Perspectives for Glyco-Engineering of Recombinant Biopharmaceuticals from Microalgae
Abstract
:1. Introduction
2. Production of Biopharmaceuticals in Alternative Hosts
3. Microalgal Biofactories
4. Post-Translational Modifications and Glycosylation
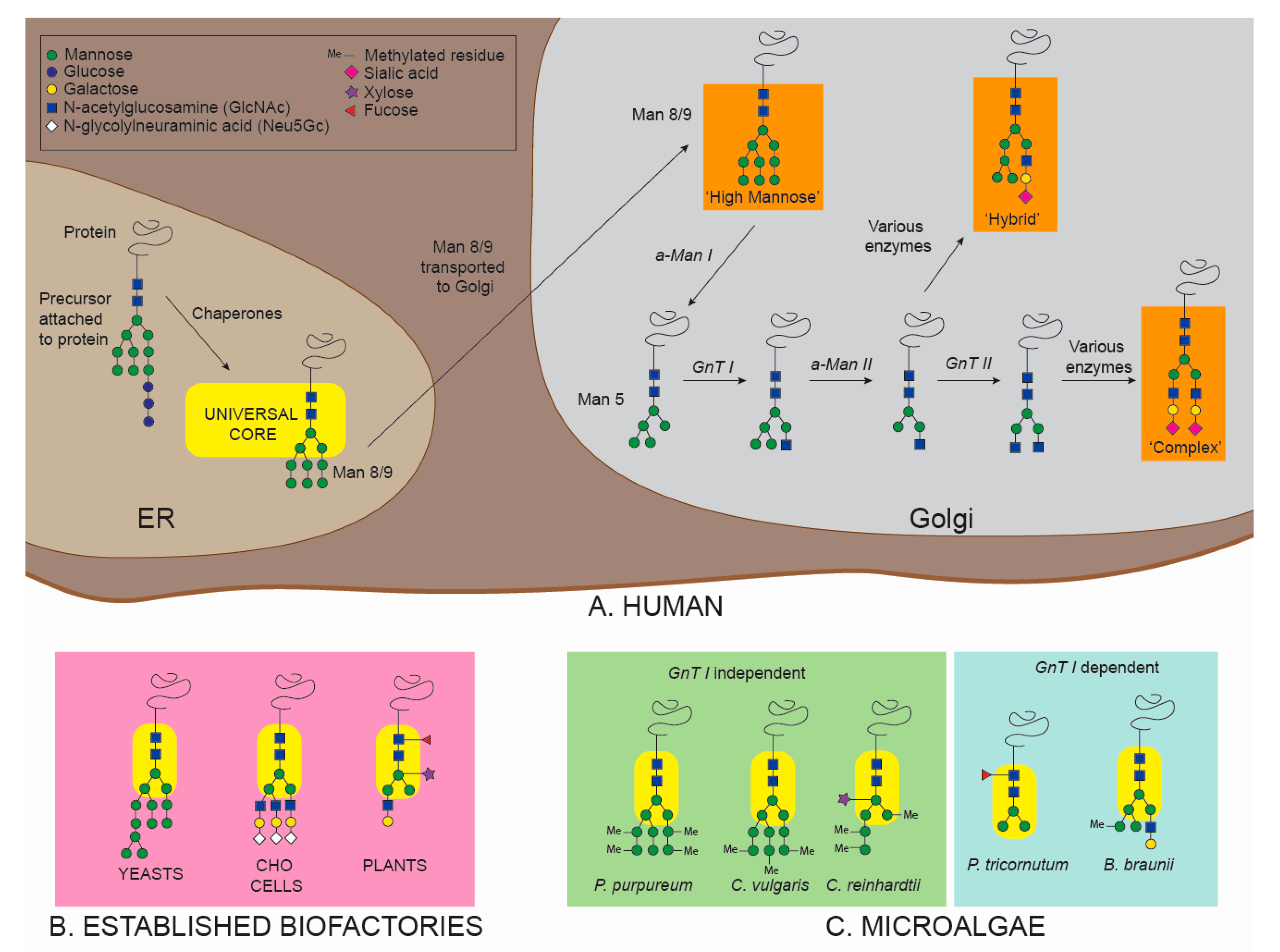
4.1. N-Glycosylation
4.2. O-Glycosylation
4.3. Computationally Predicted Distribution of Microalgal N- and O- Glycosylation Enzymes
5. Strategies for Manipulating Protein Glycosylation
5.1. Protein Engineering
5.1.1. Glycoprotein Sequence Engineering
5.1.2. Subcellular Location Engineering
5.1.3. Glycosylation Pattern Engineering
5.2. Cell Glyco-Engineering
5.2.1. Glyco-Engineering by Inhibitor Interference
5.2.2. Genetic Glyco-Engineering
6. Future Perspective for Glyco-Engineering in Microalgae
Funding
Conflicts of Interest
References
- Ghaderi, D.; Zhang, M.; Hurtado-Ziola, N.; Varki, A. Production platforms for biotherapeutic glycoproteins. Occurrence, impact, and challenges of non-human sialylation. Biotechnol. Genet. Eng. Rev. 2012, 28, 147–176. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, G. Biopharmaceutical benchmarks 2014. Nat. Biotechnol. 2014, 32, 992. [Google Scholar] [CrossRef] [PubMed]
- Jayapal, K.P.; Wlaschin, K.F.; Hu, W.; Yap, M.G. Recombinant protein therapeutics from CHO cells-20 years and counting. Chem. Eng. Prog. 2007, 103, 40. [Google Scholar]
- Lalonde, M.E.; Durocher, Y. Therapeutic glycoprotein production in mammalian cells. J. Biotechnol. 2017, 251, 128–140. [Google Scholar] [CrossRef] [PubMed]
- Hippler, M. Chlamydomonas: Biotechnology and Biomedicine; Springer: Berlin, Germany, 2017; Volume 31. [Google Scholar]
- Baeshen, M.N.; Al-Hejin, A.M.; Bora, R.S.; Ahmed, M.M.; Ramadan, H.A.; Saini, K.S.; Baeshen, N.A.; Redwan, E.M. Production of Biopharmaceuticals in E. coli: Current Scenario and Future Perspectives. J. Microbiol. Biotechnol. 2015, 25, 953–962. [Google Scholar] [CrossRef]
- Mattanovich, D.; Branduardi, P.; Dato, L.; Gasser, B.; Sauer, M.; Porro, D. Recombinant Protein Production in Yeasts. In Recombinant Gene Expression; Lorence, A., Ed.; Humana Press: Totowa, NJ, USA, 2012; pp. 329–358. [Google Scholar] [CrossRef]
- Berlec, A.; Strukelj, B. Current state and recent advances in biopharmaceutical production in Escherichia coli, yeasts and mammalian cells. J. Ind. Microbiol. Biotechnol. 2013, 40, 257–274. [Google Scholar] [CrossRef]
- Gomord, V.; Faye, L. Posttranslational modification of therapeutic proteins in plants. Curr. Opin. Plant. Biol. 2004, 7, 171–181. [Google Scholar] [CrossRef]
- Mathieu-Rivet, E.; Kiefer-Meyer, M.C.; Vanier, G.; Ovide, C.; Burel, C.; Lerouge, P.; Bardor, M. Protein N-glycosylation in eukaryotic microalgae and its impact on the production of nuclear expressed biopharmaceuticals. Front. Plant. Sci. 2014, 5, 359. [Google Scholar] [CrossRef]
- Rasala, B.A.; Mayfield, S.P. Photosynthetic biomanufacturing in green algae; production of recombinant proteins for industrial, nutritional, and medical uses. Photosynth. Res. 2015, 123, 227–239. [Google Scholar] [CrossRef]
- Specht, E.; Miyake-Stoner, S.; Mayfield, S. Micro-algae come of age as a platform for recombinant protein production. Biotechnol. Lett. 2010, 32, 1373–1383. [Google Scholar] [CrossRef] [Green Version]
- Harris, E.H. The Chlamydomonas sourcebook: Introduction to Chlamydomonas and its laboratory use; Academic press: Cambridge, MA, USA, 2009; Volume 1. [Google Scholar]
- Hawkins, R.L.; Nakamura, M. Expression of human growth hormone by the eukaryotic alga, Chlorella. Curr. Microbiol. 1999, 38, 335–341. [Google Scholar] [CrossRef] [PubMed]
- Bai, L.L.; Yin, W.B.; Chen, Y.H.; Niu, L.L.; Sun, Y.R.; Zhao, S.M.; Yang, F.Q.; Wang, R.R.; Wu, Q.; Zhang, X.Q.; et al. A new strategy to produce a defensin: Stable production of mutated NP-1 in nitrate reductase-deficient Chlorella ellipsoidea. PLoS ONE 2013, 8, e54966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Geng, D.; Wang, Y.; Wang, P.; Li, W.; Sun, Y. Stable expression of hepatitis B surface antigen gene in Dunaliella salina (Chlorophyta). J. Appl. Phycol. 2003, 15, 451–456. [Google Scholar] [CrossRef]
- Chen, H.L.; Li, S.S.; Huang, R.; Tsai, H.-J. Conditional Production Of A Functional Fish Growth Hormone In The Transgenic Line Of Nannochloropsis Oculata (Eustigmatophyceae). J. Phycol. 2008, 44, 768–776. [Google Scholar] [CrossRef] [PubMed]
- Li, S.-S.; Tsai, H.-J. Transgenic microalgae as a non-antibiotic bactericide producer to defend against bacterial pathogen infection in the fish digestive tract. Fish. Shellfish Immunol. 2009, 26, 316–325. [Google Scholar] [CrossRef]
- Hempel, F.; Maier, U.G. An engineered diatom acting like a plasma cell secreting human IgG antibodies with high efficiency. Microb. Cell Factories 2012, 11, 126. [Google Scholar] [CrossRef] [Green Version]
- Beltrao, P.; Albanese, V.; Kenner, L.R.; Swaney, D.L.; Burlingame, A.; Villen, J.; Lim, W.A.; Fraser, J.S.; Frydman, J.; Krogan, N.J. Systematic functional prioritization of protein posttranslational modifications. Cell 2012, 150, 413–425. [Google Scholar] [CrossRef] [Green Version]
- Beltrao, P.; Trinidad, J.C.; Fiedler, D.; Roguev, A.; Lim, W.A.; Shokat, K.M.; Burlingame, A.L.; Krogan, N.J. Evolution of phosphoregulation: Comparison of phosphorylation patterns across yeast species. Plos Biol. 2009, 7, e1000134. [Google Scholar] [CrossRef]
- Weinert, B.T.; Wagner, S.A.; Horn, H.; Henriksen, P.; Liu, W.R.; Olsen, J.V.; Jensen, L.J.; Choudhary, C. Proteome-Wide Mapping of the Drosophila Acetylome Demonstrates a High Degree of Conservation of Lysine Acetylation. Sci. Signal. 2011, 4, ra48. [Google Scholar] [CrossRef]
