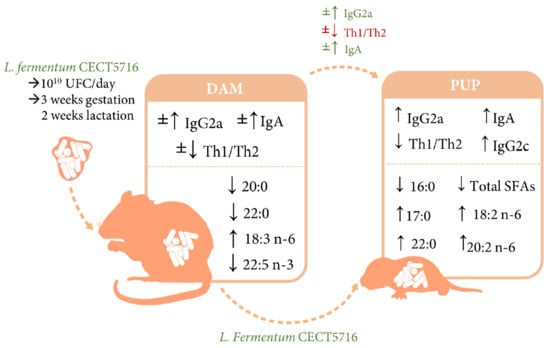
Lactobacillus fermentum CECT5716 Supplementation in Rats during Pregnancy and Lactation Impacts Maternal and Offspring Lipid Profile, Immune System and Microbiota
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Experimental Design
2.3. Extraction, Methylation, and Quantification of FA
2.4. Quantification of Immunoglobulins and Cytokines
2.5. Analysis of Microbiota Composition
2.6. Phagocytic Activity of Blood Monocytes and Granulocytes
2.7. Lymphocyte Isolation from Mesenteric Lymph Nodes (MLNs)
2.8. Immunofluorescence Staining and Phenotype of MLN Lymphocytes
2.9. Proliferative Response of MLN Lymphocytes
2.10. Quantification of Immunoglobulins in Different Compartments
2.11. Detection of Lactobacillus fermentum CECT5716
2.12. Statistical Analysis
3. Results
3.1. Transmission of Lactobacillus fermentum CECT5716
3.2. Growth, Morphometrics
3.3. Impact on the Dams’ Immune System
3.4. Impact on the Ig and Cytokines of Dams and Pups
3.5. Impact on the Plasma FA Profile of Dams and Pups
3.6. Impact on the Cecal Microbiota Composition of Dams and Pups
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Kominiarek, M.A.; Rajan, P. Nutrition recommendations in pregnancy and lactation. Med. Clin. N. Am. 2016, 100, 1199–1215. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chu, D.M.; Meyer, K.M.; Prince, A.L.; Aagaard, K.M. Impact of maternal nutrition in pregnancy and lactation on offspring gut microbial composition and function. Gut Microbes 2016, 7, 459–470. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mueller, N.T.; Bakacs, E.; Combellick, J.; Grigoryan, Z.; Dominguez-Bello, M.G. The infant microbiome development: mom matters. Trends Mol. Med. 2015, 21, 109–117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sohn, K.; Underwood, M.A. Prenatal and postnatal administration of prebiotics and probiotics. Semin. Fetal Neonatal Med. 2017, 22, 284–289. [Google Scholar] [CrossRef] [PubMed]
- Edwards, S.M.; Cunningham, S.A.; Dunlop, A.L.; Corwin, E.J. The maternal gut microbiome during pregnancy. MCN Am. J. Matern. Child. Nurs. 2017, 42, 310–316. [Google Scholar] [CrossRef]
- Rodríguez, J.M.; Murphy, K.; Stanton, C.; Ross, R.P.; Kober, O.I.; Juge, N.; Avershina, E.; Rudi, K.; Narbad, A.; Jenmalm, M.C.; et al. The composition of the gut microbiota throughout life, with an emphasis on early life. Microb. Ecol. Heal. Dis. 2015, 26, 26050. [Google Scholar] [CrossRef]
- Collado, M.C.; Rautava, S.; Aakko, J.; Isolauri, E.; Salminen, S. Human gut colonisation may be initiated in utero by distinct microbial communities in the placenta and amniotic fluid. Sci. Rep. 2016, 6, 23129. [Google Scholar] [CrossRef] [Green Version]
- Jiménez, E.; Marín, M.L.; Martín, R.; Odriozola, J.M.; Olivares, M.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Is meconium from healthy newborns actually sterile? Res. Microbiol. 2008, 159, 187–193. [Google Scholar] [CrossRef]
- Swartwout, B.; Luo, X.M. Implications of probiotics on the maternal-neonatal interface: Gut microbiota, immunomodulation, and autoimmunity. Front. Immunol. 2018, 9, 2840. [Google Scholar] [CrossRef] [Green Version]
- DiGiulio, D.B.; Romero, R.; Amogan, H.P.; Kusanovic, J.P.; Bik, E.M.; Gotsch, F.; Kim, C.J.; Erez, O.; Edwin, S.; Relman, D.A. Microbial prevalence, diversity and abundance in amniotic fluid during preterm labor: A molecular and culture-based investigation. PLoS ONE 2008, 3, e3056. [Google Scholar] [CrossRef]
