Technical Considerations in Ex Vivo Human Regulatory T Cell Migration and Suppression Assays
Abstract
:1. Introduction
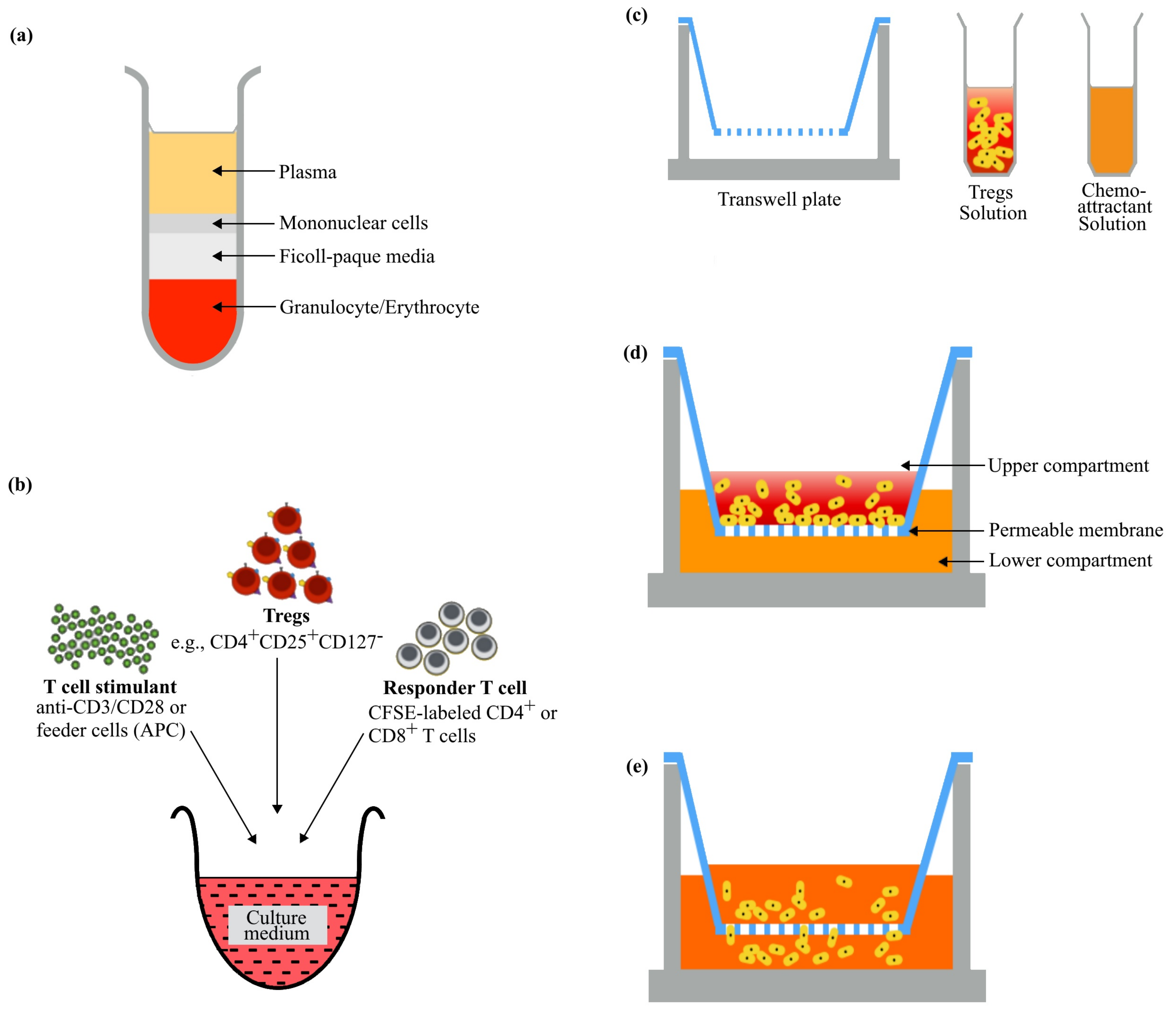
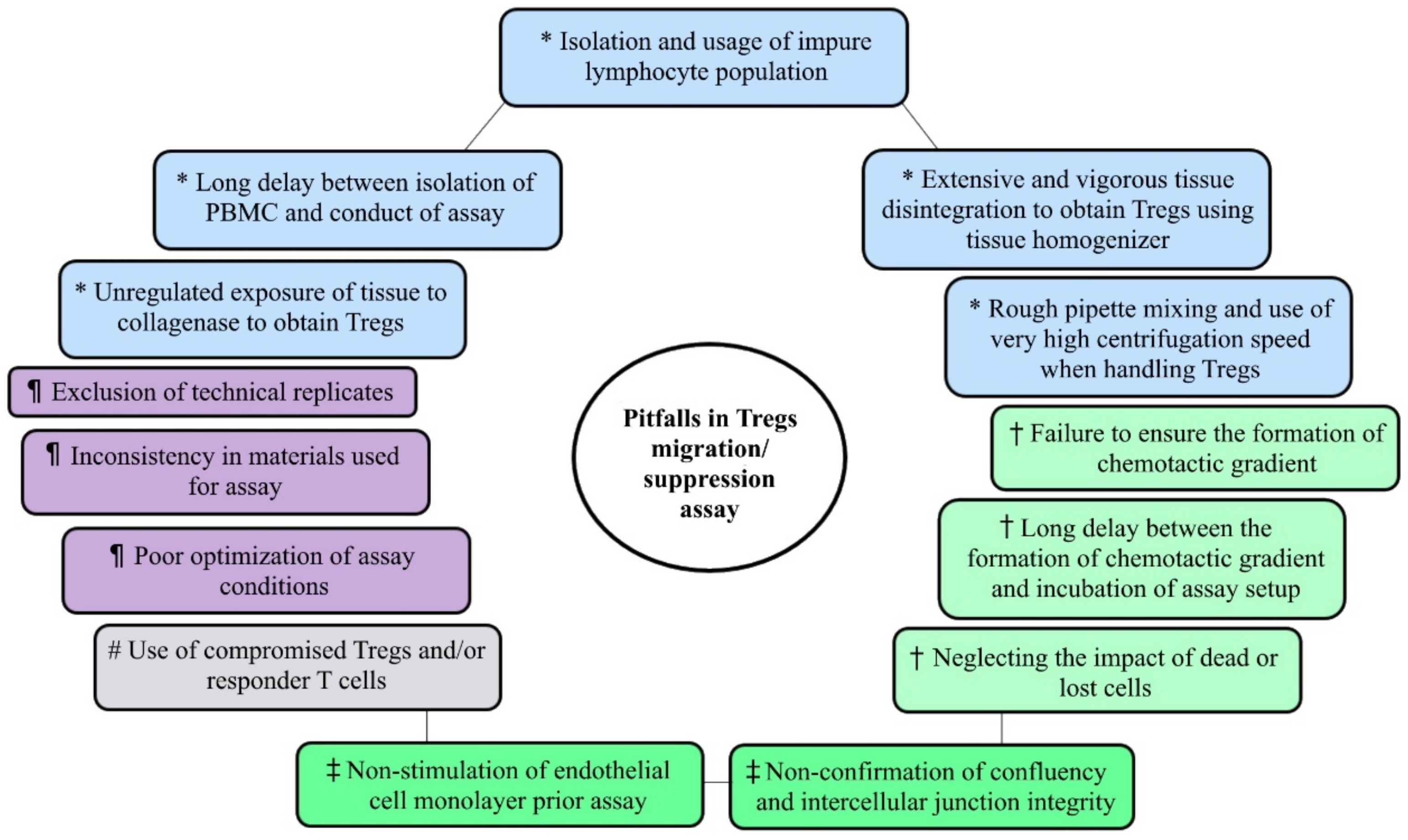
2. Sample Processing and Preparation of Single-Cell Suspension
3. Optimization of Experiment
4. Tregs Migration Assay
4.1. Choice of Transwell
4.2. Endothelial Cell Monolayer
4.3. Establishing a Chemotactic Gradient
4.4. Counting Migrated Cells
5. Tregs Suppression Assay
5.1. Choice of Responder T Cell
5.2. Tregs/Tconv Ratio
5.3. Monitoring Suppression of Proliferation
6. Concluding Remarks
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Liu, C.; Workman, C.J.; Vignali, D.A.A. Targeting regulatory T cells in tumors. FEBS J. 2016, 283, 2731–2748. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.S.; Sohn, H.; Jang, S.W.; Lee, G.R. The transcription factor NFIL3 controls regulatory T-cell function and stability. Exp. Mol. Med. 2019, 51, 80. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Srivastava, R.K.; Dar, H.Y.; Mishra, P.K. Immunoporosis: Immunology of osteoporosis-role of T cells. Front. Immunol. 2018, 9, 657. [Google Scholar] [CrossRef]
- Santegoets, S.J.A.M.; Dijkgraaf, E.M.; Battaglia, A.; Beckhove, P.; Britten, C.M.; Gallimore, A.; Godkin, A.; Gouttefangeas, C.; de Gruijl, T.D.; Koenen, H.J.P.M.; et al. Monitoring regulatory T cells in clinical samples: Consensus on an essential marker set and gating strategy for regulatory T cell analysis by flow cytometry. Cancer Immunol. Immunother. 2015, 64, 1271–1286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, F.; Du, R.; Wei, F.; Zhao, H.; Yu, J.; Wang, C.; Zhan, Z.; Ding, T.; Ren, X.; Chen, X.; et al. Expression of TNFR2 by regulatory T cells in peripheral blood is correlated with clinical pathology of lung cancer patients. Cancer Immunol. Immunother. 2015, 64, 1475–1485. [Google Scholar] [CrossRef]
- Mercadante, E.R.; Lorenz, U.M. Breaking free of control: How conventional T cells overcome regulatory T cell suppression. Front. Immunol. 2016, 7, 193. [Google Scholar] [CrossRef] [Green Version]
- Ding, Y.; Xu, J.; Bromberg, J.S. Regulatory T cell migration during an immune response. Trends Immunol. 2012, 33, 174–180. [Google Scholar] [CrossRef] [Green Version]
- Fritzsching, B.; Haas, J.; König, F.; Kunz, P.; Fritzsching, E.; Pöschl, J.; Krammer, P.H.; Brück, W.; Suri-Payer, E.; Wildemann, B. Intracerebral human regulatory T cells: Analysis of CD4+CD25+FOXP3+ T cells in brain lesions and cerebrospinal fluid of multiple sclerosis patients. PLoS ONE 2011, 6, 17988. [Google Scholar] [CrossRef] [Green Version]
- Dominguez-Villar, M.; Baecher-Allan, C.M.; Hafler, D.A. Identification of T helper type 1-like, Foxp3+ regulatory T cells in human autoimmune disease. Nat. Med. 2011, 17, 673–675. [Google Scholar] [CrossRef] [Green Version]
- Brusko, T.M.; Wasserfall, C.H.; Clare-Salzler, M.J.; Schatz, D.A.; Atkinson, M.A. Functional defects and the influence of age on the frequency of CD4 +CD25+ T-cells in type 1 diabetes. Diabetes 2005, 54, 1407–1414. [Google Scholar] [CrossRef] [Green Version]
