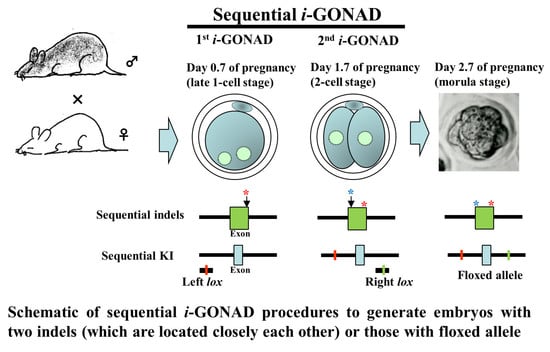
Sequential i-GONAD: An Improved In Vivo Technique for CRISPR/Cas9-Based Genetic Manipulations in Mice
Abstract
:1. Introduction
2. Materials and Methods
2.1. Mouse and Superovulation Induction
2.2. Preparation of Reagents Used for i-GONAD, si-GONAD, and simi-GONAD
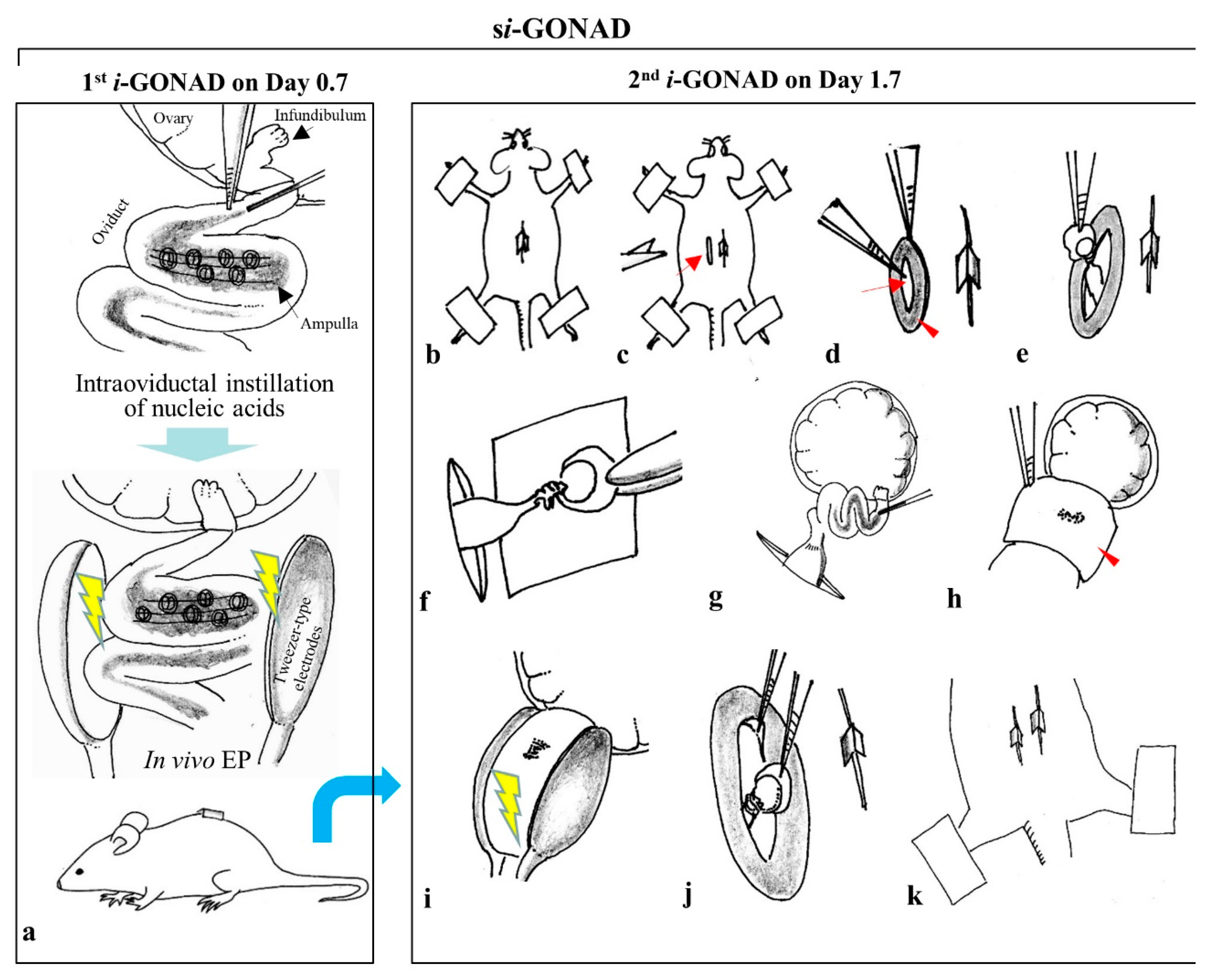
2.3. si-GONAD, simi-GONAD, and i-GONAD
2.4. Embryo Collection, ZP Removal, and Staining with Fluorescence-Labeled Lectin
2.5. Fluorescence Imaging
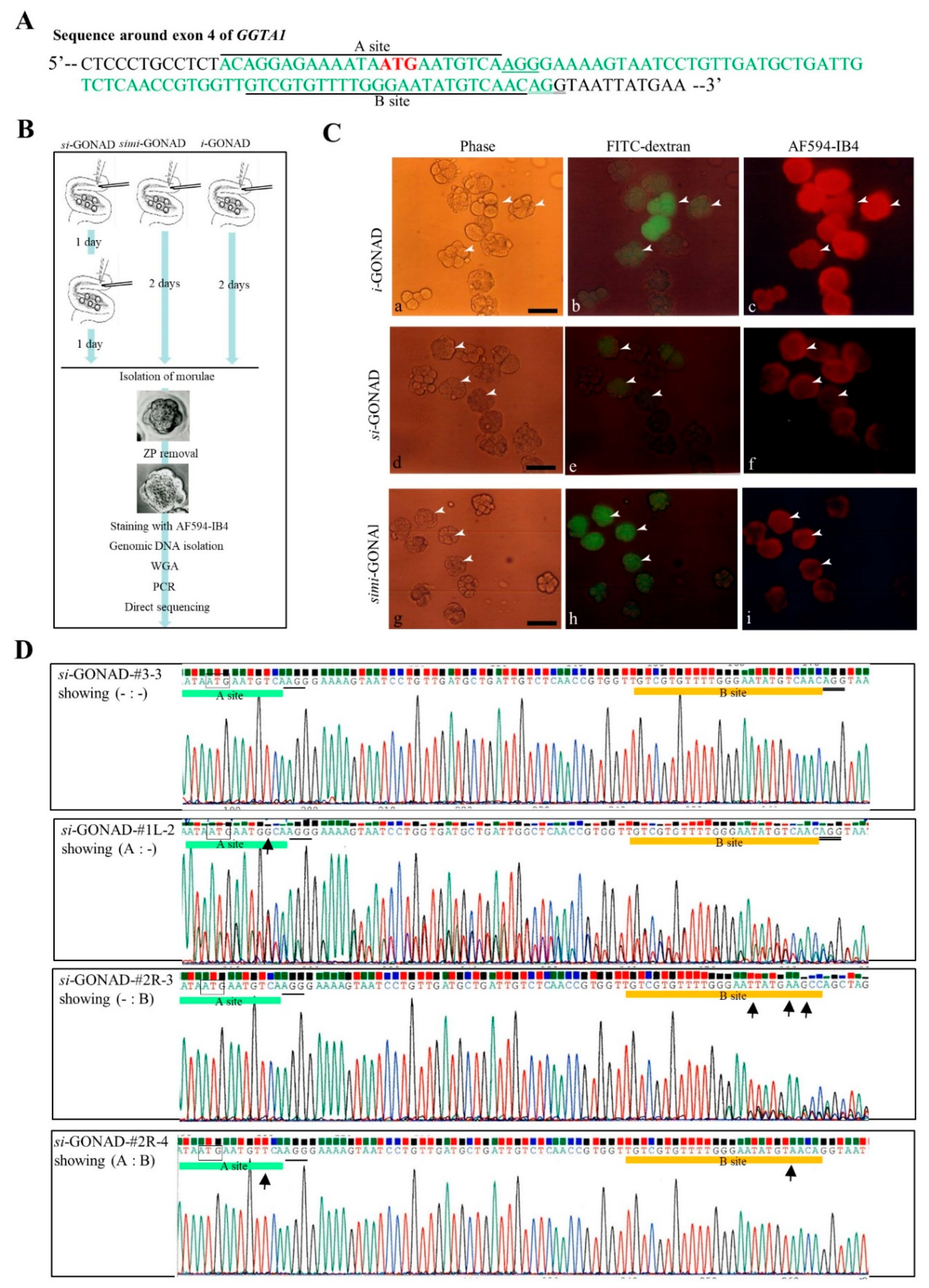
2.6. Single Embryo Analysis
2.7. Assay for the Floxed Allele
3. Results
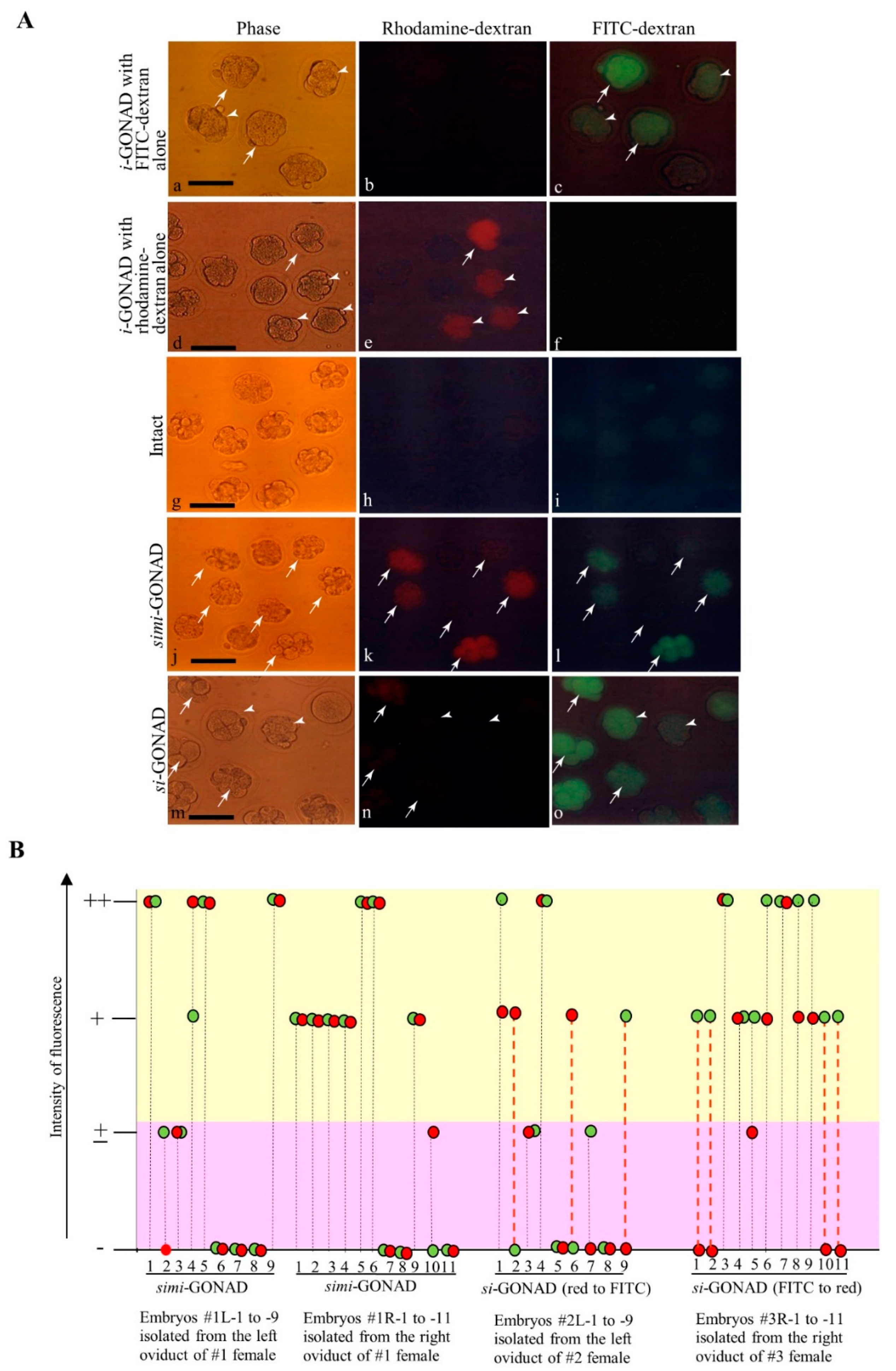
3.1. Experiment 1: si-GONAD Using Two Fluorescent Dextrans
3.2. Experiment 2: Induction of Indels in the Two Sites (Which Are Located Close to Each Other) of Mouse GGTA1 (exon 4) by si-GONAD
3.3. Experiment 3: si-GONAD-Mediated KI of Mutated lox Sites into the Intronic Sequences Interposing Exon 3 of Mouse Mecp2 Gene
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| AF594 | Alexa Fluor 594 |
| α-GalT | α-1,3-galactosyltransferase |
| CRISPR/Cas9 | Clustered regularly interspaced palindrome repeats (CRISPR)/Caspase 9 (Cas9) |
| crRNA | CRISPR RNA |
| EP | Electroporation |
| FBS | Fetal bovine serum |
| FITC | Fluorescein isothiocyanate |
| gRNA | Guide RNA |
| GOI | Gene of interest |
| HDR | Homology direct repair |
| IB4 | Isolectin BS-I-B4 lectin |
| i-GONAD | Improved genome-editing via oviductal nucleic acid delivery |
| indels | Insertion and deletion mutations |
| IP | Intraperitoneally |
| IVF | In vitro fertilization |
| KI | Knock-in |
| LD | Large deletion |
| NHEJ | Non-homologous end joining |
| ODNs | Oligodeoxynucleotides |
| PAM | Protospacer adjacent motif |
| PBS | Dulbecco’s modified Ca2+, Mg2+-free phosphate-buffered saline |
| PCR | Polymerase chain reaction |
| Pp | Poring pulse |
| rAAV-6 | Recombinant adeno-associated virus serotype 6 |
| RNP | Ribonucleoprotein |
| SCNT | Somatic cell nuclear transfer |