- Song, S.J.; Dominguez-Bello, M.G.; Knight, R. How delivery mode and feeding can shape the bacterial community in the infant gut. CMAJ 2013, 185, 373–374. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sirilun, S.; Takahashi, H.; Boonyaritichaikij, S.; Chaiyasut, C.; Lertruangpanya, P.; Koga, Y.; Mikami, K. Impact of maternal bifidobacteria and the mode of delivery on Bifidobacterium microbiota in infants. Benef. Microbes 2015, 6, 767–774. [Google Scholar] [CrossRef] [PubMed]
- Lundgren, S.N.; Madan, J.C.; Emond, J.A.; Morrison, H.G.; Christensen, B.C.; Karagas, M.R.; Hoen, A.G. Maternal diet during pregnancy is related with the infant stool microbiome in a delivery mode-dependent manner. Microbiome 2018, 6, 109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gotoh, A.; Katoh, T.; Sakanaka, M.; Ling, Y.; Yamada, C.; Asakuma, S.; Urashima, T.; Tomabechi, Y.; Katayama-Ikegami, A.; Kurihara, S.; et al. Sharing of human milk oligosaccharides degradants within bifidobacterial communities in faecal cultures supplemented with Bifidobacterium bifidum. Sci. Rep. 2018, 8, 13958. [Google Scholar] [CrossRef] [PubMed]
- Rodríguez, J.M. The origin of human milk bacteria: Is there a bacterial entero-mammary pathway during late pregnancy and lactation? Adv. Nutr. 2014, 5, 779–784. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fernández, L.; Langa, S.; Martín, V.; Maldonado, A.; Jiménez, E.; Martín, R.; Rodríguez, J.M. The human milk microbiota: origin and potential roles in health and disease. Pharmacol. Res. 2013, 69, 1–10. [Google Scholar] [CrossRef]
- Jeurink, P.V.; van Bergenhenegouwen, J.; Jiménez, E.; Knippels, L.M.J.; Fernández, L.; Garssen, J.; Knol, J.; Rodríguez, J.M.; Martín, R. Human milk: A source of more life than we imagine. Benef. Microbes 2013, 4, 17–30. [Google Scholar] [CrossRef]
- Gregory, K.E.; Walker, W.A. Immunologic factors in human milk and disease prevention in the preterm infant. Curr. Pediatr. Rep. 2013, 1, 222–228. [Google Scholar] [CrossRef] [Green Version]
- Ganapathy, S. Long chain polyunsaturated fatty acids and immunity in infants. Indian Pediatr. 2009, 46, 785–790. [Google Scholar]
- Cacho, N.T.; Lawrence, R.M. Innate immunity and breast milk. Front. Immunol. 2017, 8, 584. [Google Scholar] [CrossRef] [Green Version]
- Simon, A.K.; Hollander, G.A.; Mcmichael, A. Evolution of the immune system in humans from infancy to old age. Proc. R. Soc. B 2015, 282, 20143085. [Google Scholar] [CrossRef]
- Wickens, K.; Stanley, T.V.; Mitchell, E.A.; Barthow, C.; Fitzharris, P.; Purdie, G.; Siebers, R.; Black, P.N.; Crane, J. Early supplementation with Lactobacillus rhamnosus HN001 reduces eczema prevalence to 6 years: Does it also reduce atopic sensitization? Clin. Exp. Allergy 2013, 43, 1048–1057. [Google Scholar] [CrossRef] [PubMed]
- Rø, A.D.B.; Simpson, M.R.; Rø, T.B.; Storrø, O.; Johnsen, R.; Videm, V.; Øien, T. Reduced Th22 cell proportion and prevention of atopic dermatitis in infants following maternal probiotic supplementation. Clin. Exp. Allergy 2017, 47, 1014–1021. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rautava, S.; Kalliomäki, M.; Isolauri, E. Probiotics during pregnancy and breast-feeding might confer immunomodulatory protection against atopic disease in the infant. J. Allergy Clin. Immunol. 2002, 109, 119–121. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rautava, S.; Kainonen, E.; Salminen, S.; Isolauri, E. Maternal probiotic supplementation during pregnancy and breast-feeding reduces the risk of eczema in the infant. J. Allergy Clin. Immunol. 2012, 130, 1355–1360. [Google Scholar] [CrossRef] [PubMed]