- Haseda, F.; Imagawa, A.; Murase-Mishiba, Y.; Terasaki, J.; Hanafusa, T. CD4+ CD45RA− FoxP3high activated regulatory T cells are functionally impaired and related to residual insulin-secreting capacity in patients with type 1 diabetes. Clin. Exp. Immunol. 2013, 173, 207–216. [Google Scholar] [CrossRef] [PubMed]
- Van Roon, J.A.G.; Hartgring, S.A.Y.; van der Wurff-Jacobs, K.M.G.; Bijlsma, J.W.J.; Lafeber, F.P.J.G. Numbers of CD25+Foxp3+ T cells that lack the IL-7 receptor are increased intra-articularly and have impaired suppressive function in RA patients. Rheumatology 2010, 49, 2084–2089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fessler, J.; Raicht, A.; Husic, R.; Ficjan, A.; Schwarz, C.; Duftner, C.; Schwinger, W.; Graninger, W.B.; Stradner, M.H.; Dejaco, C. Novel senescent regulatory T-cell subset with impaired suppressive function in rheumatoid arthritis. Front. Immunol. 2017, 8, 300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Liang, J.; Tian, C.; Zeng, Y.; Yang, Q.; Liu, Y.; Liu, Y.; Wu, J.; Hu, Y.; Gu, F.; Zhang, K.; et al. FOXA1+ regulatory T cells: A novel T cell subset that suppresses antitumor immunity in lung cancer. Biochem. Biophys. Res. Commun. 2019, 514, 308–315. [Google Scholar] [CrossRef]
- Wang, D.; Yang, L.; Yu, W.; Wu, Q.; Lian, J.; Li, F.; Liu, S.; Li, A.; He, Z.; Liu, J.; et al. Colorectal cancer cell-derived CCL20 recruits regulatory T cells to promote chemoresistance via FOXO1/CEBPB/NF-κB signaling. J. Immunother. Cancer 2019, 7, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Mrizak, D.; Martin, N.; Barjon, C.; Jimenez-Pailhes, A.-S.; Mustapha, R.; Niki, T.; Guigay, J.; Pancré, V.; de Launoit, Y.; Busson, P.; et al. Effect of nasopharyngeal carcinoma-derived exosomes on human regulatory T cells. J. Natl. Cancer Inst. 2014, 107, 363. [Google Scholar]
- Liu, H.; Wang, S.H.; Chen, S.C.; Chen, C.Y.; Lin, T.M. Zoledronic acid blocks the interaction between breast cancer cells and regulatory T-cells. BMC Cancer 2019, 19, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Brinkman, C.C.; Iwami, D.; Hritzo, M.K.; Xiong, Y.; Ahmad, S.; Simon, T.; Hippen, K.L.; Blazar, B.R.; Bromberg, J.S. Treg engage lymphotoxin beta receptor for afferent lymphatic transendothelial migration. Nat. Commun. 2016, 7, 1–6. [Google Scholar] [CrossRef] [Green Version]
- Chaudhary, B.; Elkord, E. Regulatory T cells in the tumor microenvironment and cancer progression: Role and therapeutic targeting. Vaccines 2016, 4, 28. [Google Scholar] [CrossRef] [Green Version]
- Fraser, H.; Safinia, N.; Grageda, N.; Thirkell, S.; Lowe, K.; Fry, L.J.; Scottá, C.; Hope, A.; Fisher, C.; Hilton, R.; et al. A Rapamycin-Based GMP-Compatible Process for the Isolation and Expansion of Regulatory T Cells for Clinical Trials. Mol. Ther.-Methods Clin. Dev. 2018, 8, 198–209. [Google Scholar] [CrossRef] [Green Version]
- Klein, A.B.; Witonsky, S.G.; Ahmed, S.A.; Holladay, S.D.; Gogal, R.M.; Link, L.; Reilly, C.M. Impact of different cell isolation techniques on lymphocyte viability and function. J. Immunoass. Immunochem. 2006, 27, 61–76. [Google Scholar] [CrossRef] [PubMed]
