Astrocyte Infection during Rabies Encephalitis Depends on the Virus Strain and Infection Route as Demonstrated by Novel Quantitative 3D Analysis of Cell Tropism
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Lines, Primary Brain Cell Preparation, and Cultivation
2.2. Viruses
2.3. Antibodies
2.4. Infection of in Vitro Cell Cultures and Immunofluorescence Staining
2.5. Brain Samples and Mouse Infections
2.6. Archived Mouse Brains Infected with rRABV Dog and rRABV Fox
2.7. Ultimate 3D Imaging of Solvent-Cleared Organs (uDISCO)
2.8. Confocal Laser Scanning Microscopy and Image Processing
2.9. Statistical Analysis
3. Results
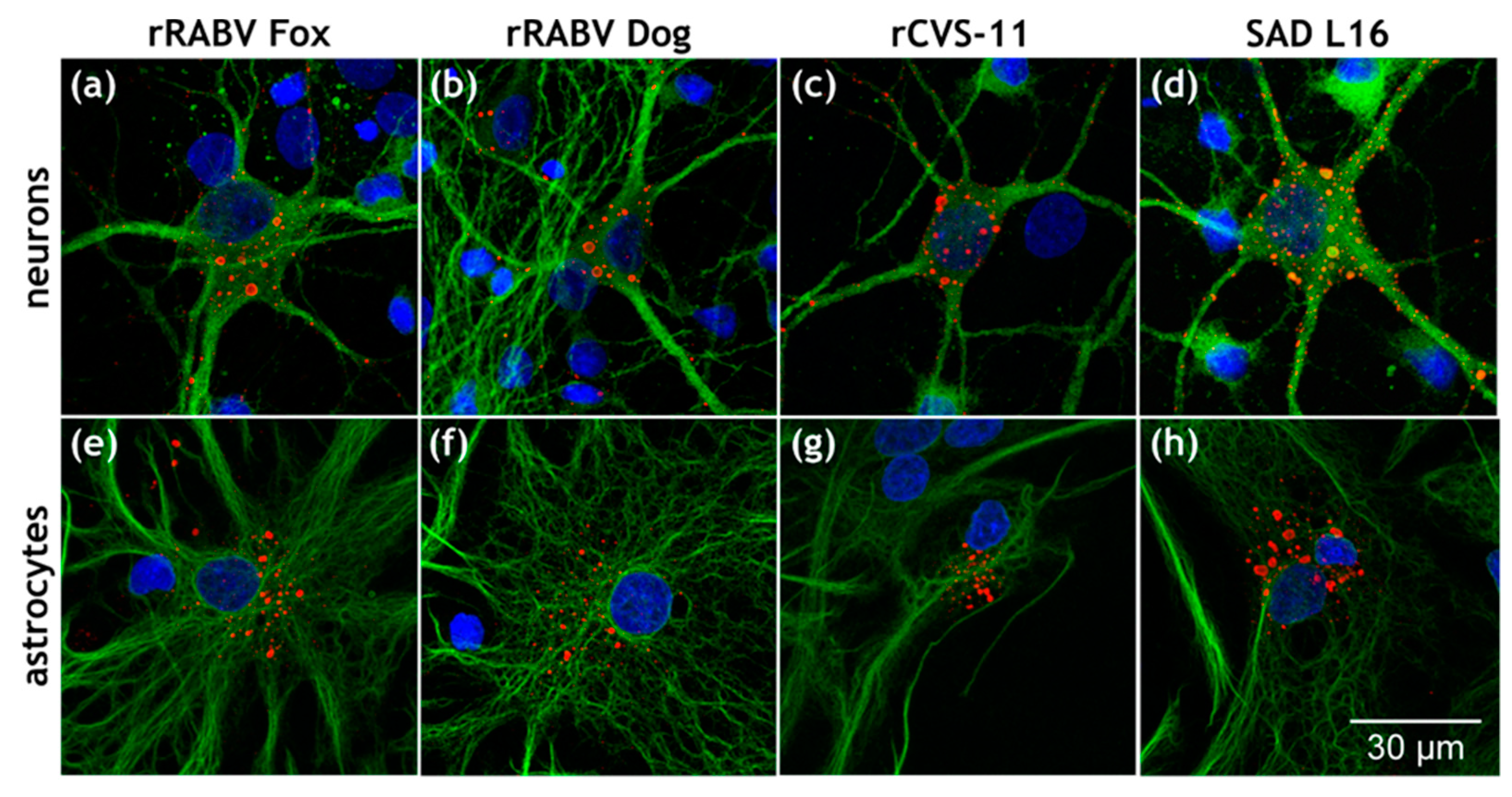
3.1. Astrocytes Are Readily Infected by both Field Viruses and Lab Strains in Mixed Primary Brain Cell Cultures
3.2. Field RABV Infects Neurons and Astrocytes In Vivo as Demonstrated by High-Resolution 3D Analysis of Infected Brain Tissue
