Sphingosine-1-Phosphate in the Tumor Microenvironment: A Signaling Hub Regulating Cancer Hallmarks
Abstract
1. Introduction
2. S1P Metabolism and Export
3. Glioblastoma
4. S1P Level and Metabolism in GBM
5. The Tumor Microenvironments and the Specific Features of the GBM Ones
6. The Cellular Contributors to S1P in the Tumor Microenvironment
6.1. S1P Secretion by GBM Cells and GSCs
6.2. S1P Secretion by Non-Cancer Cells Recruited in the GBM Microenvironment
6.2.1. Microglia and Macrophages
6.2.2. Endothelial Cells
6.2.3. Neurons and Astrocytes
7. S1P Role in Cancer Hallmarks
7.1. S1P in the Cancer Microenvironment Promotes Sustained Proliferation
7.2. S1P in the Cancer Microenvironment Promotes Invasive Behaviour
7.3. S1P in the Cancer Microenvironment Promotes Death Resistance
7.4. S1P in the Cancer Microenvironment Promotes Immune-Evasion
7.5. S1P in the Cancer Microenvironment Promotes Intense Angiogenesis
7.6. S1P in the Cancer Microenvironment Promotes Deregulated Energy Metabolism
8. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Maceyka, M.; Harikumar, K.B.; Milstien, S.; Spiegel, S. Sphingosine-1-phosphate signaling and its role in disease. Trends Cell. Biol. 2012, 22, 50–60. [Google Scholar] [CrossRef] [PubMed]
- Kostenis, E. Novel clusters of receptors for sphingosine-1-phosphate, sphingosylphosphorylcholine, and (lyso)-phosphatidic acid: New receptors for “old” ligands. J. Cell. Biochem. 2004, 92, 923–936. [Google Scholar] [CrossRef] [PubMed]
- Hait, N.C.; Allegood, J.; Maceyka, M.; Strub, G.M.; Harikumar, K.B.; Singh, S.K.; Luo, C.; Marmorstein, R.; Kordula, T.; Milstien, S.; et al. Regulation of histone acetylation in the nucleus by sphingosine-1-phosphate. Science 2009, 325, 1254–1257. [Google Scholar] [CrossRef] [PubMed]
- Strub, G.M.; Maceyka, M.; Hait, N.C.; Milstien, S.; Spiegel, S. Extracellular and intracellular actions of sphingosine-1-phosphate. Adv. Exp. Med. Biol. 2010, 688, 141–155. [Google Scholar] [PubMed]
- Nagahashi, M.; Abe, M.; Sakimura, K.; Takabe, K.; Wakai, T. The role of sphingosine-1-phosphate in inflammation and cancer progression. Cancer Sci. 2018, 109, 3671–3678. [Google Scholar] [CrossRef]
- Ogretmen, B. Sphingolipid metabolism in cancer signalling and therapy. Nat. Rev. Cancer. 2018, 18, 33–50. [Google Scholar] [CrossRef]
- Pyne, N.J.; El Buri, A.; Adams, D.R.; Pyne, S. Sphingosine 1-phosphate and cancer. Adv. Biol. Regul. 2018, 8, 97–106. [Google Scholar] [CrossRef]
- Wang, P.; Yuan, Y.; Lin, W.; Zhong, H.; Xu, K.; Qi, X. Roles of sphingosine-1-phosphate signaling in cancer. Cancer Cell. Int. 2019, 19, 295. [Google Scholar] [CrossRef]
- Abdel Hadi, L.; Di Vito, C.; Marfia, G.; Navone, S.N.; Campanella, R.; Riboni, L. The role and function of sphingolipids in glioblastoma multiforme. In Bioactive Sphingolipids in Cancer Biology and Therapy; Hannun, Y.A., Luberto, C., Mao, C., Obeid, L.M., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 259–291. [Google Scholar]
- Mahajan-Thakur, S.; Bien-Möller, S.; Marx, S.; Schroeder, H.; Rauch, B.H. Sphingosine 1-phosphate (S1P) signaling in glioblastoma multiforme - A systematic review. Int. J. Mol. Sci. 2017, 18, 2448. [Google Scholar] [CrossRef]
- Korbecki, J.; Gutowska, I.; Kojder, I.; Jeżewski, D.; Goschorska, M.; Łukomska, A.; Chlubek, D.; Baranowska-Bosiacka, I. New extracellular factors in glioblastoma multiforme development: Neurotensin, growth differentiation factor-15, sphingosine-1-phosphate and cytomegalovirus infection. Oncotarget 2018, 9, 7219–7270. [Google Scholar] [CrossRef]
- Haddadi, N.; Lin, Y.; Simpson, A.M.; Nassif, N.T.; McGowan, E.M. “Dicing and splicing” sphingosine kinase and relevance to cancer. Int. J. Mol. Sci. 2017, 18, 1891. [Google Scholar] [CrossRef]
- Merrill, A.H., Jr. Sphingolipid and glycosphingolipid metabolic pathways in the era of sphingolipidomics. Chem. Rev. 2011, 111, 6387–6422. [Google Scholar] [CrossRef] [PubMed]
- Hatoum, D.; Haddadi, N.; Lin, Y.; Nassif, N.T.; McGowan, E.M. Mammalian sphingosine kinase (SphK) isoenzymes and isoform expression: Challenges for SphK as an oncotarget. Oncotarget 2017, 8, 36898–36929. [Google Scholar] [CrossRef] [PubMed]
- Pulkoski-Gross, M.J.; Obeid, L.M. Molecular mechanisms of regulation of sphingosine kinase 1. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2018, 1863, 1413–1422. [Google Scholar] [CrossRef]
- Song, D.D.; Zhou, J.H.; Sheng, R. Regulation and function of sphingosine kinase 2 in diseases. Histol. Histopathol. 2018, 33, 433–445. [Google Scholar]
- Pitson, S.M. Regulation of sphingosine kinase and sphingolipid signaling. Trends Biochem. Sci. 2011, 36, 97–107. [Google Scholar] [CrossRef]
- Siow, D.; Wattenberg, B. The compartmentalization and translocation of the sphingosine kinases: Mechanisms and functions in cell signaling and sphingolipid metabolism. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 365–375. [Google Scholar] [CrossRef]
- Neubauer, H.A.; Pitson, S.M. Roles, regulation and inhibitors of sphingosine kinase 2. FEBS J. 2013, 280, 5317–5336. [Google Scholar] [CrossRef]
- Newton, J.; Lima, S.; Maceyka, M.; Spiegel, S. Revisiting the sphingolipid rheostat: Evolving concepts in cancer therapy. Exp. Cell Res. 2015, 333, 195–200. [Google Scholar] [CrossRef]
- Spiegel, S.; Milstien, S. Functions of the multifaceted family of sphingosine kinases and some close relatives. J. Biol. Chem. 2007, 282, 2125–2129. [Google Scholar] [CrossRef]
- Proia, R.L.; Hla, T. Emerging biology of sphingosine-1-phosphate: Its role in pathogenesis and therapy. J. Clin. Investig. 2015, 125, 1379–1387. [Google Scholar] [CrossRef] [PubMed]
- Ancellin, N.; Colmont, C.; Su, J.; Li, Q.; Mittereder, N.; Chae, S.S.; Stefansson, S.; Liau, G.; Hla, T. Extracellular export of sphingosine kinase-1 enzyme. Sphingosine 1-phosphate generation and the induction of angiogenic vascular maturation. J. Biol. Chem. 2002, 277, 6667–6675. [Google Scholar] [CrossRef] [PubMed]
- Venkataraman, K.; Thangada, S.; Michaud, J.; Oo, M.L.; Ai, Y.; Lee, Y.M.; Wu, M.; Parikh, N.S.; Khan, F.; Proia, R.L.; et al. Extracellular export of sphingosine kinase-1a contributes to the vascular S1P gradient. Biochem. J. 2006, 397, 461–471. [Google Scholar] [CrossRef] [PubMed]
- Weigert, A.; Cremer, S.; Schmidt, M.V.; von Knethen, A.; Angioni, C.; Geisslinger, G.; Brune, B. Cleavage of sphingosine kinase 2 by caspase-1 provokes its release from apoptotic cells. Blood 2010, 115, 3531–3540. [Google Scholar] [CrossRef] [PubMed]
- Heffernan-Stroud, L.A.; Obeid, L.M. Sphingosine kinase 1 in cancer. Adv. Cancer Res. 2013, 117, 201–235. [Google Scholar] [PubMed]
- Wang, Q.; Li, J.; Li, G.; Li, Y.; Xu, C.; Li, M.; Xu, G.; Fu, S. Prognostic significance of sphingosine kinase 2 expression in non-small cell lung cancer. Tumour Biol. 2014, 35, 363e368. [Google Scholar]
- Ikeda, M.; Kihara, A.; Igarashi, Y. Sphingosine-1-phosphate lyase SPL is an endoplasmic reticulum-resident, integral membrane protein with the pyridoxal 5′-phosphate binding domain exposed to the cytosol. Biochem. Biophys. Res. Commun. 2004, 325, 338–343. [Google Scholar] [CrossRef]
- Le Stunff, H.; Peterson, C.; Liu, H.; Milstien, S.; Spiegel, S. Sphingosine-1-phosphate and lipid phosphohydrolases. Biochim. Biophys. Acta 2002, 1582, 8–17. [Google Scholar] [CrossRef]
- Tang, X.; Benesch, M.G.; Brindley, D.N. Lipid phosphate phosphatases and their roles in mammalian physiology and pathology. J. Lipid Res. 2015, 56, 2048–2060. [Google Scholar] [CrossRef]
- Cuvillier, O.; Pirianov, G.; Kleuser, B.; Vanek, P.G.; Coso, O.A.; Gutkind, S.; Spiegel, S. Suppression of ceramide-mediated programmed cell death by sphingosine-1-phosphate. Nature 1996, 381, 800–803. [Google Scholar] [CrossRef]
- Olivera, A.; Allende, M.L.; Proia, R.L. Shaping the landscape: Metabolic regulation of S1P gradients. Biochim. Biophys. Acta 2013, 1831, 193–202. [Google Scholar] [CrossRef]
- Nishi, T.; Kobayashi, N.; Hisano, Y.; Kawahara, A.; Yamaguchi, A. Molecular and physiological functions of sphingosine 1-phosphate transporters. Biochim. Biophys. Acta 2014, 1841, 759–765. [Google Scholar] [CrossRef]
- Kobayashi, N.; Kawasaki-Nishi, S.; Otsuka, M.; Hisano, Y.; Yamaguchi, A.; Nishi, T. MFSD2B is a sphingosine 1-phosphate transporter in erythroid cells. Sci. Rep. 2018, 8, 4969. [Google Scholar] [CrossRef]
- Zhu, X.; Ren, K.; Zeng, Y.-Z.; Zheng, Z.; Yi, G.H. Biological function of SPNS2: From zebrafish to human. Mol. Immunol. 2018, 103, 55–62. [Google Scholar] [CrossRef] [PubMed]
- Vu, T.M.; Ishizu, A.N.; Foo, J.C.; Toh, X.R.; Zhang, F.; Whee, D.M.; Torta, F.; Cazenave-Gassiot, A.; Matsumura, T.; Kim, S.; et al. Mfsd2b is essential for the sphingosine-1-phosphate export in erythrocytes and platelets. Nature 2017, 550, 524–528. [Google Scholar] [CrossRef] [PubMed]
- Książek, M.; Chacińska, M.; Chabowski, A.; Baranowski, M. Sources, metabolism, and regulation of circulating sphingosine-1-phosphate. Lipid Res. 2015, 56, 1271–1281. [Google Scholar] [CrossRef]
- Louis, D.N.; Perry, A.; Reifenberger, G.; von Deimling, A.; Figarella-Branger, D.; Cavenee, W.K.; Ohgaki, H.; Wiestler, O.D.; Kleihues, P.; Ellison, D.W. The 2016 World Health Organization classification of tumors of the central nervous system: A summary. Acta Neuropathol. 2016, 31, 803–820. [Google Scholar] [CrossRef] [PubMed]
- Wen, P.Y.; Kesari, S. Malignant gliomas in adults. N. Engl. J. Med. 2008, 359, 492–507. [Google Scholar] [CrossRef] [PubMed]
- Rees, J. Tumors of the brain. In Neurology and Clinical Neuroscience; Schapira, A.H.V., Ed.; Mosby Elsevier: Philadelphia, PA, USA, 2007; pp. 1324–1339. [Google Scholar]
- Johnson, D.R.; O’Neill, B.P. Glioblastoma survival in the United States before and during the temozolomide era. J. Neurooncol. 2012, 107, 359–364. [Google Scholar] [CrossRef] [PubMed]
- Gallego, O. Nonsurgical treatment of recurrent glioblastoma. Curr. Oncol. 2015, 22, e273–e281. [Google Scholar] [CrossRef] [PubMed]
- Ostrom, Q.T.; Gittleman, H.; Xu, J.; Kromer, C.; Wolinsky, Y.; Kruchko, C.; Barnholtz-Sloan, JS. CBTRUS statistical report: Primary brain and other central nervous system tumors diagnosed in the United States in 2009–2013. Neuro Oncol. 2016, 18, v1–v75. [Google Scholar] [CrossRef]