- Reichard, A.; Asosingh, K. Best Practices for Preparing a Single Cell Suspension from Solid Tissues for Flow Cytometry. Cytom. Part A 2019, 95, 219–226. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kleiveland, C.R. Peripheral Blood Mononuclear Cells BT—The Impact of Food Bioactives on Health: In Vitro and Ex Vivo Models; Verhoeckx, K., Cotter, P., López-Expósito, I., Kleiveland, C., Lea, T., Mackie, A., Requena, T., Swiatecka, D., Wichers, H., Eds.; Springer: Berlin/Heidelberg, Germany, 2015; pp. 161–167. ISBN 978-3-319-16104-4. [Google Scholar]
- Tyler, C.J.; Pérez-Jeldres, T.; Ehinger, E.; Capaldo, B.; Karuppuchamy, T.; Boyer, J.D.; Patel, D.; Dulai, P.; Boland, B.S.; Lannigan, J.; et al. Implementation of mass cytometry as a tool for mechanism of action studies in inflammatory bowel disease. Inflamm. Bowel Dis. 2018, 24, 2366–2376. [Google Scholar] [CrossRef] [PubMed]
- Ward, S.T.; Li, K.K.; Hepburn, E.; Weston, C.J.; Curbishley, S.M.; Reynolds, G.M.; Hejmadi, R.K.; Bicknell, R.; Eksteen, B.; Ismail, T.; et al. The effects of CCR5 inhibition on regulatory T-cell recruitment to colorectal cancer. Br. J. Cancer 2015, 112, 319–328. [Google Scholar] [CrossRef] [Green Version]
- Plitas, G.; Konopacki, C.; Wu, K.; Bos, P.D.; Morrow, M.; Putintseva, E.V.; Chudakov, D.M.; Rudensky, A.Y. Regulatory T Cells Exhibit Distinct Features in Human Breast Cancer. Immunity 2016, 45, 1122–1134. [Google Scholar] [CrossRef] [Green Version]
- Mishalian, I.; Bayuh, R.; Eruslanov, E.; Michaeli, J.; Levy, L.; Zolotarov, L.; Singhal, S.; Albelda, S.M.; Granot, Z.; Fridlender, Z.G. Neutrophils recruit regulatory T-cells into tumors via secretion of CCL17—A new mechanism of impaired antitumor immunity. Int. J. Cancer 2014, 135, 1178–1186. [Google Scholar] [CrossRef]
- Chen, Z.; Chen, X.; Xu, Y.; Xiong, P.; Fang, M.; Tan, Z.; Gong, F.; Zheng, F. Collagenase digestion down-regulates the density of CD27 on lymphocytes. J. Immunol. Methods 2014, 413, 57–61. [Google Scholar] [CrossRef]
- Liu, Q.; Zhao, R.; Huang, C.; Zhou, K.L.; Zhang, X.L.; Pan, Q. Effects of enzymatic digestion, cell culture and preservation conditions on surface CD62L expression of primary murine CD3+CD4+T cells. Biomed. Res. 2018, 29, 2153–2159. [Google Scholar] [CrossRef] [Green Version]
- Cumba Garcia, L.M.; Huseby Kelcher, A.M.; Malo, C.S.; Johnson, A.J. Superior isolation of antigen-specific brain infiltrating T cells using manual homogenization technique. J. Immunol. Methods 2016, 439, 23–28. [Google Scholar] [CrossRef] [Green Version]