- Boekhorst, J.; van Breukelen, B.; Heck, A., Jr.; Snel, B. Comparative phosphoproteomics reveals evolutionary and functional conservation of phosphorylation across eukaryotes. Genome Biol. 2008, 9, R144. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wang, Y.; Yao, Q.; Justice, N.B.; Ahn, T.H.; Xu, D.; Hettich, R.L.; Banfield, J.F.; Pan, C. Diverse and divergent protein post-translational modifications in two growth stages of a natural microbial community. Nat. Commun. 2014, 5, 4405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bürkle, A. Posttranslational Modification. In Encyclopedia of Genetics; Brenner, S., Miller, J.H., Eds.; Academic Press: New York, NY, USA, 2001; p. 1533. [Google Scholar]
- An, H.J.; Froehlich, J.W.; Lebrilla, C.B. Determination of glycosylation sites and site-specific heterogeneity in glycoproteins. Curr. Opin. Chem. Biol. 2009, 13, 421–426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Walsh, G.; Jefferis, R. Post-translational modifications in the context of therapeutic proteins. Nat. Biotechnol. 2006, 24, 1241–1252. [Google Scholar] [CrossRef] [PubMed]
- Higgins, E. Carbohydrate analysis throughout the development of a protein therapeutic. Glycoconj. J. 2010, 27, 211–225. [Google Scholar] [CrossRef] [Green Version]
- Butler, M.; Spearman, M. The choice of mammalian cell host and possibilities for glycosylation engineering. Curr. Opin. Biotechnol. 2014, 30, 107–112. [Google Scholar] [CrossRef]
- Mizukami, A.; Caron, A.L.; Picanço-Castro, V.; Swiech, K. Platforms for Recombinant Therapeutic Glycoprotein Production. In Recombinant Glycoprotein Production: Methods and Protocols; Picanço-Castro, V., Swiech, K., Eds.; Springer: New York, NY, USA, 2018; pp. 1–14. [Google Scholar] [CrossRef]
- Hajba, L.; Szekrenyes, A.; Borza, B.; Guttman, A. On the glycosylation aspects of biosimilarity. Drug Discov. Today 2018, 23, 616–625. [Google Scholar] [CrossRef] [Green Version]
- Lagasse, H.A.; Alexaki, A.; Simhadri, V.L.; Katagiri, N.H.; Jankowski, W.; Sauna, Z.E.; Kimchi-Sarfaty, C. Recent advances in (therapeutic protein) drug development. F1000Research 2017, 6, 113. [Google Scholar] [CrossRef] [Green Version]
- Dumont, J.; Euwart, D.; Mei, B.; Estes, S.; Kshirsagar, R. Human cell lines for biopharmaceutical manufacturing: History, status, and future perspectives. Crit. Rev. Biotechnol. 2016, 36, 1110–1122. [Google Scholar] [CrossRef] [Green Version]
- Ahmad, Z.A.; Yeap, S.K.; Ali, A.M.; Ho, W.Y.; Alitheen, N.B.; Hamid, M. scFv antibody: Principles and clinical application. Clin. Dev. Immunol. 2012, 2012, 980250. [Google Scholar] [CrossRef]
- Junutula, J.R.; Raab, H.; Clark, S.; Bhakta, S.; Leipold, D.D.; Weir, S.; Chen, Y.; Simpson, M.; Tsai, S.P.; Dennis, M.S.; et al. Site-specific conjugation of a cytotoxic drug to an antibody improves the therapeutic index. Nat. Biotechnol. 2008, 26, 925–932. [Google Scholar] [CrossRef]
- Qiu, X.; Wong, G.; Audet, J.; Bello, A.; Fernando, L.; Alimonti, J.B.; Fausther-Bovendo, H.; Wei, H.; Aviles, J.; Hiatt, E.; et al. Reversion of advanced Ebola virus disease in nonhuman primates with ZMapp. Nature 2014, 514, 47–53. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Budzianowski, J. Tobacco against Ebola virus disease. Przegl. Lek. 2015, 72, 567–571. [Google Scholar] [PubMed]
- Davidson, E.; Bryan, C.; Fong, R.H.; Barnes, T.; Pfaff, J.M.; Mabila, M.; Rucker, J.B.; Doranz, B.J. Mechanism of Binding to Ebola Virus Glycoprotein by the ZMapp, ZMAb, and MB-003 Cocktail Antibodies. J. Virol. 2015, 89, 10982–10992. [Google Scholar] [CrossRef] [Green Version]
- Cadoret, J.P.; Bernard, O. Lipid biofuel production with microalgae: Potential and challenges. J. Soc. Biol. 2008, 202, 201–211. [Google Scholar] [CrossRef]
- Sasso, S.; Pohnert, G.; Lohr, M.; Mittag, M.; Hertweck, C. Microalgae in the postgenomic era: A blooming reservoir for new natural products. Fems Microbiol. Rev. 2012, 36, 761–785. [Google Scholar] [CrossRef]
- Merlin, M.; Gecchele, E.; Capaldi, S.; Pezzotti, M.; Avesani, L. Comparative evaluation of recombinant protein production in different biofactories: The green perspective. Biomed. Res. Int. 2014, 2014, 136419. [Google Scholar] [CrossRef] [Green Version]
- Eichler-Stahlberg, A.; Weisheit, W.; Ruecker, O.; Heitzer, M. Strategies to facilitate transgene expression in Chlamydomonas reinhardtii. Planta 2009, 229, 873–883. [Google Scholar] [CrossRef]
- Ramos-Martinez, E.M.; Fimognari, L.; Sakuragi, Y. High-yield secretion of recombinant proteins from the microalga Chlamydomonas reinhardtii. Plant Biotechnol. J. 2017, 15, 1214–1224. [Google Scholar] [CrossRef] [Green Version]
- Murbach, T.S.; Glavits, R.; Endres, J.R.; Hirka, G.; Vertesi, A.; Beres, E.; Szakonyine, I.P. A Toxicological Evaluation of Chlamydomonas reinhardtii, a Green Algae. Int J. Toxicol. 2018, 37, 53–62. [Google Scholar] [CrossRef] [Green Version]
- Fields, F.J.; Ostrand, J.T.; Mayfield, S.P. Fed-batch mixotrophic cultivation of Chlamydomonas reinhardtii for high-density cultures. Algal Res. 2018, 33, 109–117. [Google Scholar] [CrossRef]
- Fahad, S.; Khan, F.A.; Pandupuspitasari, N.S.; Ahmed, M.M.; Liao, Y.C.; Waheed, M.T.; Sameeullah, M.; Darkhshan; Hussain, S.; Saud, S.; et al. Recent developments in therapeutic protein expression technologies in plants. Biotechnol. Lett. 2015, 37, 265–279. [Google Scholar] [CrossRef] [PubMed]
- Scaife, M.A.; Nguyen, G.T.; Rico, J.; Lambert, D.; Helliwell, K.E.; Smith, A.G. Establishing Chlamydomonas reinhardtii as an industrial biotechnology host. Plant. J. 2015, 82, 532–546. [Google Scholar] [CrossRef] [PubMed]
- Demurtas, O.C.; Massa, S.; Ferrante, P.; Venuti, A.; Franconi, R.; Giuliano, G. A Chlamydomonas-derived Human Papillomavirus 16 E7 vaccine induces specific tumor protection. PLoS ONE 2013, 8, e61473. [Google Scholar] [CrossRef] [Green Version]
- Dreesen, I.A.; Charpin-El Hamri, G.; Fussenegger, M. Heat-stable oral alga-based vaccine protects mice from Staphylococcus aureus infection. J. Biotechnol. 2010, 145, 273–280. [Google Scholar] [CrossRef]