- Dotterud, C.K.; Storrø, O.; Johnsen, R.; Øien, T. Probiotics in pregnant women to prevent allergic disease: A randomized, double-blind trial. Br. J. Dermatol. 2010, 163, 616–623. [Google Scholar] [CrossRef] [PubMed]
- Barthow, C.; Wickens, K.; Stanley, T.; Mitchell, E.A.; Maude, R.; Abels, P.; Purdie, G.; Murphy, R.; Stone, P.; Kang, J.; et al. The Probiotics in Pregnancy Study (PiP Study): Rationale and design of a double-blind randomised controlled trial to improve maternal health during pregnancy and prevent infant eczema and allergy. BMC Pregnancy Childbirth 2016, 16, 133. [Google Scholar] [CrossRef] [Green Version]
- Azad, M.B.; Coneys, G.J.; Kozyrskyj, A.L.; Field, C.J.; Ramsey, C.D.; Becker, A.B.; Friesen, C.; Abou-Setta, A.M.; Zarychanski, R. Probiotic supplementation during pregnancy or infancy for the prevention of asthma and wheeze: systematic review and meta-analysis. BMJ 2013, 347, f6471. [Google Scholar] [CrossRef] [Green Version]
- Fiocchi, A.; Pawankar, R.; Cuello-Garcia, C.; Ahn, K.; Al-Hammadi, S.; Agarwal, A.; Beyer, K.; Burks, W.; Canonica, G.W.; Ebisawa, M.; et al. World Allergy Organization-McMaster University Guidelines for Allergic Disease Prevention (GLAD-P): Probiotics. World Allergy Organ. J. 2015, 8, 4. [Google Scholar] [CrossRef] [Green Version]
- Laitinen, K.; Poussa, T.; Isolauri, E. Probiotics and dietary counselling contribute to glucose regulation during and after pregnancy: A randomised controlled trial. Br. J. Nutr. 2009, 101, 1679–1687. [Google Scholar] [CrossRef] [Green Version]
- Luoto, R.; Laitinen, K.; Nermes, M.; Isolauri, E. Impact of maternal probiotic-supplemented dietary counselling on pregnancy outcome and prenatal and postnatal growth: A double-blind, placebo-controlled study. Br. J. Nutr. 2010, 103, 1792–1799. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wickens, K.L.; Barthow, C.A.; Murphy, R.; Abels, P.R.; Maude, R.M.; Stone, P.R.; Mitchell, E.A.; Stanley, T.V.; Purdie, G.L.; Kang, J.M.; et al. Early pregnancy probiotic supplementation with Lactobacillus rhamnosus HN001 may reduce the prevalence of gestational diabetes mellitus: A randomised controlled trial. Br. J. Nutr. 2017, 117, 804–813. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Anukam, K.C.; Osazuwa, E.; Osemene, G.I.; Ehigiagbe, F.; Bruce, A.W.; Reid, G. Clinical study comparing probiotic Lactobacillus GR-1 and RC-14 with metronidazole vaginal gel to treat symptomatic bacterial vaginosis. Microbes Infect. 2006, 8, 2772–2776. [Google Scholar] [CrossRef] [PubMed]
- Larsson, P.-G.; Stray-Pedersen, B.; Ryttig, K.R.; Larsen, S. Human lactobacilli as supplementation of clindamycin to patients with bacterial vaginosis reduce the recurrence rate; a 6-month, double-blind, randomized, placebo-controlled study. BMC Womens. Health 2008, 8, 3. [Google Scholar] [CrossRef] [PubMed]
- Mastromarino, P.; Macchia, S.; Meggiorini, L.; Trinchieri, V.; Mosca, L.; Perluigi, M.; Midulla, C. Effectiveness of Lactobacillus-containing vaginal tablets in the treatment of symptomatic bacterial vaginosis. Clin. Microbiol. Infect. 2009, 15, 67–74. [Google Scholar] [CrossRef] [Green Version]