- Henriksson, R.; Asklund, T.; Poulsen, H.S. Impact of therapy on quality of life, neurocognitive function and their correlates in glioblastoma multiforme: A review. J. Neurooncol. 2011, 104, 639–646. [Google Scholar] [CrossRef]
- Wilson, C.B. Glioblastoma: The past, the present, and the future. Clin. Neurosurg. 1992, 38, 32–48. [Google Scholar]
- Glas, M.; Rath, B.H.; Simon, M.; Reinartz, R.; Schramme, A.; Trageser, D.; Eisenreich, R.; Leinhaas, A.; Keller, M.; Schildhaus, H.-U.; et al. Residual tumor cells are unique cellular targets in glioblastoma. Ann. Neurol. 2010, 68, 264–269. [Google Scholar] [CrossRef]
- Siegel, R.; Ward, E.; Brawley, O.; Jemal, A. Cancer statistics, 2011: The impact of eliminating socioeconomic and racial disparities on premature cancer deaths. CA Cancer J. Clin. 2011, 61, 212e36. [Google Scholar] [CrossRef] [PubMed]
- Serwer, L.P.; James, C.D. Challenges in drug delivery to tumors of the central nervous system: An overview of pharmacological and surgical considerations. Adv. Drug Deliv. Rev. 2012, 64, 590–597. [Google Scholar] [CrossRef] [PubMed]
- Riboni, L.; Campanella, R.; Bassi, R.; Villani, R.; Gaini, S.M.; Martinelli-Boneschi, F.; Viani, P.; Tettamanti, G. Ceramide levels are inversely associated with malignant progression of human glial tumors. Glia 2002, 39, 105–113. [Google Scholar] [CrossRef]
- Abuhusain, H.J.; Matin, A.; Qiao, Q.; Shen, H.; Kain, N.; Day, B.W.; Stringer, B.W.; Daniels, B.; Laaksonen, M.A.; Teo, C.; et al. A metabolic shift favoring sphingosine 1-phosphate at the expense of ceramide controls glioblastoma angiogenesis. J. Biol. Chem. 2013, 288, 37355–37364. [Google Scholar] [CrossRef]
- Hannun, Y.A.; Obeid, L.M. Many ceramides. J. Biol. Chem. 2011, 286, 27855–27862. [Google Scholar] [CrossRef]
- Senkal, C.E.; Ponnusamy, S.; Rossi, M.J.; Bialewski, J.; Sinha, D.; Jiang, J.C.; Jazwinski, S.M.; Hannun, Y.A.; Ogretmen, B. Role of human longevity assurance gene 1 and C18-ceramide in chemotherapy-induced cell death in human head and neck squamous cell carcinomas. Mol. Cancer Ther. 2007, 6, 712–722. [Google Scholar] [CrossRef]
- Sullards, M.C.; Wang, E.; Peng, Q.; Merrill, A.H., Jr. Metabolomic profiling of sphingolipids in human glioma cell lines by liquid chromatography tandem mass spectrometry. Cell. Mol. Biol. 2003, 49, 789–797. [Google Scholar] [PubMed]
- Van Brocklyn, J.R.; Jackson, C.A.; Pearl, D.K.; Kotur, M.S.; Snyder, P.J.; Prior, T.W. Sphingosine kinase-1 expression correlates with poor survival of patients with glioblastoma multiforme: Roles of sphingosine kinase isoforms in growth of glioblastoma cell lines. J. Neuropathol. Exp. Neurol. 2005, 64, 695–705. [Google Scholar] [CrossRef] [PubMed]
- Li, J.; Guan, H.Y.; Gong, L.Y.; Song, L.-B.; Zhang, N.; Wu, J.; Yuan, J.; Zheng, Y.-J.; Huang, Z.-S.; Li, M. Clinical significance of sphingosine kinase-1 expression in human astrocytomas progression and overall patient survival. Clin. Cancer Res. 2008, 14, 6996–7003. [Google Scholar] [CrossRef]
- Quint, K.; Stiel, N.; Neureiter, D.; Schlicker, H.U.; Nimsky, C.; Ocker, M.; Strik, H.; Kolodziej, M.A. The role of sphingosine kinase isoforms and receptors S1P1, S1P2, S1P3, and S1P5 in primary, secondary, and recurrent glioblastomas. Tum. Biol. 2014, 35, 8979–8989. [Google Scholar] [CrossRef] [PubMed]
- Bien-Möller, S.; Lange, S.; Holm, T.; Bohm, A.; Paland, H.; Kupper, J.; Herzog, S.; Weitmann, K.; Havemann, C.; Vogelgesang, S.; et al. Expression of S1P metabolizing enzymes and receptors correlate with survival time and regulate cell migration in glioblastoma multiforme. Oncotarget 2016, 7, 13031–13046. [Google Scholar] [CrossRef] [PubMed]
- Yoshida, Y.; Nakada, M.; Harada, T.; Tanaka, S.; Furuta, T.; Hayashi, Y.; Kita, D.; Uchiyama, N.; Hayashi, Y.; Hamada, J. The expression level of sphingosine-1-phosphate receptor type 1 is related to MIB-1 labeling index and predicts survival of glioblastoma patients. J. Neurooncol. 2010, 98, 41–47. [Google Scholar] [CrossRef]
- Postepska-Igielska, A.; Giwojna, A.; Gasri-Plotnitsky, L.; Schmitt, N.; Dold, A.; Ginsberg, D.; Grummt, I. LncRNA Khps1 regulates expression of the proto-oncogene SPHK1 via triplex-mediated changes in chromatin structure. Mol. Cell. 2015, 60, 626–636. [Google Scholar] [CrossRef]
- Kiang, K.M.; Zhang, X.-Q.; Leung, G.K. Long Non-Coding RNAs: The key players in glioma pathogenesis. Cancers 2015, 7, 1406–1424. [Google Scholar] [CrossRef]
- Donaires, F.S.; Godoy, P.R.; Leandro, G.S.; Puthier, D.; Sakamoto-Hojo, E.T. E2F transcription factors associated with up-regulated genes in glioblastoma. Cancer Biomark. 2017, 18, 199–208. [Google Scholar] [CrossRef]
- Yagoub, D.; Wilkins, M.R.; Lay, A.J.; Kaczorowski, D.C.; Hatoum, D.; Bajan, S.; Hutvagner, G.; Lai, J.H.; Wu, W.; Martiniello-Wilks, R.; et al. Sphingosine kinase 1 isoform-specific interactions in breast cancer. Mol. Endocrinol. 2014, 28, 1899–1915. [Google Scholar] [CrossRef]
- Lim, K.G.; Tonelli, F.; Berdyshev, E.; Gorshkovab, I.; Leclercq, T.; Pitson, S.M.; Bittman, R.; Pyne, S.; Pyne, N.J. Inhibition kinetics and regulation of sphingosine kinase 1 expression in prostate cancer cells: Functional differences between sphingosine kinase 1a and 1b. Int. J. Biochem. Cell. Biol. 2012, 44, 1457–1464. [Google Scholar] [CrossRef] [PubMed]
- Okada, T.; Ding, G.; Sonoda, H.; Kajimoto, T.; Haga, Y.; Khosrowbeygi, A.; Gao, N.; Miwa, S.; Jahangeer, S.; Nakamura, J. Involvement of N-terminal-extended form of sphingosine kinase 2 in serum-dependent regulation of cell proliferation and apoptosis. J. Biol. Chem. 2005, 280, 36318–36325. [Google Scholar] [CrossRef] [PubMed]
- Sordillo, L.A.; Sordillo, P.P.; Helson, L. Sphingosine kinase inhibitors as maintenance therapy of glioblastoma after ceramide-induced response. Anticancer Res. 2016, 36, 2085–2095. [Google Scholar] [PubMed]
- Jensen, S.A.; Calvert, A.E.; Volpert, G.; Kouri, F.M.; Hurley, L.A.; Luciano, J.P.; Wu, Y.; Chalastanis, A.; Futerman, A.H.; Stegh, A.H. Bcl2L13 is a ceramide synthase inhibitor in glioblastoma. Proc. Natl. Acad. Sci. USA 2014, 111, 5682–5687. [Google Scholar] [CrossRef] [PubMed]
- Tettamanti, G.; Bassi, R.; Viani, P.; Riboni, L. Salvage pathways in glycosphingolipid metabolism. Biochimie 2003, 85, 423–437. [Google Scholar] [CrossRef]
- Dilillo, M.; Ait-Belkacem, R.; Esteve, C.; Pellegrini, D.; Nicolardi, S.; Costa, M.; Vannini, E.; de Graaf, E.L.; Caleo, M.; McDonnell, L.A. Ultra-High Mass Resolution MALDI Imaging Mass Spectrometry of Proteins and Metabolites in a Mouse Model of Glioblastoma. Sci. Rep. 2017, 7, 603. [Google Scholar] [CrossRef]
- Doan, N.B.; Nguyen, H.S.; Alhajala, H.S.; Jaber, B.; Al-Gizawiy, M.M.; Ahn, E.E.; Mueller, W.M.; Chitambar, C.R.; Mirza, S.P.; Schmainda, K.M. Identification of radiation responsive genes and transcriptome profiling via complete RNA sequencing in a stable radioresistant U87 glioblastoma model. Oncotarget 2018, 9, 23532–23542. [Google Scholar] [CrossRef]
- Steck, P.A.; Ligon, A.H.; Cheong, P.; Yung, W.K.; Pershouse, MA. Two tumor suppressive loci on chromosome 10 involved in human glioblastomas. Genes Chromosomes Cancer 1995, 12, 255–261. [Google Scholar] [CrossRef]
- Ogawa, C.; Kihara, A.; Gokoh, M.; Igarashi, Y. Identification and characterization of a novel human sphingosine-1-phosphate phosphohydrolase, hSPP2. J. Biol. Chem. 2003, 278, 1268–1272. [Google Scholar] [CrossRef]
- Mora, R.; Dokic, I.; Kees, T.; Hüber, C.M.; Keitel, D.; Geibig, R.; Brügge, B.; Zentgraf, H.; Brady, N.R.; Régnier-Vigouroux, A. Sphingolipid rheostat alterations related to transformation can be exploited for specific induction of lysosomal cell death in murine and human glioma. Glia 2010, 58, 1364–1383. [Google Scholar] [CrossRef]
- Qin, J.; Berdyshev, E.; Goya, J.; Natarajan, V.; Dawson, G. Neurons and oligodendrocytes recycle sphingosine 1-phosphate to ceramide: Significance for apoptosis and multiple sclerosis. J. Biol. Chem. 2010, 285, 14134–14143. [Google Scholar] [CrossRef] [PubMed]
- Brizuela, L.; Ader, I.; Mazerolles, C.; Bocquet, M.; Malavaud, B.; Cuvillier, O. First evidence of sphingosine 1-phosphate lyase protein expression and activity downregulation in human neoplasm: Implication for resistance to therapeutics in prostate cancer. Mol. Cancer Ther. 2012, 11, 1841–1851. [Google Scholar] [CrossRef] [PubMed]
- Kurek, K.; Łukaszuk, B.; Świdnicka-Siergiejko, A.; Rogalski, P.; Wróblewski , E.; Chabowski, A.; Dąbrowski, A.; Żendzian-Piotrowska, M. Sphingolipid metabolism in colorectal adenomas varies depending on histological architecture of polyps and grade of nuclear dysplasia. Lipids 2015, 50, 349–358. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, Y.; Furuya, H.; Tamashiro, P.M.; Iino, K.; Chan, O.T.M.; Goodison, S.; Pagano, I.; Hokutan, K.; Peres, R.; Loo, L.W.M.; et al. Genetic deletion of sphingosine kinase 1 suppresses mouse breast tumor development in an HER2 transgenic model. Carcinogenesis 2018, 39, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Quail, D.F.; Joyce, J.A. The microenvironmental landscape of brain tumors. Cancer Cell 2017, 31, 326–341. [Google Scholar] [CrossRef] [PubMed]
- Fang, H.; Declerck, Y.A. Targeting the tumor microenvironment: From understanding pathways to effective clinical trials. Cancer Res. 2013, 73, 4965–49774. [Google Scholar] [CrossRef]
- Charles, N.A.; Holland, E.C.; Gilbertson, R.; Glass, R.; Kettenmann, H. The brain tumor microenvironment. Glia 2011, 59, 1169–1180. [Google Scholar] [CrossRef]
- Swartling, F.J.; Hede, S.-M.; Weiss, W.A. What underlies the diversity of brain tumors? Cancer Metastasis Rev. 2013, 32, 5–24. [Google Scholar] [CrossRef]
- Broekman, M.L.; Maas, S.L.N.; Abels, E.R.; Mempel, T.R.; Krichevsky, A.M.; Breakefield, X.O. Multidimensional communication in the microenvirons of glioblastoma. Nat. Rev. Neurol. 2018, 14, 482–495. [Google Scholar] [CrossRef]