- Georgianna, D.R. Production of recombinant enzymes in the marine alga Dunaliella tertiolecta. Algal Res. 2013, 2, 2–9. [Google Scholar] [CrossRef]
- Gregory, J.A.; Li, F.; Tomosada, L.M.; Cox, C.J.; Topol, A.B.; Vinetz, J.M.; Mayfield, S. Algae-produced Pfs25 elicits antibodies that inhibit malaria transmission. PLoS ONE 2012, 7, e37179. [Google Scholar] [CrossRef] [Green Version]
- Gregory, J.A.; Topol, A.B.; Doerner, D.Z.; Mayfield, S. Alga-produced cholera toxin-Pfs25 fusion proteins as oral vaccines. Appl. Env. Microbiol. 2013, 79, 3917–3925. [Google Scholar] [CrossRef] [Green Version]
- He, D.M.; Qian, K.X.; Shen, G.F.; Zhang, Z.F.; Li, Y.N.; Su, Z.L.; Shao, H.B. Recombination and expression of classical swine fever virus (CSFV) structural protein E2 gene in Chlamydomonas reinhardtii chroloplasts. Colloids Surf. B. Biointerfaces 2007, 55, 26–30. [Google Scholar] [CrossRef]
- Jones, C.S.; Luong, T.; Hannon, M.; Tran, M.; Gregory, J.A.; Shen, Z.; Briggs, S.P.; Mayfield, S.P. Heterologous expression of the C-terminal antigenic domain of the malaria vaccine candidate Pfs48/45 in the green algae Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2013, 97, 1987–1995. [Google Scholar] [CrossRef]
- Manuell, A.L.; Beligni, M.V.; Elder, J.H.; Siefker, D.T.; Tran, M.; Weber, A.; McDonald, T.L.; Mayfield, S.P. Robust expression of a bioactive mammalian protein in Chlamydomonas chloroplast. Plant. Biotechnol. J. 2007, 5, 402–412. [Google Scholar] [CrossRef]
- Mayfield, S.P.; Franklin, S.E.; Lerner, R.A. Expression and assembly of a fully active antibody in algae. Proc. Natl. Acad. Sci. USA 2003, 100, 438–442. [Google Scholar] [CrossRef] [Green Version]
- Rasala, B.A.; Muto, M.; Lee, P.A.; Jager, M.; Cardoso, R.M.; Behnke, C.A.; Kirk, P.; Hokanson, C.A.; Crea, R.; Mendez, M.; et al. Production of therapeutic proteins in algae, analysis of expression of seven human proteins in the chloroplast of Chlamydomonas reinhardtii. Plant. Biotechnol. J. 2010, 8, 719–733. [Google Scholar] [CrossRef] [Green Version]
- Su, Z.L.; Qian, K.X.; Tan, C.P.; Meng, C.X.; Qin, S. Recombination and heterologous expression of allophycocyanin gene in the chloroplast of Chlamydomonas reinhardtii. Acta Biochim. Biophys. Sin. (Shanghai) 2005, 37, 709–712. [Google Scholar] [CrossRef] [Green Version]
- Sun, M.; Qian, K.; Su, N.; Chang, H.; Liu, J.; Shen, G. Foot-and-mouth disease virus VP1 protein fused with cholera toxin B subunit expressed in Chlamydomonas reinhardtii chloroplast. Biotechnol Lett. 2003, 25, 1087–1092. [Google Scholar] [CrossRef]
- Surzycki, R.; Greenham, K.; Kitayama, K.; Dibal, F.; Wagner, R.; Rochaix, J.D.; Ajam, T.; Surzycki, S. Factors effecting expression of vaccines in microalgae. Biologicals 2009, 37, 133–138. [Google Scholar] [CrossRef]
- Tran, M.; Zhou, B.; Pettersson, P.L.; Gonzalez, M.J.; Mayfield, S.P. Synthesis and assembly of a full-length human monoclonal antibody in algal chloroplasts. Biotechnol. Bioeng. 2009, 104, 663–673. [Google Scholar] [CrossRef]
- Tran, M.; Henry, R.E.; Siefker, D.; Van, C.; Newkirk, G.; Kim, J.; Bui, J.; Mayfield, S.P. Production of anti-cancer immunotoxins in algae: Ribosome inactivating proteins as fusion partners. Biotechnol. Bioeng. 2013, 110, 2826–2835. [Google Scholar] [CrossRef]
- Tran, M.; Van, C.; Barrera, D.J.; Pettersson, P.L.; Peinado, C.D.; Bui, J.; Mayfield, S.P. Production of unique immunotoxin cancer therapeutics in algal chloroplasts. Proc. Natl. Acad. Sci. USA 2013, 110, E15–E22. [Google Scholar] [CrossRef] [Green Version]
- Wang, X.; Brandsma, M.; Tremblay, R.; Maxwell, D.; Jevnikar, A.M.; Huner, N.; Ma, S. A novel expression platform for the production of diabetes-associated autoantigen human glutamic acid decarboxylase (hGAD65). Bmc Biotechnol 2008, 8, 87. [Google Scholar] [CrossRef] [Green Version]
- Yang, Z.; Li, y.; Chen, F.; Li, D.; Zhang, Z.; Liu, Y.; Zheng, D.; Wang, Y.; Shen, G. Expression of human soluble TRAIL in Chlamydomonas reinhardtii chloroplast. Chin. Sci. Bull. 2006, 51, 1703–1709. [Google Scholar] [CrossRef]
- Yoon, S.M.; Kim, S.Y.; Li, K.F.; Yoon, B.H.; Choe, S.; Kuo, M.M. Transgenic microalgae expressing Escherichia coli AppA phytase as feed additive to reduce phytate excretion in the manure of young broiler chicks. Appl. Microbiol. Biotechnol 2011, 91, 553–563. [Google Scholar] [CrossRef]
- Zhang, Y.K.; Shen, G.F.; Ru, B.G. Survival of human metallothionein-2 transplastomic Chlamydomonas reinhardtii to ultraviolet B exposure. Acta Biochim. Biophys. Sin. (Shanghai) 2006, 38, 187–193. [Google Scholar] [CrossRef] [Green Version]
- Baier, T.; Kros, D.; Feiner, R.C.; Lauersen, K.J.; Muller, K.M.; Kruse, O. Engineered Fusion Proteins for Efficient Protein Secretion and Purification of a Human Growth Factor from the Green Microalga Chlamydomonas reinhardtii. Acs Synth Biol 2018. [Google Scholar] [CrossRef]
- Chavez, M.N.; Schenck, T.L.; Hopfner, U.; Centeno-Cerdas, C.; Somlai-Schweiger, I.; Schwarz, C.; Machens, H.G.; Heikenwalder, M.; Bono, M.R.; Allende, M.L.; et al. Towards autotrophic tissue engineering: Photosynthetic gene therapy for regeneration. Biomaterials 2016, 75, 25–36. [Google Scholar] [CrossRef] [Green Version]
- Dauvillee, D.; Delhaye, S.; Gruyer, S.; Slomianny, C.; Moretz, S.E.; d’Hulst, C.; Long, C.A.; Ball, S.G.; Tomavo, S. Engineering the chloroplast targeted malarial vaccine antigens in Chlamydomonas starch granules. PLoS ONE 2010, 5, e15424. [Google Scholar] [CrossRef]
- Hou, Q.; Qiu, S.; Liu, Q.; Tian, J.; Hu, Z.; Ni, J. Selenoprotein-transgenic Chlamydomonas reinhardtii. Nutrients 2013, 5, 624–636. [Google Scholar] [CrossRef]
- Lauersen, K.J.; Vanderveer, T.L.; Berger, H.; Kaluza, I.; Mussgnug, J.H.; Walker, V.K.; Kruse, O. Ice recrystallization inhibition mediated by a nuclear-expressed and -secreted recombinant ice-binding protein in the microalga Chlamydomonas reinhardtii. Appl. Microbiol. Biotechnol. 2013, 97, 9763–9772. [Google Scholar] [CrossRef]