- Hanson, L.; Vandevusse, L.; Duster, M.; Warrack, S.; Safdar, N. Feasibility of oral prenatal probiotics against maternal group B Streptococcus vaginal and rectal colonization. JOGNN 2014, 43, 294–304. [Google Scholar] [CrossRef] [Green Version]
- Brantsæter, A.L.; Myhre, R.; Haugen, M.; Myking, S.; Sengpiel, V.; Magnus, P.; Jacobsson, B.; Meltzer, H.M. Intake of probiotic food and risk of preeclampsia in primiparous women. Am. J. Epidemiol. 2011, 174, 807–815. [Google Scholar] [CrossRef] [Green Version]
- Nordqvist, M.; Jacobsson, B.; Brantsæter, A.L.; Myhre, R.; Nilsson, S.; Sengpiel, V. Timing of probiotic milk consumption during pregnancy and effects on the incidence of preeclampsia and preterm delivery: A prospective observational cohort study in Norway. BMJ Open 2018, 8, e018021. [Google Scholar] [CrossRef] [Green Version]
- Gjessing, K.; Sengpiel, V.; Meltzer, H.; Myhre, R.; Brantsæter, A.; Myking, S.; Haugen, M.; Jacobsson, B. Intake of probiotic food and risk of spontaneous preterm delivery. Am. J. Clin. Nutr. 2011, 93, 151–157. [Google Scholar]
- Martín, R.; Langa, S.; Reviriego, C.; Jiménez, E.; Marín, M.L.; Xaus, J.; Fernández, L.; Rodríguez, J.M. Human milk is a source of lactic acid bacteria for the infant gut. J. Pediatr. 2003, 143, 754–758. [Google Scholar] [CrossRef]
- Bond, D.M.; Morris, J.M.; Nassar, N. Study protocol: evaluation of the probiotic Lactobacillus Fermentum CECT5716 for the prevention of mastitis in breastfeeding women: A randomised controlled trial. BMC Pregnancy Childbirth 2017, 17, 148. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Maldonado-Lobón, J.A.; Gil-Campos, M.; Maldonado, J.; López-Huertas, E.; Flores-Rojas, K.; Valero, A.D.; Rodríguez-Benítez, M.V.; Bañuelos, O.; Lara-Villoslada, F.; Fonollá, J.; et al. Long-term safety of early consumption of Lactobacillus fermentum CECT5716: A 3-year follow-up of a randomized controlled trial. Pharmacol. Res. 2015, 95–96, 12–19. [Google Scholar] [CrossRef] [PubMed]
- Gil-Campos, M.; López, M.Á.; Rodriguez-Benítez, M.V.; Romero, J.; Roncero, I.; Linares, M.D.; Maldonado, J.; López-Huertas, E.; Berwind, R.; Ritzenthaler, K.L.; et al. Lactobacillus fermentum CECT 5716 is safe and well tolerated in infants of 1-6 months of age: A randomized controlled trial. Pharmacol. Res. 2012, 65, 231–238. [Google Scholar] [CrossRef] [PubMed]
- Maldonado, J.; Cañabate, F.; Sempere, L.; Vela, F.; Sánchez, A.R.; Narbona, E.; López-Huertas, E.; Geerlings, A.; Valero, A.D.; Olivares, M.; et al. Human milk probiotic Lactobacillus fermentum CECT5716 reduces the incidence of gastrointestinal and upper respiratory tract infections in infants. J. Pediatr. Gastroenterol. Nutr. 2012, 54, 55–61. [Google Scholar] [CrossRef]
- Arroyo, R.; Martín, V.; Maldonado, A.; Jiménez, E.; Fernández, L.; Rodríguez, J.M. Treatment of infectious mastitis during lactation: Antibiotics versus oral administration of lactobacilli isolated from breast milk. Clin. Infect. Dis. 2010, 50, 1551–1558. [Google Scholar] [CrossRef] [Green Version]
- Maldonado-Lobón, J.A.; Díaz-López, M.A.; Carputo, R.; Duarte, P.; Díaz-Ropero, M.P.; Valero, A.D.; Sañudo, A.; Sempere, L.; Ruiz-López, M.D.; Bañuelos, Ó.; et al. Lactobacillus fermentum CECT 5716 reduces Staphylococcus load in the breastmilk of lactating mothers suffering breast pain: A randomized controlled trial. Breastfeed. Med. 2015, 10, 425–432. [Google Scholar] [CrossRef]
