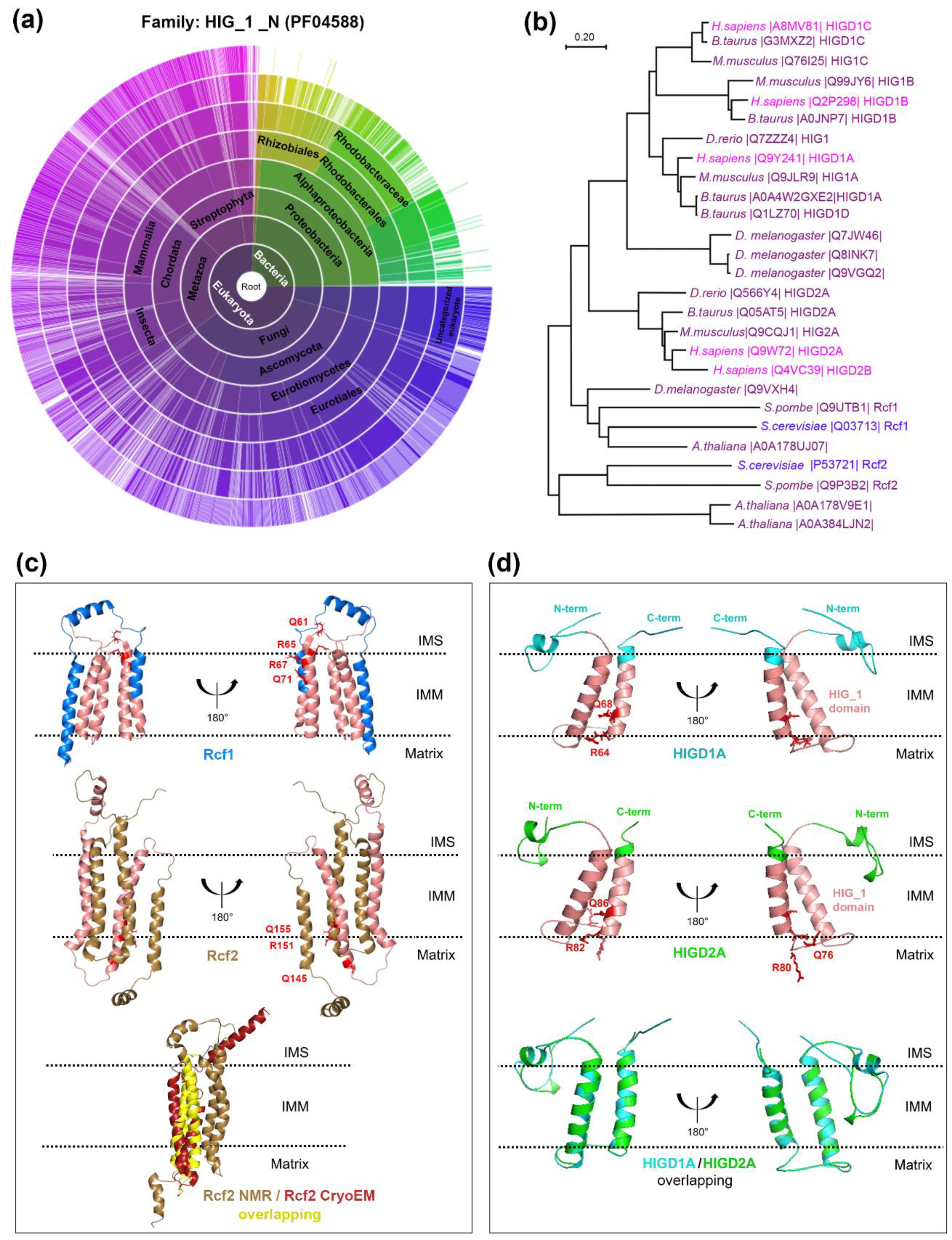
:1. Introduction
2. Role of HIGD Proteins in Yeast Cytochrome c Oxidase Biogenesis and Function
2.1. Discovery of Rcf Proteins in Saccharomyces cerevisiae
2.2. Role of Rcf Proteins in Cytochrome c Oxidase Assembly and Function
2.3. Role of Rcf Proteins in Yeast Respiratory Supercomplex Biogenesis and Function
2.4. Role of Rcf Proteins under Hypoxia and Oxidative Stress
3. Role of HIGD1A and HIGD2A in Mammalian Cytochrome c Oxidase Biogenesis and Function
3.1. Regulation of HIGD Gene Expression
3.2. Role of HIGD Proteins in the Assembly and Regulation of Mammalian Cytochrome c Oxidase
3.2.1. Role in the Assembly and Regulation of Cytochrome c Oxidase in Physiological Conditions
3.2.2. Role in the Assembly and Regulation of Cytochrome c Oxidase under Hypoxic Conditions
4. Concluding Remarks
Author Contributions
Funding
Conflicts of Interest
References
- Kadenbach, B. Complex IV—The regulatory center of mitochondrial oxidative phosphorylation. Mitochondrion 2020, in press. [Google Scholar]
- Timon-Gomez, A.; Nyvltova, E.; Abriata, L.A.; Vila, A.J.; Hosler, J.; Barrientos, A. Mitochondrial cytochrome c oxidase biogenesis: Recent developments. Semin. Cell Dev. Biol. 2018, 76, 163–178. [Google Scholar] [CrossRef] [PubMed]
- Pierron, D.; Wildman, D.E.; Huttemann, M.; Markondapatnaikuni, G.C.; Aras, S.; Grossman, L.I. Cytochrome c oxidase: Evolution of control via nuclear subunit addition. Biochim. Biophys. Acta Bioenerg. 2012, 1817, 590–597. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttemann, M.; Kadenbach, B.; Grossman, L.I. Mammalian subunit IV isoforms of cytochrome c oxidase. Gene 2001, 267, 111–123. [Google Scholar] [CrossRef]
- Poyton, R.O.; Burke, P.V. Oxygen regulated transcription of cytochrome c and cytochrome c oxidase genes in yeast. Biochim. Biophys. Acta Bioenerg. 1992, 1101, 252–256. [Google Scholar] [CrossRef]
- Liu, J.; Barrientos, A. Transcriptional regulation of yeast OXPHOS hypoxic genes by oxidative stress. Antiox. Redox Signal. 2012, 19, 1916–1927. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fukuda, R.; Zhang, H.; Kim, J.-W.; Shimoda, L.; Dang, C.V.; Semenza, G.L. HIF-1 regulates cytochrome oxidase subunits to optimize efficiency of respiration in hypoxic cells. Cell 2007, 129, 111–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bourens, M.; Fontanesi, F.; Soto, I.C.; Liu, J.; Barrientos, A. Redox and reactive oxygen species regulation of mitochondrial cytochrome c oxidase biogenesis. Antioxid. Redox Signal. 2013, 19, 1940–1952. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Poyton, R.O.; McEwen, J.E. Crosstalk between nuclear and mitochondrial genomes. Annu. Rev. Biochem. 1996, 65, 563–607. [Google Scholar] [CrossRef]
- Fontanesi, F.; Soto, I.C.; Horn, D.; Barrientos, A. Assembly of mitochondrial cytochrome c oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 2006, 291, C1129–C1147. [Google Scholar] [CrossRef] [Green Version]