- Persano, L.; Rampazzo, E.; Basso, G.; Viola, G. Glioblastoma cancer stem cells: Role of the microenvironment and therapeutic targeting. Biochem. Pharmacol. 2013, 85, 612–622. [Google Scholar] [CrossRef]
- Brat, D.J.; Van Meir, E.G. Vaso-occlusive and prothrombotic mechanisms associated with tumor hypoxia, necrosis, and accelerated growth in glioblastoma. Lab. Investig. 2004, 84, 397–405. [Google Scholar] [CrossRef]
- Peitzsch, C.; Perrin, R.; Hill, R.P.; Dubrovska, A.; Kurth, I. Hypoxia as a biomarker for radioresistant cancer stem cells. Int. J. Radiat. Biol. 2014, 90, 636–652. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.; Liu, Y. Hypoxia-Inducible Factors in Cancer Stem Cells and Inflammation. Trends Pharmacol. Sci. 2015, 36, 374–383. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Qian, B.-Z.; Pollard, J.W. Immune cell promotion of metastasis. Nat. Rev. Immunol. 2015, 15, 73–86. [Google Scholar] [CrossRef]
- Uribe, D.; Torres, Á.; Rocha, J.D.; Niechi, I.; Oyarzún, C.; Sobrevia, L.; San Martín, R.; Quezada, C. Multidrug resistance in glioblastoma stem-like cells: Role of the hypoxic microenvironment and adenosine signaling. Mol. Asp. Med. 2017, 55, 140–151. [Google Scholar] [CrossRef] [PubMed]
- Hambardzumyan, D.; Bergers, G. Glioblastoma: Defining tumor niches. Trends Cancer 2015, 1, 252–265. [Google Scholar] [CrossRef]
- Filatova, A.; Acker, T.; Garvalov, B.K. The cancer stem cell niche(s): The crosstalk between glioma stem cells and their microenvironment. Biochim. Biophys. Acta 2013, 1830, 2496–2508. [Google Scholar] [CrossRef]
- Infanger, D.W.; Cho, Y.; Lopez, B.S.; Mohanan, S.; Liu, S.C.; Gursel, D.; Boockvar, J.A.; Fischbach, C. Glioblastoma stem cells are regulated by interleukin-8 signaling in a tumoral perivascular niche. Cancer Res. 2013, 73, 7079–7089. [Google Scholar] [CrossRef]
- Riboni, L.; Giussani, P.; Viani, P. Sphingolipid transport. Adv. Exp. Med. Biol. 2010, 688, 24–45. [Google Scholar]
- Takabe, K.; Spiegel, S. Export of sphingosine-1-phosphate and cancer progression. J. Lipid Res. 2014, 55, 1839–1846. [Google Scholar] [CrossRef]
- Edsall, L.C.; Cuvillier, O.; Twitty, S.; Spiegel, S.; Milstien, S. Sphingosine kinase expression regulates apoptosis and caspase activation in PC12 cells. J. Neurochem. 2001, 76, 1573–1584. [Google Scholar] [CrossRef] [PubMed]
- Anelli, V.; Gault, C.R.; Snider, A.J.; Obeid, L.M. Role of sphingosine kinase-1 in paracrine/transcellular angiogenesis and lymphangiogenesis in vitro. FASEB J. 2010, 24, 2727–2738. [Google Scholar] [CrossRef]
- Riccitelli, E.; Giussani, P.; Di Vito, C.; Condomitti, G.; Tringali, C.; Caroli, M.; Galli, R.; Viani, P.; Riboni, L. Extracellular sphingosine-1-phosphate: A novel actor in human glioblastoma stem cell survival. PLoS ONE 2013, 8, e68229. [Google Scholar] [CrossRef] [PubMed]
- Abdel Hadi, L.; Anelli, V.; Guarnaccia, L.; Navone, S.; Beretta, M.; Moccia, F.; Tringali, C.; Urechie, V.; Campanella, R.; Marfia, G.; et al. A bidirectional crosstalk between glioblastoma and brain endothelial cells potentiates the angiogenic and proliferative signaling of sphingosine-1-phosphate in the glioblastoma microenvironment. Biochim. Biophys. Acta Mol. Cell. Biol. Lipids 2018, 1863, 1179–1192. [Google Scholar] [CrossRef] [PubMed]
- Bassi, R.; Anelli, V.; Giussani, P.; Tettamanti, G.; Viani, P.; Riboni, L. Sphingosine-1-phosphate is released by cerebellar astrocytes in response to bFGF and induces astrocyte proliferation through Gi-protein-coupled receptors. Glia 2006, 53, 621–630. [Google Scholar] [CrossRef] [PubMed]
- LaMontagne, K.; Littlewood-Evans, A.; Schnell, C.; O’Reilly, T.; Wyder, L.; Sanchez, T.; Probst, B.; Butler, J.; Wood, A.; Liau, G.; et al. Antagonism of sphingosine-1-phosphate receptors by FTY720 inhibits angiogenesis and tumor vascularization. Cancer Res. 2006, 66, 221–231. [Google Scholar] [CrossRef] [PubMed]
- Paugh, B.S.; Paugh, S.W.; Bryan, L.; Kapitonov, D.; Wilczynska, K.M.; Gopalan, S.M.; Rokita, H.; Milstien, S.; Spiegel, S.; Kordula, T. EGF regulates plasminogen activator inhibitor-1 by a pathway involving c-Src, PKCδ, and sphingosine kinase 1 in glioblastoma cells. FASEB J. 2008, 22, 455–465. [Google Scholar] [CrossRef]
- Riboni, L.; Viani, P.; Bassi, R.; Stabilini, A.; Tettamanti, G. Biomodulatory role of ceramide in basic fibroblast growth-factor induced proliferation of cerebellar astrocytes in primary culture. Glia 2000, 32, 137–145. [Google Scholar] [CrossRef]
- Sukocheva, O.; Wadham, C.; Holmes, A.; Albanese, N.; Verrier, E.; Feng, F.; Bernal, A.; Derian, C.K.; Ullrich, A.; Vadas, M.A.; et al. Estrogen transactivates EGFR via the sphingosine 1-phosphate receptor Edg-3: The role of sphingosine kinase-1. J. Cell. Biol. 2006, 173, 301–310. [Google Scholar] [CrossRef]
- Chen, C.H.; Chen, P.Y.; Lin, Y.Y.; Feng, L.Y.; Chen, S.H.; Chen, C.Y.; Huang, Y.C.; Huang, C.Y.; Jung, S.M.; Chen, L.Y.; et al. Suppression of tumor growth via IGFBP3 depletion as a potential treatment in glioma. J. Neurosurg. 2019, 11, 1–12. [Google Scholar] [CrossRef]
- Cattaneo, M.G.; Vanetti, C.; Samarani, M.; Aureli, M.; Bassi, R.; Sonnino, S.; Giussani, P. Cross-talk between sphingosine-1-phosphate and EGFR signaling pathways enhances human glioblastoma cell invasiveness. FEBS Lett. 2018, 592, 949–961. [Google Scholar] [CrossRef] [PubMed]
- Meyer zu Heringdorf, D.; Lass, H.; Kuchar, I.; Lipinski, M.; Alemany, R.; Rumenapp, U.; Jakobs, K.H. Stimulation of intracellular sphingosine-1-phosphate production by G-protein-coupled sphingosine-1-phosphate receptors. Eur. J. Pharmacol. 2001, 145–154. [Google Scholar]
- Blom, T.; Bergelin, N.; Meinander, A.; Löf, L.; Slotte, J.P.; Eriksson, J.E.; Törnquist, K. An autocrine sphingosine-1-phosphate signaling loop enhances NF-κB-activation and survival. BMC Cell. Biol. 2010, 11, 45. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, S.E.; Milstien, S.; Spiegel, S. Autocrine and paracrine roles of sphingosine-1-phosphate. Trends Endocrinol. Metab. 2007, 18, 300–307. [Google Scholar] [CrossRef] [PubMed]
- Chan, H.; Pitson, S.M. Post-translational regulation of sphingosine kinases. Biochim. Biophys. Acta 2013, 1831, 147–156. [Google Scholar] [CrossRef] [PubMed]
- Libermann, T.A.; Nusbaum, H.R.; Razon, N.; Kris, R.; Lax, I.; Soreq, H.; Whittle, N.; Waterfield, M.D.; Ullrich, A.; Schlessinger, J. Amplification, enhanced expression and possible rearrangement of EGF receptor gene in primary human brain tumours of glial origin. Nature 1985, 313, 144–147. [Google Scholar] [CrossRef]
- Bryan, L.; Kordula, T.; Spiegel, S.; Milstien, S. Regulation and functions of sphingosine kinases in the brain. Biochim. Biophys. Acta 2008, 1781, 459–466. [Google Scholar] [CrossRef]
- Lu, T.; Tian, L.; Han, Y.; Vogelbaum, M.; Stark, G.R. Dose-dependent cross-talk between the transforming growth factor-β and interleukin-1 signaling pathways. Proc. Natl. Acad. Sci. USA 2007, 104, 4365–4370. [Google Scholar] [CrossRef]
- Paugh, B.S.; Bryan, L.; Paugh, S.W.; Wilczynska, K.M.; Alvarez, S.M.; Singh, S.K.; Rokita, H.; Wright, S.; Griswold-Prenner, I.; Milstien, S.; et al. Interleukin-1 regulates the expression of sphingosine kinase 1 in glioblastoma cells. J. Biol. Chem. 2009, 284, 3408–3417. [Google Scholar] [CrossRef]
- Anelli, V.; Gault, C.R.; Cheng, A.B.; Obeid, L.M. Sphingosine kinase 1 is up-regulated during hypoxia in U87MG glioma cells. Role of hypoxia-inducible factors 1 and 2. J. Biol. Chem. 2008, 283, 3365–3375. [Google Scholar] [CrossRef]
- Zhang, H.; Li, W.; Sun, S.; Yu, S.; Zhang, M.; Zou, F. Inhibition of sphingosine kinase 1 suppresses proliferation of glioma cells under hypoxia by attenuating activity of extracellular signal-regulated kinase. Cell Prolif. 2012, 45, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Bouquerel, P.; Gstalder, C.; Müller, D.; Laurent, J.; Brizuela, L.; Sabbadini, R.A.; Malavaud, B.; Pyronnet, S.; Martineau, Y.; Ader, I.; et al. Essential role for SphK1/S1P signaling to regulate hypoxia-inducible factor 2α expression and activity in cancer. Oncogene 2016, 5, e209. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.-H.; Kim, E.-J.; Hitomi, M.; Oh, S.-Y.; Jin, X.; Jeon, H.-M.; Beck, S.; Jin, X.; Kim, J.-K.; Park, C.G.; et al. The LIM-only transcription factor LMO2 determines tumorigenic and angiogenic traits in glioma stem cells. Cell Death Differ. 2015, 22, 1517–1525. [Google Scholar] [CrossRef]
- Matrone, G.; Meng , S.; Gu, Q.; Lv, J.; Fang, L.; Chen, K.; Cooke, J.P. Lmo2 (LIM-Domain-Only 2) modulates Sphk1 (sphingosine kinase) and promotes endothelial cell migration. Arter. Thromb. Vasc. Biol. 2017, 37, 1860–1868. [Google Scholar] [CrossRef]
- Rigogliuso, S.; Donati, C.; Cassarà, D.; Taverna, S.; Salamone, M.; Bruni, P.; Vittorelli, L.M. An active form of sphingosine kinase-1 is released in the extracellular medium as component of membrane vesicles shed by two human tumor cell lines. J. Oncol. 2010. [Google Scholar] [CrossRef]
- Nojima, H.; Freeman, C.M.; Schuster, R.M.; Japtok, L.; Kleuser, B.; Edwards, M.J.; Gulbins, E.; Lentsch, A.B. Hepatocyte exosomes mediate liver repair and regeneration via sphingosine-1-phosphate. J. Hepatol. 2016, 64, 60–68. [Google Scholar] [CrossRef]
- Mohamed, N.N.B.; Okada, T.; Kajimoto, T.; Nakamura, S.-I. Essential role of sphingosine kinase 2 in the regulation of cargo contents in the exosomes from K562 cells. J. Med. Sci. 2017, 63, E123–E129. [Google Scholar]
- Neubauer, H.A.; Tea, M.N.; Zebol, J.R.; Gliddon, B.L.; Stefanidis, C.; Moretti, P.A.B.; Pitman, M.R.; Costabile, M.; Kular, J.; Stringer, B.W.; et al. Cytoplasmic dynein regulates the subcellular localization of sphingosine kinase 2 to elicit tumor-suppressive functions in glioblastoma. Oncogene 2019, 38, 1151–1165. [Google Scholar] [CrossRef]
- Bernhart, E.; Damm, S.; Wintersperger, A.; Nusshold, C.; Brunner, A.M.; Plastira, I.; Rechberger, G.; Reicher, H.; Wadsack, C.; Zimmer, A.; et al. Interference with distinct steps of sphingolipid synthesis and signaling attenuates proliferation of U87MG glioma cells. Biochem. Pharm. 2015, 96, 119–130. [Google Scholar] [CrossRef]
