Entacapone Treatment Modulates Hippocampal Proteins Related to Synaptic Vehicle Trafficking
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals
2.2. Experimental Animals and Entacapone Treatment
2.3. Entacapone Treatment
2.4. Protein Preparation for 2DE
2.5. Analysis of 2-DE Gels
2.6. Trypsin Digestion of Master Gels
2.7. Protein Identification Using MALDI-TOF MS
2.8. Validation of Selected Proteins
2.8.1. Western Blot Analyses
2.8.2. Immunohistochemistry
2.9. Statistical Analysis
3. Results
3.1. Proteomic Profiles Changed by Entacapone Treatment
3.2. Proteins Responsible for Vesicular Trafficking and Endocytosis
3.3. Proteins Responsible for Cellular Signaling and Processes
3.4. Proteins Responsible for the Regulation of Cell Structure and Morphology
3.5. Proteins Regulating Energy Metabolism
3.6. Proteins with Various Enzymatic Activities
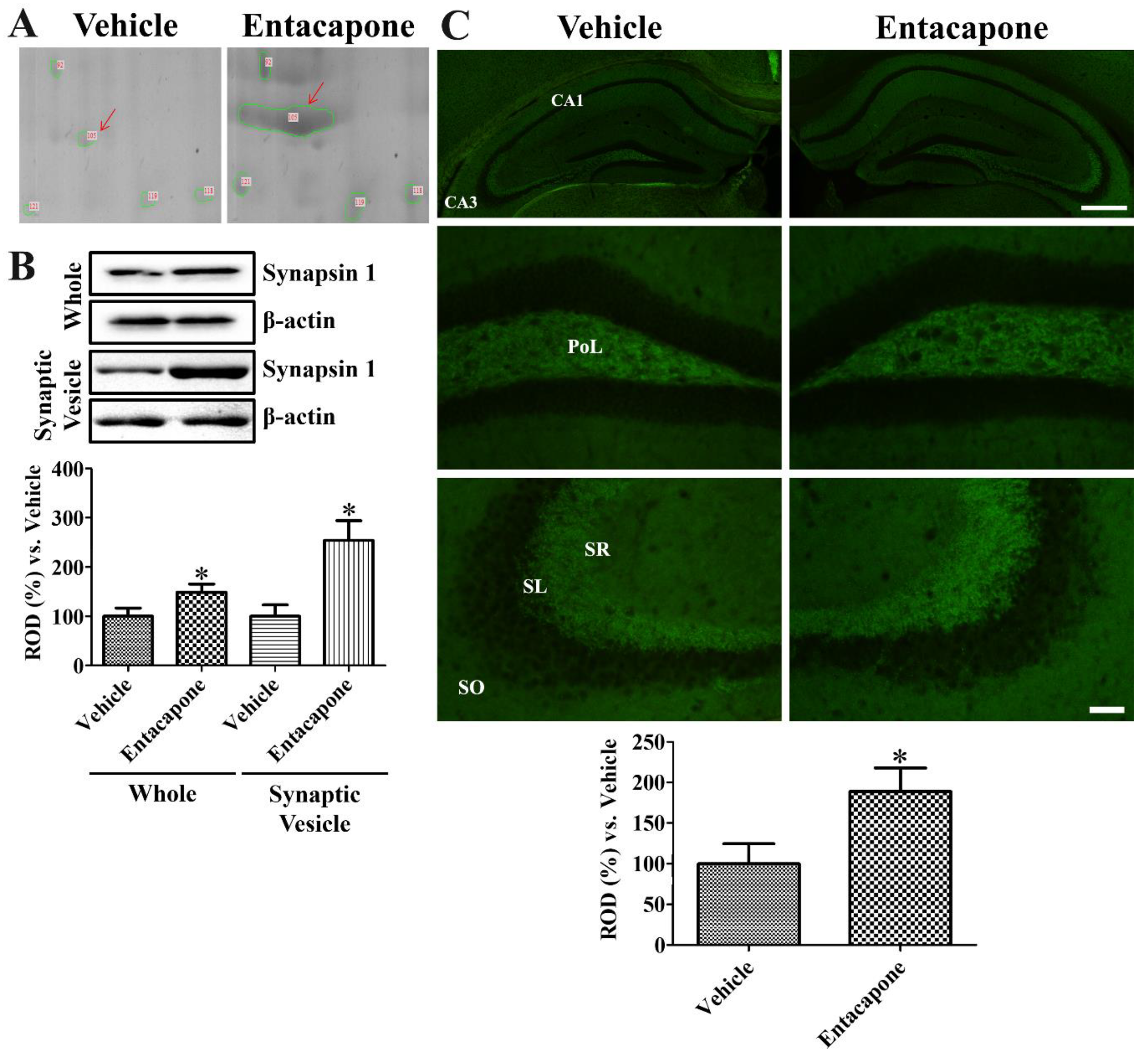
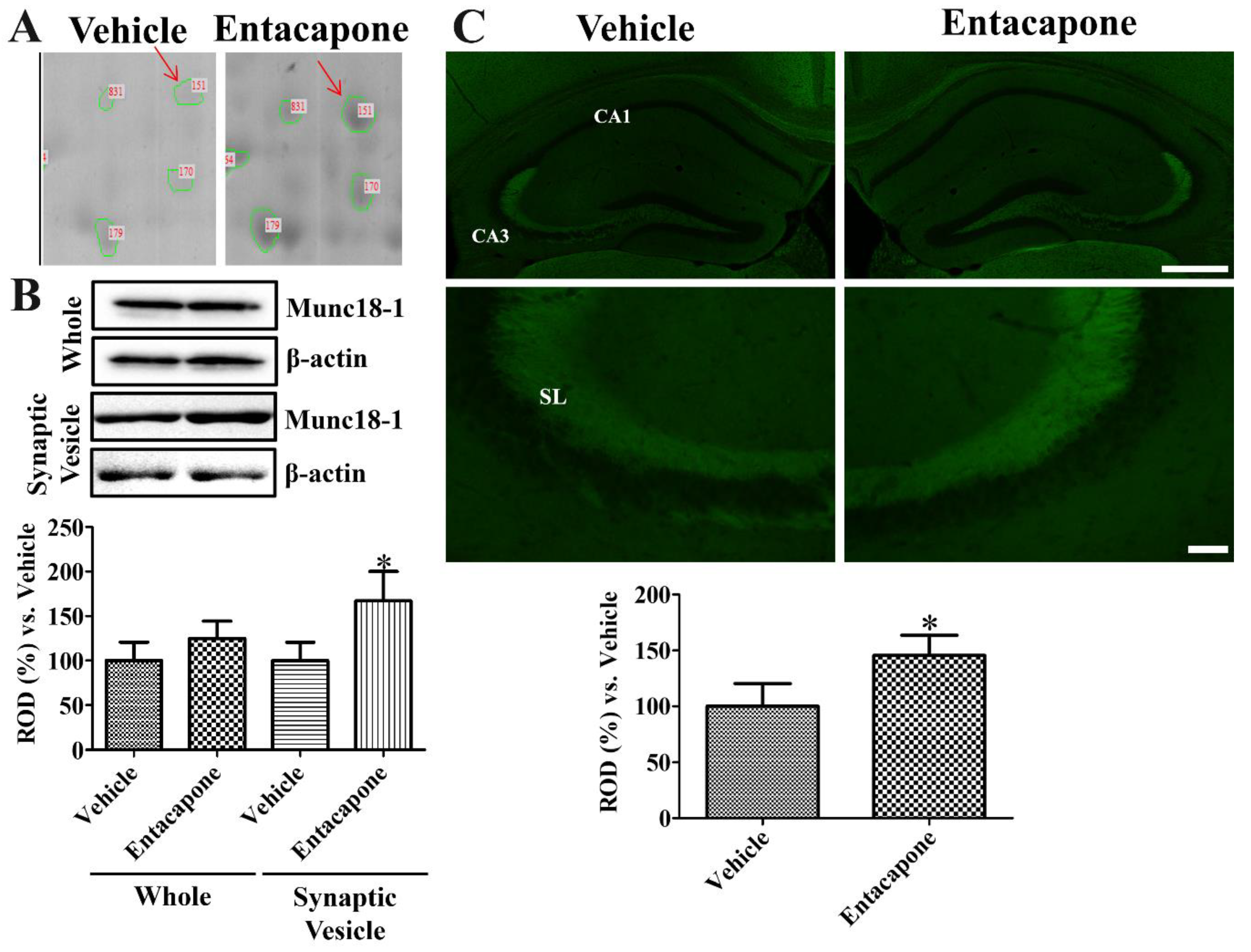
3.7. Validation of Protein Level Changes for Dynamin 1, Synapsin I, and Munc18-1
4. Discussion
4.1. Vesicular Trafficking and Endocytosis
4.2. Cellular Signaling and Various Cellular Processes Including the Cell Cycle, Mitosis, and Differentiation
4.3. Cell Structure and Morphology
4.4. Energy Metabolism
4.5. Various Enzymatic Activities
4.6. Validation of Synaptic Vesicle Trafficking-Related Proteins
Author Contributions
Funding
Conflicts of Interest
References
- Niethard, N.; Born, J. A backup of hippocampal spatial code outside the hippocampus? New light on systems memory consolidation. Neuron 2020, 106, 204–206. [Google Scholar] [CrossRef]
- Kim, W.B.; Cho, J.H. Encoding of contextual fear memory in hippocampal-amygdala circuit. Nat. Commun. 2020, 11, 1382. [Google Scholar] [CrossRef] [PubMed]
- Manning, C.E.; Eagle, A.L.; Kwiatkowski, C.C.; Achargui, R.; Woodworth, H.; Potter, E.; Ohnishi, Y.; Leinninger, G.M.; Robison, A.J. Hippocampal subgranular zone FosB expression is critical for neurogenesis and learning. Neuroscience 2019, 406, 225–233. [Google Scholar] [CrossRef]
- Wood, E.R.; Mumby, D.G.; Pinel, J.P.; Phillips, A.G. Impaired object recognition memory in rats following ischemia-induced damage to the hippocampus. Behav. Neurosci. 1993, 107, 51–62. [Google Scholar] [CrossRef]
- Cole, E.; Ziadé, J.; Simundic, A.; Mumby, D.G. Effects of perirhinal cortex and hippocampal lesions on rats’ performance on two object-recognition tasks. Behav. Brain Res. 2020, 381, 112450. [Google Scholar] [CrossRef] [PubMed]
- Jessberger, S.; Clark, R.E.; Broadbent, N.J.; Clemenson, G.D., Jr.; Consiglio, A.; Lie, D.C.; Squire, L.R.; Gage, F.H. Dentate gyrus-specific knockdown of adult neurogenesis impairs spatial and object recognition memory in adult rats. Learn. Mem. 2009, 16, 147–154. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ohara, S.; Sato, S.; Tsutsui, K.; Witter, M.P.; Iijima, T. Organization of multisynaptic inputs to the dorsal and ventral dentate gyrus: Retrograde trans-synaptic tracing with rabies virus vector in the rat. PLoS ONE 2013, 8, e78928. [Google Scholar] [CrossRef] [Green Version]
- Kempadoo, K.A.; Mosharov, E.V.; Choi, S.J.; Sulzer, D.; Kandel, E.R. Dopamine release from the locus coeruleus to the dorsal hippocampus promotes spatial learning and memory. Proc. Natl. Acad. Sci. USA 2016, 113, 14835–14840. [Google Scholar] [CrossRef] [Green Version]
- Strehl, A.; Galanis, C.; Radic, T.; Schwarzacher, S.W.; Deller, T.; Vlachos, A. Dopamine modulates homeostatic excitatory synaptic plasticity of immature dentate granule cells in entorhino-hippocampal slice cultures. Front. Mol. Neurosci. 2018, 11, 303. [Google Scholar] [CrossRef]