- Franco, L.V.R.; Su, C.H.; Tzagoloff, A. Modular assembly of yeast mitochondrial ATP synthase and cytochrome oxidase. Biol. Chem. 2020, 401, 835–853. [Google Scholar] [CrossRef]
- McStay, G.P.; Su, C.H.; Tzagoloff, A. Modular assembly of yeast cytochrome oxidase. Mol. Biol. Cell 2013, 24, 440–452. [Google Scholar] [CrossRef] [PubMed]
- Cobine, P.A.; Pierrel, F.; Winge, D.R. Copper trafficking to the mitochondrion and assembly of copper metalloenzymes. Biochim. Biophys. Acta Mol. Cell Res. 2006, 1763, 759–772. [Google Scholar] [CrossRef] [PubMed]
- Swenson, S.A.; Moore, C.M.; Marcero, J.R.; Medlock, A.E.; Reddi, A.R.; Khalimonchuk, O. From synthesis to utilization: The ins and outs of mitochondrial heme. Cells 2020, 9, 579. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Timón-Gómez, A.; Garlich, J.; Stuart, R.A.; Ugalde, C.; Barrientos, A. Distinct roles of mitochondrial HIGD1A and HIGD2A in respiratory complex and supercomplex biogenesis. Cell Rep. 2020, 31, 107607. [Google Scholar] [CrossRef] [PubMed]
- Lobo-Jarne, T.; Pérez-Pérez, R.; Fontanesi, F.; Timón-Gómez, A.; Wittig, I.; Peñas, A.; Serrano-Lorenzo, P.; García-Consuegra, I.; Arenas, J.; Martín, M.A.; et al. Multiple pathways coordinate assembly of human mitochondrial complex IV and stabilization of respiratory supercomplexes. EMBO J. 2020, 39, e103912. [Google Scholar] [CrossRef] [PubMed]
- Vidoni, S.; Harbour, M.E.; Guerrero-Castillo, S.; Signes, A.; Ding, S.; Fearnley, I.M.; Taylor, R.W.; Tiranti, V.; Arnold, S.; Fernandez-Vizarra, E.; et al. MR-1S interacts with PET100 and PET117 in module-based assembly of human cytochrome c oxidase. Cell Rep. 2017, 18, 1727–1738. [Google Scholar] [CrossRef] [Green Version]
- Hock, D.H.; Reljic, B.; Ang, C.-S.; Muellner-Wong, L.; Mountford, H.S.; Compton, A.G.; Ryan, M.T.; Thorburn, D.R.; Stroud, D.A. HIGD2A is required for assembly of the COX3 module of human mitochondrial complex IV. Mol. Cell Proteom. 2020, 19, 1145–1160. [Google Scholar] [CrossRef] [Green Version]
- Strogolova, V.; Furness, A.; Robb-McGrath, M.; Garlich, J.; Stuart, R.A. Rcf1 and Rcf2, members of the hypoxia induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol. Cell Biol. 2012, 32, 1363–1373. [Google Scholar] [CrossRef] [Green Version]
- Vukotic, M.; Oeljeklaus, S.; Wiese, S.; Vogtle, F.N.; Meisinger, C.; Meyer, H.E.; Zieseniss, A.; Katschinski, D.M.; Jans, D.C.; Jakobs, S.; et al. Rcf1 Mediates Cytochrome Oxidase Assembly and Respirasome Formation, Revealing Heterogeneity of the Enzyme Complex. Cell Metab. 2012, 7, 336–347. [Google Scholar] [CrossRef] [Green Version]
- Su, C.H.; McStay, G.P.; Tzagoloff, A. The Cox3p assembly module of yeast cytochrome oxidase. Mol. Biol. Cell 2014, 25, 965–976. [Google Scholar] [CrossRef]