- Eibinger, G.; Fauler, G.; Bernhart, E.; Frank, S.; Hammer, A.; Wintersperger, A.; Eder, H.; Heinemann, A.; Mischel, P.S.; Malle, E.; et al. On the role of 25-hydroxycholesterol synthesis by glioblastoma cell lines. Implications for chemotactic monocyte recruitment. Exp. Cell Res. 2013, 319, 1828–1838. [Google Scholar] [CrossRef]
- Marfia, G.; Campanella, R.; Navone, S.E.; Di Vito, C.; Riccitelli, E.; Abdel Hadi, L.; Bornati, A.; de Rezende, G.; Giussani, P.; Tringali, C.; et al. Autocrine/paracrine sphingosine-1-phosphate fuels proliferative and stemness qualities of glioblastoma stem cells. Glia 2014, 62, 1968–1981. [Google Scholar] [CrossRef] [PubMed]
- Kawamura, Y.; Takouda, J.; Yoshimoto, K.; Nakashima, K. New aspects of glioblastoma multiforme revealed by similarities between neural and glioblastoma stem cells. Cell Biol. Toxicol. 2018, 34, 425–440. [Google Scholar] [CrossRef] [PubMed]
- Kimura, A.; Ohmori, T.; Ohkawa, R.; Madoiwa, S.; Mimuro, J.; Murakami, T.; Kobayashi, E.; Hoshino, Y.; Yatomi, Y.; Sakata, Y. Essential roles of sphingosine 1-phosphate/S1P1 receptor axis in the migration of neural stem cells toward a site of spinal cord injury. Stem Cells 2007, 25, 115–124. [Google Scholar] [CrossRef] [PubMed]
- Seidel, S.; Garvalov, B.K.; Wirta, V.; von Stechow, L.; Schänzer, A.; Meletis, K.; Wolter, M.; Sommerlad, D.; Henze, A.-T.; Nistér, M.; et al. A hypoxic niche regulates glioblastoma stem cells through hypoxia inducible factor 2α. Brain 2010, 133, 983–995. [Google Scholar] [CrossRef] [PubMed]
- Bleau, A.M.; Hambardzumyan, D.; Ozawa, T.; Fomchenko, E.I.; Huse, J.T.; Brennan, C.W.; Holland, E.C. PTEN/PI3K/Akt pathway regulates the side population phenotype and ABCG2 activity in glioma tumor stem-like cells. Cell Stem Cell 2009, 4, 226–235. [Google Scholar] [CrossRef]
- Quezada, C.; Peigñan, L.; Segura, R.; Riquelme, F.; Melo, R.; Rojas, Z.D.; Ayach, F.; San Martín, R.; Cárcamo, J.G. Study of resistance to chemotherapy mediated by ABC transporters in biopsies of glioblastoma multiforme. Rev. Med. Chil. 2011, 139, 415–424. [Google Scholar] [CrossRef]
- Begicevic, R.-R.; Falasca, M. ABC Transporters in cancer stem cells: Beyond chemoresistance. Int. J. Mol. Sci. 2017, 18, 2362. [Google Scholar] [CrossRef]
- Dréan, A.; Rosenberg, S.; Lejeune, F.-X.; Goli, L.; Nadaradjane, A.A.; Guehennec, J.; Schmitt, C.; Verreault, M.; Bielle, F.; Mokhtari, K.; et al. ATP binding cassette (ABC) transporters: Expression and clinical value in glioblastoma. J. Neurooncol. 2018, 138, 479–486. [Google Scholar] [CrossRef]
- Riboni, L. Cross-talk between tumor stem cells and tumor cells: A glioblastoma strategy to promote malignancy. Stem Cell Investig. 2018, 5. [Google Scholar] [CrossRef]
- Wthamang, X.; Prager, B.C.; Wu, Q.; Kim, L.J.Y.; Gimple, R.C.; Shi, Y.; Yang, K.; Morton, A.R.; Zhou, W.; Zhu, Z.; et al. Reciprocal signaling between glioblastoma stem cells and differentiated tumor cells promotes malignant progression. Cell Stem Cell 2018, 22, 514–528. [Google Scholar]
- Bougnaud, S.; Golebiewska, A.; Oudin, A.; Keunen, O.; Harter, P.N.; Mäder, L.; Azuaje, F.; Fritah, S.; Stieber, D.; Kaoma, T.; et al. Molecular crosstalk between tumour and brain parenchyma instructs histopathological features in glioblastoma. Oncotarget 2016, 7, 31955–31971. [Google Scholar] [CrossRef]
- Codrici, E.; Enciu, A.M.; Popescu, I.D.; Mihat, S.; Tanase, C. Glioma stem cells and their microenvironments: Providers of challenging therapeutic targets. Stem Cells Int. 2016, 2016, 5728438. [Google Scholar] [CrossRef] [PubMed]
- Ho, I.A.W.; Shim, W.S.N. Contribution of the microenvironmental niche to glioblastoma heterogeneity. BioMed Res. Int. 2017, 9634172. [Google Scholar] [CrossRef] [PubMed]
- Roos, A.; Ding, Z.; Loftus, J.C.; Tran, N.L. Molecular and microenvironmental determinants of glioma stem-like cell survival and invasion. Front. Oncol. 2017, 7, 120. [Google Scholar] [CrossRef] [PubMed]
- Hambardzumyan, D.; Gutmann, D.H.; Kettenmann, H. The role of microglia and macrophages in glioma maintenance and progression. Nat. Neurosci. 2016, 19, 20–27. [Google Scholar] [CrossRef] [PubMed]
- Arcuri, C.; Fioretti, B.; Bianchi, R.; Mecca, C.; Tubaro, C.; Beccari, T.; Franciolini, F.; Giambanco, I.; Donato, R. Microglia-glioma cross-talk: A two way approach to new strategies against glioma. Front. Biosci. 2017, 22, 268–309. [Google Scholar] [CrossRef]
- Dello Russo, C.; Lisi, L.; Tentori, L.; Navarra, P.; Graziani, G.; Combs, C.K. Exploiting microglial functions for the treatment of glioblastoma. Curr. Cancer Drug Targets 2017, 17, 267–281. [Google Scholar] [CrossRef]
- Glass, R.; Synowitz, M. CNS macrophages and peripheral myeloid cells in brain tumours. Acta Neuropathol. 2014, 128, 347–362. [Google Scholar] [CrossRef]
- Roesch, S.; Rapp, C.; Dettling, S.; Herold-Mende, C. When immune cells turn bad-tumor-associated microglia/macrophages in glioma. Int. J. Mol. Sci. 2018, 19, 436. [Google Scholar] [CrossRef]
- Maceyka, M.; Spiegel, S. Sphingolipid metabolites in inflammatory disease. Nature 2014, 510, 58–67. [Google Scholar] [CrossRef]
- Aoki, M.; Aoki, H.; Ramanathan, R.; Hait, N.C.; Takabe, K. sphingosine-1-phosphate signaling in immune cells and inflammation: Roles and therapeutic potential. Mediat. Inflamm. 2016, 2016, 8606878. [Google Scholar] [CrossRef]
- Di Vito, C.; Abdel Hadi, L.; Navone, S.E.; Marfia, G.; Campanella, R.; Mancuso, M.E.; Riboni, L. Platelet-derived sphingosine-1-phosphate and inflammation: From basic mechanisms to clinical implications. Platelets 2016, 27, 393–401. [Google Scholar] [CrossRef] [PubMed]
- Kitatani, K.; Iwabuchi, K.; Snider, A.; Riboni, L. Sphingolipids in inflammation: From bench to bedside. Mediat. Inflamm. 2016, 7602526. [Google Scholar] [CrossRef] [PubMed]
- Gualtierotti, R.; Guarnaccia, L.; Beretta, M.; Navone, S.E.; Campanella, R.; Riboni, L.; Rampini, P.; Marfia, G. Modulation of neuroinflammation in the central nervous system: Role of chemokines and sphingolipids. Adv. Ther. 2017, 34, 396–420. [Google Scholar] [CrossRef]
- Hammad, S.M.; Crellin, H.G.; Wu, B.X.; Melton, J.; Anelli, V.; Obeid, L.M. Dual and distinct roles for sphingosine kinase 1 and sphingosine 1 phosphate in the response to inflammatory stimuli in raw macrophages. Prostaglandins Other Lipid Mediat. 2008, 85, 107–114. [Google Scholar] [CrossRef]
- Nayak, D.; Huo, Y.; Kwang, W.X.; Pushparaj, P.N.; Kumar, S.D.; Ling, E.A.; Dheen, S.T. Sphingosine kinase 1 regulates the expression of proinflammatory cytokines and nitric oxide in activated microglia. Neuroscience 2010, 166, 132–144. [Google Scholar] [CrossRef]
- Marfia, G.; Navone, S.E.; Abdel Hadi, L.; Paroni, M.; Berno, V.; Beretta, M.; Gualtierotti, R.; Ingegnoli, F.; Levi, V.; Miozzo, M.; et al. The adipose mesenchymal stem cell secretome inhibits inflammatory responses of microglia: Evidence for an involvement of sphingosine-1-phosphate signalling. Stem Cells Dev. 2016, 25, 1095–1107. [Google Scholar] [CrossRef]
- Gibson, C.J.; Hossain, M.M.; Richardson, J.R.; Aleksunes, L.M. Inflammatory regulation of ATP binding cassette efflux transporter expression and function in microglia. J. Pharm. Exp. Ther. 2012, 343, 650–660. [Google Scholar] [CrossRef]
- Cole, S.P.C. Multidrug resistance protein 1 (MRP1, ABCC1), a “multitasking” ATP-binding cassette (ABC) transporter. J. Biol. Chem. 2014, 289, 30880–30888. [Google Scholar] [CrossRef]
- Matias, D.; Balça-Silva, J.; da Graça, G.C.; Wanjiru, C.M.; Macharia, L.W.; Nascimento, C.P.; Roque, N.R.; Coelho-Aguiar, J.M.; Pereira, C.M.; Dos Santos, M.F.; et al. Microglia/astrocytes-glioblastoma crosstalk: Crucial molecular mechanisms and microenvironmental factors. Front. Cell. Neurosci. 2018, 12, 235. [Google Scholar] [CrossRef]
- Zhong, L.; Jiang, X.; Zhu, Z.; Qin, H.; Dinkins, M.B.; Kong, J.N.; Leanhart, S.; Wang, R.; Elsherbini, A.; Bieberich, E.; et al. Lipid transporter Spns2 promotes microglia pro-inflammatory activation in response to amyloid-beta peptide. Glia 2019, 67, 498–511. [Google Scholar] [CrossRef] [PubMed]
- Pilorget, A.; Demeule, M.; Barakat, S.; Marvaldi, J.; Luis, J.; Beliveau, R. Modulation of P-glycoprotein function by sphingosine kinase-1 in brain endothelial cells. J. Neurochem. 2007, 100, 1203–1210. [Google Scholar] [CrossRef]
- Hisano, Y.; Kobayashi, N.; Yamaguchi, A.; Nishi, T. Mouse SPNS2 functions as a spingosine-1-phosphate transporter in vascular endothelial cells. PLoS ONE 2012, 7, e38941. [Google Scholar] [CrossRef]
- Anelli, V.; Bassi, R.; Tettamanti, G.; Viani, P.; Riboni, L. Extracellular release of newly synthesized sphingosine-1-phosphate by cerebellar granule cells and astrocytes. J. Neurochem. 2005, 92, 1204–1215. [Google Scholar] [CrossRef] [PubMed]
- Kajimoto, T.; Okada, Y.; Yu, H.; Goparaju, S.K.; Jahangeer, S.; Nakamura, S.-I. Involvement of sphingosine-1-phosphate in glutamate secretion in hippocampal neurons. Mol. Cell. Biol. 2007, 27, 3429–3440. [Google Scholar] [CrossRef] [PubMed]
- Brown, P.D.; Jensen, A.W.; Felten, S.J.; Ballman, K.V.; Schaefer, P.L.; Jaeckle, K.A.; Cerhan, J.H.; Buckner, J.C. Detrimental effects of tumor progression on cognitive function of patients with high-grade glioma. J. Clin. Oncol. 2006, 24, 5427–5433. [Google Scholar] [CrossRef] [PubMed]
- Buckingham, S.C.; Campbell, S.L.; Haas, B.R.; Montana, V.; Robel, S.; Ogunrinu, T.; Sontheimer, H. Glutamate release by primary brain tumors induces epileptic activity. Nat. Med. 2011, 17, 1269–1274. [Google Scholar] [CrossRef]
- Strong, A.D.; Indart, M.C.; Hill, N.R.; Daniels, R.L. GL261 glioma tumor cells respond to ATP with an intracellular calcium rise and glutamate release. Mol. Cell. Biochem. 2018, 446, 53–62. [Google Scholar] [CrossRef]
- Marcus, H.J.; Carpenter, K.L.; Price, S.J.; Hutchinson, P.J. In vivo assessment of high-grade glioma biochemistry using microdialysis: A study of energy-related molecules, growth factors and cytokines. J. Neurooncol. 2010, 97, 11–23. [Google Scholar] [CrossRef]
- Haley, E.-M.; Kim, Y. The role of basic fibroblast growth factor in glioblastoma multiforme and glioblastoma stem cells and in their in vitro culture. Cancer Lett. 2014, 346, 1–5. [Google Scholar] [CrossRef]