| si-GONAD | Sequential i-GONAD |
| simi-GONAD | Simultaneous i-GONAD |
| ss | Single-stranded |
| Tp | Transfer pulse |
| tracrRNA | Trans-activating small RNA |
| WGA | Whole genome amplification |
| ZP | Zona pellucida |
References
- Harrison, M.M.; Jenkins, B.V.; O’Connor-Giles, K.M.; Wildonger, J.A. CRISPR view of development. Genes Dev. 2014, 28, 1859–1872. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hsu, P.D.; Lander, E.S.; Zhang, F. Development and applications of CRISPR-Cas9 for genome engineering. Cell 2014, 157, 1262–1278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Qiu, Z.; Shao, Y.; Chen, Y.; Guan, Y.; Liu, M.; Li, Y.; Gao, N.; Wang, L.; Lu, X.; et al. Heritable gene targeting in the mouse and rat using a CRISPR-Cas system. Nat. Biotechnol. 2013, 31, 681–683. [Google Scholar] [CrossRef]
- Wang, H.; Yang, H.; Shivalila, C.S.; Dawlaty, M.M.; Cheng, A.W.; Zhang, F.; Jaenisch, R. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell 2013, 153, 910–918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, H.; Wang, H.; Shivalila, C.S.; Cheng, A.W.; Shi, L.; Jaenisch, R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell 2013, 154, 1370–1379. [Google Scholar] [CrossRef] [Green Version]
- Mashiko, D.; Fujihara, Y.; Satouh, Y.; Miyata, H.; Isotani, A.; Ikawa, M. Generation of mutant mice by pronuclear injection of circular plasmid expressing Cas9 and single guided RNA. Sci. Rep. 2013, 3, 3355. [Google Scholar] [CrossRef]
- Fujii, W.; Kawasaki, K.; Sugiura, K.; Naito, K. Efficient generation of large-scale genome-modified mice using gRNA and CAS9 endonuclease. Nucleic Acids Res. 2013, 41, e187. [Google Scholar] [CrossRef]
- Shen, B.; Zhang, J.; Wu, H.; Wang, J.; Ma, K.; Li, Z.; Zhang, X.; Zhang, P.; Huang, X. Generation of gene-modified mice via Cas9/RNA-mediated gene targeting. Cell Res. 2013, 23, 720–723. [Google Scholar] [CrossRef]