3.3. Similar Levels of Field Virus Infection in both Neurons and Astrocytes in the Infected Mouse Brain
3.4. Astrocyte Infection by RABV Depends on the Type of Virus (Field vs. Lab-Adapted).
3.5. Confirmation of the Specific Astrocyte Tropism of Field RABV, ERA, and SAD L16 after i.c. Inoculation and Route-Dependent Astrocyte Infection by rCVS-11
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fooks, A.R.; Cliquet, F.; Finke, S.; Freuling, C.; Hemachudha, T.; Mani, R.S.; Müller, T.; Nadin-Davis, S.; Picard-Meyer, E.; Wilde, H.; et al. Rabies. Nat. Rev. Dis. Primers 2017, 3, 17091. [Google Scholar] [CrossRef]
- Walker, P.J.; Blasdell, K.R.; Calisher, C.H.; Dietzgen, R.G.; Kondo, H.; Kurath, G.; Longdon, B.; Stone, D.M.; Tesh, R.B.; Tordo, N.; et al. ICTV Virus Taxonomy Profile: Rhabdoviridae. J. Gen. Virol. 2018, 99, 447–448. [Google Scholar] [CrossRef] [PubMed]
- Finke, S.; Conzelmann, K.-K. Replication strategies of rabies virus. Virus Res. 2005, 111, 120–131. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Wang, H.; Mahmood, F.; Fu, Z.F. Rabies virus glycoprotein is an important determinant for the induction of innate immune responses and the pathogenic mechanisms. Vet. Microbiol. 2013, 162, 601–613. [Google Scholar] [CrossRef] [PubMed]
- Besson, B.; Sonthonnax, F.; Duchateau, M.; Ben Khalifa, Y.; Larrous, F.; Eun, H.; Hourdel, V.; Matondo, M.; Chamot-Rooke, J.; Grailhe, R.; et al. Regulation of NF-κB by the p105-ABIN2-TPL2 complex and RelAp43 during rabies virus infection. PLoS Pathog. 2017, 13, e1006697. [Google Scholar] [CrossRef] [PubMed]
- Masatani, T.; Ito, N.; Shimizu, K.; Ito, Y.; Nakagawa, K.; Sawaki, Y.; Koyama, H.; Sugiyama, M. Rabies virus nucleoprotein functions to evade activation of the RIG-I-mediated antiviral response. J. Virol. 2010, 84, 4002–4012. [Google Scholar] [CrossRef] [PubMed]
- Ben Khalifa, Y.; Luco, S.; Besson, B.; Sonthonnax, F.; Archambaud, M.; Grimes, J.M.; Larrous, F.; Bourhy, H. The matrix protein of rabies virus binds to RelAp43 to modulate NF-κB-dependent gene expression related to innate immunity. Sci. Rep. 2016, 6, 39420. [Google Scholar] [CrossRef]
- Brzózka, K.; Finke, S.; Conzelmann, K.-K. Identification of the rabies virus alpha/beta interferon antagonist: Phosphoprotein P interferes with phosphorylation of interferon regulatory factor 3. J. Virol. 2005, 79, 7673–7681. [Google Scholar] [CrossRef]
- Brzózka, K.; Finke, S.; Conzelmann, K.-K. Inhibition of interferon signaling by rabies virus phosphoprotein P: Activation-dependent binding of STAT1 and STAT2. J. Virol. 2006, 80, 2675–2683. [Google Scholar] [CrossRef]
- Ito, N.; Moseley, G.W.; Blondel, D.; Shimizu, K.; Rowe, C.L.; Ito, Y.; Masatani, T.; Nakagawa, K.; Jans, D.A.; Sugiyama, M. Role of interferon antagonist activity of rabies virus phosphoprotein in viral pathogenicity. J. Virol. 2010, 84, 6699–6710. [Google Scholar] [CrossRef]