- García-Poyatos, C.; Cogliati, S.; Calvo, E.; Hernansanz-Agustín, P.; Lagarrigue, S.; Magni, R.; Botos, M.; Langa, X.; Amati, F.; Vázquez, J.; et al. Scaf1 promotes respiratory supercomplexes and metabolic efficiency in zebrafish. EMBO Rep. 2020, 21, e50287. [Google Scholar] [CrossRef] [PubMed]
- Viscomi, C.; Bottani, E.; Civiletto, G.; Cerutti, R.; Moggio, M.; Fagiolari, G.; Schon, E.A.; Lamperti, C.; Zeviani, M. In vivo correction of COX deficiency by activation of the AMPK/PGC-1alpha axis. Cell Metab. 2011, 14, 80–90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, S.; Kadenbach, B. Cell respiration is controlled by ATP, an allosteric inhibitor of cytochrome-c oxidase. Eur. J. Biochem. 1997, 249, 350–354. [Google Scholar] [CrossRef] [PubMed]
- Babcock, G.T.; Wikstrom, M. Oxygen activation and the conservation of energy in cell respiration. Nature 1992, 356, 301–309. [Google Scholar] [CrossRef]
- Bender, E.; Kadenbach, B. The allosteric ATP-inhibition of cytochrome c oxidase activity is reversibly switched on by cAMP-dependent phosphorylation. FEBS Lett. 2000, 466, 130–134. [Google Scholar] [CrossRef] [Green Version]
- Shinzawa-Itoh, K.; Sugimura, T.; Misaki, T.; Tadehara, Y.; Yamamoto, S.; Hanada, M.; Yano, N.; Nakagawa, T.; Uene, S.; Yamada, T.; et al. Monomeric structure of an active form of bovine cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2019, 116, 19945–19951. [Google Scholar] [CrossRef] [Green Version]
- Ramzan, R.; Rhiel, A.; Weber, P.; Kadenbach, B.; Vogt, S. Reversible dimerization of cytochrome c oxidase regulates mitochondrial respiration. Mitochondrion 2019, 49, 149–155. [Google Scholar] [CrossRef]
- Salazar, C.; Elorza, A.A.; Cofre, G.; Ruiz-Hincapie, P.; Shirihai, O.; Ruiz, L.M. The OXPHOS supercomplex assembly factor HIG2A responds to changes in energetic metabolism and cell cycle. J. Cell. Physiol. 2019, 234, 17405–17419. [Google Scholar] [CrossRef]
- Ameri, K.; Jahangiri, A.; Rajah, A.M.; Tormos, K.V.; Nagarajan, R.; Pekmezci, M.; Nguyen, V.; Wheeler, M.L.; Murphy, M.P.; Sanders, T.A.; et al. HIGD1A regulates oxygen consumption, ROS production, and AMPK activity during glucose deprivation to modulate cell survival and tumor growth. Cell Rep. 2015, 10, 891–899. [Google Scholar] [CrossRef] [Green Version]
- Guerra-Castellano, A.; Díaz-Quintana, A.; Pérez-Mejías, G.; Elena-Real, C.A.; González-Arzola, K.; García-Mauriño, S.M.; De la Rosa, M.A.; Díaz-Moreno, I. Oxidative stress is tightly regulated by cytochrome c phosphorylation and respirasome factors in mitochondria. Proc. Natl. Acad. Sci. USA 2018, 115, 7955–7960. [Google Scholar] [CrossRef] [Green Version]
- Nagao, T.; Shintani, Y.; Hayashi, T.; Kioka, H.; Kato, H.; Nishida, Y.; Yamazaki, S.; Tsukamoto, O.; Yashirogi, S.; Yazawa, I.; et al. Higd1a improves respiratory function in the models of mitochondrial disorder. FASEB J. 2020, 34, 1859–1871. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Strogolova, V.; Hoang, N.H.; Hosler, J.; Stuart, R.A. The yeast mitochondrial proteins Rcf1 and Rcf2 support the enzymology of the cytochrome c oxidase complex and generation of the proton motive force. J. Biol. Chem. 2019, 294, 4867–4877. [Google Scholar] [CrossRef] [Green Version]