- Stubbendorff, C.; Stevenson, C.W. Dopamine regulation of contextual fear and associated neural circuit function. Eur. J. Neurosci. 2020. [Google Scholar] [CrossRef]
- Müller, T. Entacapone. Expert Opin. Drug Metab. Toxicol. 2010, 6, 983–993. [Google Scholar] [CrossRef]
- Bertolini, F.; Novaroli, L.; Carrupt, P.A.; Reist, M. Novel screening assay for antioxidant protection against peroxyl radical-induced loss of protein function. J. Pharm. Sci. 2007, 96, 2931–2944. [Google Scholar] [CrossRef]
- Chen, A.Y.; Lü, J.M.; Yao, Q.; Chen, C. Entacapone is an antioxidant more potent than vitamin C and vitamin E for scavenging of hypochlorous acid and peroxynitrite, and the inhibition of oxidative stress-induced cell death. Med. Sci. Monit. 2016, 22, 687–696. [Google Scholar] [CrossRef] [Green Version]
- Di Giovanni, S.; Eleuteri, S.; Paleologou, K.E.; Yin, G.; Zweckstetter, M.; Carrupt, P.A.; Lashuel, H.A. Entacapone and tolcapone, two catechol O-methyltransferase inhibitors, block fibril formation of alpha-synuclein and beta-amyloid and protect against amyloid-induced toxicity. J. Biol. Chem. 2010, 285, 14941–14954. [Google Scholar] [CrossRef] [Green Version]
- Karlsson, M.; Wikberg, T. Liquid chromatographic determination of a new catechol-O-methyltransferase inhibitor, entacapone, and its Z-isomer in human plasma and urine. J. Pharm. Biomed. Anal. 1992, 10, 593–600. [Google Scholar] [CrossRef]
- Heikkinen, H.; Varhe, A.; Laine, T.; Puttonen, J.; Kela, M.; Kaakkola, S.; Reinikainen, K. Entacapone improves the availability of L-dopa in plasma by decreasing its peripheral metabolism independent of L-dopa/carbidopa dose. Br. J. Clin. Pharmacol. 2002, 54, 363–371. [Google Scholar] [CrossRef] [Green Version]
- Brannan, T.; Prikhojan, A.; Yahr, M.D. Peripheral and central inhibitors of catechol-O-methyl transferase: Effects on liver and brain COMT activity and L-DOPA metabolism. J. Neural Transm. 1997, 104, 77–87. [Google Scholar] [CrossRef]
- Kaakkola, S.; Wurtman, R.J. Effects of COMT inhibitors on striatal dopamine metabolism: A microdialysis study. Brain Res. 1992, 587, 241–249. [Google Scholar] [CrossRef]
- Polak, P.E.; Lin, S.X.; Pelligrino, D.; Feinstein, D.L. The blood-brain barrier-permeable catechol-O-methyltransferase inhibitor dinitrocatechol suppresses experimental autoimmune encephalomyelitis. J. Neuroimmunol. 2014, 276, 135–141. [Google Scholar] [CrossRef]
- Yoo, D.Y.; Jung, H.Y.; Kim, W.; Hahn, K.R.; Kwon, H.J.; Nam, S.M.; Chung, J.Y.; Yoon, Y.S.; Kim, D.W.; Hwang, I.K. Entacapone increases proliferating cells and immature neurons in the mouse hippocampus via up-regulation of the BDNF/TrkB/pCREB pathway. Neural Reg. Res. 2021, 16, 1005–1010. [Google Scholar]
- Yoo, D.Y.; Cho, S.B.; Jung, H.Y.; Kim, W.; Choi, G.M.; Won, M.H.; Kim, D.W.; Hwang, I.K.; Choi, S.Y.; Moon, S.M. Tat-protein disulfide-isomerase A3: A possible candidate for preventing ischemic damage in the spinal cord. Cell Death Dis. 2017, 8, e3075. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.Y.; Kim, W.; Hahn, K.R.; Kwon, H.J.; Nam, S.M.; Chung, J.Y.; Yoon, Y.S.; Kim, D.W.; Yoo, D.Y.; Hwang, I.K. Effects of pyridoxine deficiency on hippocampal function and its possible association with V-type proton ATPase subunit B2 and heat shock cognate protein 70. Cells 2020, 9, 1067. [Google Scholar] [CrossRef]
- Paxinos, G.; Franklin, K.B.J. The Mouse Brain in Stereotaxic Coordinates; Academic Press: San Diego, CA, USA, 2001. [Google Scholar]
- Reis, C.R.; Chen, P.H.; Bendris, N.; Schmid, S.L. TRAIL-death receptor endocytosis and apoptosis are selectively regulated by dynamin-1 activation. Proc. Natl. Acad. Sci. USA 2017, 114, 504–509. [Google Scholar] [CrossRef] [Green Version]