- Faber, M.; Pulmanausahakul, R.; Hodawadekar, S.S.; Spitsin, S.; McGettigan, J.P.; Schnell, M.J.; Dietzschold, B. Overexpression of the rabies virus glycoprotein results in enhancement of apoptosis and antiviral immune response. J. Virol. 2002, 76, 3374–3381. [Google Scholar] [CrossRef] [PubMed]
- Jackson, A.C.; Rasalingam, P.; Weli, S.C. Comparative pathogenesis of recombinant rabies vaccine strain SAD-L16 and SAD-D29 with replacement of Arg333 in the glycoprotein after peripheral inoculation of neonatal mice: Less neurovirulent strain is a stronger inducer of neuronal apoptosis. Acta Neuropathol. 2006, 111, 372–378. [Google Scholar] [CrossRef] [PubMed]
- Morimoto, K.; Hooper, D.C.; Spitsin, S.; Koprowski, H.; Dietzschold, B. Pathogenicity of different rabies virus variants inversely correlates with apoptosis and rabies virus glycoprotein expression in infected primary neuron cultures. J. Virol. 1999, 73, 510–518. [Google Scholar] [CrossRef] [PubMed]
- Sarmento, L.; Li, X.-q.; Howerth, E.; Jackson, A.C.; Fu, Z.F. Glycoprotein-mediated induction of apoptosis limits the spread of attenuated rabies viruses in the central nervous system of mice. J. Neurovirol. 2005, 11, 571–581. [Google Scholar] [CrossRef] [PubMed]
- Dietzschold, B.; Li, J.; Faber, M.; Schnell, M. Concepts in the pathogenesis of rabies. Future Virol. 2008, 3, 481–490. [Google Scholar] [CrossRef] [PubMed]
- Tsiang, H.; Lycke, E.; Ceccaldi, P.E.; Ermine, A.; Hirardot, X. The anterograde transport of rabies virus in rat sensory dorsal root ganglia neurons. J. Gen. Virol. 1989, 70, 2075–2085. [Google Scholar] [CrossRef]
- Astic, L.; Saucier, D.; Coulon, P.; Lafay, F.; Flamand, A. The CVS strain of rabies virus as transneuronal tracer in the olfactory system of mice. Brain Res. 1993, 619, 146–156. [Google Scholar] [CrossRef]
- Lycke, E.; Tsiang, H. Rabies virus infection of cultured rat sensory neurons. J. Virol. 1987, 61, 2733–2741. [Google Scholar] [CrossRef]
- Ceccaldi, P.E.; Gillet, J.P.; Tsiang, H. Inhibition of the transport of rabies virus in the central nervous system. J. Neuropathol. Exp. Neurol. 1989, 48, 620–630. [Google Scholar] [CrossRef]
- Gluska, S.; Zahavi, E.E.; Chein, M.; Gradus, T.; Bauer, A.; Finke, S.; Perlson, E. Rabies Virus Hijacks and accelerates the p75NTR retrograde axonal transport machinery. PLoS Pathog. 2014, 10, e1004348. [Google Scholar] [CrossRef]
- Bauer, A.; Nolden, T.; Schröter, J.; Römer-Oberdörfer, A.; Gluska, S.; Perlson, E.; Finke, S. Anterograde glycoprotein-dependent transport of newly generated rabies virus in dorsal root ganglion neurons. J. Virol. 2014, 88, 14172–14183. [Google Scholar] [CrossRef] [PubMed]
- Wickersham, I.R.; Finke, S.; Conzelmann, K.-K.; Callaway, E.M. Retrograde neuronal tracing with a deletion-mutant rabies virus. Nat. Methods 2007, 4, 47–49. [Google Scholar] [CrossRef] [PubMed]