- Horii, T.; Arai, Y.; Yamazaki, M.; Morita, S.; Kimura, M.; Itoh, M.; Abe, Y.; Hatada, I. Validation of microinjection methods for generating knockout mice by CRISPR/Cas-mediated genome engineering. Sci. Rep. 2014, 4, 4513. [Google Scholar] [CrossRef]
- Kaneko, T.; Sakuma, T.; Yamamoto, T.; Mashimo, T. Simple knockout by electroporation of engineered endonucleases into intact rat embryos. Sci. Rep. 2014, 4, 6382. [Google Scholar] [CrossRef] [Green Version]
- Hashimoto, M.; Takemoto, T. Electroporation enables the efficient mRNA delivery into the mouse zygotes and facilitates CRISPR/Cas9-based genome editing. Sci. Rep. 2015, 5, 11315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, W.; Dion, S.L.; Kutny, P.M.; Zhang, Y.; Cheng, A.W.; Jillette, N.L.; Malhotra, A.; Geurts, A.M.; Chen, Y.G.; Wang, H. Efficient CRISPR/Cas9-mediated genome editing in mice by zygote electroporation of nuclease. Genetics 2015, 200, 423–430. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, S.; Lee, B.; Lee, A.Y.; Modzelewski, A.J.; He, L. Highly efficient mouse genome editing by CRISPR ribonucleoprotein electroporation of zygotes. J. Biol. Chem. 2016, 291, 14457–14467. [Google Scholar] [CrossRef] [Green Version]
- Tröder, S.E.; Ebert, L.K.; Butt, L.; Assenmacher, S.; Schermer, S.B.; Zevnik, B. An optimized electroporation approach for efficient CRISPR/Cas9 genome editing in murine zygotes. PLoS ONE 2018, 13, e0196891. [Google Scholar] [CrossRef] [Green Version]
- Teixeira, M.; Py, B.F.; Bosc, C.; Laubreton, D.; Moutin, M.J.; Marvel, J.; Flamant, F.; Markossian, S. Electroporation of mice zygotes with dual guide RNA/Cas9 complexes for simple and efficient cloning-free genome editing. Sci. Rep. 2018, 8, 474. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yoon, Y.; Wang, D.; Tai, P.W.L.; Riley, J.; Gao, G.; Rivera-Pérez, J.A. Streamlined ex vivo and in vivo genome editing in mouse embryos using recombinant adeno-associated viruses. Nat. Commun. 2018, 9, 412. [Google Scholar] [CrossRef]