- Hartley, A.M.; Meunier, B.; Pinotsis, N.; Maréchal, A. Rcf2 revealed in cryo-EM structures of hypoxic isoforms of mature mitochondrial III-IV supercomplexes. Proc. Natl. Acad. Sci. USA 2020, 117, 9329–9337. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schäfer, J.; Dawitz, H.; Ott, M.; Ädelroth, P.; Brzezinski, P. Regulation of cytochrome c oxidase activity by modulation of the catalytic site. Sci. Rep. 2018, 8, 11397. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dawitz, H.; Schäfer, J.; Schaart, J.M.; Magits, W.; Brzezinski, P.; Ott, M. Rcf1 modulates cytochrome c oxidase activity especially under energy-demanding conditions. Front. Physiol. 2019, 10, 1555. [Google Scholar] [CrossRef]
- Hoang, N.H.; Strogolova, V.; Mosley, J.J.; Stuart, R.A.; Hosler, J. Hypoxia-inducible gene domain 1 proteins in yeast mitochondria protect against proton leak through complex IV. J. Biol. Chem. 2019, 294, 17669–17677. [Google Scholar] [CrossRef] [PubMed]
- Rydstrom Lundin, C.; von Ballmoos, C.; Ott, M.; Adelroth, P.; Brzezinski, P. Regulatory role of the respiratory supercomplex factors in Saccharomyces cerevisiae. Proc. Natl. Acad. Sci. USA 2016, 113, E4476–E4485. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Cao, Y.; Chen, Y.; Chen, Y.; Gardner, P.; Steiner, D.F. Pancreatic beta cells lack a low glucose and O2-inducible mitochondrial protein that augments cell survival. Proc. Natl. Acad. Sci. USA 2006, 103, 10636–10641. [Google Scholar] [CrossRef] [Green Version]
- Bedo, G.; Vargas, M.; Ferreiro, M.J.; Chalar, C.; Agrati, D. Characterization of hypoxia induced gene 1: Expression during rat central nervous system maturation and evidence of antisense RNA expression. Int. J. Dev. Biol. 2005, 49, 431–436. [Google Scholar] [CrossRef] [Green Version]
- Ameri, K.; Rajah, A.M.; Nguyen, V.; Sanders, T.A.; Jahangiri, A.; Delay, M.; Donne, M.; Choi, H.J.; Tormos, K.V.; Yeghiazarians, Y.; et al. Nuclear localization of the mitochondrial factor HIGD1A during metabolic stress. PLoS ONE 2013, 8, e62758. [Google Scholar] [CrossRef] [Green Version]
- Stecher, G.; Tamura, K.; Kumar, S. Molecular evolutionary genetics analysis (MEGA) for macOS. Mol. Biol. Evol. 2020, 37, 1237–1239. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Pettersson, P.; Huang, J.; Brzezinski, P.; Pomès, R.; Mäler, L.; Ädelroth, P. NMR Structure and dynamics studies of yeast Respiratory Supercomplex Factor 2. Structure 2020, in press. [Google Scholar]
- Bienert, S.; Waterhouse, A.; de Beer, T.A.P.; Tauriello, G.; Studer, G.; Bordoli, L.; Schwede, T. The SWISS-MODEL Repository-new features and functionality. Nucleic Acids Res. 2017, 45, D313–D319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guex, N.; Peitsch, M.C.; Schwede, T. Automated comparative protein structure modeling with SWISS-MODEL and Swiss-PdbViewer: A historical perspective. Electrophoresis 2009, 30, S162–S173. [Google Scholar] [CrossRef] [PubMed]
