Tissue-Specific Metabolic Regulation of FOXO-Binding Protein: FOXO Does Not Act Alone
Abstract
:1. Introduction
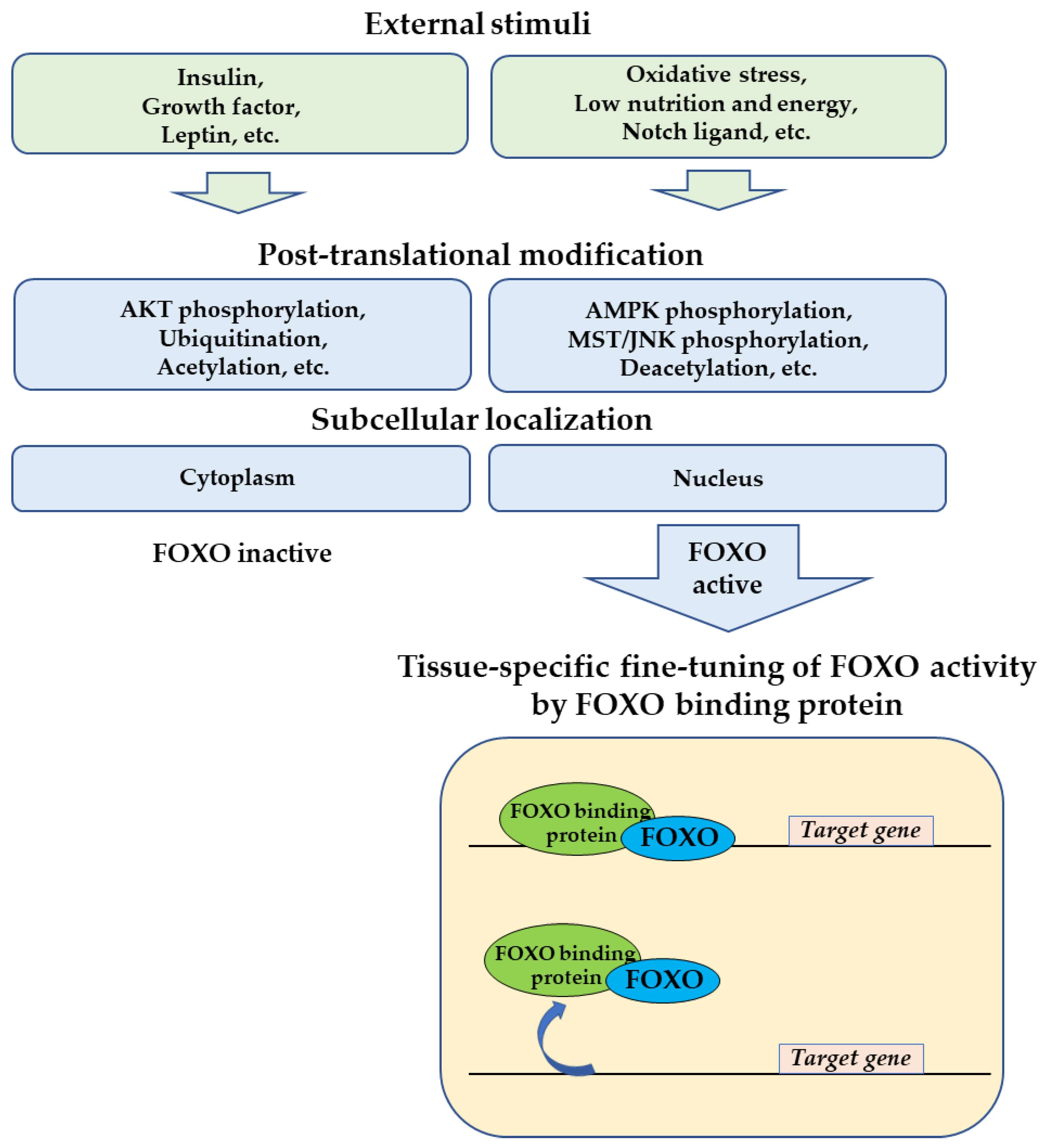
2. General Regulation of FOXOs by FOXO-Binding Proteins
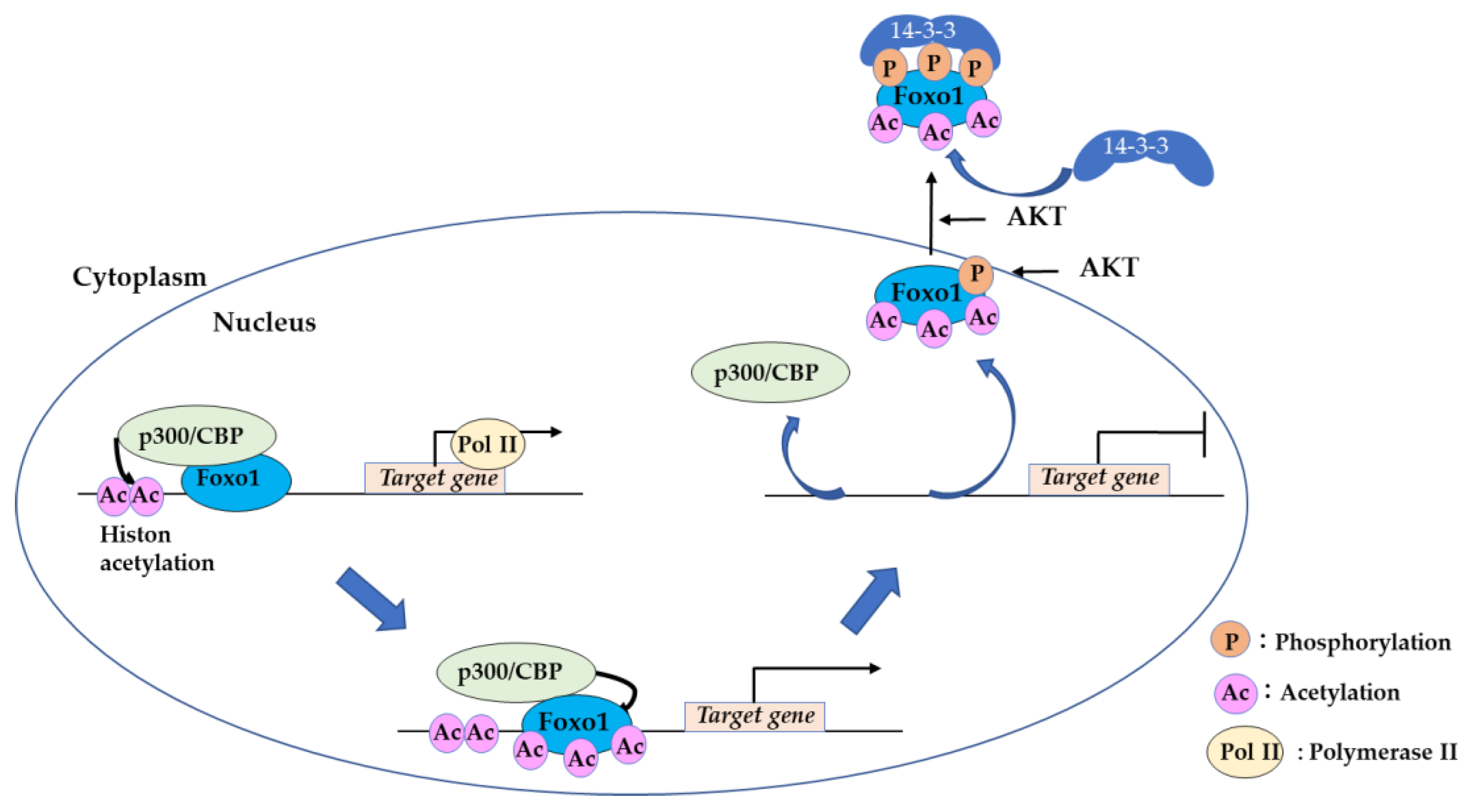