- Bahl, K.; Xie, S.; Spagnol, G.; Sorgen, P.; Naslavsky, N.; Caplan, S. EHD3 protein is required for tubular recycling endosome stabilization, and an asparagine-glutamic acid residue pair within its Eps15 homology (EH) domain dictates its selective binding to NPF peptides. J. Biol. Chem. 2016, 291, 13465–13478. [Google Scholar] [CrossRef] [Green Version]
- Ritter, B.; Philie, J.; Girard, M.; Tung, E.C.; Blondeau, F.; McPherson, P.S. Identification of a family of endocytic proteins that define a new alpha-adaptin ear-binding motif. EMBO Rep. 2003, 4, 1089–1095. [Google Scholar] [CrossRef]
- Tapias, A.; Zhou, Z.W.; Shi, Y.; Chong, Z.; Wang, P.; Groth, M.; Platzer, M.; Huttner, W.; Herceg, Z.; Yang, Y.G.; et al. Trrap-dependent histone acetylation specifically regulates cell-cycle gene transcription to control neural progenitor fate decisions. Cell Stem Cell 2014, 14, 632–643. [Google Scholar] [CrossRef] [Green Version]
- Zinin, N.; Adameyko, I.; Wilhelm, M.; Fritz, N.; Uhlén, P.; Ernfors, P.; Henriksson, M.A. MYC proteins promote neuronal differentiation by controlling the mode of progenitor cell division. EMBO Rep. 2014, 15, 383–391. [Google Scholar] [CrossRef]
- Lucio-Eterovic, A.K.; Singh, M.M.; Gardner, J.E.; Veerappan, C.S.; Rice, J.C.; Carpenter, P.B. Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc. Natl. Acad. Sci. USA 2010, 107, 16952–16957. [Google Scholar] [CrossRef] [Green Version]
- Huang, Y.; Thathiah, A. Regulation of neuronal communication by G protein-coupled receptors. FEBS Lett. 2015, 589, 1607–1619. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eguren, M.; Porlan, E.; Manchado, E.; García-Higuera, I.; Cañamero, M.; Fariñas, I.; Malumbres, M. The APC/C cofactor Cdh1 prevents replicative stress and p53-dependent cell death in neural progenitors. Nat. Commun. 2013, 4, 2880. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Betsch, L.; Boltz, V.; Brioudes, F.; Pontier, G.; Girard, V.; Savarin, J.; Wipperman, B.; Chambrier, P.; Tissot, N.; Benhamed, M.; et al. TCTP and CSN4 control cell cycle progression and development by regulating CULLIN1 neddylation in plants and animals. PLoS Genet. 2019, 15, e1007899. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, S.Y.; Oh, M.; Lee, K.S.; Kim, W.K.; Oh, K.J.; Lee, S.C.; Bae, K.H.; Han, B.S. Profiling analysis of protein tyrosine phosphatases during neuronal differentiation. Neurosci. Lett. 2016, 612, 219–224. [Google Scholar] [CrossRef] [PubMed]
- Tomita, H.; Cornejo, F.; Aranda-Pino, B.; Woodard, C.L.; Rioseco, C.C.; Neel, B.G.; Alvarez, A.R.; Kaplan, D.R.; Miller, F.D.; Cancino, G.I. The protein tyrosine phosphatase receptor delta regulates developmental neurogenesis. Cell Rep. 2020, 30, 215–228.e5. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cornell, B.; Toyo-Oka, K. 14-3-3 Proteins in brain development: Neurogenesis, neuronal migration and neuromorphogenesis. Front. Mol. Neurosci. 2017, 10, 318. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ding, H.; Underwood, R.; Lavalley, N.; Yacoubian, T.A. 14-3-3 inhibition promotes dopaminergic neuron loss and 14-3-3θ overexpression promotes recovery in the MPTP mouse model of Parkinson’s disease. Neuroscience 2015, 307, 73–82. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Foote, M.; Qiao, H.; Graham, K.; Wu, Y.; Zhou, Y. Inhibition of 14-3-3 proteins leads to Schizophrenia-related behavioral phenotypes and synaptic defects in mice. Biol. Psychiatry 2015, 78, 386–395. [Google Scholar] [CrossRef] [Green Version]
- Takács-Kollár, V.; Nyitrai, M.; Hild, G. The effect of mouse twinfilin-1 on the structure and dynamics of monomeric actin. Biochim. Biophys. Acta 2016, 1864, 840–846. [Google Scholar] [CrossRef]