- Rasala, B.A.; Lee, P.A.; Shen, Z.; Briggs, S.P.; Mendez, M.; Mayfield, S.P. Robust expression and secretion of Xylanase1 in Chlamydomonas reinhardtii by fusion to a selection gene and processing with the FMDV 2A peptide. PLoS ONE 2012, 7, e43349. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Wang, Y.; Sun, Y.; Zhang, L.; Li, W. Highly efficient expression of rabbit neutrophil peptide-1 gene in Chlorella ellipsoidea cells. Curr Genet. 2001, 39, 365–370. [Google Scholar] [CrossRef]
- Kim, D.H.; Kim, Y.T.; Cho, J.J.; Bae, J.H.; Hur, S.B.; Hwang, I.; Choi, T.J. Stable integration and functional expression of flounder growth hormone gene in transformed microalga, Chlorella ellipsoidea. Mar. Biotechnol (NY) 2002, 4, 63–73. [Google Scholar] [CrossRef]
- Feng, S.; Feng, W.; Zhao, L.; Gu, H.; Li, Q.; Shi, K.; Guo, S.; Zhang, N. Preparation of transgenic Dunaliella salina for immunization against white spot syndrome virus in crayfish. Arch. Virol 2014, 159, 519–525. [Google Scholar] [CrossRef]
- Hempel, F.; Maurer, M.; Brockmann, B.; Mayer, C.; Biedenkopf, N.; Kelterbaum, A.; Becker, S.; Maier, U.G. From hybridomas to a robust microalgal-based production platform: Molecular design of a diatom secreting monoclonal antibodies directed against the Marburg virus nucleoprotein. Microb. Cell Fact. 2017, 16, 131. [Google Scholar] [CrossRef]
- Walsh, G. Biopharmaceutical benchmarks 2018. Nat. Biotechnol 2018, 36, 1136–1145. [Google Scholar] [CrossRef]
- Shaaltiel, Y.; Bartfeld, D.; Hashmueli, S.; Baum, G.; Brill-Almon, E.; Galili, G.; Dym, O.; Boldin-Adamsky, S.A.; Silman, I.; Sussman, J.L.; et al. Production of glucocerebrosidase with terminal mannose glycans for enzyme replacement therapy of Gaucher’s disease using a plant cell system. Plant. Biotechnol. J. 2007, 5, 579–590. [Google Scholar] [CrossRef]
- New antibodies best ZMapp in Ebola trial. Nat. Biotechnol. 2019, 37, 1105. [CrossRef]
- Walsh, C.T.; Garneau-Tsodikova, S.; Gatto, G.J., Jr. Protein posttranslational modifications: The chemistry of proteome diversifications. Angew. Chem. Int. Ed. Engl. 2005, 44, 7342–7372. [Google Scholar] [CrossRef]
- Amoresano, A.; Carpentieri, A.; Giangrande, C.; Palmese, A.; Chiappetta, G.; Marino, G.; Pucci, P. Technical advances in proteomics mass spectrometry: Identification of post-translational modifications. Clin. Chem. Lab. Med. 2009, 47, 647–665. [Google Scholar] [CrossRef]
- Deribe, Y.L.; Pawson, T.; Dikic, I. Post-translational modifications in signal integration. Nat. Struct. Mol. Biol. 2010, 17, 666–672. [Google Scholar] [CrossRef]
- Nikov, G.; Bhat, V.; Wishnok, J.S.; Tannenbaum, S.R. Analysis of nitrated proteins by nitrotyrosine-specific affinity probes and mass spectrometry. Anal. Biochem. 2003, 320, 214–222. [Google Scholar] [CrossRef]
- Swaney, D.L.; Beltrao, P.; Starita, L.; Guo, A.; Rush, J.; Fields, S.; Krogan, N.J.; Villen, J. Global analysis of phosphorylation and ubiquitylation cross-talk in protein degradation. Nat. Methods 2013, 10, 676–682. [Google Scholar] [CrossRef]
- Uversky, V.N. Wrecked regulation of intrinsically disordered proteins in diseases: Pathogenicity of deregulated regulators. Front. Mol. Biosci. 2014, 1, 6. [Google Scholar] [CrossRef] [PubMed]
- Ohtsubo, K.; Marth, J.D. Glycosylation in cellular mechanisms of health and disease. Cell 2006, 126, 855–867. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jefferis, R. Glycosylation as a strategy to improve antibody-based therapeutics. Nat. Rev. Drug Discov. 2009, 8, 226–234. [Google Scholar] [CrossRef]
- Shields, R.L.; Lai, J.; Keck, R.; O’Connell, L.Y.; Hong, K.; Meng, Y.G.; Weikert, S.H.; Presta, L.G. Lack of fucose on human IgG1 N-linked oligosaccharide improves binding to human FcγRIII and antibody-dependent cellular toxicity. J. Biol. Chem. 2002, 277, 26733–26740. [Google Scholar] [CrossRef] [Green Version]
- Niwa, R.; Natsume, A.; Uehara, A.; Wakitani, M.; Iida, S.; Uchida, K.; Satoh, M.; Shitara, K. IgG subclass-independent improvement of antibody-dependent cellular cytotoxicity by fucose removal from Asn297-linked oligosaccharides. J. Immunol. Methods 2005, 306, 151–160. [Google Scholar] [CrossRef]
- Shinkawa, T.; Nakamura, K.; Yamane, N.; Shoji-Hosaka, E.; Kanda, Y.; Sakurada, M.; Uchida, K.; Anazawa, H.; Satoh, M.; Yamasaki, M. The absence of fucose but not the presence of galactose or bisecting N-acetylglucosamine of human IgG1 complex-type oligosaccharides shows the critical role of enhancing antibody-dependent cellular cytotoxicity. J. Biol. Chem. 2003, 278, 3466–3473. [Google Scholar] [CrossRef] [Green Version]
- Kessler, M.; Goldsmith, D.; Schellekens, H. Immunogenicity of biopharmaceuticals. Nephrol Dial. Transpl. 2006, 21 (Suppl. 5), v9–v12. [Google Scholar] [CrossRef]
- van Beers, M.M.C.; Bardor, M. Minimizing immunogenicity of biopharmaceuticals by controlling critical quality attributes of proteins. Biotechnol. J. 2012, 7, 1473–1484. [Google Scholar] [CrossRef]
- Vanier, G.; Lucas, P.L.; Loutelier-Bourhis, C.; Vanier, J.; Plasson, C.; Walet-Balieu, M.L.; Tchi-Song, P.C.; Remy-Jouet, I.; Richard, V.; Bernard, S.; et al. Heterologous expression of the N-acetylglucosaminyltransferase I dictates a reinvestigation of the N-glycosylation pathway in Chlamydomonas reinhardtii. Sci. Rep. 2017, 7, 10156. [Google Scholar] [CrossRef] [Green Version]
- Marshall, R.D. The nature and metabolism of the carbohydrate-peptide linkages of glycoproteins. Biochem. Soc. Symp. 1974, 17–26. [Google Scholar]
- Zielinska, D.F.; Gnad, F.; Wisniewski, J.R.; Mann, M. Precision mapping of an in vivo N-glycoproteome reveals rigid topological and sequence constraints. Cell 2010, 141, 897–907. [Google Scholar] [CrossRef] [Green Version]