- Benck, C.J.; Martinov, T.; Fife, B.T.; Chatterjea, D. Isolation of infiltrating leukocytes from mouse skin using enzymatic digest and gradient separation. J. Vis. Exp. 2016, 2016, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Leelatian, N.; Doxie, D.B.; Greenplate, A.R.; Mobley, B.C.; Lehman, J.M.; Sinnaeve, J.; Kauffmann, R.M.; Werkhaven, J.A.; Mistry, A.M.; Weaver, K.D.; et al. Single cell analysis of human tissues and solid tumors with mass cytometry. Cytom. Part B-Clin. Cytom. 2017, 92, 68–78. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akimova, T.; Levine, M.H.; Beier, U.H.; Hancock, W.W. Standardization, Evaluation, and Area-Under-Curve Analysis of Human and Murine Treg Suppressive Function. In Suppression and Regulation of Immune Responses: Methods and Protocols; Cuturi, M.C., Anegon, I., Eds.; Springer: New York, NY, USA, 2016; Volume 2, pp. 43–78. ISBN 978-1-4939-3139-2. [Google Scholar]
- Su, S.; Liao, J.; Liu, J.; Huang, D.; He, C.; Chen, F.; Yang, L.B.; Wu, W.; Chen, J.; Lin, L.; et al. Blocking the recruitment of naive CD4+ T cells reverses immunosuppression in breast cancer. Cell Res. 2017, 27, 461–482. [Google Scholar] [CrossRef] [PubMed]
- Kishore, M.; Cheung, K.C.P.; Fu, H.; Bonacina, F.; Wang, G.; Coe, D.; Ward, E.J.; Colamatteo, A.; Jangani, M.; Baragetti, A.; et al. Regulatory T Cell Migration Is Dependent on Glucokinase-Mediated Glycolysis. Immunity 2017, 47, 875–889. [Google Scholar] [CrossRef] [PubMed]
- Zitzer, N.C.; Snyder, K.; Meng, X.; Taylor, P.A.; Efebera, Y.A.; Devine, S.M.; Blazar, B.R.; Garzon, R.; Ranganathan, P. MicroRNA-155 Modulates Acute Graft-versus-Host Disease by Impacting T Cell Expansion, Migration, and Effector Function. J. Immunol. 2018, 200, 4170–4179. [Google Scholar] [CrossRef]
- Hoeppli, R.E.; MacDonald, K.N.; Leclair, P.; Fung, V.C.W.; Mojibian, M.; Gillies, J.; Rahavi, S.M.R.; Campbell, A.I.M.; Gandhi, S.K.; Pesenacker, A.M.; et al. Tailoring the homing capacity of human Tregs for directed migration to sites of Th1-inflammation or intestinal regions. Am. J. Transplant. 2019, 19, 62–76. [Google Scholar] [CrossRef] [Green Version]
- Fletcher, D.A.; Mullins, R.D. Cell mechanics and the cytoskeleton. Nature 2010, 463, 485–492. [Google Scholar] [CrossRef] [Green Version]
- Zhang, N.N.; Chen, J.N.; Xiao, L.; Tang, F.; Zhang, Z.G.; Zhang, Y.W.; Feng, Z.Y.; Jiang, Y.; Shao, C.K. Accumulation Mechanisms of CD4 + CD25 + FOXP3 + Regulatory T Cells in EBV-associated Gastric Carcinoma. Sci. Rep. 2015, 5, 18057. [Google Scholar] [CrossRef] [Green Version]
- Ghaffarian, R.; Muro, S. Models and methods to evaluate transport of drug delivery systems across cellular barriers. J. Vis. Exp. 2013, 7–9. [Google Scholar] [CrossRef]
- Molestina, R.E.; Miller, R.D.; Ramirez, J.A.; Summersgill, J.T. Infection of human endothelial cells with Chlamydia pneumoniae stimulates transendothelial migration of neutrophils and monocytes. Infect. Immun. 1999, 67, 1323–1330. [Google Scholar] [CrossRef] [Green Version]
- Prager, G.W.; Lackner, E.M.; Krauth, M.T.; Unseld, M.; Poettler, M.; Laffer, S.; Cerny-Reiterer, S.; Lamm, W.; Kornek, G.V.; Binder, B.R.; et al. Targeting of VEGF-dependent transendothelial migration of cancer cells by bevacizumab. Mol. Oncol. 2010, 4, 150–160. [Google Scholar] [CrossRef] [Green Version]
- Dejana, E. Endothelial cell-cell junctions: Happy together. Nat. Rev. Mol. Cell Biol. 2004, 5, 261–270. [Google Scholar] [CrossRef] [PubMed]