- Li, Y.; Wang, P.S.; Lucas, G.; Li, R.; Yao, L. ARP2/3 complex is required for directional migration of neural stem cell-derived oligodendrocyte precursors in electric fields. Stem Cell Res. Ther. 2015, 6, 41. [Google Scholar] [CrossRef] [Green Version]
- Wang, D.; Zhang, L.; Zhao, G.; Wahlström, G.; Heino, T.I.; Chen, J.; Zhang, Y.Q. Drosophila twinfilin is required for cell migration and synaptic endocytosis. J. Cell Sci. 2010, 123, 1546–1556. [Google Scholar] [CrossRef] [Green Version]
- Wahlström, G.; Vartiainen, M.; Yamamoto, L.; Mattila, P.K.; Lappalainen, P.; Heino, T.I. Twinfilin is required for actin-dependent developmental processes in Drosophila. J. Cell Biol. 2001, 155, 787–796. [Google Scholar] [CrossRef] [Green Version]
- Takaya, R.; Nagai, J.; Piao, W.; Niisato, E.; Nakabayashi, T.; Yamazaki, Y.; Nakamura, F.; Yamashita, N.; Kolattukudy, P.; Goshima, Y.; et al. CRMP1 and CRMP4 are required for proper orientation of dendrites of cerebral pyramidal neurons in the developing mouse brain. Brain Res. 2017, 1655, 161–167. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Wang, G.; Xiao, L.; Wei, Y.; Wang, H.; Zhou, L.; Sun, L. Resilience in the LPS-induced acute depressive-like behaviors: Increase of CRMP2 neuroprotection and microtubule dynamics in hippocampus. Brain Res. Bull. 2020, 162, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Frappier, T.; Derancourt, J.; Pradel, L.A. Actin and neurofilament binding domain of brain spectrin beta subunit. Eur. J. Biochem. 1992, 205, 85–91. [Google Scholar] [CrossRef] [PubMed]
- Rafalski, V.A.; Brunet, A. Energy metabolism in adult neural stem cell fate. Prog. Neurobiol. 2011, 93, 182–203. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Shen, W.B.; Yang, P.; Dong, D.; Sun, W.; Yang, P. High glucose inhibits neural stem cell differentiation through oxidative stress and endoplasmic reticulum stress. Stem Cells Dev. 2018, 27, 745–755. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.Y.; Yim, H.S.; Yoo, D.Y.; Kim, J.W.; Chung, J.Y.; Seong, J.K.; Yoon, Y.S.; Kim, D.W.; Hwang, I.K. Postnatal changes in glucose transporter 3 expression in the dentate gyrus of the C57BL/6 mouse model. Lab. Anim. Res. 2016, 32, 1–7. [Google Scholar] [CrossRef] [Green Version]
- Jung, H.Y.; Kwon, H.J.; Kim, W.; Nam, S.M.; Kim, J.W.; Hahn, K.R.; Yoo, D.Y.; Won, M.H.; Yoon, Y.S.; Kim, D.W.; et al. Phosphoglycerate mutase 1 promotes cell proliferation and neuroblast differentiation in the dentate gyrus by facilitating the phosphorylation of cAMP response element-binding protein. Neurochem. Res. 2019, 44, 323–332. [Google Scholar] [CrossRef]
- Peng, S.; Xiao, W.; Ju, D.; Sun, B.; Hou, N.; Liu, Q.; Wang, Y.; Zhao, H.; Gao, C.; Zhang, S.; et al. Identification of entacapone as a chemical inhibitor of FTO mediating metabolic regulation through FOXO1. Sci. Transl. Med. 2019, 11, eaau7116. [Google Scholar] [CrossRef]
- Adeniyi, P.A.; Shrestha, A.; Ogundele, O.M. Distribution of VTA glutamate and dopamine terminals, and their significance in CA1 neural network activity. Neuroscience 2020, 446, 171–198. [Google Scholar] [CrossRef]
- Barnes, J.R.; Mukherjee, B.; Rogers, B.C.; Nafar, F.; Gosse, M.; Parsons, M.P. The relationship between glutamate dynamics and activity-dependent synaptic plasticity. J. Neurosci. 2020, 40, 2793–2807. [Google Scholar] [CrossRef]
- Palacios-Filardo, J.; Mellor, J.R. Neuromodulation of hippocampal long-term synaptic plasticity. Curr. Opin. Neurobiol. 2019, 54, 37–43. [Google Scholar] [CrossRef] [PubMed]