- Waterhouse, A.; Bertoni, M.; Bienert, S.; Studer, G.; Tauriello, G.; Gumienny, R.; Heer, F.T.; de Beer, T.A.P.; Rempfer, C.; Bordoli, L.; et al. SWISS-MODEL: Homology modelling of protein structures and complexes. Nucleic Acids Res. 2018, 46, W296–W303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hess, D.C.; Myers, C.L.; Huttenhower, C.; Hibbs, M.A.; Hayes, A.P.; Paw, J.; Clore, J.J.; Mendoza, R.M.; Luis, B.S.; Nislow, C.; et al. Computationally driven, quantitative experiments discover genes required for mitochondrial biogenesis. PLoS Genet. 2009, 5, e1000407. [Google Scholar] [CrossRef] [Green Version]
- Helbig, A.O.; de Groot, M.J.; van Gestel, R.A.; Mohammed, S.; de Hulster, E.A.; Luttik, M.A.; Daran-Lapujade, P.; Pronk, J.T.; Heck, A.J.; Slijper, M. A three-way proteomics strategy allows differential analysis of yeast mitochondrial membrane protein complexes under anaerobic and aerobic conditions. Proteomics 2009, 9, 4787–4798. [Google Scholar] [CrossRef]
- Chen, Y.C.; Taylor, E.B.; Dephoure, N.; Heo, J.M.; Tonhato, A.; Papandreou, I.; Nath, N.; Denko, N.C.; Gygi, S.P.; Rutter, J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012, 15, 348–360. [Google Scholar] [CrossRef] [Green Version]
- Garlich, J.; Strecker, V.; Wittig, I.; Stuart, R.A. Mutational analysis of the QRRQ motif in the yeast Hig1 type 2 protein Rcf1 reveals a regulatory role for the cytochrome c oxidase complex. J. Biol. Chem. 2017, 292, 5216–5226. [Google Scholar] [CrossRef] [Green Version]
- Ghosh, A.; Pratt, A.T.; Soma, S.; Theriault, S.G.; Griffin, A.T.; Trivedi, P.P.; Gohil, V.M. Mitochondrial disease genes COA6, COX6B and SCO2 have overlapping roles in COX2 biogenesis. Hum. Mol. Genet. 2016, 25, 660–671. [Google Scholar] [CrossRef] [Green Version]
- Varanasi, L.; Mills, D.; Murphree, A.; Gray, J.; Purser, C.; Baker, R.; Hosler, J. Altering conserved lipid binding sites in cytochrome c oxidase of Rhodobacter sphaeroides perturbs the interaction between subunits I and III and promotes suicide inactivation of the enzyme. Biochemistry 2006, 45, 14896–14907. [Google Scholar] [CrossRef] [PubMed]
- Shinzawa-Itoh, K.; Aoyama, H.; Muramoto, K.; Terada, H.; Kurauchi, T.; Tadehara, Y.; Yamasaki, A.; Sugimura, T.; Kurono, S.; Tsujimoto, K.; et al. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. EMBO J. 2007, 26, 1713–1725. [Google Scholar] [CrossRef] [PubMed]
- Gilderson, G.; Salomonsson, L.; Aagaard, A.; Gray, J.; Brzezinski, P.; Hosler, J. Subunit III of cytochrome c oxidase of Rhodobacter sphaeroides is required to maintain rapid proton uptake through the D pathway at physiologic pH. Biochemistry 2003, 42, 7400–7409. [Google Scholar] [CrossRef] [PubMed]