- Robles-Vera, I.; Toral, M.; de la Visitación, N.; Sánchez, M.; Romero, M.; Olivares, M.; Jiménez, R.; Duarte, J. The probiotic Lactobacillus fermentum prevents dysbiosis and vascular oxidative stress in rats with hypertension induced by chronic nitric oxide blockade. Mol. Nutr. Food Res. 2018, 62, 1800298. [Google Scholar] [CrossRef]
- Díaz-Ropero, M.P.; Martín, R.; Sierra, S.; Lara-Villoslada, F.; Rodríguez, J.M.; Xaus, J.; Olivares, M. Two Lactobacillus strains, isolated from breast milk, differently modulate the immune response. J. Appl. Microbiol. 2007, 102, 337–343. [Google Scholar] [CrossRef]
- Pérez-Cano, F.J.; Dong, H.; Yaqoob, P. In vitro immunomodulatory activity of Lactobacillus fermentum CECT5716 and Lactobacillus salivarius CECT5713: two probiotic strains isolated from human breast milk. Immunobiology 2010, 215, 996–1004. [Google Scholar] [CrossRef]
- Azagra-Boronat, I.; Tres, A.; Massot-Cladera, M.; Franch, À.; Castell, M.; Guardiola, F.; Pérez-Cano, F.J.; Rodríguez-Lagunas, M.J. Lactobacillus fermentum CECT5716 supplementation in rats during pregnancy and lactation impacts breastmilk composition. J. Dairy Sci. 2020. In press. [Google Scholar] [CrossRef]
- Reeves, P.G.; Nielsen, F.H.; Fahey, G.C. AIN-93 purified diets for laboratory rodents: Final report of the American Institute of Nutrition ad hoc Writing Committee on the reformulation of the AIN-76A rodent diet. J. Nutr. 1993, 123, 1939–1951. [Google Scholar] [CrossRef] [PubMed]
- Novákova, V.; Babický, A. Coprophagy in young laboratory rat. Physiol Bohemoslov 1989, 38, 21–28. [Google Scholar] [PubMed]
- Azagra-Boronat, I.; Massot-Cladera, M.; Knipping, K.; van‘t Land, B.; Stahl, B.; Garssen, J.; Rodríguez-Lagunas, M.J.; Franch, À.; Castell, M.; Pérez-Cano, F.J. Supplementation with 2’-FL and scGOS/lcFOS ameliorates rotavirus-induced diarrhea in suckling rats. Front. Cell. Infect. Microbiol. 2018, 8, 372. [Google Scholar] [CrossRef]
- Azagra-Boronat, I.; Massot-Cladera, M.; Knipping, K.; van’t Land, B.; Tims, S.; Stahl, B.; Knol, J.; Garssen, J.; Franch, À.; Castell, M.; et al. Oligosaccharides modulate rotavirus-associated dysbiosis and TLR gene expression in neonatal rats. Cells 2019, 8, 876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Grases-Pintó, B.; Torres-Castro, P.; Abril-Gil, M.; Castell, M.; Rodríguez-Lagunas, M.J.; Pérez-Cano, F.J.; Franch, À. A preterm rat model for immunonutritional studies. Nutrients 2019, 11, 999. [Google Scholar] [CrossRef] [Green Version]
- Torres-Castro, P.; Abril-Gil, M.; Rodríguez-Lagunas, M.J.; Castell, M.; Pérez-Cano, F.J.; Franch, À. TGF-β2, EGF, and FGF21 growth factors present in breast milk promote mesenteric lymph node lymphocytes maturation in suckling rats. Nutrients 2018, 10, 1171. [Google Scholar] [CrossRef] [Green Version]
- Azagra-Boronat, I.; Massot-Cladera, M.; Mayneris-Perxachs, J.; Knipping, K.; van’t Land, B.; Tims, S.; Stahl, B.; Garssen, J.; Franch, À.; Castell, M.; et al. Immunomodulatory and prebiotic effects of 2′-fucosyllactose in suckling rats. Front. Immunol. 2019, 10, 1773. [Google Scholar] [CrossRef] [Green Version]
- Grases-Pintó, B.; Abril-Gil, M.; Rodríguez-Lagunas, M.J.; Castell, M.; Pérez-Cano, F.J.; Franch, À. Leptin and adiponectin supplementation modifies mesenteric lymph node lymphocyte composition and functionality in suckling rats. Br. J. Nutr. 2018, 119, 486–495. [Google Scholar] [CrossRef] [Green Version]