- Mizuno, N.; Mizutani, E.; Sato, H.; Kasai, M.; Ogawa, A.; Suchy, F.; Yamaguchi, T.; Nakauchi, H. Intra-embryo gene cassette knockin by CRISPR/Cas9-mediated genome editing with adeno-associated viral vector. iScience 2018, 9, 286–297. [Google Scholar] [CrossRef] [Green Version]
- Takahashi, G.; Gurumurthy, C.B.; Wada, K.; Miura, H.; Sato, M.; Ohtsuka, M. GONAD: Genome-editing via Oviductal Nucleic Acids Delivery system: A novel microinjection independent genome engineering method in mice. Sci. Rep. 2015, 5, 11406. [Google Scholar] [CrossRef] [Green Version]
- Ohtsuka, M.; Sato, M.; Miura, H.; Takabayashi, S.; Matsuyama, M.; Koyano, T.; Arifin, N.; Nakamura, S.; Wada, K.; Gurumurthy, C.B. i-GONAD: A robust method for in situ germline genome engineering using CRISPR nucleases. Genome Biol. 2018, 19, 25. [Google Scholar] [CrossRef] [Green Version]
- Kobayashi, T.; Namba, M.; Koyano, T.; Fukushima, M.; Sato, M.; Ohtsuka, M.; Matsuyama, M. Successful production of genome-edited rats by the rGONAD method. BMC Biotechnol. 2018, 18, 19. [Google Scholar] [CrossRef] [Green Version]
- Takabayashi, S.; Aoshima, T.; Kabashima, K.; Aoto, K.; Ohtsuka, M.; Sato, M. i-GONAD (improved genome-editing via oviductal nucleic acids delivery), a convenient in vivo tool to produce genome-edited rats. Sci. Rep. 2018, 8, 12059. [Google Scholar] [CrossRef] [PubMed]
- Horii, T.; Morita, S.; Kimura, M.; Terawaki, N.; Shibutani, M.; Hatada, I. Efficient generation of conditional knockout mice via sequential introduction of lox sites. Sci. Rep. 2017, 7, 7891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, M.; Ohtsuka, M.; Nakamura, S. Intraoviductal instillation of a solution as an effective route for manipulating preimplantation mammalian embryos in vivo. In New Insights into Theriogenology; Payan-Carreira, R., Ed.; InTechOpen: Rijeka, Croatia, 2018; pp. 135–150. [Google Scholar] [CrossRef] [Green Version]
- Gurumurthy, C.B.; Sato, M.; Nakamura, A.; Inui, M.; Kawano, N.; Islam, M.; Ogiwara, S.; Takabayashi, S.; Matsuyama, M.; Nakagawa, S.; et al. Creation of CRISPR-based germline-genome-engineered mice without ex vivo handling of zygotes by i-GONAD. Nat. Protoc. 2019, 14, 2452–2482. [Google Scholar] [CrossRef]