- Wickersham, I.R.; Lyon, D.C.; Barnard, R.J.O.; Mori, T.; Finke, S.; Conzelmann, K.-K.; Young, J.A.T.; Callaway, E.M. Monosynaptic restriction of transsynaptic tracing from single, genetically targeted neurons. Neuron 2007, 53, 639–647. [Google Scholar] [CrossRef] [PubMed]
- Reardon, T.R.; Murray, A.J.; Turi, G.F.; Wirblich, C.; Croce, K.R.; Schnell, M.J.; Jessell, T.M.; Losonczy, A. Rabies Virus CVS-N2c(ΔG) Strain Enhances Retrograde Synaptic Transfer and Neuronal Viability. Neuron 2016, 89, 711–724. [Google Scholar] [CrossRef] [PubMed]
- Thoulouze, M.I.; Lafage, M.; Schachner, M.; Hartmann, U.; Cremer, H.; Lafon, M. The neural cell adhesion molecule is a receptor for rabies virus. J. Virol. 1998, 72, 7181–7190. [Google Scholar] [CrossRef] [PubMed]
- Tuffereau, C.; Bénéjean, J.; Blondel, D.; Kieffer, B.; Flamand, A. Low-affinity nerve-growth factor receptor (P75NTR) can serve as a receptor for rabies virus. EMBO J. 1998, 17, 7250–7259. [Google Scholar] [CrossRef]
- Lentz, T.; Burrage, T.; Smith, A.; Crick, J.; Tignor, G. Is the acetylcholine receptor a rabies virus receptor? Science 1982, 215, 182–184. [Google Scholar] [CrossRef]
- Wang, J.; Wang, Z.; Liu, R.; Shuai, L.; Wang, X.; Luo, J.; Wang, C.; Chen, W.; Wang, X.; Ge, J.; et al. Metabotropic glutamate receptor subtype 2 is a cellular receptor for rabies virus. PLoS Pathog. 2018, 14, e1007189. [Google Scholar] [CrossRef]
- Ray, N.B.; Power, C.; Lynch, W.P.; Ewalt, L.C.; Lodmell, D.L. Rabies viruses infect primary cultures of murine, feline, and human microglia and astrocytes. Arch. Virol. 1997, 142, 1011–1019. [Google Scholar] [CrossRef]
- Davis, B.M.; Rall, G.F.; Schnell, M.J. Everything You Always Wanted to Know About Rabies Virus (But Were Afraid to Ask). Annu. Rev. Virol. 2015, 2, 451–471. [Google Scholar] [CrossRef]
- Jackson, A.C.; Phelan, C.C.; Rossiter, J.P. Infection of Bergmann glia in the cerebellum of a skunk experimentally infected with street rabies virus. Can. J. Vet. Res. 2000, 64, 226–228. [Google Scholar] [PubMed]
- Tsiang, H.; Koulakoff, A.; Bizzini, B.; Berwald-Netter, Y. Neurotropism of rabies virus. An in vitro study. J. Neuropathol. Exp. Neurol. 1983, 42, 439–452. [Google Scholar] [CrossRef] [PubMed]
- Pfefferkorn, C.; Kallfass, C.; Lienenklaus, S.; Spanier, J.; Kalinke, U.; Rieder, M.; Conzelmann, K.-K.; Michiels, T.; Staeheli, P. Abortively Infected Astrocytes Appear to Represent the Main Source of Interferon Beta in the Virus-Infected Brain. J. Virol. 2016, 90, 2031–2038. [Google Scholar] [CrossRef] [PubMed]
- Tian, B.; Zhou, M.; Yang, Y.; Yu, L.; Luo, Z.; Tian, D.; Wang, K.; Cui, M.; Chen, H.; Fu, Z.F.; et al. Lab-Attenuated Rabies Virus Causes Abortive Infection and Induces Cytokine Expression in Astrocytes by Activating Mitochondrial Antiviral-Signaling Protein Signaling Pathway. Front. Immunol. 2018, 8, 2011. [Google Scholar] [CrossRef] [PubMed]