2.1. 14-3-3 Proteins Bind to Phosphorylated FOXO and Suppress DNA Binding
2.2. Acetylation of FOXO by CREB-Binding Protein (CBP) is a “Hit and Run” Regulation
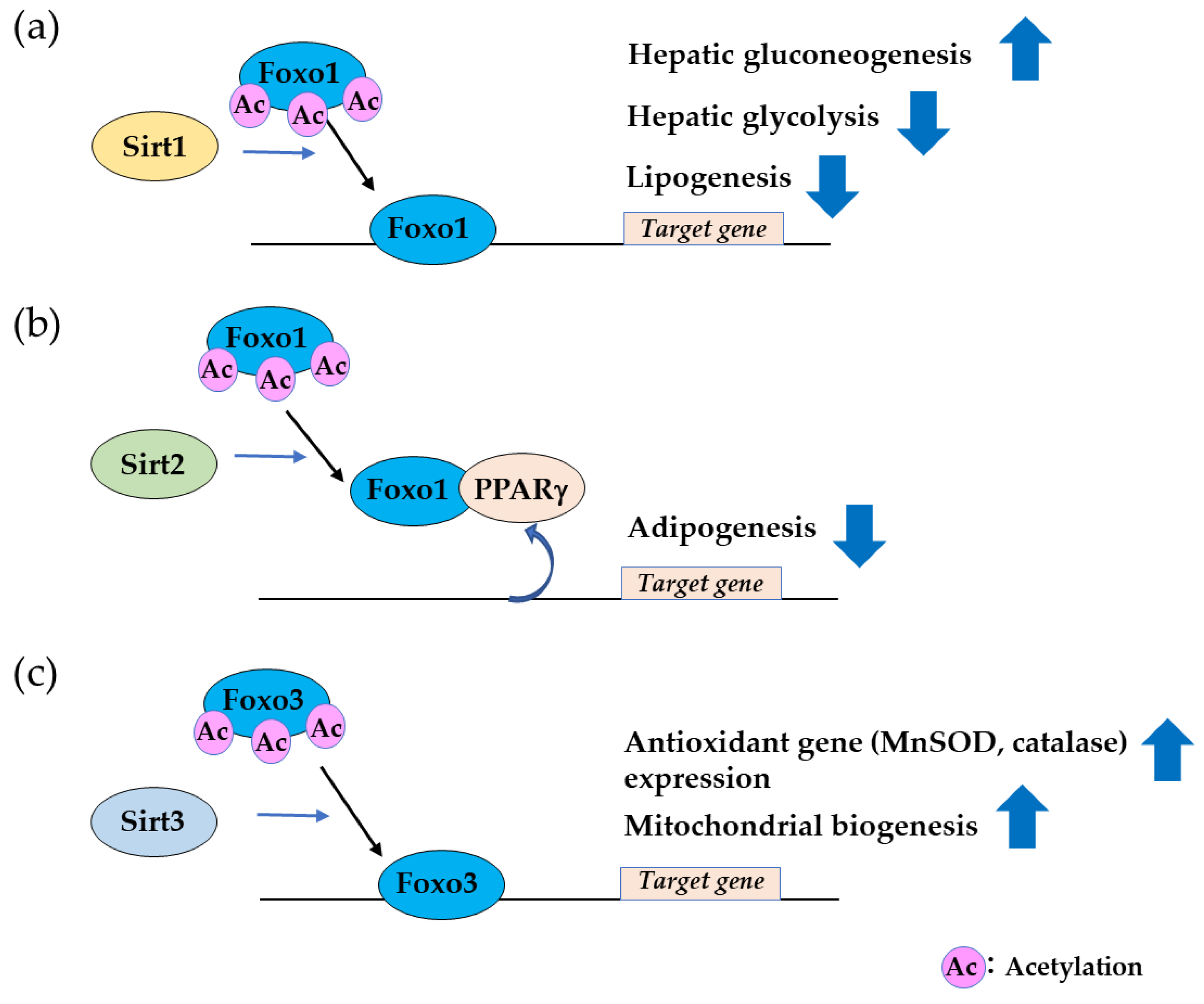
2.3. Sirtuin Family Proteins Bind and Deacetylate FOXO, Leading to Nuclear Localization.