- Ohtsuka, M.; Sato, M. i-GONAD: A method for generating genome-edited animals without ex vivo handling of embryos. Dev. Growth Differ. 2019, 61, 306–315. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sato, M.; Akasaka, E.; Saitoh, I.; Ohtsuka, M.; Nakamura, S.; Sakurai, T.; Watanabe, S. Targeted toxin-based selectable drug-free enrichment of mammalian cells with high transgene expression. Biology 2013, 2, 341–355. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Akasaka, E.; Ozawa, A.; Mori, H.; Mizobe, Y.; Yoshida, M.; Miyoshi, K.; Sato, M. Whole-genome amplification-based GenomiPhi for multiple genomic analysis of individual early porcine embryos. Theriogenology 2011, 75, 1543–1549. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Koriyama, M.; Watanabe, S.; Ohtsuka, M.; Sakurai, T.; Inada, E.; Saitoh, I.; Nakamura, S.; Miyoshi, K. Direct injection of CRISPR/Cas9-related mRNA into cytoplasm of parthenogenetically activated porcine oocytes causes frequent mosaicism for indel mutations. Int. J. Mol. Sci. 2015, 16, 17838–17856. [Google Scholar] [CrossRef] [Green Version]
- Vaughan, H.A.; Loveland, B.E.; Sandrin, M.S. Gal(α1-3)Gal is the major xenoepitope expressed on pig endothelial cells recognized by naturally occurring cytotoxic human antibodies. Transplantation 1994, 58, 879–882. [Google Scholar] [CrossRef]
- Akasaka, E.; Watanabe, S.; Himaki, T.; Ohtsuka, M.; Yoshida, M.; Miyoshi, K.; Sato, M. Enrichment of xenograft-competent genetically modified pig cells using a targeted toxin, isolectin BS-I-B4 conjugate. Xenotransplantation 2010, 17, 81–89. [Google Scholar] [CrossRef] [PubMed]
- Sato, M.; Miyoshi, K.; Nagao, Y.; Nishi, Y.; Ohtsuka, M.; Nakamura, S.; Sakurai, T.; Watanabe, S. The combinational use of CRISPR/Cas9-based gene editing and targeted toxin technology enables efficient biallelic knockout of the α-1,3-galactosyltransferase gene in porcine embryonic fibroblasts. Xenotransplantation 2014, 21, 291–300. [Google Scholar] [CrossRef]