- Fessler, E.; Borovski, T.; Medema, J.P. Endothelial cells induce cancer stem cell features in differentiated glioblastoma cells via bFGF. Mol. Cancer. 2015, 14, 157. [Google Scholar] [CrossRef]
- Okamoto, S.; Nitta, M.; Maruyama, T.; Sawada, T.; Komori, T.; Okada, Y.; Muragaki, Y. Bevacizumab changes vascular structure and modulates the expression of angiogenic factors in recurrent malignant gliomas. Brain Tumor Pathol. 2016, 33, 129–136. [Google Scholar] [CrossRef]
- Sato, K.; Malchinkhuu, E.; Horiuchi, Y.; Mogi, C.; Tomura, H.; Tosaka, M.; Yoshimoto, Y.; Kuwabara, A.; Okajima, F. Critical role of ABCA1 transporter in sphingosine 1-phosphate release from astrocytes. J. Neurochem. 2007, 103, 2610–2619. [Google Scholar] [CrossRef] [PubMed]
- Hanahan, D.; Weinberg, R.A. Hallmarks of cancer: The next generation. Cell 2011, 144, 646–674. [Google Scholar] [CrossRef]
- Nørøxe, D.S.; Poulsen, H.S.; Lassen, U. Hallmarks of glioblastoma: A systematic review. ESMO Open 2016. [Google Scholar] [CrossRef]
- Mendelson, K.; Evans, T.; Hla, T. Sphingosine 1-phosphate signalling. Development 2014. [Google Scholar] [CrossRef] [PubMed]
- Patmanathan, S.N.; Wang, W.; Yap, L.F.; Herr, D.R.; Paterson, I.C. Mechanisms of sphingosine 1-phosphate receptor signalling in cancer. Cell Signal 2017, 34, 66–75. [Google Scholar] [CrossRef]
- Blaho, V.A.; Hla, T. Regulation of mammalian physiology, development, and disease by the sphingosine 1-phosphate and lysophosphatidic acid receptors. Chem. Rev. 2011, 111, 6299–6320. [Google Scholar] [CrossRef]
- Yoshida, Y.; Nakada, M.; Sugimoto, N.; Harada, T.; Hayashi, Y.; Kita, D.; Uchiyama, N.; Hayashi, Y.; Yachie, A.; Takuwa, Y.; et al. Sphingosine-1-phosphate receptor type 1 regulates glioma cell proliferation and correlates with patient survival. Int. J. Cancer. 2010, 126, 2341–2352. [Google Scholar] [CrossRef]
- Marfe, G.; Mirone, G.; Shukla, A.; Di Stefano, C. Sphingosine kinases signalling in carcinogenesis. Mini Rev. Med. Chem. 2015, 15, 300–314. [Google Scholar] [CrossRef]
- El-Shewy, H.M.; Johnson, K.R.; Lee, M.H.; Jaffa, A.A.; Obeid, L.M.; Luttrell, L.M. Insulin-like growth factors mediate heterotrimeric G protein-dependent ERK1/2 activation by transactivating sphingosine 1-phosphate receptors. J. Biol. Chem. 2006, 281, 31399–313407. [Google Scholar] [CrossRef] [PubMed]
- Lebman, D.A.; Spiegel, S. Cross-talk at the crossroads of sphingosine-1-phosphate, growth factors, and cytokine signaling. J. Lipid Res. 2008, 49, 1388–1394. [Google Scholar] [CrossRef] [PubMed]
- El Buri, A.; Adams, D.R.; Smith, D.; Tate, R.J.; Mullin, M.; Pyne, S.; Pyne, N.J. The sphingosine 1-phosphate receptor 2 is shed in exosomes from breast cancer cells and is N-terminally processed to a short constitutively active form that promotes extracellular signal regulated kinase activation and DNA synthesis in fibroblasts. Oncotarget 2018, 9, 29453–29467. [Google Scholar] [PubMed]
- Radeff-Huang, J.; Seasholtz, T.M.; Chang, J.W.; Smith, J.M.; Walsh, C.T.; Brown, J.H. Tumor necrosis factor-α-stimulated cell proliferation is mediated through sphingosine kinase-dependent Akt activation and cyclin D expression. J. Biol. Chem. 2007, 282, 863–870. [Google Scholar] [CrossRef]
- Kapitonov, D.; Allegood, J.C.; Mitchell, C.; Hait, N.C.; Almenara, J.A.; Adams, J.K.; Zipkin, R.E.; Dent, P.; Kordula, T.; Milstien, S.; et al. Targeting sphingosine kinase 1 inhibits Akt signaling, induces apoptosis, and suppresses growth of human glioblastoma cells and xenografts. Cancer Res. 2009, 69, 6915–6923. [Google Scholar] [CrossRef]
- Yu, O.M.; Miyamoto, S.; Brown, J.H. Myocardin-related transcription factor A and Yes-associated protein exert dual control in G protein-coupled receptor- and RhoA-mediated transcriptional regulation and cell proliferation. Mol. Cell. Biol. 2016, 36, 39–49. [Google Scholar]
- Santosh, V.; Arivazhagan, A.; Sreekanthreddy, P.; Srinivasan, H.; Thota, B.; Srividya, M.R.; Vrinda, M.; Sridevi, S.; Shailaja, B.C.; Samuel, C.; et al. Grade-specific expression of insulin-like growth factor-binding proteins-2, -3, and -5 in astrocytomas: IGFBP-3 emerges as a strong predictor of survival in patients with newly diagnosed glioblastoma. Cancer Epidemiol Biomark. Prev. 2010, 19, 1399–1408. [Google Scholar] [CrossRef]
- Martin, J.L.; Lin, M.Z.; McGowan, E.M.; Baxter, R.C. Potentiation of growth factor signaling by insulin-like growth factor-binding protein-3 in breast epithelial cells requires sphingosine kinase activity. J. Biol Chem. 2009, 284, 25542–25552. [Google Scholar] [CrossRef]
- Usui, S.; Sugimoto, N.; Takuwa, N.; Sakagami, S.; Takata, S.; Kaneko, S.; Takuwa, Y. Blood lipid mediator sphingosine 1-phosphate potently stimulates platelet-derived growth factor-A and -B chain expression through S1P1-Gi-Ras-MAPK-dependent induction of Kruppel-like factor. J. Biol. Chem. 2004, 279, 12300–12311. [Google Scholar] [CrossRef]
- Van Brocklyn, J.R.; Letterle, C.A.; Snyder, P.J.; Prior, T.W. Sphingosine-1-phosphate stimulates human glioma cell proliferation through Gi-coupled receptors: Role of ERK MAP kinase and phosphatidylinositol 3-kinase β. Cancer Lett. 2002, 181, 195–204. [Google Scholar] [CrossRef]
- Van Brocklyn, J.R.; Young, N.; Roof, R. Sphingosine-1-phosphate stimulates motility and invasiveness of human glioblastoma multiforme cells. Cancer Lett. 2003, 199, 53–60. [Google Scholar] [CrossRef]
- Sato, K.; Ui, M.; Okajima, F. Differential roles of Edg-1 and Edg-5, sphingosine 1-phosphate receptors, in the signaling pathways in C6 glioma cells. Mol. Brain Res. 2000, 85, 151–160. [Google Scholar] [CrossRef]
- Young, N.; Van Brocklyn, J.R. Roles of sphingosine-1-phosphate (S1P) receptors in malignant behavior of glioma cells. Differential effects of S1P2 on cell migration and invasiveness. Exp. Cell. Res. 2007, 313, 1615–1627. [Google Scholar] [CrossRef] [PubMed]
- Estrada-Bernal, A.; Lawler, S.E.; Nowicki, M.O.; Chaudhury, A.R.; Van Brocklyn, J.R. The role of sphingosine kinase-1 in EGFRvIII-regulated growth and survival of glioblastoma cells. J. Neurooncol. 2011, 102, 353–366. [Google Scholar] [CrossRef] [PubMed]
- Oliver, T.G.; Wechsler-Reya, R.J. Getting at the root and stem of brain tumours. Neuron 2004, 42, 885–888. [Google Scholar] [CrossRef]
- Ganguly, D.; Fan, M.; Yang, C.H.; Zbytek, B.; Finkelstein, D.; Roussel, M.F.; Pfeffer, L.M. The critical role that STAT3 plays in glioma-initiating cells: STAT3 addiction in glioma. Oncotarget 2018, 9, 22095–22112. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.; Deng, J.; Kujawski, M.; Yang, C.; Liu, Y.; Herrmann, A.; Kortylewski, M.; Horne, D.; Somlo, G.; Forman, S.; et al. STAT3-induced S1PR1 expression is crucial for persistent STAT3 activation in tumors. Nat. Med. 2010, 16, 1421–1428. [Google Scholar] [CrossRef] [PubMed]
- Hirata, N.; Yamada, S.; Shoda, T.; Kurihara, M.; Sekino, Y.; Kanda, Y. Sphingosine-1-phosphate promotes expansion of cancer stem cells via S1PR3 by a ligand-independent Notch activation. Nat. Commun. 2014, 5, 4806. [Google Scholar] [CrossRef]
- Yu, O.M.; Benitez, J.A.; Plouffe, S.W.; Ryback, D.; Klein, A.; Smith, J.; Greenbaum, J.; Delatte, B.; Rao, A.; Guan, K.L.; et al. YAP and MRTF-A, transcriptional co-activators of RhoA-mediated gene expression, are critical for glioblastoma tumorigenicity. Oncogene 2018, 37, 5492–5507. [Google Scholar] [CrossRef]
- Hatoum, A.; Mohammed, R.; Zakieh, O. The unique invasiveness of glioblastoma and possible drug targets on extracellular matrix. Cancer Manag. Res. 2019, 11, 1843–1855. [Google Scholar] [CrossRef]
- Kalokhe, G.; Grimm, S.A.; Chandler, J.P.; Helenowski, I.; Rademaker, A.; Raizer, J.J. Metastatic glioblastoma: Case presentations and a review of the literature. J. Neurooncol. 2012, 107, 21–27. [Google Scholar] [CrossRef] [PubMed]
- Lun, M.; Lok, E.; Gautam, S.; Wu, E.; Wong, E.T. The natural history of extracranial metastasis from glioblastoma multiforme. J. Neurooncol. 2011, 105, 261–273. [Google Scholar] [CrossRef] [PubMed]
- Hoelzinger, D.B.; Demuth, T.; Berens, M.E. Autocrine factors that sustain glioma invasion and paracrine biology in the brain microenvironment. J. Natl. Cancer Inst. 2007, 99, 1583–1593. [Google Scholar] [CrossRef] [PubMed]
- Van Brocklyn, J.R. Regulation of cancer cell migration and invasion by sphingosine-1-phosphate. World J. Biol. Chem. 2010, 1, 307–312. [Google Scholar] [CrossRef] [PubMed]
- Lepley, D.; Paik, J.H.; Hla, T.; Ferrer, F. The G protein-coupled receptor S1P2 regulates Rho/Rho kinase pathway to inhibit tumor cell migration. Cancer Res. 2005, 65, 3788–3795. [Google Scholar] [CrossRef] [PubMed]
- Malchinkhuu, E.; Sato, K.; Maehama, T.; Mogi, C.; Tomura, H.; Ishiuchi, S.; Saito, N.; Kurose, H.; Tomura, H.; Okajima, F. S1P2 receptors mediate inhibition of glioma cell migration through Rho signaling pathways independent of PTEN. Biochem. Biophys. Res. Commun. 2008, 366, 963–968. [Google Scholar] [CrossRef]
- Sugimoto, N.; Takuwa, N.; Okamoto, H.; Sakurada, S.; Takuwa, Y. Inhibitory and stimulatory regulation of Rac and cell motility by the G12/13-Rho and Gi pathways integrated downstream of a single G protein-coupled sphingosine-1-phosphate receptor isoform. Mol. Cell. Biol. 2003, 23, 1534–1545. [Google Scholar] [CrossRef]
- Young, N.; Pearl, D.K.; Van Brocklyn, J.R. Sphingosine-1-phosphate regulates glioblastoma cell invasiveness through the urokinase plasminogen activator system and CCN1/Cyr61. Mol. Cancer Res. 2009, 7, 23–32. [Google Scholar] [CrossRef]
- Bryan, L.; Paugh, B.S.; Kapitonov, D.; Wilczynska, K.M.; Alvarez, S.M.; Singh, S.K.; Milstien, S.; Spiegel, S.; Kordula, T. Sphingosine-1-phosphate and interleukin-1 independently regulate plasminogen activator inhibitor-1 and urokinase-type plasminogen activator receptor expression in glioblastoma cells: Implications for invasiveness. Mol. Cancer Res. 2008, 6, 1469–1477. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, H.; Ding, K.; Xu, J. FTY720 reduces migration and invasion of human glioblastoma cell lines via inhibiting the PI3K/AKT/mTOR/p70S6K signaling pathway. Tumour Biol. 2014, 35, 10707–10714. [Google Scholar] [CrossRef]