- Camps-Bossacoma, M.; Pérez-Cano, F.J.; Franch, À.; Castell, M. Gut microbiota in a rat oral sensitization model: effect of a cocoa-enriched diet. Oxid. Med. Cell. Longev. 2017, 2017, 7417505. [Google Scholar] [CrossRef]
- Andreasson, U.; Perret-Liaudet, A.; van Waalwijk van Doorn, L.J.C.; Blennow, K.; Chiasserini, D.; Engelborghs, S.; Fladby, T.; Genc, S.; Kruse, N.; Kuiperij, H.B.; et al. A practical guide to immunoassay method validation. Front. Neurol. 2015, 6, 179. [Google Scholar] [CrossRef]
- Makino, H.; Kushiro, A.; Ishikawa, E.; Muylaert, D.; Kubota, H.; Sakai, T.; Oishi, K.; Martin, R.; Ben Amor, K.; Oozeer, R.; et al. Transmission of intestinal Bifidobacterium longum subsp. longum strains from mother to infant, determined by multilocus sequencing typing and amplified fragment length polymorphism. Appl. Environ. Microbiol. 2011, 77, 6788–6793. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Martín, V.; Maldonado-Barragán, A.; Moles, L.; Rodriguez-Baños, M.; Del Campo, R.; Fernández, L.; Rodríguez, J.M.; Jiménez, E. Sharing of bacterial strains between breast milk and infant feces. J. Hum. Lact. 2012, 28, 36–44. [Google Scholar] [CrossRef] [PubMed]
- Jiménez, E.; Delgado, S.; Maldonado, A.; Arroyo, R.; Albújar, M.; García, N.; Jariod, M.; Fernández, L.; Gómez, A.; Rodríguez, J.M. Staphylococcus epidermidis: A differential trait of the fecal microbiota of breast-fed infants. BMC Microbiol. 2008, 8, 143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Andrés, J.; Jiménez, E.; Chico-Calero, I.; Fresno, M.; Fernández, L.; Rodríguez, J.M. Physiological translocation of lactic acid bacteria during pregnancy contributes to the composition of the milk microbiota in mice. Nutrients 2018, 10, 14. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lara-Villoslada, F.; Sierra, S.; Díaz-Ropero, M.P.; Rodríguez, J.M.; Xaus, J.; Olivares, M. Safety assessment of Lactobacillus fermentum CECT5716, a probiotic strain isolated from human milk. J. Dairy Res. 2009, 76, 216–221. [Google Scholar] [CrossRef]
- Vanhaecke, T.; Aubert, P.; Grohard, P.A.; Durand, T.; Hulin, P.; Paul-Gilloteaux, P.; Fournier, A.; Docagne, F.; Ligneul, A.; Fressange-Mazda, C.; et al. L. fermentum CECT5716 prevents stress-induced intestinal barrier dysfunction in newborn rats. Neurogastroenterol. Motil. 2017, 29, e13069. [Google Scholar] [CrossRef]
- Pramanik, S.S.; Pramanik, T.; Mondal, S.C.; Chanda, R. Number, maturity and phagocytic activity of neutrophils in the three trimesters of pregnancy. East. Mediterr. Heal. J. 2007, 13, 862–867. [Google Scholar]
- Luzía França, E.; Pernet Hara, C.C.; Fagundes, D.L.G.; Peixoto Lima, N.A.; Bilotti Ratto, S.H.; Honorio-Franca, A.C. Fluctuation in the functional activity of human colostrum phagocytes to Streptococcus pneumoniae and Enteropathogenic Escherichia coli. J. Med. Microbiol. Diagnosis 2012, 1, 1000104. [Google Scholar] [CrossRef]
- Maldonado Galdeano, C.; De Moreno De Leblanc, A.; Vinderola, G.; Bibas Bonet, M.E.; Perdigón, G. Proposed model: mechanisms of immunomodulation induced by probiotic bacteria. Clin. Vaccine Immunol. 2007, 14, 485–492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kemgang, T.S.; Kapila, S.; Shanmugam, V.P.; Kapila, R. Cross-talk between probiotic lactobacilli and host immune system. J. Appl. Microbiol. 2014, 117, 303–319. [Google Scholar] [CrossRef]