- Costa, A.R.; Rodrigues, M.E.; Henriques, M.; Oliveira, R.; Azeredo, J. Glycosylation: Impact, control and improvement during therapeutic protein production. Crit. Rev. Biotechnol. 2014, 34, 281–299. [Google Scholar] [CrossRef]
- Parodi, A.J. Role of N-oligosaccharide endoplasmic reticulum processing reactions in glycoprotein folding and degradation. Biochem. J. 2000, 348, 1. [Google Scholar] [CrossRef]
- Dell, A.; Morris, H.R. Glycoprotein Structure Determination by Mass Spectrometry. Science 2001, 291, 2351. [Google Scholar] [CrossRef]
- Harvey, D.J. Analysis of carbohydrates and glycoconjugates by matrix-assisted laser desorption/ionization mass spectrometry: An update for 2011-2012. Mass Spectrom. Rev. 2015. [Google Scholar] [CrossRef]
- Varelas, X.; Bouchie, M.P.; Kukuruzinska, M.A. Protein N-glycosylation in oral cancer: Dysregulated cellular networks among DPAGT1, E-cadherin adhesion and canonical Wnt signaling. Glycobiology 2014, 24, 579–591. [Google Scholar] [CrossRef] [Green Version]
- Wacker, M.; Linton, D.; Hitchen, P.G.; Nita-Lazar, M.; Haslam, S.M.; North, S.J.; Panico, M.; Morris, H.R.; Dell, A.; Wren, B.W.; et al. N-linked glycosylation in Campylobacter jejuni and its functional transfer into E. coli. (Reports). Science 2002, 298, 1790–1793. [Google Scholar] [CrossRef]
- Linton, D.; Dorrell, N.; Hitchen, P.G.; Amber, S.; Karlyshev, A.V.; Morris, H.R.; Dell, A.; Valvano, M.A.; Aebi, M.; Wren, B.W. Functional analysis of the Campylobacter jejuni N-linked protein glycosylation pathway. Mol. Microbiol 2005, 55, 1695–1703. [Google Scholar] [CrossRef]
- Tang, H.; Wang, S.; Wang, J.; Song, M.; Xu, M.; Zhang, M.; Shen, Y.; Hou, J.; Bao, X. N-hypermannose glycosylation disruption enhances recombinant protein production by regulating secretory pathway and cell wall integrity in Saccharomyces cerevisiae. Sci Rep. 2016, 6, 25654. [Google Scholar] [CrossRef]
- Parsaie Nasab, F.; Aebi, M.; Bernhard, G.; Frey, A.D. A combined system for engineering glycosylation efficiency and glycan structure in Saccharomyces cerevisiae. Appl Env. Microbiol 2013, 79, 997–1007. [Google Scholar] [CrossRef] [Green Version]
- Gomord, V.; Fitchette, A.C.; Menu-Bouaouiche, L.; Saint-Jore-Dupas, C.; Plasson, C.; Michaud, D.; Faye, L. Plant-specific glycosylation patterns in the context of therapeutic protein production. Plant. Biotechnol. J. 2010, 8, 564–587. [Google Scholar] [CrossRef] [PubMed]
- Mathieu-Rivet, E.; Scholz, M.; Arias, C.; Dardelle, F.; Schulze, S.; Le Mauff, F.; Teo, G.; Hochmal, A.K.; Blanco-Rivero, A.; Loutelier-Bourhis, C.; et al. Exploring the N-glycosylation pathway in Chlamydomonas reinhardtii unravels novel complex structures. Mol. Cell Proteom. 2013, 12, 3160–3183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Levy-Ontman, O.; Arad, S.M.; Harvey, D.J.; Parsons, T.B.; Fairbanks, A.; Tekoah, Y. Unique N-glycan moieties of the 66-kDa cell wall glycoprotein from the red microalga Porphyridium sp. J. Biol. Chem. 2011, 286, 21340–21352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Baiet, B.; Burel, C.; Saint-Jean, B.; Louvet, R.; Menu-Bouaouiche, L.; Kiefer-Meyer, M.C.; Mathieu-Rivet, E.; Lefebvre, T.; Castel, H.; Carlier, A.; et al. N-glycans of Phaeodactylum tricornutum diatom and functional characterization of its N-acetylglucosaminyltransferase I enzyme. J. Biol. Chem. 2011, 286, 6152–6164. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schulze, S.; Urzica, E.; Reijnders, M.; van de Geest, H.; Warris, S.; Bakker, L.V.; Fufezan, C.; Martins Dos Santos, V.A.P.; Schaap, P.J.; Peters, S.A.; et al. Identification of methylated GnTI-dependent N-glycans in Botryococcus brauni. New Phytol. 2017, 215, 1361–1369. [Google Scholar] [CrossRef] [Green Version]
- Mocsai, R.; Figl, R.; Troschl, C.; Strasser, R.; Svehla, E.; Windwarder, M.; Thader, A.; Altmann, F. N-glycans of the microalga Chlorella vulgaris are of the oligomannosidic type but highly methylated. Sci. Rep. 2019, 9, 331. [Google Scholar] [CrossRef] [Green Version]
- Lerouge, P.; Cabanes-Macheteau, M.; Rayon, C.; Fischette-Lainé, A.-C.; Gomord, V.; Faye, L. N-Glycoprotein biosynthesis in plants: Recent developments and future trends. Plant. Mol. Biol. 1998, 38, 31–48. [Google Scholar] [CrossRef]
- Zhang, P.; Burel, C.; Plasson, C.; Kiefer-Meyer, M.C.; Ovide, C.; Gugi, B.; Wan, C.; Teo, G.; Mak, A.; Song, Z.; et al. Characterization of a GDP-Fucose Transporter and a Fucosyltransferase Involved in the Fucosylation of Glycoproteins in the Diatom Phaeodactylum tricornutum. Front. Plant. Sci 2019, 10, 610. [Google Scholar] [CrossRef]
- Nakayama, Y.; Nakamura, N.; Tsuji, D.; Itoh, K.; Kurosak, A. Genetic Diseases Associated with Protein Glycosylation Disorders in Mammals. Genetic Disorders 2013. [Google Scholar] [CrossRef]
- Yusibov, V.; Kushnir, N.; Streatfield, S.J. Antibody Production in Plants and Green Algae. Annu Rev. Plant. Biol 2016, 67, 669–701. [Google Scholar] [CrossRef]
- Mumm, J.S.; Kopan, R. Notch signaling: From the outside in. Dev. Biol 2000, 228, 151–165. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rana, N.A.; Haltiwanger, R.S. Fringe benefits: Functional and structural impacts of O-glycosylation on the extracellular domain of Notch receptors. Curr. Opin. Struct. Biol. 2011, 21, 583–589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dobson, C.M.; Hempel, S.J.; Stalnaker, S.H.; Stuart, R.; Wells, L. O-Mannosylation and human disease. Cell Mol. Life Sci. 2013, 70, 2849–2857. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- De Pourcq, K.; De Schutter, K.; Callewaert, N. Engineering of glycosylation in yeast and other fungi: Current state and perspectives. Appl. Microbiol. Biotechnol. 2010, 87, 1617–1631. [Google Scholar] [CrossRef]
- Liu, J.; Jin, C.; Cherian, R.M.; Karlsson, N.G.; Holgersson, J. O-glycan repertoires on a mucin-type reporter protein expressed in CHO cell pools transiently transfected with O-glycan core enzyme cDNAs. J. Biotechnol. 2015, 199, 77–89. [Google Scholar] [CrossRef]