- Natarajan, J.; Berrar, D.; Dubitzky, W.; Hack, C.; Zhang, Y.; DeSesa, C.; Van Brocklyn, J.R.; Bremer, E.G. Text mining of full-text journal articles combined with gene expression analysis reveals a relationship between sphingosine-1-phosphate and invasiveness of a glioblastoma cell line. BMC Bioinform. 2006, 7. [Google Scholar] [CrossRef] [PubMed]
- Fortier, S.; Labelle, D.; Sina, A.; Moreau, R.; Annabi, B. Silencing of the MT1-MMP/G6PT axis suppresses calcium mobilization by sphingosine-1-phosphate in glioblastoma cells. FEBS Lett. 2008, 582, 799–804. [Google Scholar] [CrossRef] [PubMed]
- Leclerc, C.; Haeich, J.; Aulestia, F.J.; Kilhoffer, M.C.; Miller, A.L.; Néant, I.; Webb, S.E.; Schaeffer, E.; Junier, M.P.; Chneiweiss, H.; et al. Calcium signaling orchestrates glioblastoma development: Facts and conjunctures. Biochim. Biophys. Acta 2016, 1863, 1447–1459. [Google Scholar] [CrossRef] [PubMed]
- Toman, R.E.; Spiegel, S. Lysophospholipid receptors in the nervous system. Neurochem. Res. 2002, 27, 619–627. [Google Scholar] [CrossRef] [PubMed]
- Pulli, I.; Asghar, M.Y.; Kemppainen, K.; Törnquist, K. Sphingolipid-mediated calcium signaling and its pathological effects. Biochim. Biophys. Acta Mol. Cell. Res. 2018, 1865, 1668–1677. [Google Scholar] [CrossRef]
- Jang, H.S.; Lal, S.; Greenwood, J.A. Calpain 2 is required for glioblastoma cell invasion: Regulation of matrix metalloproteinase 2. Neurochem. Res. 2010, 35, 1796–1804. [Google Scholar] [CrossRef]
- Chen, P.Y.; Csutora, P.; Veyna-Burke, N.A.; Marchase, R.B. Glucose-6-phosphate and Ca2+ sequestration are mutually enhanced in microsomes from liver, brain, and heart. Diabetes 1998, 47, 874–881. [Google Scholar] [CrossRef]
- Belkaid, A.; Currie, J.C.; Desgagnés, J.; Annabi, B. The chemopreventive properties of chlorogenic acid reveal a potential new role for the microsomal glucose-6-phosphate translocase in brain tumor progression. Cancer Cell Int. 2006, 6, 7. [Google Scholar] [CrossRef]
- Lepannetier, S.; Zanou, N.; Yerna, X.; Emeriau, N.; Dufour, I.; Masquelier, J.; Muccioli, G.; Tajeddine, N.; Gailly, P. Sphingosine-1-phosphate-activated TRPC1 channel controls chemotaxis of glioblastoma cells. Cell Calcium 2016, 60, 373–383. [Google Scholar] [CrossRef]
- Annabi, B.; Lachambre, M.P.; Plouffe, K.; Sartelet, H.; Béliveau, R. Modulation of invasive properties of CD133+ glioblastoma stem cells: A role for MT1-MMP in bioactive lysophospholipid signaling. Mol. Carcinog. 2009, 48, 910–919. [Google Scholar] [CrossRef]
- Giussani, P.; Tringali, C.; Riboni, L.; Viani, P.; Venerando, B. Sphingolipids: Key regulators of apoptosis and pivotal players in cancer drug resistance. Int. J. Mol. Sci. 2014, 15, 4356–4392. [Google Scholar] [CrossRef]
- Giussani, P.; Bassi, R.; Anelli, V.; Brioschi, L.; de Zen, F.; Riccitelli, E.; Caroli, M.; Campanella, R.; Gaini, S.M.; Viani, P.; et al. Glucosylceramide synthase protects glioblastoma cells against autophagic and apoptotic death induced by temozolomide and Paclitaxel. Cancer Investig. 2012, 30, 27–37. [Google Scholar]
- Vadas, M.; Xia, P.; McCaughan, G.; Gamble, J. The role of sphingosine kinase 1 in cancer: Oncogene or non-oncogene addiction? Biochim. Biophys. Acta 2008, 1781, 442–447. [Google Scholar] [CrossRef] [PubMed]
- Gault, C.R.; Obeid, L.M. Still benched on its way to the bedside: Sphingosine kinase 1 as an emerging target in cancer chemotherapy. Crit. Rev. Biochem. Mol. Biol. 2011, 46, 342–351. [Google Scholar] [CrossRef] [PubMed][Green Version]
- Noack, J.; Choi, J.; Richter, K.; Kopp-Schneider, A.; Régnier-Vigouroux, A. A sphingosine kinase inhibitor combined with temozolomide induces glioblastoma cell death through accumulation of dihydrosphingosine and dihydroceramide, endoplasmic reticulum stress and autophagy. Cell Death Dis. 2014, 5, e1425. [Google Scholar] [CrossRef] [PubMed]
- Oancea-Castillo, L.R.; Klein, C.; Abdollahi, A.; Weber, K.J.; Régnier-Vigouroux, A.; Dokic, I. Comparative analysis of the effects of a sphingosine kinase inhibitor to temozolomide and radiation treatment on glioblastoma cell lines. Cancer Biol. Ther. 2017, 18, 400–406. [Google Scholar] [CrossRef] [PubMed]
- Bektas, M.; Johnson, S.P.; Poe, WE.; Bigmar, D.D.; Friedman, H.S. A sphingosine kinase inhibitor induces cell death in temozolomide resistant glioblastoma cells. Cancer Chemother. Pharm. 2009, 64, 1053–1058. [Google Scholar] [CrossRef]
- Doan, N.B.; Alhajala, H.; Al-Gizawiy, M.M.; Mueller, W.M.; Rand, S.D.; Connelly, J.M.; Cochran, E.J.; Chitambar, C.R.; Clark, P.; Kuo, J.; et al. Acid ceramidase and its inhibitors: A de novo drug target and a new class of drugs for killing glioblastoma cancer stem cells with high efficiency. Oncotarget 2017, 8, 112662–112674. [Google Scholar] [CrossRef]
- Radeff-Huang, J.; Seasholtz, T.M.; Matteo, R.G.; Brown, J.H. G protein mediated signaling pathways in lysophospholipid induced cell proliferation and survival. J. Cell. Biochem. 2004, 92, 949–966. [Google Scholar] [CrossRef]
- Koul, D. PTEN Signaling pathways in glioblastoma. Cancer Biol. Ther. 2008, 7, 1321–1325. [Google Scholar] [CrossRef]
- Sami, A.; Karsy, M. Targeting the PI3K/AKT/mTOR signaling pathway in glioblastoma: Novel therapeutic agents and advances in understanding. Tumour Biol. 2013, 34, 1991–2002. [Google Scholar] [CrossRef] [PubMed]
- Mantamadiotis, T. Towards targeting PI3K-dependent regulation of gene expression in brain cancer. Cancers 2017, 9, 60. [Google Scholar] [CrossRef] [PubMed]
- Brazil, D.P.; Yang, Z.Z.; Hemmings, B.A. Advances in protein kinase B signalling: AKTion on multiple fronts. Trends Biochem. Sci. 2001, 26, 657–664. [Google Scholar] [CrossRef]
- Guan, H.; Song, L.; Cai, J.; Huang, Y.; Wu, J.; Yuan, J.; Li, J.; Li, M. Sphingosine kinase 1 regulates the Akt/FOXO3a/Bim pathway and contributes to apoptosis resistance in glioma cells. PLoS ONE 1994, 6, e19946. [Google Scholar] [CrossRef]
- Sonoda, Y.; Yamamoto, D.; Sakurai, S.; Hasegawa, M.; Aizu-Yokota, E.; Momoi, T.; Kasahara, T. FTY720, a novel immunosuppressive agent, induces apoptosis in human glioma cells. Biochem. Biophys. Res. Commun. 2001, 281, 282–288. [Google Scholar] [CrossRef]
- Zhang, L.; Wang, H.; Ding, K.; Xu, J. FTY720 induces autophagy-related apoptosis and necroptosis in human glioblastoma cells. Toxicol. Lett. 2015, 236, 43–59. [Google Scholar] [CrossRef]
- Giussani, P.; Brioschi, L.; Bassi, R.; Riboni, L.; Viani, P. Phosphatidylinositol 3-kinase/AKT pathway regulates the endoplasmic reticulum to Golgi traffic of ceramide in glioma cells: A link between lipid signaling pathways involved in the control of cell survival. J. Biol. Chem. 2009, 284, 5088–5096. [Google Scholar] [CrossRef]
- Imran, M.; Rauf, A.; Abu-Izneid, T.; Nadeem, M.; Shariati, M.A.; Khan, I.A.; Imran, A.; Orhan, I.E.; Rizwan, M.; Atif, M.; et al. Luteolin, a flavonoid, as an anticancer agent: A review. Biomed. Pharm. 2019, 112, 108612. [Google Scholar] [CrossRef]
- Abdel Hadi, L.; Di Vito, C.; Marfia, G.; Ferraretto, A.; Tringali, C.; Viani, P.; Riboni, L. Sphingosine kinase 2 and ceramide transport as key targets of the natural flavonoid luteolin to induce apoptosis in colon cancer cells. PLoS ONE 2015, 10, e0143384. [Google Scholar] [CrossRef]
- Estrada-Bernal, A.; Palanichamy, K.; Ray Chaudhury, A.; Van Brocklyn, J.R. Induction of brain tumor stem cell apoptosis by FTY720: A potential therapeutic agent for glioblastoma. Neuro-Oncol. 2012, 14, 405–415. [Google Scholar] [CrossRef]
- Brandenburg, S.; Müller, A.; Turkowski, K.; Radev, Y.T.; Rot, S.; Schmidt, C.; Bungert, A.D.; Acker, G.; Schorr, A.; Hippe, A.; et al. Resident microglia rather than peripheral macrophages promote vascularization in brain tumors and are source of alternative pro-angiogenic factors. Acta Neuropathol. 2016, 131, 365–378. [Google Scholar] [CrossRef]
- Watters, J.J.; Schartner, J.M.; Badie, B. Microglia function in brain tumors. J. Neurosci. Res. 2005, 81, 447–455. [Google Scholar] [CrossRef] [PubMed]
- Lisi, L.; Ciotti, G.M.; Braun, D.; Kalinin, S.; Currò, D.; Dello Russo, C.; Coli, A.; Mangiola, A.; Anile, C.; Feinstein, D.L.; et al. Expression of iNOS, CD163 and ARG-1 taken as M1 and M2 markers of microglial polarization in human glioblastoma and the surrounding normal parenchyma. Neurosci. Lett. 2017, 645, 106–112. [Google Scholar] [CrossRef] [PubMed]
- Zhai, H.; Heppner, F.L.; Tsirka, S.E. Microglia/macrophages promote glioma progression. Glia 2011, 59, 472–485. [Google Scholar] [CrossRef]
- Ye, X.-Z.; Xu, S.-L.; Xin, Y.-H.; Yu, S.-C.; Ping, Y.-F.; Chen, L.; Xiao, H.-L.; Wang, B.; Yi, L.; Wang, Q.-L.; et al. Tumor-associated microglia/macrophages enhance the invasion of glioma stem-like cells via TGF-β1 signaling pathway. J. Immunol. 2012, 189, 444–453. [Google Scholar] [CrossRef] [PubMed]
- Gabrusiewicz, K.; Ellert-Miklaszewska, A.; Lipko, M.; Sielska, M.; Frankowska, M.; Kaminska, B. Characteristics of the alternative phenotype of microglia/macrophages and its modulation in experimental gliomas. PLoS ONE 2011, 6, e23902. [Google Scholar] [CrossRef] [PubMed]
- Gabrilovich, D.I.; Ostrand-Rosenberg, S.; Bronte, V. Coordinated regulation of myeloid cells by tumours. Nat. Rev. Immunol. 2012, 12, 253–268. [Google Scholar] [CrossRef] [PubMed]
- Komohara, Y.; Ohnishi, K.; Kuratsu, J.; Takeya, M. Possible involvement of the M2 anti-inflammatory macrophage phenotype in growth of human gliomas. J. Pathol. 2008, 216, 15–24. [Google Scholar] [CrossRef]
- Prosniak, M.; Harshyne, L.A.; Andrews, D.W.; Kenyon, L.C.; Bedelbaeva, K.; Apanasovich, T.V.; Heber-Katz, E.; Curtis, M.T.; Cotzia, P.; Hooper, D.C. Glioma grade is associated with the accumulation and activity of cells bearing M2 monocyte markers. Clin. Cancer Res. 2013, 19, 3776–3786. [Google Scholar] [CrossRef]
- Poon, C.C.; Sarkar, S.; Yong, V.W.; Kelly, J.J.P. Glioblastoma-associated microglia and macrophages: Targets for therapies to improve prognosis. Brain 2017, 140, 1548–1560. [Google Scholar] [CrossRef]
- Gordon, S. Alternative activation of macrophages. Nat. Rev. Immunol. 2003, 3, 23–35. [Google Scholar] [CrossRef] [PubMed]