- Berndtsson, J.; Aufschnaiter, A.; Rathore, S.; Marin-Buera, L.; Dawitz, H.; Diessl, J.; Kohler, V.; Barrientos, A.; Büttner, S.; Fontanesi, F.; et al. Respiratory supercomplexes enhance electron transport by decreasing cytochrome c diffusion distance. EMBO Rep. 2020, 32, e51015. [Google Scholar] [CrossRef] [PubMed]
- Ogunjimi, E.O.; Pokalsky, C.N.; Shroyer, L.A.; Prochaska, L.J. Evidence for a conformational change in subunit III of bovine heart mitochondrial cytochrome c oxidase. J. Bioenerg. Biomembr. 2000, 32, 617–626. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Pettersson, P.; Huang, J.; Sjöholm, J.; Sjöstrand, D.; Pomès, R.; Högbom, M.; Brzezinski, P.; Mäler, L.; Ädelroth, P. Solution NMR structure of yeast Rcf1, a protein involved in respiratory supercomplex formation. Proc. Natl. Acad. Sci. USA 2018, 115, 3048–3053. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalpage, H.A.; Wan, J.; Morse, P.T.; Zurek, M.P.; Turner, A.A.; Khobeir, A.; Yazdi, N.; Hakim, L.; Liu, J.; Vaishnav, A.; et al. Cytochrome c phosphorylation: Control of mitochondrial electron transport chain flux and apoptosis. Int. J. Biochem. Cell Biol. 2020, 121, 105704. [Google Scholar] [CrossRef]
- Moreno-Beltrán, B.; Guerra-Castellano, A.; Díaz-Quintana, A.; Del Conte, R.; García-Mauriño, S.M.; Díaz-Moreno, S.; González-Arzola, K.; Santos-Ocaña, C.; Velázquez-Campoy, A.; De la Rosa, M.A.; et al. Structural basis of mitochondrial dysfunction in response to cytochrome c phosphorylation at tyrosine 48. Proc. Natl. Acad. Sci. USA 2017, 114, E3041–E3050. [Google Scholar]
- Römpler, K.; Müller, T.; Juris, L.; Wissel, M.; Vukotic, M.; Hofmann, K.; Deckers, M. Overlapping role of Respiratory Supercomplex Factor Rcf2 and Its N-terminal homolog Rcf3 in Saccharomyces cerevisiae. J. Biol. Chem. 2016, 291, 23769–23778. [Google Scholar] [CrossRef] [Green Version]
- Milenkovic, D.; Blaza, J.N.; Larsson, N.G.; Hirst, J. The Enigma of the Respiratory Chain Supercomplex. Cell Metab. 2017, 25, 765–776. [Google Scholar] [CrossRef] [Green Version]
- Lobo-Jarne, T.; Ugalde, C. Respiratory chain supercomplexes: Structures, function and biogenesis. Semin. Cell Dev. Biol. 2017, 76, 179–190. [Google Scholar] [CrossRef] [PubMed]
- Acin-Perez, R.; Enriquez, J.A. The function of the respiratory supercomplexes: The plasticity model. Biochim. Biophys. Acta Bioenerg. 2014, 1837, 444–450. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stuchebrukhov, A.; Schäfer, J.; Berg, J.; Brzezinski, P. Kinetic advantage of forming respiratory supercomplexes. Biochim. Biophys. Acta Bioenerg. 2020, 1861, 148193. [Google Scholar] [CrossRef] [PubMed]
- Maranzana, E.; Barbero, G.; Falasca, A.I.; Lenaz, G.; Genova, M.L. Mitochondrial Respiratory Supercomplex Association Limits Production of Reactive Oxygen Species from Complex, I. Antioxid. Redox Signal. 2013, 19, 1469–1480. [Google Scholar] [CrossRef] [Green Version]