- Sato, M.; Miyoshi, M.; Nakamura, S.; Ohtsuka, M.; Sakurai, T.; Watanabe, S.; Kawaguchi, H.; Tanimoto, A. Efficient generation of somatic cell nuclear transfer-competent porcine cells with mutated alleles at multiple target loci by using CRISPR/Cas9 combined with targeted toxin-based selection system. Int. J. Mol. Sci. 2017, 18, 2610. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chi, H.; Sato, M.; Yoshida, M.; Miyoshi, K. Expression analysis of a α-1, 3-galactosyltransferase, an enzyme that creates xenotransplantation-related α-Gal epitope, in pig preimplantation embryos. Anim. Sci. J. 2012, 83, 88–93. [Google Scholar] [CrossRef] [PubMed]
- Maruyama, T.; Dougan, S.K.; Truttmann, M.C.; Bilate, A.M.; Ingram, J.R.; Ploegh, H.L. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat. Biotechnol. 2015, 33, 538–542. [Google Scholar] [CrossRef] [PubMed]

| Method 1 | Name of Oviducts Examined 2 | Rate of Normal Morulae (%) 3 | Total No. of Morulae Examined | Mode of Mutations 4 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (A : -) (%) | (- : B) (%) | (A : B) (%) | LD (%) | Complex (%) | (- : -) (%) | ||||
| si-GONAD | #1L | 11/11 (100) | 11 | 1 (9) | 5 (45) | 1 (9) | 0 (0) | 2 (18) | 2 (18) |
| #2R | 11/13 (85) | 11 | 0 (0) | 5 (45) | 4 (36) | 1 (9) | 1 (9) | 0 (0) | |
| #3R | 6/6 (100) | 6 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 1 (17) | 5 (83) | |
| Total | 28/30 (93) | 28 | 1 (4) | 10 (36) | 5 (18) | 1 (4) | 4 (14) | 7 (25) | |
| simi-GONAD | #1L | 10/12 (83) | 10 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 2 (20) | 8 (80) |
| #1R | 12/12 (100) | 12 | 0 (0) | 2 (17) | 0 (0) | 0 (0) | 2 (17) | 8 (67) | |
| Total | 22/24 (92) | 22 | 0 (0) | 2 (9) | 0 (0) | 0 (0) | 4 (18) | 16 (73) | |
| i-GONAD | #1R | 10/11 (91) | 10 | 0 (0) | 8 (80) | 0 (0) | 1 (10) | 1 (10) | 0 (0) |
| Method 1 | No. Oviducts Examined 2 | Rate of Normal Morulae (%) 3 | Total no. of Morulae Examined | Mode of KI 5 | |||||