- Ito, N.; Moseley, G.W.; Sugiyama, M. The importance of immune evasion in the pathogenesis of rabies virus. J. Vet. Med. Sci. 2016, 78, 1089–1098. [Google Scholar] [CrossRef]
- Suja, M.S.; Mahadevan, A.; Madhusudana, S.N.; Shankar, S.K. Role of apoptosis in rabies viral encephalitis: A comparative study in mice, canine, and human brain with a review of literature. Patholog. Res. Int. 2011, 2011, 374286. [Google Scholar] [CrossRef]
- Wang, Z.W.; Sarmento, L.; Wang, Y.; Li, X.-q.; Dhingra, V.; Tseggai, T.; Jiang, B.; Fu, Z.F. Attenuated Rabies Virus Activates, while Pathogenic Rabies Virus Evades, the Host Innate Immune Responses in the Central Nervous System. J. Virol. 2005, 79, 12554–12565. [Google Scholar] [CrossRef]
- Renier, N.; Wu, Z.; Simon, D.J.; Yang, J.; Ariel, P.; Tessier-Lavigne, M. iDISCO: A simple, rapid method to immunolabel large tissue samples for volume imaging. Cell 2014, 159, 896–910. [Google Scholar] [CrossRef]
- Pan, C.; Cai, R.; Quacquarelli, F.P.; Ghasemigharagoz, A.; Lourbopoulos, A.; Matryba, P.; Plesnila, N.; Dichgans, M.; Hellal, F.; Ertürk, A. Shrinkage-mediated imaging of entire organs and organisms using uDISCO. Nat. Methods 2016, 13, 859–867. [Google Scholar] [CrossRef]
- Zaeck, L.; Potratz, M.; Freuling, C.M.; Müller, T.; Finke, S. High-Resolution 3D Imaging of Rabies Virus Infection in Solvent-Cleared Brain Tissue. J. Vis. Exp. 2019, 30. [Google Scholar] [CrossRef]
- Brewer, G.J.; Torricelli, J.R. Isolation and culture of adult neurons and neurospheres. Nat. Protoc. 2007, 2, 1490–1498. [Google Scholar] [CrossRef] [PubMed]
- Nolden, T.; Pfaff, F.; Nemitz, S.; Freuling, C.M.; Höper, D.; Müller, T.; Finke, S. Reverse genetics in high throughput: Rapid generation of complete negative strand RNA virus cDNA clones and recombinant viruses thereof. Sci. Rep. 2016, 6, 23887. [Google Scholar] [CrossRef] [PubMed]
- Schnell, M.J.; Mebatsion, T.; Conzelmann, K.K. Infectious rabies viruses from cloned cDNA. EMBO J. 1994, 4195–4203. [Google Scholar] [CrossRef]
- Abelseth, M.K. An attenuated rabies vaccine for domestic animals produced in tissue culture. Can. Vet. J. 1964, 5, 279–286. [Google Scholar]
- Höper, D.; Freuling, C.M.; Müller, T.; Hanke, D.; von Messling, V.; Duchow, K.; Beer, M.; Mettenleiter, T.C. High definition viral vaccine strain identity and stability testing using full-genome population data--The next generation of vaccine quality control. Vaccine 2015, 33, 5829–5837. [Google Scholar] [CrossRef]
- Vos, A.; Nolden, T.; Habla, C.; Finke, S.; Freuling, C.M.; Teifke, J.; Müller, T. Raccoons (Procyon lotor) in Germany as potential reservoir species for Lyssaviruses. Eur J. Wildl Res. 2013, 59, 637–643. [Google Scholar] [CrossRef]
- Fu, C.; Donovan, W.P.; Shikapwashya-Hasser, O.; Ye, X.; Cole, R.H. Hot Fusion: An efficient method to clone multiple DNA fragments as well as inverted repeats without ligase. PLoS ONE 2014, 9, e115318. [Google Scholar] [CrossRef]