- Schneider-Hohendorf, T.; Stenner, M.P.; Weidenfeller, C.; Zozulya, A.L.; Simon, O.J.; Schwab, N.; Wiendl, H. Regulatory T cells exhibit enhanced migratory characteristics, a feature impaired in patients with multiple sclerosis. Eur. J. Immunol. 2010, 40, 3581–3590. [Google Scholar] [CrossRef] [PubMed]
- Munro, J.M.; Pober, J.S.; Cotran, R.S. Tumor necrosis factor and interferon-γ induced distinct patterns of endothelial activation and associated leukocyte accumulation in skin of Papio anubis. Am. J. Pathol. 1989, 135, 121–133. [Google Scholar]
- Shaw, S.K.; Perkins, B.N.; Lim, Y.C.; Liu, Y.; Nusrat, A.; Schnell, F.J.; Parkos, C.A.; Luscinskas, F.W. Reduced expression of junctional adhesion molecule and platelet/endothelial cell adhesion molecule-1 (CD31) at human vascular endothelial junctions by cytokines tumor necrosis factor-α plus interferon-γ does not reduce leukocyte transmigration under flow. Am. J. Pathol. 2001, 159, 2281–2291. [Google Scholar] [CrossRef]
- Mohan, K.; Ding, Z.; Hanly, J.; Issekutz, T.B. IFN-γ-Inducible T Cell α Chemoattractant Is a Potent Stimulator of Normal Human Blood T Lymphocyte Transendothelial Migration: Differential Regulation by IFN-γ and TNF-α. J. Immunol. 2002, 168, 6420–6428. [Google Scholar] [CrossRef] [Green Version]
- Fu, H.; Kishore, M.; Gittens, B.; Wang, G.; Coe, D.; Komarowska, I.; Infante, E.; Ridley, A.J.; Cooper, D.; Perretti, M.; et al. Self-recognition of the endothelium enables regulatory T-cell trafficking and defines the kinetics of immune regulation. Nat. Commun. 2014, 5, 3436. [Google Scholar] [CrossRef] [Green Version]
- Ehirchiou, D.; Muller, Y.D.; Chicheportiche, R.; Nobari, R.H.; Madelon, N.; Schneider, M.K.J.; Seebach, J.D. Chemoattractant signals and adhesion molecules promoting human regulatory T cell recruitment to porcine endothelium. Transplantation 2016, 100, 753–762. [Google Scholar] [CrossRef]
- Curiel, T.J.; Coukos, G.; Zou, L.; Alvarez, X.; Cheng, P.; Mottram, P.; Evdemon-Hogan, M.; Conejo-Garcia, J.R.; Zhang, L.; Burow, M.; et al. Specific recruitment of regulatory T cells in ovarian carcinoma fosters immune privilege and predicts reduced survival. Nat. Med. 2004, 10, 942–949. [Google Scholar] [CrossRef]
- Nummer, D.; Suri-Payer, E.; Schmitz-Winnenthal, H.; Bonertz, A.; Galindo, L.; Antolovich, D.; Koch, M.; Büchler, M.; Weitz, J.; Schirrmacher, V.; et al. Role of tumor endothelium in CD4+CD25+ regulatory T cell infiltration of human pancreatic carcinoma. J. Natl. Cancer Inst. 2007, 99, 1188–1199. [Google Scholar] [CrossRef] [Green Version]
- Chen, D.; Bromberg, J.S. T regulatory cells and migration. Am. J. Transplant. 2006, 6, 1518–1523. [Google Scholar] [CrossRef]