- Mitoma, J.; Petryniak, B.; Hiraoka, N.; Yeh, J.C.; Lowe, J.B.; Fukuda, M. Extended core 1 and core 2 branched O-glycans differentially modulate sialyl Lewis X-type L-selectin ligand activity. J. Biol. Chem. 2003, 278, 9953–9961. [Google Scholar] [CrossRef] [Green Version]
- Nguema-Ona, E.; Vicre-Gibouin, M.; Gotte, M.; Plancot, B.; Lerouge, P.; Bardor, M.; Driouich, A. Cell wall O-glycoproteins and N-glycoproteins: Aspects of biosynthesis and function. Front. Plant. Sci. 2014, 5, 499. [Google Scholar] [CrossRef] [Green Version]
- Xu, J.; Tan, L.; Goodrum, K.J.; Kieliszewski, M.J. High-yields and extended serum half-life of human interferon alpha2b expressed in tobacco cells as arabinogalactan-protein fusions. Biotechnol. Bioeng. 2007, 97, 997–1008. [Google Scholar] [CrossRef]
- Bollig, K.; Lamshoft, M.; Schweimer, K.; Marner, F.J.; Budzikiewicz, H.; Waffenschmidt, S. Structural analysis of linear hydroxyproline-bound O-glycans of Chlamydomonas reinhardtii--conservation of the inner core in Chlamydomonas and land plants. Carbohydr. Res. 2007, 342, 2557–2566. [Google Scholar] [CrossRef]
- Rice, P.; Longden, I.; Bleasby, A. EMBOSS: The European Molecular Biology Open Software Suite. Trends Genet. 2000, 16, 276–277. [Google Scholar] [CrossRef]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic local alignment search tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- El-Gebali, S.; Mistry, J.; Bateman, A.; Eddy, S.R.; Luciani, A.; Potter, S.C.; Qureshi, M.; Richardson, L.J.; Salazar, G.A.; Smart, A.; et al. The Pfam protein families database in 2019. Nucleic Acids Res. 2019, 47, D427–D432. [Google Scholar] [CrossRef] [PubMed]
- Johnson, L.S.; Eddy, S.R.; Portugaly, E. Hidden Markov model speed heuristic and iterative HMM search procedure. BMC Bioinform. 2010, 11, 431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lucas, P.-L.; Mathieu-Rivet, E.; Chan Tchi Song, P.; Oltmanns, A.; Loutelier-Bourhis, C.; Plasson, C.; Afonso, C.; Hippler, M.; Lerouge, P.; Mati-Baouche, N.; et al. Multiple xylosyltransferases heterogeneously xylosylate protein N-linked glycans in Chlamydomonas reinhardtii. Plant. J. 2019. [Google Scholar] [CrossRef]
- Kowarik, M.; Young, N.M.; Numao, S.; Schulz, B.L.; Hug, I.; Callewaert, N.; Mills, D.C.; Watson, D.C.; Hernandez, M.; Kelly, J.F.; et al. Definition of the bacterial N-glycosylation site consensus sequence. EMBO J. 2006, 25, 1957–1966. [Google Scholar] [CrossRef] [Green Version]
- Schwarz, D.; Daley, D.; Beckhaus, T.; Dötsch, V.; Bernhard, F. Cell-free expression profiling of E. coli inner membrane proteins. Proteomics 2010, 10, 1762–1779. [Google Scholar] [CrossRef]
- Hamilton, S.R.; Davidson, R.C.; Sethuraman, N.; Nett, J.H.; Jiang, Y.; Rios, S.; Bobrowicz, P.; Stadheim, T.A.; Li, H.; Choi, B.K.; et al. Humanization of yeast to produce complex terminally sialylated glycoproteins. Science 2006, 313, 1441–1443. [Google Scholar] [CrossRef] [Green Version]
- Malphettes, L.; Freyvert, Y.; Chang, J.; Liu, P.Q.; Chan, E.; Miller, J.C.; Zhou, Z.; Nguyen, T.; Tsai, C.; Snowden, A.W.; et al. Highly efficient deletion of FUT8 in CHO cell lines using zinc-finger nucleases yields cells that produce completely nonfucosylated antibodies. Biotechnol. Bioeng. 2010, 106, 774–783. [Google Scholar] [CrossRef]
- Ronda, C.; Pedersen, L.E.; Hansen, H.G.; Kallehauge, T.B.; Betenbaugh, M.J.; Nielsen, A.T.; Kildegaard, H.F. Accelerating genome editing in CHO cells using CRISPR Cas9 and CRISPy, a web-based target finding tool. Biotechnol. Bioeng. 2014, 111, 1604–1616. [Google Scholar] [CrossRef] [Green Version]
- Piron, R.; Santens, F.; De Paepe, A.; Depicker, A.; Callewaert, N. Using GlycoDelete to produce proteins lacking plant-specific N-glycan modification in seeds. Nat. Biotechnol. 2015, 33, 1135. [Google Scholar] [CrossRef]
- Castilho, A.; Steinkellner, H. Glyco-engineering in plants to produce human-like N-glycan structures. Biotechnol J. 2012, 7, 1088–1098. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Q.; Shankara, S.; Roy, A.; Qiu, H.; Estes, S.; McVie-Wylie, A.; Culm-Merdek, K.; Park, A.; Pan, C.; Edmunds, T. Development of a simple and rapid method for producing non-fucosylated oligomannose containing antibodies with increased effector function. Biotechnol. Bioeng. 2008, 99, 652–665. [Google Scholar] [CrossRef] [PubMed]
- Wang, L.X.; Lomino, J.V. Emerging technologies for making glycan-defined glycoproteins. ACS Chem. Biol. 2012, 7, 110–122. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chalker, J.M.; Bernardes, G.J.L.; Davis, B.G. A “Tag-and-Modify” Approach to Site-Selective Protein Modification. Acc. Chem. Res. 2011, 44, 730–741. [Google Scholar] [CrossRef] [PubMed]
- Narasimhan, S. Control of glycoprotein synthesis. UDP-GlcNAc:glycopeptide beta 4-N-acetylglucosaminyltransferase III, an enzyme in hen oviduct which adds GlcNAc in beta 1-4 linkage to the beta-linked mannose of the trimannosyl core of N-glycosyl oligosaccharides. J. Biol. Chem. 1982, 257, 10235–10242. [Google Scholar]
- Sizova, I.; Greiner, A.; Awasthi, M.; Kateriya, S.; Hegemann, P. Nuclear gene targeting in Chlamydomonas using engineered zinc-finger nucleases. Plant J. 2013, 73, 873–882. [Google Scholar] [CrossRef]
- Daboussi, F.; Leduc, S.; Marechal, A.; Dubois, G.; Guyot, V.; Perez-Michaut, C.; Amato, A.; Falciatore, A.; Juillerat, A.; Beurdeley, M.; et al. Genome engineering empowers the diatom Phaeodactylum tricornutum for biotechnology. Nat. Commun. 2014, 5, 3831. [Google Scholar] [CrossRef]
- Jiang, W.; Bikard, D.; Cox, D.; Zhang, F.; Marraffini, L.A. RNA-guided editing of bacterial genomes using CRISPR-Cas systems. Nat. Biotechnol 2013, 31, 233–239. [Google Scholar] [CrossRef]