- Finke, S.; Granzow, H.; Hurst, J.; Pollin, R.; Mettenleiter, T.C. Intergenotypic replacement of lyssavirus matrix proteins demonstrates the role of lyssavirus M proteins in intracellular virus accumulation. J. Virol. 2010, 84, 1816–1827. [Google Scholar] [CrossRef]
- Buchholz, U.J.; Finke, S.; Conzelmann, K.K. Generation of bovine respiratory syncytial virus (BRSV) from cDNA: BRSV NS2 is not essential for virus replication in tissue culture, and the human RSV leader region acts as a functional BRSV genome promoter. J. Virol. 1999, 73, 251–259. [Google Scholar] [CrossRef]
- Orbanz, J.; Finke, S. Generation of recombinant European bat lyssavirus type 1 and inter-genotypic compatibility of lyssavirus genotype 1 and 5 antigenome promoters. Arch. Virol. 2010, 155, 1631–1641. [Google Scholar] [CrossRef]
- Eggerbauer, E.; Pfaff, F.; Finke, S.; Höper, D.; Beer, M.; Mettenleiter, T.C.; Nolden, T.; Teifke, J.-P.; Müller, T.; Freuling, C.M. Comparative analysis of European bat lyssavirus 1 pathogenicity in the mouse model. PLoS Negl. Trop. Dis. 2017, 11, e0005668. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Bolte, S.; Cordelières, F.P. A guided tour into subcellular colocalization analysis in light microscopy. J. Microsc. 2006, 224, 213–232. [Google Scholar] [CrossRef] [PubMed]
- de Chaumont, F.; Dallongeville, S.; Chenouard, N.; Hervé, N.; Pop, S.; Provoost, T.; Meas-Yedid, V.; Pankajakshan, P.; Lecomte, T.; Le Montagner, Y.; et al. Icy: An open bioimage informatics platform for extended reproducible research. Nat. Methods 2012, 9, 690–696. [Google Scholar] [CrossRef]
- Jackson, A.C.; Reimer, D.L. Pathogenesis of experimental rabies in mice: An immunohistochemical study. Acta Neuropathol. 1989, 78, 159–165. [Google Scholar] [CrossRef]
- Conzelmann, K.K.; Cox, J.H.; Schneider, L.G.; Thiel, H.J. Molecular cloning and complete nucleotide sequence of the attenuated rabies virus SAD B19. Virology 1990, 175, 485–499. [Google Scholar] [CrossRef]
- Weiland, F.; Cox, J.H.; Meyer, S.; Dahme, E.; Reddehase, M.J. Rabies Virus Neuritic Paralysis: Immunopathogenesis of Nonfatal Paralytic Rabies. J. Virol. 1992, 5096–5099. [Google Scholar] [CrossRef]
- Zlotnik, I. The reaction of astrocytes to acute virus infections of the central nervous system. Br. J. Exp. Pathol 1968, 49, 555–564. [Google Scholar]
- Jackson, A.C. Rabies. Scientific Basis of the Disease and Its Management, 3rd ed.; Elsevier Science: San Diego, CA, USA, 2013; ISBN 9780123965479. [Google Scholar]
- Detje, C.N.; Lienenklaus, S.; Chhatbar, C.; Spanier, J.; Prajeeth, C.K.; Soldner, C.; Tovey, M.G.; Schlüter, D.; Weiss, S.; Stangel, M.; et al. Upon intranasal vesicular stomatitis virus infection, astrocytes in the olfactory bulb are important interferon Beta producers that protect from lethal encephalitis. J. Virol. 2015, 89, 2731–2738. [Google Scholar] [CrossRef]