- Rigas, D.; Lewis, G.; Aron, J.L.; Wang, B.; Banie, H.; Sankaranarayanan, I.; Galle-Treger, L.; Maazi, H.; Lo, R.; Freeman, G.J.; et al. Type 2 innate lymphoid cell suppression by regulatory T cells attenuates airway hyperreactivity and requires inducible T-cell costimulator–inducible T-cell costimulator ligand interaction. J. Allergy Clin. Immunol. 2017, 139, 1468–1477. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yan, M.; Jene, N.; Byrne, D.; Millar, E.K.A.; O’Toole, S.A.; McNeil, C.M.; Bates, G.J.; Harris, A.L.; Banham, A.H.; Sutherland, R.L.; et al. Recruitment of regulatory T cells is correlated with hypoxia-induced CXCR4 expression, and is associated with poor prognosis in basal-like breast cancers. Breast Cancer Res. 2011, 13, R47. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Facciabene, A.; Peng, X.; Hagemann, I.S.; Balint, K.; Barchetti, A.; Wang, L.P.; Gimotty, P.A.; Gilks, C.B.; Lal, P.; Zhang, L.; et al. Tumour hypoxia promotes tolerance and angiogenesis via CCL28 and T reg cells. Nature 2011, 475, 226–230. [Google Scholar] [CrossRef] [PubMed]
- Sugiyama, D.; Nishikawa, H.; Maeda, Y.; Nishioka, M.; Tanemura, A.; Katayama, I.; Ezoe, S.; Kanakura, Y.; Sato, E.; Fukumori, Y.; et al. Anti-CCR4 mAb selectively depletes effector-Type FoxP3+CD4+ regulatory T cells, evoking antitumor immune responses in humans. Proc. Natl. Acad. Sci. USA 2013, 110, 17945–17950. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, P.; Li, Q.J.; Feng, Y.; Zhang, Y.; Markowitz, G.J.; Ning, S.; Deng, Y.; Zhao, J.; Jiang, S.; Yuan, Y.; et al. TGF-β-miR-34a-CCL22 Signaling-Induced Treg Cell Recruitment Promotes Venous Metastases of HBV-Positive Hepatocellular Carcinoma. Cancer Cell 2012, 22, 291–303. [Google Scholar] [CrossRef] [Green Version]
- Xia, S.; Wei, J.; Wang, J.; Sun, H.; Zheng, W.; Li, Y.; Sun, Y.; Zhao, H.; Zhang, S.; Wen, T.; et al. A requirement of dendritic cell-derived interleukin-27 for the tumor infiltration of regulatory T cells. J. Leukoc. Biol. 2014, 95, 733–742. [Google Scholar] [CrossRef]
- Wiedemann, G.M.; Knott, M.M.L.; Vetter, V.K.; Rapp, M.; Haubner, S.; Fesseler, J.; Kühnemuth, B.; Layritz, P.; Thaler, R.; Kruger, S.; et al. Cancer cell-derived IL-1α induces CCL22 and the recruitment of regulatory T cells. Oncoimmunology 2016, 5, e1175794. [Google Scholar] [CrossRef] [Green Version]
- Cook, K.W.; Letley, D.P.; Ingram, R.J.M.; Staples, E.; Skjoldmose, H.; Atherton, J.C.; Robinson, K. CCL20/CCR6-mediated migration of regulatory T cells to the Helicobacter pylori-infected human gastric mucosa. Gut 2014, 63, 1550–1559. [Google Scholar] [CrossRef] [Green Version]
- Liu, W.; Wei, X.; Li, L.; Wu, X.; Yan, J.; Yang, H.; Song, F. CCR4 mediated chemotaxis of regulatory T cells suppress the activation of T cells and NK cells via TGF-β pathway in human non-small cell lung cancer. Biochem. Biophys. Res. Commun. 2017, 488, 196–203. [Google Scholar] [CrossRef]
- Scharschmidt, T.C.; Vasquez, K.S.; Pauli, M.L.; Leitner, E.G.; Chu, K.; Truong, H.A.; Lowe, M.M.; Sanchez Rodriguez, R.; Ali, N.; Laszik, Z.G.; et al. Commensal Microbes and Hair Follicle Morphogenesis Coordinately Drive Treg Migration into Neonatal Skin. Cell Host Microbe 2017, 21, 467–477. [Google Scholar] [CrossRef] [Green Version]
- Kim, Y.U.; Kim, B.S.; Lim, H.; Wetsel, R.A.; Chung, Y. Enforced expression of CXCR5 drives T follicular regulatory-like features in Foxp3+T cells. Biomol. Ther. 2017, 25, 130–139. [Google Scholar] [CrossRef] [Green Version]