- Nymark, M.; Sharma, A.K.; Sparstad, T.; Bones, A.M.; Winge, P. A CRISPR/Cas9 system adapted for gene editing in marine algae. Sci. Rep. 2016, 6, 24951. [Google Scholar] [CrossRef]
- Wang, Q.; Lu, Y.; Xin, Y.; Wei, L.; Huang, S.; Xu, J. Genome editing of model oleaginous microalgae Nannochloropsis spp. by CRISPR/Cas9. Plant. J. 2016, 88, 1071–1081. [Google Scholar] [CrossRef]
- Elliott, S.; Lorenzini, T.; Asher, S.; Aoki, K.; Brankow, D.; Buck, L.; Busse, L.; Chang, D.; Fuller, J.; Grant, J.; et al. Enhancement of therapeutic protein in vivo activities through glycoengineering. Nat. Biotechnol. 2003, 21, 414–421. [Google Scholar] [CrossRef] [PubMed]
- Perlman, S.; van den Hazel, B.; Christiansen, J.; Gram-Nielsen, S.; Jeppesen, C.B.; Andersen, K.V.; Halkier, T.; Okkels, S.; Schambye, H.T. Glycosylation of an N-Terminal Extension Prolongs the Half-Life and Increases the in Vivo Activity of Follicle Stimulating Hormone. J. Clin. Endocrinol. Metab. 2003, 88, 3227–3235. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Runkel, L.; Meier, W.; Pepinsky, R.B.; Karpusas, M.; Whitty, A.; Kimball, K.; Brickelmaier, M.; Muldowney, C.; Jones, W.; Goelz, S.E. Structural and Functional Differences Between Glycosylated and Non-glycosylated Forms of Human Interferon-β (IFN-β). Pharm. Res. 1998, 15, 641–649. [Google Scholar] [CrossRef] [PubMed]
- Sola, R.J.; Griebenow, K. Effects of glycosylation on the stability of protein pharmaceuticals. J. Pharm. Sci. 2009, 98, 1223–1245. [Google Scholar] [CrossRef] [Green Version]
- Shebanova, A.; Ismagulova, T.; Solovchenko, A.; Baulina, O.; Lobakova, E.; Ivanova, A.; Moiseenko, A.; Shaitan, K.; Polshakov, V.; Nedbal, L.; et al. Versatility of the green microalga cell vacuole function as revealed by analytical transmission electron microscopy. Protoplasma 2017, 254, 1323–1340. [Google Scholar] [CrossRef]
- Schreiber, V.; Dersch, J.; Puzik, K.; Backer, O.; Liu, X.; Stork, S.; Schulz, J.; Heimerl, T.; Klingl, A.; Zauner, S.; et al. The Central Vacuole of the Diatom Phaeodactylum tricornutum: Identification of New Vacuolar Membrane Proteins and of a Functional Di-leucine-based Targeting Motif. Protist 2017, 168, 271–282. [Google Scholar] [CrossRef]
- Wei, C.; Zhou, X.; Zhang, Y. Improving intracellular production of recombinant protein in Pichia pastoris using an optimized preinduction glycerol-feeding scheme. Appl. Microbiol. Biotechnol. 2008, 78, 257–264. [Google Scholar] [CrossRef]
- Zou, G.; Ochiai, H.; Huang, W.; Yang, Q.; Li, C.; Wang, L.-X. Chemoenzymatic Synthesis and Fcγ Receptor Binding of Homogeneous Glycoforms of Antibody Fc Domain. Presence of a Bisecting Sugar Moiety Enhances the Affinity of Fc to FcγIIIa Receptor. J. Am. Chem. Soc. 2011, 133, 18975–18991. [Google Scholar] [CrossRef] [Green Version]
- Fabris, M.; Matthijs, M.; Carbonelle, S.; Moses, T.; Pollier, J.; Dasseville, R.; Baart, G.J.; Vyverman, W.; Goossens, A. Tracking the sterol biosynthesis pathway of the diatom Phaeodactylum tricornutum. New Phytol. 2014, 204, 521–535. [Google Scholar] [CrossRef]
- Pollier, J.; Vancaester, E.; Kuzhiumparambil, U.; Vickers, C.E.; Vandepoele, K.; Goossens, A.; Fabris, M. A widespread alternative squalene epoxidase participates in eukaryote steroid biosynthesis. Nat. Microbiol 2019, 4, 226–233. [Google Scholar] [CrossRef]
- Schuster, M.; Umana, P.; Ferrara, C.; Brunker, P.; Gerdes, C.; Waxenecker, G.; Wiederkum, S.; Schwager, C.; Loibner, H.; Himmler, G.; et al. Improved effector functions of a therapeutic monoclonal Lewis Y-specific antibody by glycoform engineering. Cancer Res. 2005, 65, 7934–7941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cameron, F.; McCormack, P.L. Obinutuzumab: First Global Approval. Drugs 2014, 74, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Cheng, J.K.; Lewis, A.M.; Kim, D.S.; Dyess, T.; Alper, H.S. Identifying and retargeting transcriptional hot spots in the human genome. Biotechnol. J. 2016, 11, 1100–1109. [Google Scholar] [CrossRef] [PubMed]
- Lee, J.S.; Kallehauge, T.B.; Pedersen, L.E.; Kildegaard, H.F. Site-specific integration in CHO cells mediated by CRISPR/Cas9 and homology-directed DNA repair pathway. Sci. Rep. 2015, 5, 8572. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Papapetrou, E.P.; Schambach, A. Gene Insertion Into Genomic Safe Harbors for Human Gene Therapy. Mol. Therapy 2016, 24, 678–684. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Clausen, H.; Wandall, H.H.; Steentoft, C.; Stanley, P.; Schnaar, R.L. Glycosylation engineering. In Essentials of Glycobiology [Internet], 3rd ed.; Cold Spring Harbor Laboratory Press: Cold Spring Harbor, NY, USA, 2017. [Google Scholar]
- Omasa, T.; Tanaka, R.; Doi, T.; Ando, M.; Kitamoto, Y.; Honda, K.; Kishimoto, M.; Ohtake, H. Decrease in antithrombin III fucosylation by expressing GDP-fucose transporter siRNA in Chinese hamster ovary cells. J. Biosci. Bioeng. 2008, 106, 168–173. [Google Scholar] [CrossRef]
- Myrbraten, I.S.; Wiull, K.; Salehian, Z.; Havarstein, L.S.; Straume, D.; Mathiesen, G.; Kjos, M. CRISPR Interference for Rapid Knockdown of Essential Cell Cycle Genes in Lactobacillus plantarum. mSphere 2019, 4. [Google Scholar] [CrossRef] [Green Version]
- Cheng, X.; Liu, G.; Ke, W.; Zhao, L.; Lv, B.; Ma, X.; Xu, N.; Xia, X.; Deng, X.; Zheng, C.; et al. Building a multipurpose insertional mutant library for forward and reverse genetics in Chlamydomonas. Plant. Methods 2017, 13, 36. [Google Scholar] [CrossRef]
- Schulze, S.; Oltmanns, A.; Machnik, N.; Liu, G.; Xu, N.; Jarmatz, N.; Scholz, M.; Sugimoto, K.; Fufezan, C.; Huang, K.; et al. N-Glycoproteomic Characterization of Mannosidase and Xylosyltransferase Mutant Strains of Chlamydomonasreinhardtii. Plant. Physiol. 2018, 176, 1952–1964. [Google Scholar] [CrossRef] [Green Version]