- Broussard, J.I.; Yang, K.; Levine, A.T.; Tsetsenis, T.; Jenson, D.; Cao, F.; Garcia, I.; Arenkiel, B.R.; Zhou, F.M.; De Biasi, M.; et al. Dopamine regulates aversive contextual learning and associated in vivo synaptic plasticity in the hippocampus. Cell Rep. 2016, 14, 1930–1939. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tominaga-Yoshino, K.; Urakubo, T.; Ueno, Y.; Kawaai, K.; Saito, S.; Tashiro, T.; Ogura, A. Transient appearance of Ca2+-permeable AMPA receptors is crucial for the production of repetitive LTP-induced synaptic enhancement (RISE) in cultured hippocampal slices. Hippocampus 2020, 30, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Lisman, J. Glutamatergic synapses are structurally and biochemically complex because of multiple plasticity processes: Long-term potentiation, long-term depression, short-term potentiation and scaling. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2017, 372, 20160260. [Google Scholar] [CrossRef] [PubMed]
- Michaelis, E.K.; Wang, X.; Pal, R.; Bao, X.; Hascup, K.N.; Wang, Y.; Wang, W.T.; Hui, D.; Agbas, A.; Choi, I.Y.; et al. Neuronal Glud1 (glutamate dehydrogenase 1) over-expressing mice: Increased glutamate formation and synaptic release, loss of synaptic activity, and adaptive changes in genomic expression. Neurochem. Int. 2011, 59, 473–481. [Google Scholar] [CrossRef] [Green Version]
- Lander, S.S.; Chornyy, S.; Safory, H.; Gross, A.; Wolosker, H.; Gaisler-Salomon, I. Glutamate dehydrogenase deficiency disrupts glutamate homeostasis in hippocampus and prefrontal cortex and impairs recognition memory. Genes Brain Behav. 2020, 19, e12636. [Google Scholar] [CrossRef]
- Hertz, L.; Rothman, D.L. Glutamine-Glutamate Cycle Flux Is Similar in Cultured Astrocytes and Brain and Both Glutamate Production and Oxidation Are Mainly Catalyzed by Aspartate Aminotransferase. Biol. 2017, 6, 17. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, R.; Tohda, C. Extracellular cytosolic aspartate aminotransferase promotes axonal growth and object recognition memory. Neurochem. Res. 2017, 42, 3465–3473. [Google Scholar] [CrossRef]
- VanGuilder, H.D.; Farley, J.A.; Yan, H.; Van Kirk, C.A.; Mitschelen, M.; Sonntag, W.E.; Freeman, W.M. Hippocampal dysregulation of synaptic plasticity-associated proteins with age-related cognitive decline. Neurobiol. Dis. 2011, 43, 201–212. [Google Scholar] [CrossRef] [Green Version]
- Yoo, D.Y.; Jung, H.Y.; Kim, J.W.; Yim, H.S.; Kim, D.W.; Nam, H.; Suh, J.G.; Choi, J.H.; Won, M.H.; Yoon, Y.S.; et al. Reduction of dynamin 1 in the hippocampus of aged mice is associated with the decline in hippocampal-dependent memory. Mol. Med. Rep. 2016, 14, 4755–4760. [Google Scholar] [CrossRef]
- Kelly, B.L.; Vassar, R.; Ferreira, A. Beta-amyloid-induced dynamin 1 depletion in hippocampal neurons. A potential mechanism for early cognitive decline in Alzheimer disease. J. Biol. Chem. 2005, 280, 31746–31753. [Google Scholar] [CrossRef] [Green Version]
- Ciavardelli, D.; Silvestri, E.; Del Viscovo, A.; Bomba, M.; De Gregorio, D.; Moreno, M.; Di Ilio, C.; Goglia, F.; Canzoniero, L.M.; Sensi, S.L. Alterations of brain and cerebellar proteomes linked to Aβ and tau pathology in a female triple-transgenic murine model of Alzheimer’s disease. Cell Death Dis. 2010, 1, e90. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zilly, F.E.; Sørensen, J.B.; Jahn, R.; Lang, T. Munc18-bound syntaxin readily forms SNARE complexes with synaptobrevin in native plasma membranes. PLoS Biol. 2006, 4, e330. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Broeke, J.H.; Roelandse, M.; Luteijn, M.J.; Boiko, T.; Matus, A.; Toonen, R.F.; Verhage, M. Munc18 and Munc13 regulate early neurite outgrowth. Biol. Cell 2010, 102, 479–488. [Google Scholar] [CrossRef]
- Rizo, J.; Südhof, T.C. The membrane fusion enigma: SNAREs, Sec1/Munc18 proteins, and their accomplices--guilty as charged? Annu. Rev. Cell Dev. Biol. 2012, 28, 279–308. [Google Scholar] [CrossRef] [PubMed]