- Wang, Y.; Jiang, X.; He, J.; Diraviyam, T.; Zhang, X. Quantitative investigation on correlation between IgG and FcRn during gestation and lactating periods in rat. Am. J. Reprod. Immunol. 2016, 75, 81–85. [Google Scholar] [CrossRef] [PubMed]
- Hurley, W.L.; Theil, P.K. Perspectives on immunoglobulins in colostrum and milk. Nutrients 2011, 3, 442–474. [Google Scholar] [CrossRef] [PubMed]
- Ya, T.; Zhang, Q.; Chu, F.; Merritt, J.; Bilige, M.; Sun, T.; Du, R.; Zhang, H. Immunological evaluation of Lactobacillus casei Zhang: A newly isolated strain from koumiss in Inner Mongolia, China. BMC Immunol. 2008, 9, 68. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tsai, Y.T.; Cheng, P.C.; Fan, C.K.; Pan, T.M. Time-dependent persistence of enhanced immune response by a potential probiotic strain Lactobacillus paracasei subsp. paracasei NTU 101. Int. J. Food Microbiol. 2008, 128, 219–225. [Google Scholar] [CrossRef]
- Maassen, C.B.M.; Boersma, W.J.A.; Van Holten-Neelen, C.; Claassen, E.; Laman, J.D. Growth phase of orally administered Lactobacillus strains differentially affects IgG1/IgG2a ratio for soluble antigens: Implications for vaccine development. Vaccine 2003, 21, 2751–2757. [Google Scholar] [CrossRef] [Green Version]
- Bauer, P.V.; Duca, F.A.; Waise, T.M.Z.; Dranse, H.J.; Rasmussen, B.A.; Puri, A.; Rasti, M.; O’Brien, C.A.; Lam, T.K.T. Lactobacillus gasseri in the upper small intestine impacts an ACSL3-dependent fatty acid-sensing pathway regulating whole-body glucose homeostasis. Cell Metab. 2018, 27, 572–587. [Google Scholar] [CrossRef] [Green Version]
- Jang, H.R.; Park, H.-J.; Kang, D.; Chung, H.; Nam, M.H.; Lee, Y.; Park, J.-H.; Lee, H.-Y. A protective mechanism of probiotic Lactobacillus against hepatic steatosis via reducing host intestinal fatty acid absorption. Exp. Mol. Med. 2019, 51, 95. [Google Scholar] [CrossRef] [Green Version]
- Cerdó, T.; García-Santos, J.A.; Bermúdez, M.G.; Campoy, C. The role of probiotics and prebiotics in the prevention and treatment of obesity. Nutrients 2019, 11, 635. [Google Scholar] [CrossRef] [Green Version]






| REF | LF | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | 12 | |
| Dams’ CC | ||||||||||||
| Pups’ CC | 50% | 25% | 25% | |||||||||
| Dams | Pups | ||||
|---|---|---|---|---|---|
| REF | LF | REF | LF | ||
| * Body Weight | G21/L4 | 406.24 ± 18.26 | 395.47 ± 12.12 | 9.91 ± 0.20 | 9.90 ± 0.16 |
| L14 | 345.57 ± 7.96 | 325.38 ± 11.80 | 28.94 ± 0.40 | 28.85 ± 0.59 | |
| Size | BMI (g/cm2) | 0.71 ± 0.04 | 0.68 ± 0.03 | 0.34 ± 0.01 | 0.34 ± 0.01 |
| Lee Index (g0.33/cm) | 318.04 ± 8.19 | 315.19 ± 5.69 | 331.83 ± 1.30 | 332.90 ± 1.42 | |
| * Organ Weights | Stomach | 0.94 ± 0.25 | 0.67 ± 0.02 | 0.65 ± 0.01 | 0.69 ± 0.05 |
| Small intestine | 3.93 ± 0.10 | 3.85 ± 0.06 | 3.06 ± 0.03 | 3.16 ± 0.03 | |
| Cecum | 0.45 ± 0.02 | 0.48 ± 0.02 | 0.15 ± 0.01 | 0.14 ± 0.01 | |
| Large intestine | 0.63 ± 0.05 | 0.57± 0.03 | 0.33 ± 0.05 | 0.28 ± 0.01 | |
| Spleen | 0.21 ± 0.01 | 0.24 ± 0.01 | 0.49 ± 0.01 | 0.52 ± 0.01 | |
| Thymus | 0.13 ± 0.01 | 0.13 ± 0.01 | 0.46 ± 0.01 | 0.57 ± 0.09 | |
| Liver | 4.02 ± 0.15 | 4.16 ± 0.10 | 3.19 ± 0.04 | 3.02 ± 0.07 | |
| Kidneys | 0.57 ± 0.01 | 0.60 ± 0.02 | 1.15 ± 0.01 | 1.16 ± 0.01 | |
| Brain | 0.42 ± 0.06 | 0.41 ± 0.07 | 3.55 ± 0.06 | 3.72 ± 0.07 | |
| Uterus | 0.10 ± 0.01 | 0.10 ± 0.01 | - | - | |
| Ovaries | 0.05 ± 0.01 | 0.05 ± 0.01 | - | - | |