- Thorsson, V.; Gibbs, D.L.; Brown, S.D.; Wolf, D.; Bortone, D.S.; Ou Yang, T.H.; Porta-Pardo, E.; Gao, G.F.; Plaisier, C.L.; Eddy, J.A.; et al. Cancer Genome Atlas Research Network, Lazar AJ, Serody JS, Demicco, E.G.; Disis ML, Vincent BG, Shmulevich, I. The immune landscape of cancer. Immunity 2018, 48, 812–830. [Google Scholar] [CrossRef] [PubMed]
- Bryan, A.M.; Del Poeta, M. Sphingosine-1-phosphate receptors and innate immunity. Cell. Microbiol. 2018, 20, e12836. [Google Scholar] [CrossRef] [PubMed]
- Tham, C.S.; Lin, F.F.; Rao, T.S.; Yu, N.; Webb, M. Microglial activation state and lysophospholipid acid receptor expression. Int. J. Dev. Neurosci. 2003, 21, 431–443. [Google Scholar] [CrossRef] [PubMed]
- Müller, J.; von Bernstorff, W.; Heidecke, C.D.; Schulze, T. Differential S1P receptor profiles on M1- and M2-polarized macrophages affect macrophage cytokine production and migration. Biomed. Res. Int. 2017, 2017, 7584621. [Google Scholar] [CrossRef]
- Awojoodu, A.O.; Ogle, M.E.; Sefcik, L.S.; Bowers, D.T.; Martin, K.; Brayman, K.L.; Lynch, K.R.; Peirce-Cottler, S.M.; Botchwey, E. Sphingosine 1-phosphate receptor 3 regulates recruitment of anti-inflammatory monocytes to microvessels during implant arteriogenesis. Proc. Natl. Acad. Sci. USA 2013, 110, 13785–13790. [Google Scholar] [CrossRef]
- Weichand, B.; Weis, N.; Weigert, A.; Grossmann, N.; Levkau, B.; Brune, B. Apoptotic cells enhance sphingosine-1-phosphate receptor 1 dependent macrophage migration. Eur. J. Immunol. 2013, 43, 3306–3313. [Google Scholar] [CrossRef]
- Weichand, B.; Popp, R.; Dziumbla, S.; Mora, J.; Strack, E.; Elwakeel, E.; Frank, A.C.; Scholich, K.; Pierre, S.; Syed, S.N.; et al. S1PR1 on tumor-associated macrophages promotes lymphangiogenesis and metastasis via NLRP3/IL-1beta. J. Exp. Med. 2017, 214, 2695–2713. [Google Scholar] [CrossRef]
- Marx, S.; Splittstöhser, M.; Kinnen, F.; Moritz, E.; Joseph, C.; Paul, S.; Paland, H.; Seifert, C.; Marx, M.; Böhm, A.; et al. Platelet activation parameters and platelet-leucocyte-conjugate formation in glioblastoma multiforme patients. Oncotarget 2018, 9, 25860–25876. [Google Scholar] [CrossRef]
- Weigert, A.; Olesch, C.; Brüne, B. Sphingosine-1-phosphate and macrophage biology-how the sphinx tames the big eater. Front. Immunol. 2019, 10, 1706. [Google Scholar] [CrossRef]
- Gaire, B.P.; Lee, C.H.; Sapkota, A.; Lee, S.Y.; Chun, J.; Cho, H.J.; Nam, T.G.; Choi, J.W. Identification of sphingosine 1-phosphate receptor subtype 1 (S1P1) as a pathogenic factor in transient focal cerebral ischemia. Mol. Neurobiol. 2018, 55, 2320–2332. [Google Scholar] [CrossRef] [PubMed]
- Kim, G.S.; Yang, L.; Zhang, G.; Zhao, H.; Selim, M.; McCullough, L.D.; Kluk, M.J.; Sanchez, T. Critical role of sphingosine-1-phosphate receptor-2 in the disruption of cerebrovascular integrity in experimental stroke. Nat. Commun. 2015, 6, 7893. [Google Scholar] [CrossRef] [PubMed]
- Gaire, B.P.; Song, M.R.; Choi, J.W. Sphingosine 1-phosphate receptor subtype 3 (S1P3) contributes to brain injury after transient focal cerebral ischemia via modulating microglial activation and their M1 polarization. J. Neuroinflamm. 2018, 15, 284. [Google Scholar] [CrossRef] [PubMed]
- Sapkota, A.; Gaire, B.P.; Kang, M.G.; Choi, J.W. S1P2 contributes to microglial activation and M1 polarization following cerebral ischemia through ERK1/2 and JNK. Sci. Rep. 2019, 9, 12106. [Google Scholar] [CrossRef]
- Gaire, B.P.; Bae, Y.J.; Choi, J.W. S1P1 Regulates M1/M2 polarization toward brain injury after transient focal cerebral ischemia. Biomol. Ther. (Seoul) 2019, 27, 522–529. [Google Scholar] [CrossRef]
- Hughes, J.E.; Srinivasan, S.; Lynch, K.R.; Proia, R.L.; Ferdek, P.; Hedrick, C.C. Sphingosine-1-phosphate induces an anti-inflammatory phenotype in macrophages. Circ. Res. 2008, 102, 950–958. [Google Scholar] [CrossRef]
- Park, S.J.; Lee, K.P.; Kang, S.; Lee, J.; Sato, K.; Chung, H.Y.; Okajima, F.; Im, D.S. Sphingosine 1-phosphate induced anti-atherogenic and atheroprotective M2 macrophage polarization through IL-4. Cell. Signal. 2014, 26, 2249–2258. [Google Scholar] [CrossRef]
- Rodriguez, Y.I.; Campos, L.E.; Castro, M.G.; Aladhami, A.; Oskeritzian, C.A.; Alvarez, S.E. Sphingosine-1 phosphate: A new modulator of immune plasticity in the tumor microenvironment. Front. Oncol. 2016, 6, 218. [Google Scholar] [CrossRef]
- Weigert, A.; Tzieply, N.; von Knethen, A.; Johann, A.M.; Schmidt, H.; Geisslinger, G.; Brüne, B. Tumor cell apoptosis polarizes macrophages role of sphingosine-1-phosphate. Mol. Biol. Cell. 2007, 18, 3810–3819. [Google Scholar] [CrossRef]
- Weigert, A.; Schiffmann, S.; Sekar, D.; Ley, S.; Menrad, H.; Werno, C.; Grosch, S.; Geisslinger, G.; Brüne, B. Sphingosine kinase 2 deficient tumor xenografts show impaired growth and fail to polarize macrophages towards an anti-inflammatory phenotype. Int. J. Cancer. 2009, 125, 2114–2121. [Google Scholar] [CrossRef]
- Weis, N.; Weigert, A.; von Knethen, A.; Brüne, B. Heme oxygenase-1 contributes to an alternative macrophage activation profile induced by apoptotic cell supernatants. Mol. Biol. Cell. 2009, 20, 1280–1288. [Google Scholar] [CrossRef] [PubMed]
- Mrad, M.; Imbert, C.; Garcia, V.; Rambow, F.; Therville, N.; Carpentier, S.; Ségui, B.; Levade, T.; Azar, R.; Marine, J.C.; et al. Downregulation of sphingosine kinase-1 induces protective tumor immunity by promoting M1 macrophage response in melanoma. Oncotarget 2016, 7, 71873–71886. [Google Scholar] [CrossRef]
- Weigert, A.; von Knethen, A.; Thomas, D.; Faria, I.; Namgaladze, D.; Zezina, E.; Fuhrmann, D.; Petcherski, A.; Heringdorf, D.M.Z.; Radeke, H.H.; et al. Sphingosine kinase 2 is a negative regulator of inflammatory macrophage activation. Biochim. Biophys. Acta Mol. Cell Biol. Lipids 2019, 1864, 1235–1246. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Zhou, Q.; Wu, C.P.; Xu, Y.W.; Liu, W.L.; Zhao, H.F.; Li, W.P. SPHK2 protein expression, Ki-67 index and infiltration of tumor-associated macrophages (TAMs) in human glioma. Histol. Histopathol. 2018, 33, 987–994. [Google Scholar] [PubMed]
- Luo, B.; Gan, W.; Liu, Z.; Shen, Z.; Wang, J.; Shi, R.; Liu, Y.; Liu, Y.; Jiang, M.; Zhang, Z.; et al. Erythropoietin signaling in macrophages promotes dying cell clearance and immune tolerance. Oncotarget 2015, 6, 2101–2119. [Google Scholar]
- Gielen, P.R.; Schulte, B.M.; Kers-Rebel, E.D.; Verrijp, K.; Petersen-Baltussen, H.M.; ter Laan, M.; Wesseling, P.; Adema, G.J. Increase in both CD14-positive a nd CD15-positive myeloid-derived suppressor cell subpopulations in the blood of patients with glioma but predominance of CD15-positive myeloid-derived suppressor cells in glioma tissue. J. Neuropathol. Exp. Neurol. 2015, 74, 390–400. [Google Scholar] [CrossRef]
- Gabrusiewicz, K.; Rodriguez, B.; Wei, J.; Hashimoto, Y.; Healy, L.M.; Maiti, S.N.; Thomas, G.; Zhou, S.; Wang, Q.; Elakkad, A.; et al. Glioblastoma-infiltrated innate immune cells resemble M0 macrophage phenotype. JCI Insight 2016, 1, 85841. [Google Scholar] [CrossRef]
- Li, Y.; Zhou, T.; Wang, Y.; Ning, C.; Lv, Z.; Han, G.; Morris, J.C.; Taylor, E.N.; Wang, R.; Xiao, H.; et al. The protumorigenic potential of FTY720 by promoting extramedullary hematopoiesis and MDSC accumulation. Oncogene 2017, 36, 3760–3771. [Google Scholar] [CrossRef]
- Woroniecka, K.I.; Rhodin, K.E.; Chongsathidkiet, P.; Keith, K.A.; Fecci, P.E. T-Cell Dysfunction in glioblastoma: Applying a new framework. Clin. Cancer Res. 2018, 24, 3792–3802. [Google Scholar] [CrossRef]
- Matloubian, M.; Lo, C.G.; Cinamon, G.; Lesneski, M.J.; Xu, Y.; Brinkmann, V.; Allende, M.L.; Proia, R.L.; Cyster, J.G. Lymphocyte egress from thymus and peripheral lymphoid organs is dependent on S1P receptor 1. Nature 2004, 427, 355–360. [Google Scholar] [CrossRef]
- Schwab, S.R.; Cyster, J.G. Finding a way out: Lymphocyte egress from lymphoid organs. Nat. Immunol. 2007, 8, 1295–1301. [Google Scholar] [CrossRef] [PubMed]
- Chongsathidkiet, P.; Jackson, C.; Koyama, S.; Loebel, F.; Cui, X.; Farber, S.H.; Woroniecka, K.; Elsamadicy, A.A.; Dechant, C.A.; Kemeny, H.R.; et al. Sequestration of T cells in bone marrow in the setting of glioblastoma and other intracranial tumors. Nat. Med. 2018, 24, 1459–1468. [Google Scholar] [CrossRef]
- Linkous, A.G.; Yazlovitskaya, E.M. Angiogenesis in glioblastoma multiforme: Navigating the maze. Anticancer Agents Med. Chem. 2011, 11, 712–718. [Google Scholar] [CrossRef] [PubMed]
- Calabrese, C.; Poppleton, H.; Kocak, M.; THogg, L.; Fuller, C.; Hamner, B.; Oh, E.Y.; Gaber, M.W.; Finklestein, D.; Allen, M.; et al. A perivascular niche for brain tumor stem cells. Cancer Cell 2007, 11, 69–82. [Google Scholar] [CrossRef] [PubMed]
- Gilbertson, R.J.; Rich, J.N. Making a tumour’s bed: Glioblastoma stem cells and the vascular niche. Nat. Rev. Cancer 2007, 7, 733–736. [Google Scholar] [CrossRef] [PubMed]
- Charles, N.; Holland, E.C. The perivascular niche microenvironment in brain tumor progression. Cell Cycle 2010, 9, 3012–3021. [Google Scholar] [CrossRef]
- Krock, B.L.; Skuli, N.; Simon, M.C. Hypoxia-induced angiogenesis: Good and evil. Genes Cancer 2011, 2, 1117–1133. [Google Scholar] [CrossRef]
- Onishi, M.; Ichikawa, T.; Kurozumi, K.; Date, I. Angiogenesis and invasion in glioma. Brain Tumor Pathol. 2011, 28, 13–24. [Google Scholar] [CrossRef]
- Takano, S. Glioblastoma angiogenesis: VEGF resistance solutions and new strategies based on molecular mechanisms of tumor vessel formation. Brain Tumor Pathol. 2012, 29, 73–86. [Google Scholar] [CrossRef]
- Jain, R.K.; di Tomaso, E.; Duda, D.G.; Loeffler, J.S.; Sorensen, A.G.; Batchelor, T.T. Angiogenesis in brain tumours. Nat. Rev. Neurosci. 2007, 8, 610–622. [Google Scholar] [CrossRef]
- Chen, W.; He, D.; Li, Z.; Zhang, X.; Pan, D.; Chen, G. Overexpression of vascular endothelial growth factor indicates poor outcomes of glioma: A systematic review and meta-analysis. Int. J. Clin. Exp. Med. 2015, 8, 8709–8719. [Google Scholar] [PubMed]