- Daraio, T.; Valladolid-Acebes, I.; Brismar, K.; Bark, C. SNAP-25a and SNAP-25b differently mediate interactions with Munc18-1 and Gβγ subunits. Neurosci. Lett. 2018, 674, 75–80. [Google Scholar] [CrossRef] [PubMed]
- Shen, C.; Rathore, S.S.; Yu, H.; Gulbranson, D.R.; Hua, R.; Zhang, C.; Schoppa, N.E.; Shen, J. The trans-SNARE-regulating function of Munc18-1 is essential to synaptic exocytosis. Nat. Commun. 2015, 6, 8852. [Google Scholar] [CrossRef] [Green Version]
- Hamada, N.; Iwamoto, I.; Tabata, H.; Nagata, K.I. MUNC18-1 gene abnormalities are involved in neurodevelopmental disorders through defective cortical architecture during brain development. Acta Neuropathol. Commun. 2017, 5, 92. [Google Scholar] [CrossRef]
- Lee, Y.I.; Kim, Y.G.; Pyeon, H.J.; Ahn, J.C.; Logan, S.; Orock, A.; Joo, K.M.; Lőrincz, A.; Deák, F. Dysregulation of the SNARE-binding protein Munc18-1 impairs BDNF secretion and synaptic neurotransmission: A novel interventional target to protect the aging brain. Geroscience 2019, 41, 109–123. [Google Scholar] [CrossRef]
- Bykhovskaia, M. Synapsin regulation of vesicle organization and functional pools. Semin. Cell Dev. Biol. 2011, 22, 387–392. [Google Scholar] [CrossRef]
- Rocchi, A.; Sacchetti, S.; De Fusco, A.; Giovedi, S.; Parisi, B.; Cesca, F.; Höltje, M.; Ruprecht, K.; Ahnert-Hilger, G.; Benfenati, F. Autoantibodies to synapsin I sequestrate synapsin I and alter synaptic function. Cell Death Dis. 2019, 10, 864. [Google Scholar] [CrossRef] [PubMed]
- Mazzocco, M.T.; Guarnieri, F.C.; Monzani, E.; Benfenati, F.; Valtorta, F.; Comai, S. Dysfunction of the serotonergic system in the brain of synapsin triple knockout mice is associated with behavioral abnormalities resembling synapsin-related human pathologies. Prog. Neuropsychopharmacol. Biol. Psychiatry 2020, 12, 110135. [Google Scholar]
- Goetzl, E.J.; Kapogiannis, D.; Schwartz, J.B.; Lobach, I.V.; Goetzl, L.; Abner, E.L.; Jicha, G.A.; Karydas, A.M.; Boxer, A.; Miller, B.L. Decreased synaptic proteins in neuronal exosomes of frontotemporal dementia and Alzheimer's disease. FASEB J. 2016, 30, 4141–4148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Pascual, O.; Achour, S.B.; Rostaing, P.; Triller, A.; Bessis, A. Microglia activation triggers astrocyte-mediated modulation of excitatory neurotransmission. Proc. Natl. Acad. Sci. USA 2012, 109, 197–205. [Google Scholar] [CrossRef] [Green Version]
- Cockerham, R.; Liu, S.; Cachope, R.; Kiyokage, E.; Cheer, J.F.; Shipley Puche, A.C. Subsecond regulation of synaptically released dopamine by COMT in the olfactory bulb. J. Neurosci. 2016, 36, 7779–7785. [Google Scholar] [CrossRef] [PubMed]
- Lapish, C.; Ahn, S.; Evangelista, L.; So, K.; Seamans, J.; Phillips, A. Tolcapone enhances food-evoked dopamine efflux and executive memory processes mediated by the rat prefrontal cortex. Psychopharmacology 2009, 202, 521–530. [Google Scholar] [CrossRef]
- Edelmann, E.; Lessmann, V. Dopaminergic innervation and modulation of hippocampal networks. Cell Tissue Res. 2018, 373, 711–727. [Google Scholar] [CrossRef]
- Hernández, V.S.; Luquín, S.; Jáuregui-Huerta, F.; Corona-Morales, A.A.; Medina, M.P.; Ruíz-Velasco, S.; Zhang, L. Dopamine receptor dysregulation in hippocampus of aged rats underlies chronic pulsatile L-Dopa treatment induced cognitive and emotional alterations. Neuropharmacology 2014, 82, 88–100. [Google Scholar] [CrossRef] [Green Version]
- Popova, D.; Castren, E.; Taira, T. Chronic fluoxetine administration enhances synaptic plasticity and increases functional dynamics in hippocampal CA3-CA1 synapses. Neuropharmacology 2017, 126, 250–256. [Google Scholar] [CrossRef] [Green Version]