| Heart | 0.34 ± 0.01 | 0.36 ± 0.01 | - | - | |
| # Organ Length | Small intestine | 25.32 ± 2.43 | 23.25 ± 3.24 | 5.55 ± 0.18 | 135.88 ± 2.36 |
| Cecum | 1.79 ± 0.06 | 1.92 ± 0.13 | - | - | |
| Large intestine | 5.63 ± 0.45 | 5.55 ± 0.18 | - | - | |
| REF | LF | |
|---|---|---|
| %B cells (CD45RA+) | 26.93 ± 3.31 | 32.75 ± 5.02 |
| CD62L+ (%) | 78.62 ± 4.35 | 78.82 ± 3.87 |
| CD25+ (%) | 0.70 ± 0.12 | 0.86 ± 0.31 |
| %Th cells (CD4+ TCRαβ+) | 43.37 ± 1.36 | 40.23 ± 1.87 |
| CD62L+ (%) | 70.91 ± 1.53 | 70.98 ± 1.89 |
| CD25+ (%) | 6.81 ± 0.34 | 8.00 ± 0.43* |
| αE integrin (%) | 4.06 ± 0.76 | 5.70 ± 1.63 |
| %Tc cells (CD8+ TCRαβ+ and TCRγδ+) | 20.54 ± 0.51 | 17.15 ± 1.14* |
| CD62L+ (%) | 85.28 ± 1.70 | 86.81 ± 0.89 |
| CD25+ (%) | 2.58 ± 0.18 | 3.38 ± 0.68* |
| αE integrin (%) | 5.13 ± 1.26 | 7.07 ± 2.60 |
| TCRαβ+ (CD8+ TCRαβ+ NK-) (%) | 19.09 ± 0.49 | 15.57 ± 1.12* |
| TCRγδ+ (%) | 1.69 ± 0.14 | 1.89 ± 0.15 |
| CD8- TCRγδ+ (%) | 0.25 ± 0.02 | 0.31 ± 0.03 |
| CD8+ TCRγδ+ (%) | 1.45 ± 0.13 | 1.58 ± 0.13 |
| CD8αα+ TCRγδ+ (%) | 0.14 ± 0.02 | 0.16 ± 0.02 |
| CD8αβ+ TCRγδ+ (%) | 1.31 ± 0.11 | 1.42 ± 0.12 |
| %NKT cells (TCRαβ+ NK+) | 2.49 ± 0.16 | 2.45 ± 0.26 |
| %NK cells (TCRαβ- NK+) | 2.11 ± 0.11 | 2.42 ± 0.19 |
| G0 | G7 | G14 | G21 | L7 | L14 | ||
|---|---|---|---|---|---|---|---|
| Fecal pH | REF | 5.88 ± 0.05 | 6.20 ± 0.10 | 6.09 ± 0.12 | 6.08 ± 0.06 | 6.48 ± 0.08 | 6.47 ± 0.05 |
| LF | 6.07 ± 0.14 | 6.48 ± 0.19 | 6.40 ± 0.26 | 6.43 ± 0.14 | 6.50 ± 0.16 | 6.11 ± 0.12 | |
| Fecal humidity (%) | REF | - | 50.59 ± 3.33 | 55.91 ± 1.24 | 57.87 ± 2.10 | 62.97 ± 1.40 | 65.41 ± 0.78 |
| LF | - | 53.44 ± 5.02 | 58.68 ± 2.81 | 58.53 ± 1.07 | 65.19 ± 2.39 | 62.07 ± 4.36 | |
| Fecal weight (mg) | REF | - | 259.4 ± 39.6 | 144.0 ± 19.2 | 249.3 ± 40.1 | 421.8 ± 53.9 | 452.6 ± 49.0 |
| LF | - | 306.83 ± 50.2 | 278.0 ± 59.6 | 185.5 ± 16.5 | 369.3 ± 16.6 | 329.8 ± 32.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Azagra-Boronat, I.; Tres, A.; Massot-Cladera, M.; Franch, À.; Castell, M.; Guardiola, F.; Pérez-Cano, F.J.; Rodríguez-Lagunas, M.J. Lactobacillus fermentum CECT5716 Supplementation in Rats during Pregnancy and Lactation Impacts Maternal and Offspring Lipid Profile, Immune System and Microbiota. Cells 2020, 9, 575. https://doi.org/10.3390/cells9030575
Azagra-Boronat I, Tres A, Massot-Cladera M, Franch À, Castell M, Guardiola F, Pérez-Cano FJ, Rodríguez-Lagunas MJ. Lactobacillus fermentum CECT5716 Supplementation in Rats during Pregnancy and Lactation Impacts Maternal and Offspring Lipid Profile, Immune System and Microbiota. Cells. 2020; 9(3):575. https://doi.org/10.3390/cells9030575
Chicago/Turabian StyleAzagra-Boronat, Ignasi, Alba Tres, Malén Massot-Cladera, Àngels Franch, Margarida Castell, Francesc Guardiola, Francisco J. Pérez-Cano, and Maria J. Rodríguez-Lagunas. 2020. "Lactobacillus fermentum CECT5716 Supplementation in Rats during Pregnancy and Lactation Impacts Maternal and Offspring Lipid Profile, Immune System and Microbiota" Cells 9, no. 3: 575. https://doi.org/10.3390/cells9030575
APA StyleAzagra-Boronat, I., Tres, A., Massot-Cladera, M., Franch, À., Castell, M., Guardiola, F., Pérez-Cano, F. J., & Rodríguez-Lagunas, M. J. (2020). Lactobacillus fermentum CECT5716 Supplementation in Rats during Pregnancy and Lactation Impacts Maternal and Offspring Lipid Profile, Immune System and Microbiota. Cells, 9(3), 575. https://doi.org/10.3390/cells9030575