|---|---|---|---|---|---|---|---|---|---|
| (KI : Indels) (%) | (Indels : -) (%) | (- : Indels) (%) | (Indels : Indels) (%) | LD (%) | (- : -) (%) | ||||
| si-GONAD | #1R | 10/11 (91) | 10 | 0 (0) | 1 (10) | 2 (20) | 1 (10) | 0 (0) | 6 (60) |
| #1L | 3/3 (100) | 3 4 | 1 (33) | 0 (0) | 0 (0) | 1 (33) | 1 (33) | 0 (0) | |
| #2L | 8/9 (89) | 8 | 0 (0) | 0 (0) | 3 (38) | 1 (13) | 0 (0) | 4 (50) | |
| #2R | 1/1 (100) | 1 | 0 (0) | 1 (100) | 0 (0) | 0 (0) | 0 (0) | 0 (0) | |
| Total | 22/24 (92) | 22 | 1 (5) | 2 (9) | 5 (23) | 3 (14) | 1 (5) | 10 (45) | |
| simi-GONAD | #3L | 7/9 (78) | 7 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 6 (86) | 1 (14) |
| #3R | 17/18 (94) | 17 | 0 (0) | 2 (12) | 0 (0) | 1 (6) | 14 (82) | 0 (0) | |
| #4L | 4/4 (100) | 4 | 0 (0) | 0 (0) | 0 (0) | 0 (0) | 3 (75) | 1 (25) | |
| Total | 28/31 (90) | 28 | 0 (0) | 2 (7) | 0 (0) | 1 (4) | 23 (82) | 1 (4) |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, M.; Miyagasako, R.; Takabayashi, S.; Ohtsuka, M.; Hatada, I.; Horii, T. Sequential i-GONAD: An Improved In Vivo Technique for CRISPR/Cas9-Based Genetic Manipulations in Mice. Cells 2020, 9, 546. https://doi.org/10.3390/cells9030546
Sato M, Miyagasako R, Takabayashi S, Ohtsuka M, Hatada I, Horii T. Sequential i-GONAD: An Improved In Vivo Technique for CRISPR/Cas9-Based Genetic Manipulations in Mice. Cells. 2020; 9(3):546. https://doi.org/10.3390/cells9030546
Chicago/Turabian StyleSato, Masahiro, Rico Miyagasako, Shuji Takabayashi, Masato Ohtsuka, Izuho Hatada, and Takuro Horii. 2020. "Sequential i-GONAD: An Improved In Vivo Technique for CRISPR/Cas9-Based Genetic Manipulations in Mice" Cells 9, no. 3: 546. https://doi.org/10.3390/cells9030546
APA StyleSato, M., Miyagasako, R., Takabayashi, S., Ohtsuka, M., Hatada, I., & Horii, T. (2020). Sequential i-GONAD: An Improved In Vivo Technique for CRISPR/Cas9-Based Genetic Manipulations in Mice. Cells, 9(3), 546. https://doi.org/10.3390/cells9030546