- Geng, S.; Zhong, Y.; Zhou, X.; Zhao, G.; Xie, X.; Pei, Y.; Liu, H.; Zhang, H.; Shi, Y.; Wang, B. Induced regulatory T cells superimpose their suppressive capacity with effector T cells in lymph nodes via antigen-specific S1p1-dependent egress blockage. Front. Immunol. 2017, 8, 663. [Google Scholar] [CrossRef] [Green Version]
- Xiong, Y.; Ahmad, S.; Iwami, D.; Brinkman, C.C.; Bromberg, J.S. T-bet Regulates Natural Regulatory T Cell Afferent Lymphatic Migration and Suppressive Function. J. Immunol. 2016, 196, 2526–2540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wan, Y.Y.; Flavell, R.A. How diverse-CD4 effector T cells and their functions. J. Mol. Cell Biol. 2009, 1, 20–36. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, A.; Oberle, N.; Weiß, E.M.; Vobis, D.; Frischbutter, S.; Baumgrass, R.; Falk, C.S.; Haag, M.; Brügger, B.; Lin, H.; et al. Human regulatory T cells rapidly suppress T cell receptor-induced Ca 2+, NF-κB, and NFAT signaling in conventional T cells. Sci. Signal. 2011, 4, RA90. [Google Scholar] [CrossRef]
- Buckner, J.H. Mechanisms of impaired regulation by CD4+ CD25+ FOXP3+ regulatory T cells in human autoimmune diseases. Nat. Rev. Immunol. 2010, 10, 849–859. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baecher-Allan, C.; Brown, J.A.; Freeman, G.J.; Hafler, D.A. CD4 + CD25 high Regulatory Cells in Human Peripheral Blood. J. Immunol. 2001, 167, 1245–1253. [Google Scholar] [CrossRef] [Green Version]
- Thornton, A.M.; Shevach, E.M. Interleukin 2 Production. J. Exp. Med. 1998, 188, 287–296. [Google Scholar] [CrossRef] [Green Version]
- Sojka, D.K.; Hughson, A.; Sukiennicki, T.L.; Fowell, D.J. Early Kinetic Window of Target T Cell Susceptibility to CD25 + Regulatory T Cell Activity. J. Immunol. 2005, 175, 7274–7280. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, T.; Kuniyasu, Y.; Toda, M.; Sakaguchi, N.; Itoh, M.; Iwata, M.; Shimizu, J.; Sakaguchi, S. Immunologic self-tolerance maintained by CD25 + CD4 + naturally anergic and suppressive T cells: Induction of autoimmune disease by breaking their anergic/suppressive state. Int. Immunol. 1998, 10, 1969–1980. [Google Scholar] [CrossRef] [Green Version]
- Sakaguchi, S.; Wing, K.; Onishi, Y.; Prieto-Martin, P.; Yamaguchi, T. Regulatory T cells: How do they suppress immune responses? Int. Immunol. 2009, 21, 1105–1111. [Google Scholar] [CrossRef] [PubMed]
- Shevach, E.M. Mechanisms of Foxp3+ T Regulatory Cell-Mediated Suppression. Immunity 2009, 30, 636–645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Taams, L.S.; Smith, J.; Rustin, M.H.; Salmon, M.; Poulter, L.W.; Akbar, A.N. Human anergic/suppressive CD4 + CD25 + T cells: A highly differentiated and apoptosis-prone population. Eur. J. Immunol. 2001, 31, 1122–1131. [Google Scholar] [CrossRef]
- Levings, M.K.; Sangregorio, R.; Roncarolo, M.G. Human CD25+CD4+ T regulatory cells suppress naive and memory T cell proliferation and can be expanded in vitro without loss of function. J. Exp. Med. 2001, 193, 1295–1301. [Google Scholar] [CrossRef]