- Klammt, C.; Maslennikov, I.; Bayrhuber, M.; Eichmann, C.; Vajpai, N.; Chiu, E.J.; Blain, K.Y.; Esquivies, L.; Kwon, J.H.; Balana, B.; et al. Facile backbone structure determination of human membrane proteins by NMR spectroscopy. Nat. Methods 2012, 9, 834–839. [Google Scholar] [CrossRef] [Green Version]
- Lindert, S.; Maslennikov, I.; Chiu, E.J.; Pierce, L.C.; McCammon, J.A.; Choe, S. Drug screening strategy for human membrane proteins: From NMR protein backbone structure to in silica- and NMR-screened hits. Biochem. Biophys. Res. Commun. 2014, 445, 724–733. [Google Scholar] [CrossRef] [Green Version]
- An, H.J.; Shin, H.; Jo, S.G.; Kim, Y.J.; Lee, J.O.; Paik, S.G.; Lee, H. The survival effect of mitochondrial Higd-1a is associated with suppression of cytochrome c release and prevention of caspase activation. Biochim. Biophys. Acta Bioenerg. 2011, 1813, 2088–2098. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, T.; Asano, Y.; Shintani, Y.; Aoyama, H.; Kioka, H.; Tsukamoto, O.; Hikita, M.; Shinzawa-Itoh, K.; Takafuji, K.; Higo, S.; et al. Higd1a is a positive regulator of cytochrome c oxidase. Proc. Natl. Acad. Sci. USA 2015, 112, 1553–1558. [Google Scholar] [CrossRef] [Green Version]
- Denko, N.; Schindler, C.; Koong, A.; Laderoute, K.; Green, C.; Giaccia, A. Epigenetic regulation of gene expression in cervical cancer cells by the tumor microenvironment. Clin. Cancer Res. 2000, 6, 480–487. [Google Scholar]
- Kasper, L.H.; Brindle, P.K. Mammalian gene expression program resiliency: The roles of multiple coactivator mechanisms in hypoxia-responsive transcription. Cell Cycle 2006, 5, 142–146. [Google Scholar] [CrossRef] [Green Version]
- Denechaud, P.D.; Fajas, L.; Giralt, A. E2F1, a Novel Regulator of Metabolism. Front. Endocrinol. 2017, 8, 311. [Google Scholar] [CrossRef] [PubMed]
- Zhao, Y.X.; Liu, H.C.; Ying, W.Y.; Wang, C.Y.; Yu, Y.J.; Sun, W.J.; Liu, J.F. microRNA-372 inhibits proliferation and induces apoptosis in human breast cancer cells by directly targeting E2F1. Mol. Med. Rep. 2017, 16, 8069–8075. [Google Scholar] [CrossRef] [PubMed]
- Agarwal, V.; Bell, G.W.; Nam, J.W.; Bartel, D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife 2015, 4, e05005. [Google Scholar] [CrossRef] [PubMed]
- Guo, J.; Yang, C.; Zhang, S.; Liang, M.; Qi, J.; Wang, Z.; Peng, Y.; Sun, B. MiR-375 induces ROS and apoptosis in ST cells by targeting the HIGD1A gene. Gene 2019, 685, 136–142. [Google Scholar] [CrossRef] [PubMed]
- Li, T.; Xian, W.J.; Gao, Y.; Jiang, S.; Yu, Q.H.; Zheng, Q.C.; Zhang, Y. Higd1a protects cells from lipotoxicity under high-fat exposure. Oxidative Med. Cell. Longev. 2019, 2019, 6051262. [Google Scholar] [CrossRef] [PubMed]
- An, H.J.; Cho, G.; Lee, J.O.; Paik, S.G.; Kim, Y.S.; Lee, H. Higd-1a interacts with Opa1 and is required for the morphological and functional integrity of mitochondria. Proc. Natl. Acad. Sci. USA 2013, 110, 13014–13019. [Google Scholar] [CrossRef] [Green Version]