- Ader, I.; Brizuela, L.; Bouquerel, P.; Malavaud, B.; Cuvillier, O. Sphingosine kinase 1: A new modulator of hypoxia inducible factor 1α during hypoxia in human cancer cells. Cancer Res. 2008, 68, 8635–8642. [Google Scholar] [CrossRef]
- Bulnes, S.; Bengoetxea, H.; Ortuzar, N.; Argandoña, E.G.; Garcia-Blanco, A.I.; Rico-Barrio, I.; Lafuente, J.V. Angiogenic signalling pathways altered in gliomas: Selection mechanisms for more aggressive neoplastic subpopulations with invasive phenotype. J. Signal. Trans. 2012, 2012, 597915. [Google Scholar] [CrossRef] [PubMed]
- Limaye, V. The role of sphingosine kinase and sphingosine-1-phosphate in the regulation of endothelial cell biology. Endothelium 2008, 15, 101–112. [Google Scholar] [CrossRef]
- Argraves, K.M.; Wilkerson, B.A.; Argraves, W.S. Sphingosine-1-phosphate signalling in vasculogenesis and angiogenesis. World J. Biol. Chem. 2010, 1, 291–297. [Google Scholar] [CrossRef]
- Lucke, S.; Levkau, B. Endothelial functions of sphingosine-1-phosphate. Cell Physiol. Biochem. 2010, 26, 87–96. [Google Scholar] [CrossRef]
- Sun, H.Y.; Wei, S.P.; Xu, R.C.; Xu, P.X.; Zhang, W.C. Sphingosine-1-phosphate induces human endothelial VEGF and MMP-2 production via transcription factor ZNF580: Novel insights into angiogenesis. Biochem. Biophys. Res. Commun. 2010, 395, 361–366. [Google Scholar] [CrossRef]
- Liu, Y.; Wada, R.; Yamashita, T.; Mi, Y.; Deng, C.X.; Hobson, J.P.; Rosenfeldt, H.M.; Nava, V.E.; Chae, S.S.; Lee, M.J.; et al. Edg-1, the G protein-coupled receptor for sphingosine-1-phosphate, is essential for vascular maturation. J. Clin. Investig. 2000, 106, 951–961. [Google Scholar] [CrossRef]
- Osada, M.; Yatomi, Y.; Ohmori, T.; Ikeda, H.; Ozaki, Y. Enhancement of sphingosine 1-phosphate-induced migration of vascular endothelial cells and smooth muscle cells by an EDG-5 antagonist. Biochem. Biophys. Res. Commun. 2002, 299, 483–487. [Google Scholar] [CrossRef]
- Igarashi, J.; Erwin, P.A.; Dantas, A.P.; Chen, H.; Michel, T. VEGF induces S1P1 receptors in endothelial cells: Implications for cross-talk between sphingolipid and growth factor receptors. Proc. Natl. Acad. Sci. USA 2003, 100, 10664–10669. [Google Scholar] [CrossRef]
- Justus, C.R.; Sanderlin, E.J.; Yang, L.V. Molecular connections between cancer cell metabolism and the tumor microenvironment. Int. J. Mol. Sci. 2015, 16, 11055–11086. [Google Scholar] [CrossRef] [PubMed]
- Cloughesy, T.F.; Cavenee, W.K.; Mischel, P.S. Glioblastoma: From molecular pathology to targeted treatment. Annu. Rev. Pathol. 2014, 9, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Kucharzewska, P.; Christianson, H.C.; Belting, M. Global profiling of metabolic adaptation to hypoxic stress in human glioblastoma cells. PLoS ONE 2015, 10, 10–1371. [Google Scholar] [CrossRef] [PubMed]
- Clark, P.M.; Mai, W.X.; Cloughesy, T.F.; Nathanson, D.A. Emerging approaches for targeting metabolic vulnerabilities in malignant glioma. Curr. Neurol. Neurosci. Rep. 2016, 16. [Google Scholar] [CrossRef] [PubMed]
- Jiang, W.; Finniss, S.; Cazacu, S.; Xiang, C.; Brodie, Z.; Mikkelsen, T.; Poisson, L.; Shackelford, D.B.; Brodie, C. Repurposing phenformin for the targeting of glioma stem cells and the treatment of glioblastoma. Oncotarget 2016, 7, 56456–56470. [Google Scholar] [CrossRef] [PubMed]
- Talasila, K.M.; Rosland, G.V.; Hagland, H.R.; Eskilsson, E.; Flones, I.H.; Fritah, S.; Azuaje, F.; Atai, N.; Harter, P.N.; Mittelbronn, M.; et al. The angiogenic switch leads to a metabolic shift in human glioblastoma. Neuro-Oncol. 2017, 19, 383–393. [Google Scholar] [CrossRef] [PubMed]
- Lunt, S.Y.; Vander Heiden, M.G. Aerobic glycolysis: Meeting the metabolic requirements of cell proliferation. Annu. Rev. Cell. Dev. Biol. 2011, 27, 441–464. [Google Scholar] [CrossRef]
- Lu, J.; Tan, M.; Cai, Q. The Warburg effect in tumor progression: Mitochondrial oxidative metabolism as an anti-metastasis mechanism. Cancer Lett. 2015, 356, 156–164. [Google Scholar] [CrossRef]
- Altman, B.J.; Stine, Z.E.; Dang, C.V. From Krebs to clinic: Glutamine metabolism to cancer therapy. Nat. Rev. Cancer 2016, 16, 619–634. [Google Scholar] [CrossRef]
- Libby, C.J.; Tran, A.N.; Scott, S.E.; Griguer, C.; Hjelmeland, A.B. The pro-tumorigenic effects of metabolic alterations in glioblastoma including brain tumor initiating cells. Biochim. Biophys. Acta Rev. Cancer 2018, 1869, 175–188. [Google Scholar] [CrossRef]
- Watson, D.G.; Tonelli, F.; Alossaimi, M.; Williamson, L.; Chan, E.; Gorshkova, I.; Berdyshev, E.; Bittman, R.; Pyne, N.J.; Pyne, S. The roles of sphingosine kinases 1 and 2 in regulating the Warburg effect in prostate cancer cells. Cell Signal 2013, 25, 1011–1017. [Google Scholar] [CrossRef] [PubMed]
- Bernacchioni, C.; Ghini, V.; Cencetti, F.; Japtok, L.; Donati, C.; Bruni, P.; Turano, P. NMR metabolomics highlights sphingosine kinase-1 as a new molecular switch in the orchestration of aberrant metabolic phenotype in cancer cells. Mol. Oncol. 2017, 11, 517–533. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhao, S.; Wang, S.; Pan, X.; Zhang, Y.; Xu, J.; Jiang, Y.; Li, H.; Zhang, Q.; Gao, J.; et al. S1P/S1PR3 axis promotes aerobic glycolysis by YAP/c-MYC/PGAM1 axis in osteosarcoma. EBioMedicine 2019, 40, 210–223. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Wang, J.J.; Fu, X.L.; Guang, R.; To, S.T. Advances in the targeting of HIF-1α and future therapeutic strategies for glioblastoma multiforme. Oncol. Rep. 2017, 37, 657–670. [Google Scholar] [CrossRef] [PubMed]
- Quinones, A.; Le, A. The multifaceted metabolism of glioblastoma. Adv. Exp. Med. Biol. 2018, 1063, 59–72. [Google Scholar] [PubMed]
- Garnier, D.; Renoult, O.; Alves-Guerra, M.C.; Paris, F.; Pecqueur, C. Glioblastoma stem-like cells, metabolic strategy to kill a challenging target. Front. Oncol. 2019, 9. [Google Scholar] [CrossRef] [PubMed]
- Ng, W.; Pébay, A.; Drummond, K.; Burgess, A.; Kaye, A.H.; Morokoff, A. Complexities of lysophospholipid signalling in glioblastoma. J. Clin. Neurosci. 2014, 21, 893–898. [Google Scholar] [CrossRef]
- Tabuchi, S. The autotaxin-lysophosphatidic acid-lysophosphatidic acid receptor cascade: Proposal of a novel potential therapeutic target for treating glioblastoma multiforme. Lipids Health Dis. 2015, 14, 56. [Google Scholar] [CrossRef]
- Clair, T.; Aoki, J.; Koh, E.; Bandle, R.W.; Nam, S.W.; Ptaszynska, M.M.; Mills, G.B.; Schiffmann, E.; Liotta, L.A.; Stracke, M.L. Autotaxin hydrolyzes sphingosylphosphorylcholine to produce the regulator of migration, sphingosine-1-phosphate. Cancer Res. 2003, 63, 5446–5453. [Google Scholar]
- van Meeteren, L.A.; Ruurs, P.; Christodoulou, E.; Goding, J.W.; Takakusa, H.; Kikuchi, K.; Perrakis, A.; Nagano, T.; Moolenaar, W.H. Inhibition of autotaxin by lysophosphatidic acid and sphingosine 1-phosphate. J. Biol. Chem. 2005, 280, 21155–21161. [Google Scholar] [CrossRef]
- Benesch, M.G.; Zhao, Y.Y.; Curtis, J.M.; McMullen, T.P.; Brindley, D.N. Regulation of autotaxin expression and secretion by lysophosphatidate and sphingosine 1-phosphate. J. Lipid Res. 2015, 56, 1134–1144. [Google Scholar] [CrossRef]
- Shida, D.; Fang, X.; Kordula, T.; Takabe, K.; Lepine, S.; Alvarez, S.E.; Milstien, S.; Spiegel, S. Cross-talk between LPA1 and epidermal growth factor receptors mediates up-regulation of sphingosine kinase 1 to promote gastric cancer cell motility and invasion. Canc. Res. 2008, 68, 6569–6577. [Google Scholar] [CrossRef] [PubMed]
- Kunkel, G.T.; Maceyka, M.; Milstien, S.; Spiegel, S. Targeting the sphingosine-1-phosphate axis in cancer, inflammation and beyond. Nat. Rev. Drug Discov. 2013, 12, 688–702. [Google Scholar] [CrossRef] [PubMed]
- Lewis, C.S.; Voelkel-Johnson, C.; Smith, C.D. Targeting sphingosine kinases for the treatment of cancer. Adv. Cancer Res. 2018, 140, 295–325. [Google Scholar]
- Zheng, X.; Li, W.; Ren, L.; Liu, J.; Pang, X.; Chen, X.; Kang, D.; Wang, J.; Du, G. The sphingosine kinase-1/sphingosine-1-phosphate axis in cancer: Potential target for anticancer therapy. Pharmacol. Ther. 2019, 195, 85–99. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, H.S.; Awad, A.J.; Shabani, S.; Doan, N. Molecular targeting of acid ceramidase in glioblastoma: A review of its role, potential treatment, and challenges. Pharmaceutics 2018, 10, 45. [Google Scholar] [CrossRef] [PubMed]
- Nagahashi, M.; Takabe, K.; Terracina, K.P.; Soma, D.; Hirose, Y.; Kobayashi, T.; Matsuda, Y.; Wakai, T. Sphingosine-1-phosphate transporters as targets for cancer therapy. Biomed. Res. Int. 2014, 2014, 651727. [Google Scholar] [CrossRef]
- O’Sullivan, S.; Dev, K.K. Sphingosine-1-phosphate receptor therapies: Advances in clinical trials for CNS-related diseases. Neuropharmacology 2017, 113, 597–607. [Google Scholar] [CrossRef]
- Xie, Z.; Liu, H.; Geng, M. Targeting sphingosine-1-phosphate signaling for cancer therapy. Sci. China Life Sci. 2017, 60, 585–600. [Google Scholar] [CrossRef]

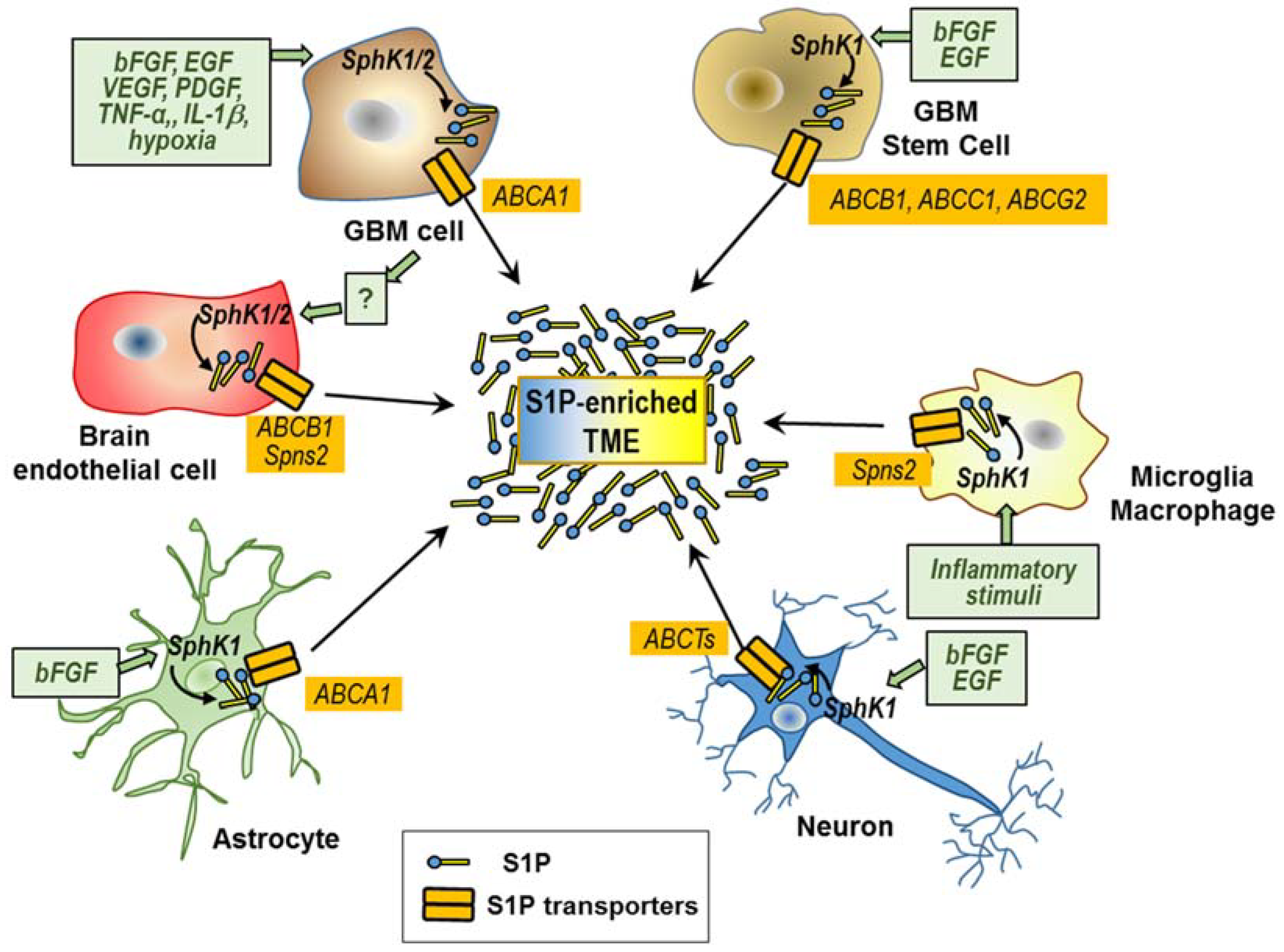
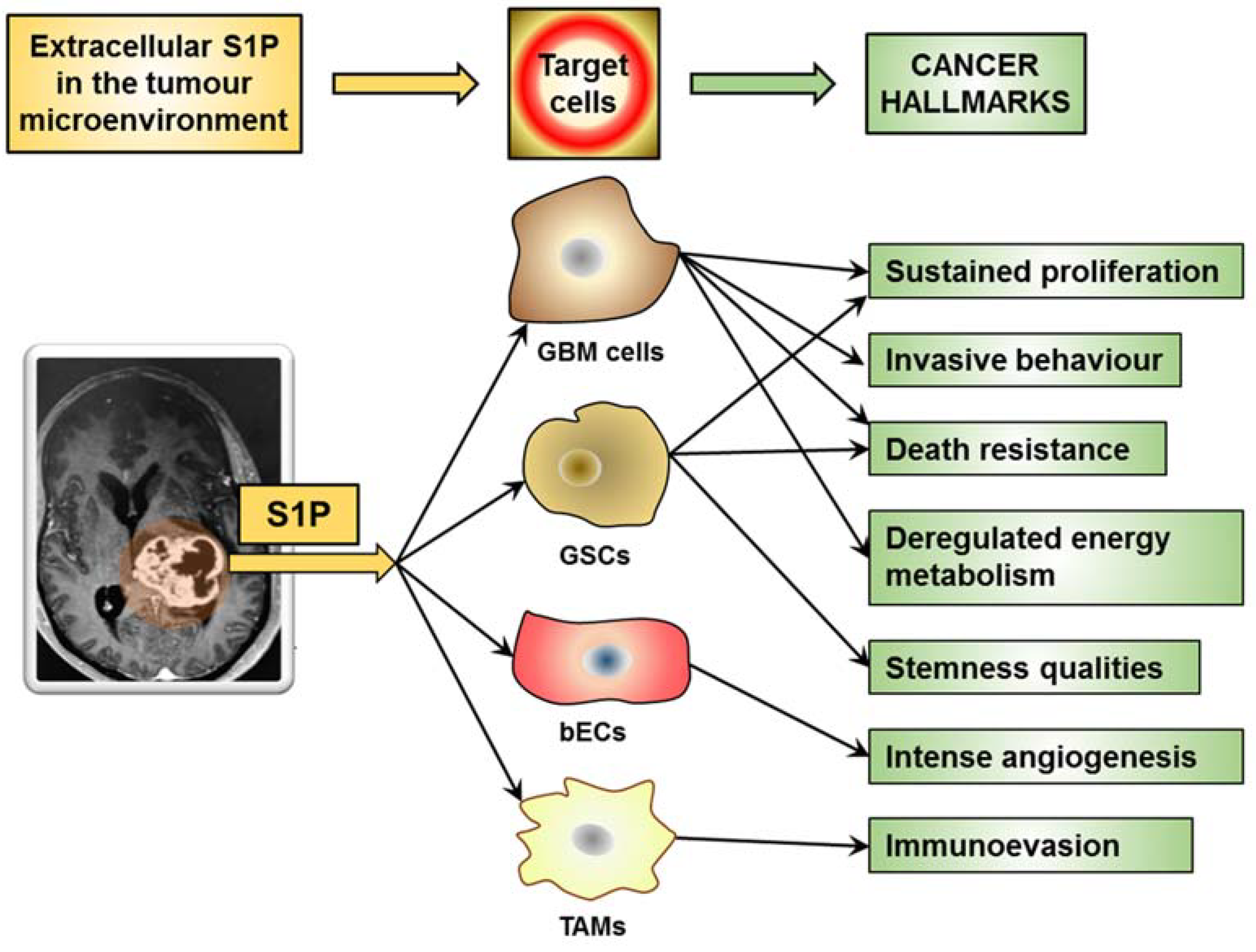
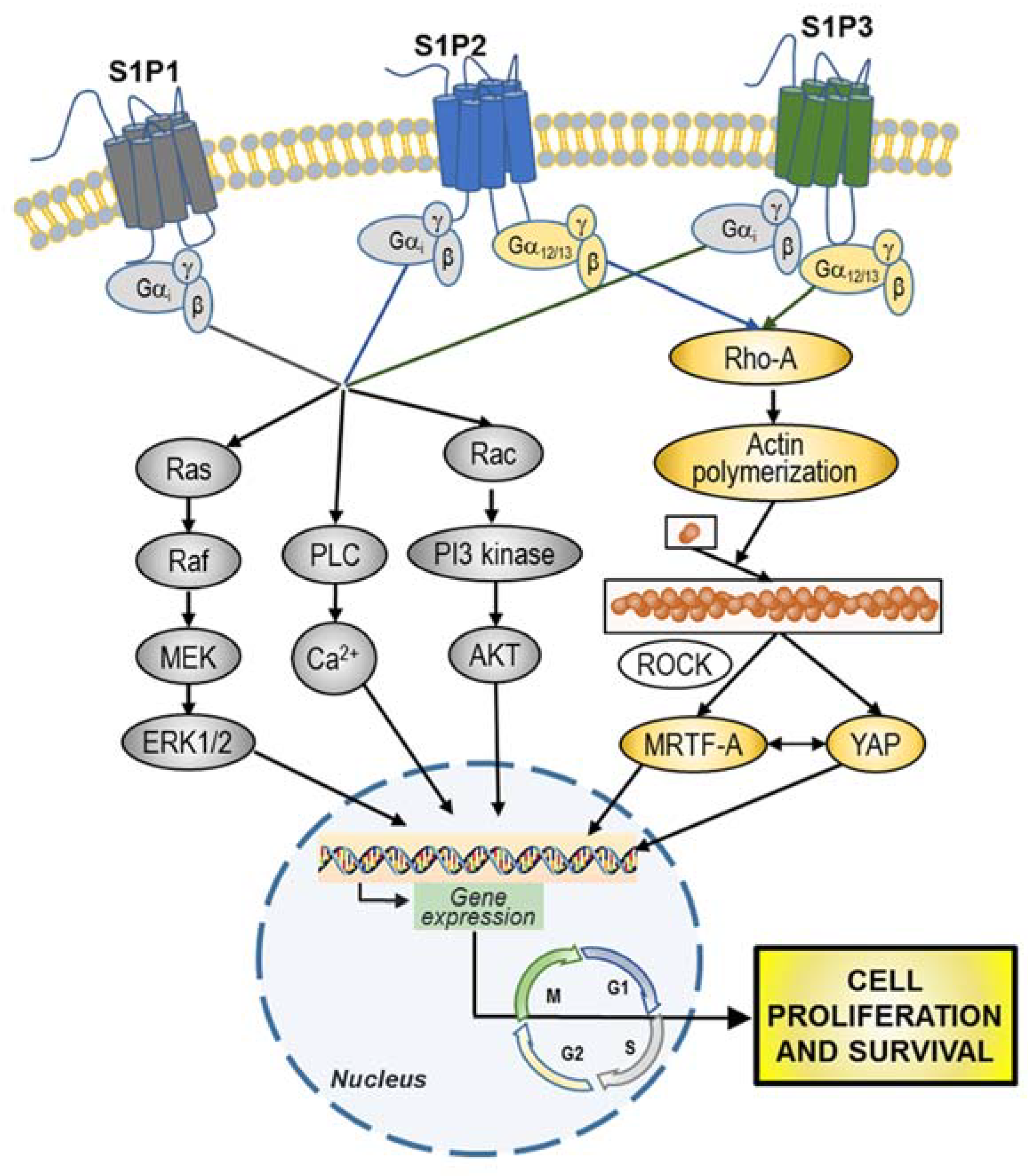
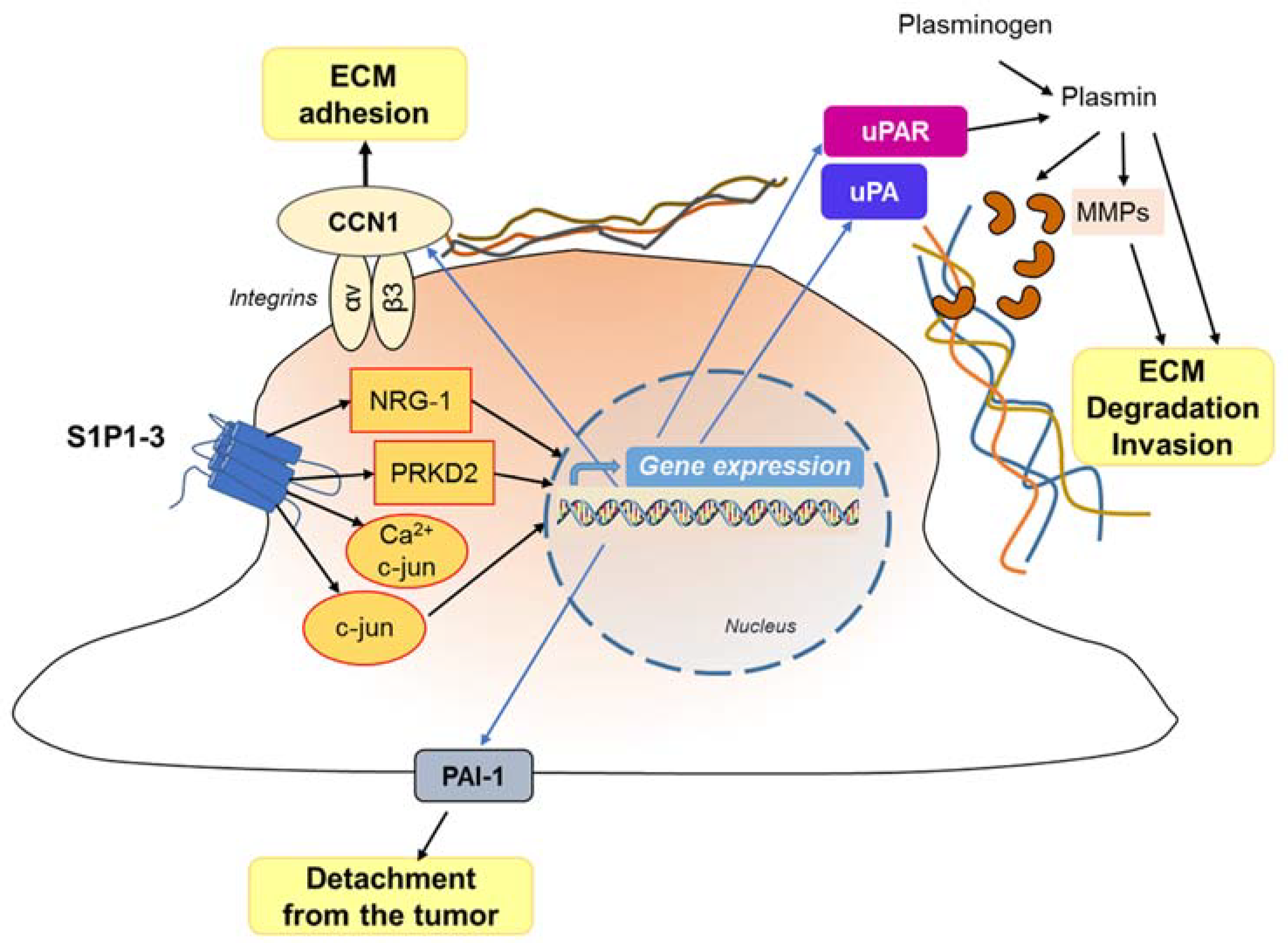
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Riboni, L.; Abdel Hadi, L.; Navone, S.E.; Guarnaccia, L.; Campanella, R.; Marfia, G. Sphingosine-1-Phosphate in the Tumor Microenvironment: A Signaling Hub Regulating Cancer Hallmarks. Cells 2020, 9, 337. https://doi.org/10.3390/cells9020337
Riboni L, Abdel Hadi L, Navone SE, Guarnaccia L, Campanella R, Marfia G. Sphingosine-1-Phosphate in the Tumor Microenvironment: A Signaling Hub Regulating Cancer Hallmarks. Cells. 2020; 9(2):337. https://doi.org/10.3390/cells9020337
Chicago/Turabian StyleRiboni, Laura, Loubna Abdel Hadi, Stefania Elena Navone, Laura Guarnaccia, Rolando Campanella, and Giovanni Marfia. 2020. "Sphingosine-1-Phosphate in the Tumor Microenvironment: A Signaling Hub Regulating Cancer Hallmarks" Cells 9, no. 2: 337. https://doi.org/10.3390/cells9020337
APA StyleRiboni, L., Abdel Hadi, L., Navone, S. E., Guarnaccia, L., Campanella, R., & Marfia, G. (2020). Sphingosine-1-Phosphate in the Tumor Microenvironment: A Signaling Hub Regulating Cancer Hallmarks. Cells, 9(2), 337. https://doi.org/10.3390/cells9020337