- Mcmurchy, A.N.; Levings, M.K. Suppression assays with human T regulatory cells: A technical guide. Eur. J. Immunol. 2012, 42, 27–34. [Google Scholar] [CrossRef]
- Walker, L.S.K.; Chodos, A.; Eggena, M.; Dooms, H.; Abbas, A.K. Antigen-dependent proliferation of CD4+ CD25+ regulatory T cells in vivo. J. Exp. Med. 2003, 198, 249–258. [Google Scholar] [CrossRef]
- Schneider, A.; Buckner, J.H. Assessment of Suppressive Capacity by Human Regulatory T Cells Using a Reproducible, Bi-Directional CFSE-Based In Vitro Assay BT—Regulatory T Cells: Methods and Protocols; Kassiotis, G., Liston, A., Eds.; Humana Press: Totowa, NJ, USA, 2011; pp. 233–241. ISBN 978-1-61737-979-6. [Google Scholar]
| Transwell Type (T) & Pore Size (S) | Number (N) & Volume (V)† of Cells in Transwell | Concentration (C) & Volume (V)† of Chemoattractant in Receiver Well | Incubation Condition | Enumeration of Migrated Cells | Reference |
|---|---|---|---|---|---|
| T: 24-well S: 5 µm | N: 5 × 104 V: NS | C: 100 ng/mLV: NS | 37 °C, 4 h | Hemocytometer | [58] |
| T: 24-well* S: 5 µm* | N: 5 × 105 *V: NS | C: 20 ng/mL V: NS | NS, 6 h | Flow cytometer | [34] |
| T: NS S: 3 µm | N: 1 × 106 V: NS | C: NS V: NS | 37 °C, 6 h | Flow cytometer | [59] |
| T: 96-well S: 5 µm | N: 1 × 105 V: 100 µL | C: 500 ng/mL V: 150 µl | NS, 5 h | Flow cytometer | [60] |
| T: 24-well S: 5 µm | N: 1 × 106 V: NS | C: 1 ng/mL V: 500 µL | 37 °C, 4 h | Flow cytometer | [61] |
| T: 24-well S: 5 µm* | N: 2.5 × 105 V: 100 µL | C: vary V: 600 µL | 37 °C, 3 h | Flow cytometer | [62] |
| T: NS S: 5 µm | N: 3 × 105 V: 100 µL | C: vary V: 600 µL | 37oC, 4 h | Flow cytometer | [63] |
| T: 96-well S: NS | N: 1 × 105 V: NS | C: 20 nM V: NS | 37 °C, 4 h | Flow cytometer | [64] |
| T: NS S: 5 µm | N: 1 × 105 V: 300 µl | C: 200 ng/mL V: NS | NS, Vary | Hemocytometer | [35] |
| T: 24-well S: 8 µm | N: NS V: NS | C: 100 ng/mL V: NS | 37 °C, 3 h | Flow cytometer | [26] |
| T: 24-well S: 5 µm* | N: 2 × 105 V: 100 µL* | C: vary V: 600 µL* | NS, 4 h | Hemocytometer | [65] |
| T: 96-well S: 5 µm | N: 1 × 105 V: 50 µL* | C: vary* V: 150 µL* | 37 °C, 90 min | Flow cytometer | [37] |
| T: 96-well S: NS | N: NS V: NS | C: 20 ng/mL V: NS | 37 °C, 5% CO2 4 h | Flow cytometer | [25] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Irekeola, A.A.; E. A. R., E.N.S.; Mat Lazim, N.; Mohamud, R.; Yean, C.Y.; Shueb, R.H. Technical Considerations in Ex Vivo Human Regulatory T Cell Migration and Suppression Assays. Cells 2020, 9, 487. https://doi.org/10.3390/cells9020487
Irekeola AA, E. A. R. ENS, Mat Lazim N, Mohamud R, Yean CY, Shueb RH. Technical Considerations in Ex Vivo Human Regulatory T Cell Migration and Suppression Assays. Cells. 2020; 9(2):487. https://doi.org/10.3390/cells9020487
Chicago/Turabian StyleIrekeola, Ahmad Adebayo, Engku Nur Syafirah E. A. R., Norhafiza Mat Lazim, Rohimah Mohamud, Chan Yean Yean, and Rafidah Hanim Shueb. 2020. "Technical Considerations in Ex Vivo Human Regulatory T Cell Migration and Suppression Assays" Cells 9, no. 2: 487. https://doi.org/10.3390/cells9020487