- Pereira, N.A.; Chan, K.F.; Lin, P.C.; Song, Z. The “less-is-more” in therapeutic antibodies: Afucosylated anti-cancer antibodies with enhanced antibody-dependent cellular cytotoxicity. MAbs 2018, 10, 693–711. [Google Scholar] [CrossRef]
- Bobrowicz, P.; Davidson, R.C.; Li, H.; Potgieter, T.I.; Nett, J.H.; Hamilton, S.R.; Stadheim, T.A.; Miele, R.G.; Bobrowicz, B.; Mitchell, T.; et al. Engineering of an artificial glycosylation pathway blocked in core oligosaccharide assembly in the yeast Pichia pastoris: Production of complex humanized glycoproteins with terminal galactose. Glycobiology 2004, 14, 757–766. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cox, K.M.; Sterling, J.D.; Regan, J.T.; Gasdaska, J.R.; Frantz, K.K.; Peele, C.G.; Black, A.; Passmore, D.; Moldovan-Loomis, C.; Srinivasan, M.; et al. Glycan optimization of a human monoclonal antibody in the aquatic plant Lemna minor. Nat. Biotechnol. 2006, 24, 1591–1597. [Google Scholar] [CrossRef] [PubMed]
- Strasser, R.; Altmann, F.; Mach, L.; Glössl, J.; Steinkellner, H. Generation of Arabidopsis thaliana plants with complex N-glycans lacking β1,2-linked xylose and core α1,3-linked fucose. Febs Lett. 2004, 561, 132–136. [Google Scholar] [CrossRef] [Green Version]
- Strasser, R.; Stadlmann, J.; Schahs, M.; Stiegler, G.; Quendler, H.; Mach, L.; Glossl, J.; Weterings, K.; Pabst, M.; Steinkellner, H. Generation of glyco-engineered Nicotiana benthamiana for the production of monoclonal antibodies with a homogeneous human-like N-glycan structure. Plant. Biotechnol. J. 2008, 6, 392–402. [Google Scholar] [CrossRef]
- Castilho, A.; Strasser, R.; Stadlmann, J.; Grass, J.; Jez, J.; Gattinger, P.; Kunert, R.; Quendler, H.; Pabst, M.; Leonard, R.; et al. In planta protein sialylation through overexpression of the respective mammalian pathway. J. Biol. Chem. 2010, 285, 15923–15930. [Google Scholar] [CrossRef] [Green Version]
- Castilho, A.; Gattinger, P.; Grass, J.; Jez, J.; Pabst, M.; Altmann, F.; Gorfer, M.; Strasser, R.; Steinkellner, H. N-glycosylation engineering of plants for the biosynthesis of glycoproteins with bisected and branched complex N-glycans. Glycobiology 2011, 21, 813–823. [Google Scholar] [CrossRef] [Green Version]
- Weikert, S.; Papac, D.; Briggs, J.; Cowfer, D.; Tom, S.; Gawlitzek, M.; Lofgren, J.; Mehta, S.; Chisholm, V.; Modi, N.; et al. Engineering Chinese hamster ovary cells to maximize sialic acid content of recombinant glycoproteins. Nat. Biotechnol. 1999, 17, 1116–1121. [Google Scholar] [CrossRef]
- Bork, K.; Reutter, W.; Weidemann, W.; Horstkorte, R. Enhanced sialylation of EPO by overexpression of UDP-GlcNAc 2-epimerase/ManAc kinase containing a sialuria mutation in CHO cells. Febs Lett. 2007, 581, 4195–4198. [Google Scholar] [CrossRef] [Green Version]
- Goh, J.S.Y.; Liu, Y.; Liu, H.; Chan, K.F.; Wan, C.; Teo, G.; Zhou, X.; Xie, F.; Zhang, P.; Zhang, Y.; et al. Highly sialylated recombinant human erythropoietin production in large-scale perfusion bioreactor utilizing CHO-gmt4 (JW152) with restored GnT I function. Biotechnol. J. 2014, 9, 100–109. [Google Scholar] [CrossRef]



| Organism | Organelle | Protein | Reference |
|---|---|---|---|
| C. reinhardtii | Chloroplast | E7 of HPV-16 | [48] |
| D2-CTB | [49] | ||
| α-galactosidase | [50] | ||
| Phytase | [50] | ||
| Xylanase | [50] | ||
| Pfs25 | [51] | ||
| Pfs28 | [51] | ||
| Pfs25-CTB | [52] | ||
| E2 | [53] | ||
| Pfs48/45 | [54] | ||
| M-SAA | [55] | ||
| Anti-HSV glycoprotein D Isc | [56] | ||
| 12FN3 | [57] | ||
| Erythropoietin | [57] | ||
| HMGB1 | [57] | ||
| Interferon β | [57] | ||
| Proinsulin | [57] | ||
| SAA-10FN3 | [57] | ||
| VEGF | [57] | ||
| Allophycocyanin | [58] | ||
| VP1-CTB | [59] | ||
| V28 | [60] | ||
| Anti-PA 83 anthrax IgG1 | [61] | ||
| Anti-CD22-gelonin sc | [62] | ||
| Anti-CD22-ETA sc | [63] | ||
| GAD65 | [64] | ||
| TRAIL | [65] | ||
| Phytase (AppA) | [66] | ||
| Metallothionein-2 | [67] | ||
| C. reinhardtii | Nucleus | Human Epidermal Growth Factor | [68] |
| VEGF-165 | [69] | ||
| GBSS-AMA1 | [70] | ||
| GBSS-MSP1 | [70] | ||
| Erythropoietin | [42] | ||
| Sep-15 | [71] | ||
| Lolium Perenme IBP | [72] | ||
| β-1,4-endoxylanase | [73] | ||
| C. vulgaris | Nucleus | Human growth hormone | [14] |
| C. sorokiniana | |||
| C. ellipsoidea | Nucleus | mNP-1 | [15] |
| NP-1 | [74] | ||
| Flounder growth hormone | [75] | ||
| D. salina | Chloroplast | α-galactosidase | [50] |
| Phytase | [50] | ||
| Xylanase | [50] | ||
| D. salina | Nucleus | V28 | [76] |
| HBsAg | [16] | ||
| P. tricornutum | Nucleus | Anti-Hepatitis B IgG | [19] |
| Anti-MARV NP IgG | [77] | ||
| N. oculata | Nucleus | Bovine lactoferricin (LFB) | [18] |
| Nucleus | Flounder growth hormone | [17] |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Barolo, L.; Abbriano, R.M.; Commault, A.S.; George, J.; Kahlke, T.; Fabris, M.; Padula, M.P.; Lopez, A.; Ralph, P.J.; Pernice, M. Perspectives for Glyco-Engineering of Recombinant Biopharmaceuticals from Microalgae. Cells 2020, 9, 633. https://doi.org/10.3390/cells9030633
Barolo L, Abbriano RM, Commault AS, George J, Kahlke T, Fabris M, Padula MP, Lopez A, Ralph PJ, Pernice M. Perspectives for Glyco-Engineering of Recombinant Biopharmaceuticals from Microalgae. Cells. 2020; 9(3):633. https://doi.org/10.3390/cells9030633
Chicago/Turabian StyleBarolo, Lorenzo, Raffaela M. Abbriano, Audrey S. Commault, Jestin George, Tim Kahlke, Michele Fabris, Matthew P. Padula, Angelo Lopez, Peter J. Ralph, and Mathieu Pernice. 2020. "Perspectives for Glyco-Engineering of Recombinant Biopharmaceuticals from Microalgae" Cells 9, no. 3: 633. https://doi.org/10.3390/cells9030633
APA StyleBarolo, L., Abbriano, R. M., Commault, A. S., George, J., Kahlke, T., Fabris, M., Padula, M. P., Lopez, A., Ralph, P. J., & Pernice, M. (2020). Perspectives for Glyco-Engineering of Recombinant Biopharmaceuticals from Microalgae. Cells, 9(3), 633. https://doi.org/10.3390/cells9030633