- Kallfass, C.; Ackerman, A.; Lienenklaus, S.; Weiss, S.; Heimrich, B.; Staeheli, P. Visualizing production of beta interferon by astrocytes and microglia in brain of La Crosse virus-infected mice. J. Virol. 2012, 86, 11223–11230. [Google Scholar] [CrossRef]






| Virus | GFAP (−) | GFAP (+) | Uncertain | |||
|---|---|---|---|---|---|---|
| Count | % | Count | % | Count | % | |
| SAD L16 | 256 | 48.3 | 30 | 5.7 | 244 | 46 |
| rCVS-11 | 58 | 64.4 | 6 | 6.7 | 26 | 28.9 |
| rRABV Dog | 252 | 46.8 | 29 | 5.4 | 258 | 47.9 |
| rRABV Fox | 1002 | 55.2 | 89 | 4.9 | 712 | 39.5 |
| Virus | MAP2 (−) | MAP2 (+) | Uncertain | |||
| Count | % | Count | % | Count | % | |
| SAD L16 | 18 | 3.5 | 387 | 74.2 | 116 | 22.3 |
| rCVS-11 | 1 | 1.0 | 44 | 43.2 | 57 | 55.9 |
| rRABV Dog | 27 | 5.3 | 347 | 73.3 | 109 | 21.4 |
| rRABV Fox | 190 | 7.3 | 2096 | 80.3 | 324 | 12.4 |
| Neurons | Astrocytes | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Region | Counted | Infected | % | Mean | SD | Counted | Infected | % | Mean | SD |
| 1 | 485 | 21 | 4.3 | 3.9 | 1.4 | 362 | 11 | 3.0 | 7.0 | 4.9 |
| 2 | 762 | 16 | 2.1 | 272 | 18 | 6.6 | ||||
| 3 | 448 | 13 | 2.9 | 106 | 18 | 17.0 | ||||
| 4 | 2518 | 132 | 5.2 | 488 | 9 | 1.8 | ||||
| 5 | 3029 | 91 | 3.0 | 273 | 20 | 7.3 | ||||
| 6 | 3847 | 234 | 6.1 | 541 | 32 | 5.9 | ||||
| ∑ | 11098 | 507 | 2042 | 108 | ||||||
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Potratz, M.; Zaeck, L.; Christen, M.; te Kamp, V.; Klein, A.; Nolden, T.; Freuling, C.M.; Müller, T.; Finke, S. Astrocyte Infection during Rabies Encephalitis Depends on the Virus Strain and Infection Route as Demonstrated by Novel Quantitative 3D Analysis of Cell Tropism. Cells 2020, 9, 412. https://doi.org/10.3390/cells9020412
Potratz M, Zaeck L, Christen M, te Kamp V, Klein A, Nolden T, Freuling CM, Müller T, Finke S. Astrocyte Infection during Rabies Encephalitis Depends on the Virus Strain and Infection Route as Demonstrated by Novel Quantitative 3D Analysis of Cell Tropism. Cells. 2020; 9(2):412. https://doi.org/10.3390/cells9020412
Chicago/Turabian StylePotratz, Madlin, Luca Zaeck, Michael Christen, Verena te Kamp, Antonia Klein, Tobias Nolden, Conrad M. Freuling, Thomas Müller, and Stefan Finke. 2020. "Astrocyte Infection during Rabies Encephalitis Depends on the Virus Strain and Infection Route as Demonstrated by Novel Quantitative 3D Analysis of Cell Tropism" Cells 9, no. 2: 412. https://doi.org/10.3390/cells9020412
APA StylePotratz, M., Zaeck, L., Christen, M., te Kamp, V., Klein, A., Nolden, T., Freuling, C. M., Müller, T., & Finke, S. (2020). Astrocyte Infection during Rabies Encephalitis Depends on the Virus Strain and Infection Route as Demonstrated by Novel Quantitative 3D Analysis of Cell Tropism. Cells, 9(2), 412. https://doi.org/10.3390/cells9020412