- Hayashi, H.; Nakagami, H.; Takeichi, M.; Shimamura, M.; Koibuchi, N.; Oiki, E.; Sato, N.; Koriyama, H.; Mori, M.; Gerardo Araujo, R.; et al. HIG1, a novel regulator of mitochondrial γ-secretase, maintains normal mitochondrial function. FASEB J. 2012, 26, 2306–2317. [Google Scholar] [CrossRef]
- Rieger, B.; Shalaeva, D.N.; Sohnel, A.C.; Kohl, W.; Duwe, P.; Mulkidjanian, A.Y.; Busch, K.B. Lifetime imaging of GFP at CoxVIIIa reports respiratory supercomplex assembly in live cells. Sci. Rep. 2017, 7, 46055. [Google Scholar] [CrossRef]
- Letts, J.A.; Fiedorczuk, K.; Sazanov, L.A. The architecture of respiratory supercomplexes. Nature 2016, 537, 644–648. [Google Scholar] [CrossRef]
- McStay, G.P.; Su, C.H.; Thomas, S.M.; Xu, J.T.; Tzagoloff, A. Characterization of assembly intermediates containing subunit 1 of yeast cytochrome oxidase. J. Biol. Chem. 2013, 288, 26546–26556. [Google Scholar] [CrossRef] [Green Version]
- Emsley, P.; Lohkamp, B.; Scott, W.G.; Cowtan, K. Features and development of Coot. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 486–501. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huttemann, M.; Lee, I.; Gao, X.; Pecina, P.; Pecinova, A.; Liu, J.; Aras, S.; Sommer, N.; Sanderson, T.H.; Tost, M.; et al. Cytochrome c oxidase subunit 4 isoform 2-knockout mice show reduced enzyme activity, airway hyporeactivity, and lung pathology. FASEB J. 2012, 26, 3916–3930. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferrari, M.; Jain, I.H.; Goldberger, O.; Rezoagli, E.; Thoonen, R.; Cheng, K.H.; Sosnovik, D.E.; Scherrer-Crosbie, M.; Mootha, V.K.; Zapol, W.M. Hypoxia treatment reverses neurodegenerative disease in a mouse model of Leigh syndrome. Proc. Natl. Acad. Sci. USA 2017, 114, E4241–E4250. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ast, T.; Meisel, J.D.; Patra, S.; Wang, H.; Grange, R.M.H.; Kim, S.H.; Calvo, S.E.; Orefice, L.L.; Nagashima, F.; Ichinose, F.; et al. Hypoxia Rescues Frataxin Loss by Restoring Iron Sulfur Cluster Biogenesis. Cell 2019, 177, 1507–1521. [Google Scholar] [CrossRef]



Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Timón-Gómez, A.; Bartley-Dier, E.L.; Fontanesi, F.; Barrientos, A. HIGD-Driven Regulation of Cytochrome c Oxidase Biogenesis and Function. Cells 2020, 9, 2620. https://doi.org/10.3390/cells9122620
Timón-Gómez A, Bartley-Dier EL, Fontanesi F, Barrientos A. HIGD-Driven Regulation of Cytochrome c Oxidase Biogenesis and Function. Cells. 2020; 9(12):2620. https://doi.org/10.3390/cells9122620
Chicago/Turabian StyleTimón-Gómez, Alba, Emma L. Bartley-Dier, Flavia Fontanesi, and Antoni Barrientos. 2020. "HIGD-Driven Regulation of Cytochrome c Oxidase Biogenesis and Function" Cells 9, no. 12: 2620. https://doi.org/10.3390/cells9122620
APA StyleTimón-Gómez, A., Bartley-Dier, E. L., Fontanesi, F., & Barrientos, A. (2020). HIGD-Driven Regulation of Cytochrome c Oxidase Biogenesis and Function. Cells, 9(12), 2620. https://doi.org/10.3390/cells9122620