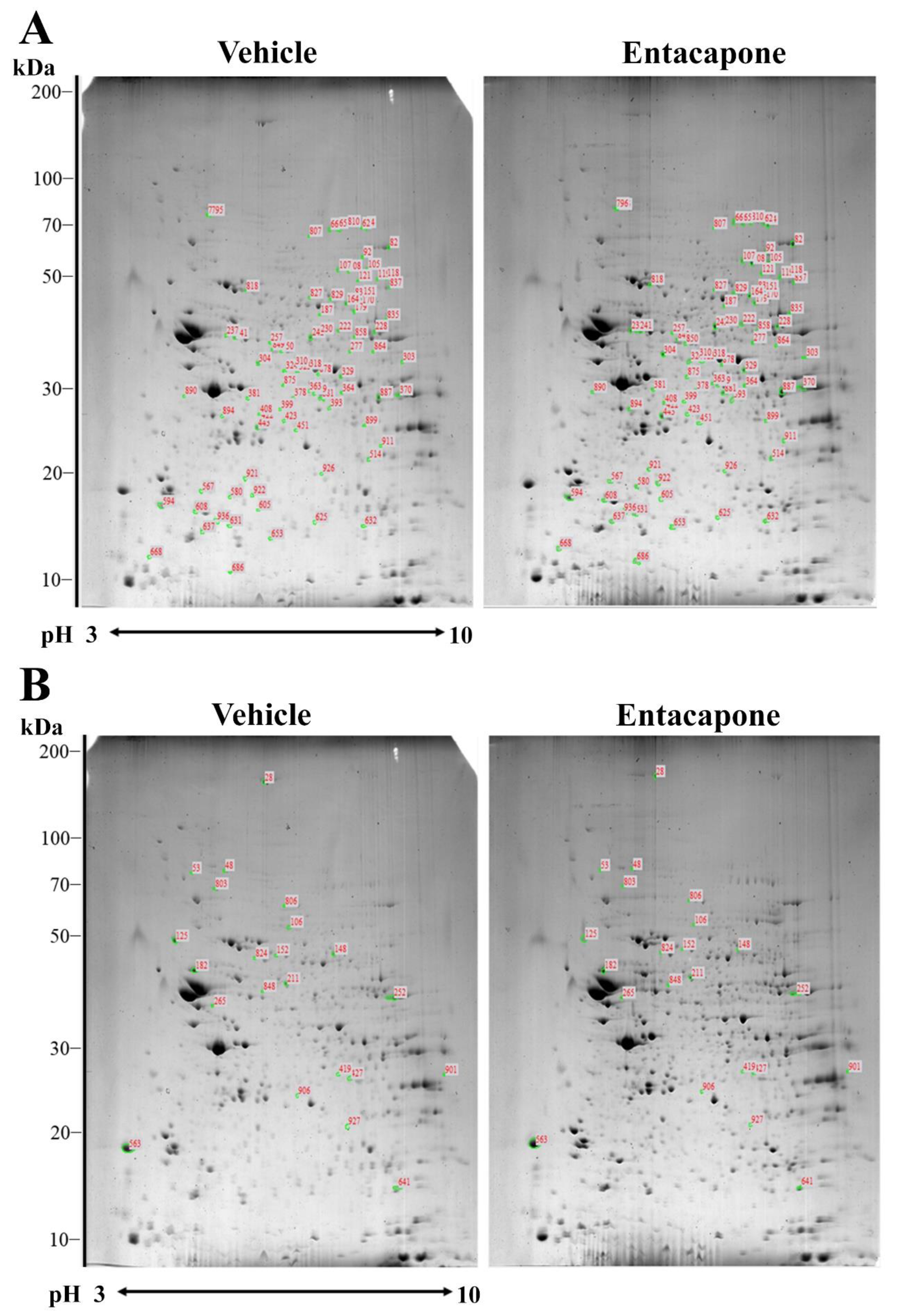

| Spot no. | Accession no. | Protein Name | Score | Nominal Mass (Da) | pI Value | Matched Peptide | Sequence Coverage (%) | Average Ratio |
|---|---|---|---|---|---|---|---|---|
| 62 | gi|148676592 | Dynamin 1 | 73 | 97,035 | 7.6 | 20 | 23 | 4.0 |
| 65 | gi|32172431 | Dynamin 1 | 195 | 98,140 | 7.61 | 39 | 42 | 2.7 |
| 810 | gi|148676592 | Dynamin 1 | 225 | 97,035 | 7.6 | 40 | 43 | 25.9 |
| 105 | gi|148668411 | Synapsin I | 137 | 69,798 | 9.84 | 22 | 44 | 20.2 |
| 118 | gi|55154394 | Synapsin II | 67 | 63,487 | 8.59 | 12 | 29 | 2.6 |
| 107 | gi|148702256 | N-Ethylmaleimide sensitive fusion protein | 109 | 78,329 | 6.68 | 25 | 35 | 2.1 |
| 108 | gi|146345470 | Vesicle-fusing ATPase | 71 | 83,131 | 6.52 | 18 | 28 | 3.1 |
| 151 | gi|3810884 | Munc18-1 | 77 | 67,906 | 6.41 | 22 | 38 | 4.0 |
| 187 | gi|172044688 | Eps15 homology (EH) domain containing protein 3 | 109 | 60,840 | 6.03 | 24 | 50 | 2.3 |
| 310 | gi|13626886 | Rab GDP dissociation inhibitor beta | 146 | 51,018 | 5.93 | 26 | 51 | 2.2 |
| 423 | gi|62287163 | Adaptin ear-binding coat-associated protein 1 | 83 | 29,678 | 5.97 | 11 | 47 | 2.5 |
| 594 | gi|46397725 | Synaptosomal-associated protein 25 | 68 | 23,528 | 4.66 | 13 | 65 | 2.5 |
| 106 | gi|73920802 | Synapsin I | 155 | 74,223 | 9.81 | 24 | 46 | −4.0 |
| Spot no. | Accession no. | Protein Name | Score | Nominal Mass (Da) | pI Value | Matched Peptide | Sequence Coverage (%) | Average Ratio |
|---|---|---|---|---|---|---|---|---|
| 277 | gi|124486949 | Transformation/transcription domain-associated protein | 70 | 439,911 | 8.51 | 37 | 12 | 3.9 |
| 324 | gi|148709230 | Nuclear receptor-binding SET-domain protein 1 | 71 | 267,700 | 8.94 | 30 | 16 | 4.7 |
| 381 | gi|55976221 | COP9 signalosome complex subunit 4 | 113 | 46,541 | 5.57 | 24 | 55 | 2.6 |
| 399 | gi|15029890 | Guanine nucleotide binding protein, alpha 11 | 79 | 42,189 | 5.7 | 15 | 48 | 2.8 |
| 422 | gi|148699469 | Fizzy/cell division cycle 20 related 1 | 67 | 46,888 | 9.74 | 12 | 27 | 2.0 |
| 803 | gi|148670554 | Valosin containing protein | 103 | 91,675 | 5.26 | 26 | 30 | −2.9 |
| 48 | gi|148698569 | Protein tyrosine phosphatase | 68 | 147,039 | 6.52 | 19 | 20 | −2.2 |
| 563 | gi|148676868 | Tyrosine 3-monooxygenase/tryptophan 5-monooxygenase activation protein, zeta polypeptide | 92 | 29,240 | 4.71 | 16 | 42 | −2.1 |
| Spot No. | Accession no. | Protein Name | Score | Nominal Mass (Da) | pI Value | Matched Peptide | Sequence Coverage (%) | Average Ratio |
|---|---|---|---|---|---|---|---|---|
| 92 | gi|77812699 | Titin isoform N2-B | 76 | 3,004,899 | 6.24 | 85 | 4 | 2.1 |
| 179 | gi|3122030 | Dihydropyrimidinase-related protein 1 | 124 | 62,471 | 6.63 | 21 | 51 | 2.2 |
| 237 | gi|146345529 | Tubulin beta-4A chain | 119 | 50,010 | 4.78 | 22 | 46 | 4.7 |
| 242 | gi|270309132 | Tetratricopeptide repeat protein 23 isoform 2 | 67 | 50,391 | 8.36 | 13 | 34 | 2.0 |
| 849 | gi|9957546 | Septin 2 | 94 | 49,050 | 5.74 | 18 | 39 | 2.0 |
| 393 | gi|30421118 | Actin monomer-binding protein twinfilin 1 | 67 | 40,368 | 6.33 | 10 | 38 | 2.3 |
| 451 | gi|67460975 | Formin-like protein 1 | 69 | 122,724 | 5.62 | 20 | 21 | 3.0 |
| 514 | gi|110825706 | Actin-related protein 2/3 complex subunit 2 | 74 | 34,450 | 6.84 | 14 | 47 | 2.0 |
| 567 | gi|18449111 | Axonemal dynein heavy chain 5 | 68 | 531,101 | 5.81 | 50 | 15 | 2.0 |
| 850 | gi|81871557 | Myosin 7 | 70 | 223,539 | 5.59 | 27 | 20 | 2.4 |
| 28 | gi|295054271 | Spectrin alpha chain | 87 | 283,519 | 5.2 | 36 | 18 | −2.1 |
| 125 | gi|116283387 | Neurofilament light polypeptide | 140 | 57,848 | 4.78 | 24 | 40 | −5.8 |
| 152 | gi|94730376 | Dihydropyrimidinase-related protein 2 (CRMP2) | 78 | 62,638 | 5.95 | 14 | 26 | −3.7 |
| 848 | gi|55977482 | Tubulin alpha-1C chain | 76 | 50,562 | 4.96 | 16 | 48 | −2.9 |
| Spot No. | Accession no. | Protein Name | Score | Nominal Mass (Da) | pI Value | Matched Peptide | Sequence Coverage (%) | Average Ratio |
|---|---|---|---|---|---|---|---|---|
| 82 | gi|63101587 | Aconitase 2, mitochondrial | 184 | 86,185 | 8.08 | 30 | 41 | 5.1 |
| 303 | gi|13543801 | Fumarate hydratase 1 | 110 | 54,564 | 9.12 | 24 | 50 | 2.3 |
| 318 | gi|13637776 | Alpha-enolase | 98 | 47,453 | 6.37 | 20 | 50 | 2.2 |
| 323 | gi|13637776 | Alpha-enolase | 79 | 47,453 | 6.37 | 15 | 38 | 2.1 |
| 364 | gi|22122625 | 3-Hydroxyisobutyryl-CoA hydrolase, mitochondrial precursor | 68 | 43,295 | 8.16 | 13 | 35 | 2.7 |
| 408 | gi|46396056 | 3′(2′),5′-Bisphosphate nucleotidase 1 | 70 | 33,517 | 5.54 | 12 | 43 | 3.3 |
| 653 | gi|23821758 | UMP-CMP kinase | 74 | 22,379 | 5.68 | 12 | 61 | 2.3 |
| 835 | gi|359807367 | Pyruvate kinase, muscle isoform M1 | 75 | 58,461 | 6.69 | 21 | 40 | 2.6 |
| 837 | gi|148692809 | Transketolase | 75 | 56,273 | 7.15 | 15 | 42 | 2.4 |
| 899 | gi|148708308 | Calcium binding protein 39 | 68 | 40,831 | 6.23 | 15 | 35 | 2.7 |
| 824 | gi|16580128 | Dihydrolipoamide S-acetyltransferase precursor | 69 | 59,389 | 5.71 | 20 | 31 | −2.2 |
| 906 | gi|148680234 | Pyrophosphatase (inorganic) 2 | 68 | 35,754 | 8.8 | 12 | 47 | −2.1 |
| Spot No. | Accession no. | Protein Name | Score | Nominal Mass (Da) | pI Value | Matched Peptide | Sequence Coverage (%) | Average Ratio |
|---|---|---|---|---|---|---|---|---|
| 119 | gi|81875980 | Isoaspartyl peptidase/L-asparaginase | 69 | 32,385 | 7.56 | 12 | 37 | 3.6 |
| 632 | gi|6137391 | mGSTA4-4 | 74 | 25,428 | 7 | 14 | 60 | 2.6 |
| 827 | gi|341941138 | Leukotriene-A4 hydrolase | 105 | 69,634 | 5.98 | 22 | 39 | 2.0 |
| 829 | gi|118600953 | Ccdc80 protein | 71 | 57,797 | 10.26 | 14 | 40 | 2.3 |
| 858 | gi|148692928 | Glutamate dehydrogenase 1 | 144 | 54,527 | 7.66 | 24 | 44 | 4.0 |
| 875 | gi|108935875 | Protein phosphatase methylesterase 1 | 69 | 42,628 | 5.67 | 15 | 38 | 3.2 |
| 887 | gi|338817898 | Aspartate aminotransferase, cytoplasmic | 165 | 46,504 | 6.68 | 21 | 60 | 2.0 |
| 911 | gi|148666837 | Monoglyceride lipase | 82 | 37,330 | 7.77 | 19 | 45 | 2.2 |
| 921 | gi|44888293 | Pyridoxal phosphate phosphatase | 70 | 31,891 | 5.53 | 13 | 48 | 2.2 |
| 926 | gi|81914662 | Carbonyl reductase [NADPH] 3 | 67 | 31,333 | 6.15 | 11 | 45 | 2.0 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yoo, D.Y.; Jung, H.Y.; Kim, W.; Hahn, K.R.; Kwon, H.J.; Nam, S.M.; Chung, J.Y.; Yoon, Y.S.; Kim, D.W.; Hwang, I.K. Entacapone Treatment Modulates Hippocampal Proteins Related to Synaptic Vehicle Trafficking. Cells 2020, 9, 2712. https://doi.org/10.3390/cells9122712
Yoo DY, Jung HY, Kim W, Hahn KR, Kwon HJ, Nam SM, Chung JY, Yoon YS, Kim DW, Hwang IK. Entacapone Treatment Modulates Hippocampal Proteins Related to Synaptic Vehicle Trafficking. Cells. 2020; 9(12):2712. https://doi.org/10.3390/cells9122712
Chicago/Turabian StyleYoo, Dae Young, Hyo Young Jung, Woosuk Kim, Kyu Ri Hahn, Hyun Jung Kwon, Sung Min Nam, Jin Young Chung, Yeo Sung Yoon, Dae Won Kim, and In Koo Hwang. 2020. "Entacapone Treatment Modulates Hippocampal Proteins Related to Synaptic Vehicle Trafficking" Cells 9, no. 12: 2712. https://doi.org/10.3390/cells9122712
APA StyleYoo, D. Y., Jung, H. Y., Kim, W., Hahn, K. R., Kwon, H. J., Nam, S. M., Chung, J. Y., Yoon, Y. S., Kim, D. W., & Hwang, I. K. (2020). Entacapone Treatment Modulates Hippocampal Proteins Related to Synaptic Vehicle Trafficking. Cells, 9(12), 2712. https://doi.org/10.3390/cells9122712