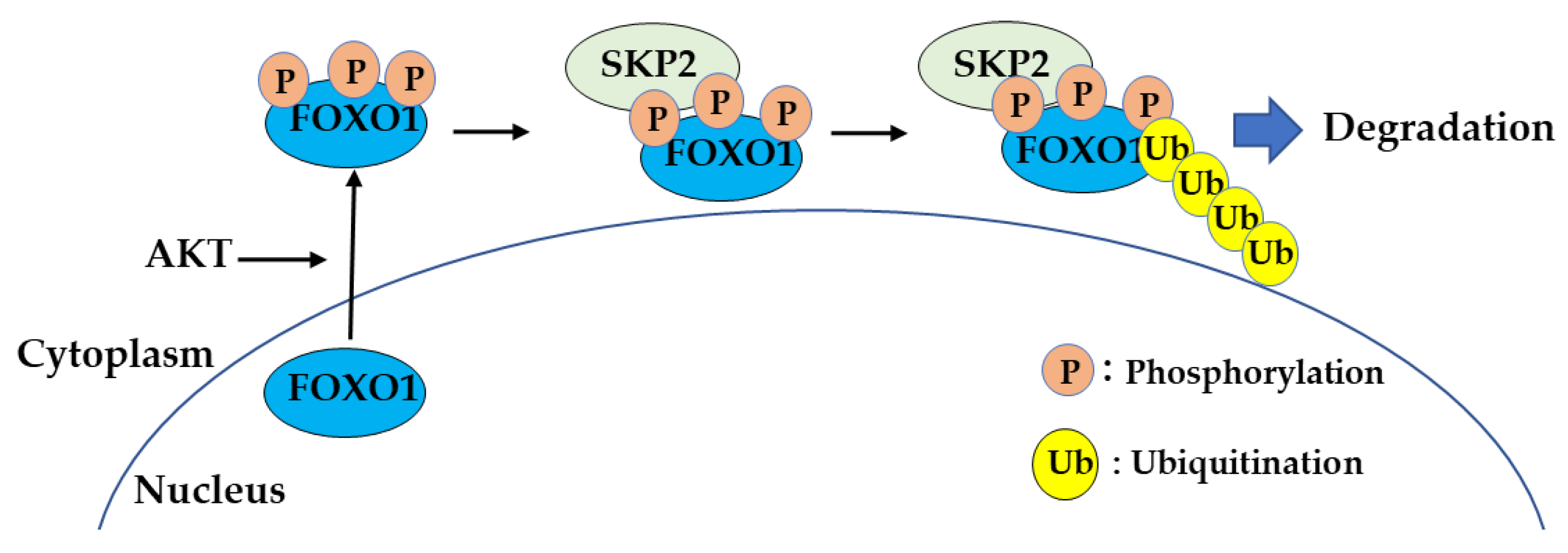
2.4. Degradation of FOXO1 by Ubiquitination is Controlled by SKP2 Binding
3. Tissue-Specific Function of FOXO1-Binding Protein in Insulin Responsive Tissues
3.1. Adipocyte
3.1.1. FOXO1 Binding to PPARγ Antagonizes Its Function in Adipocytes
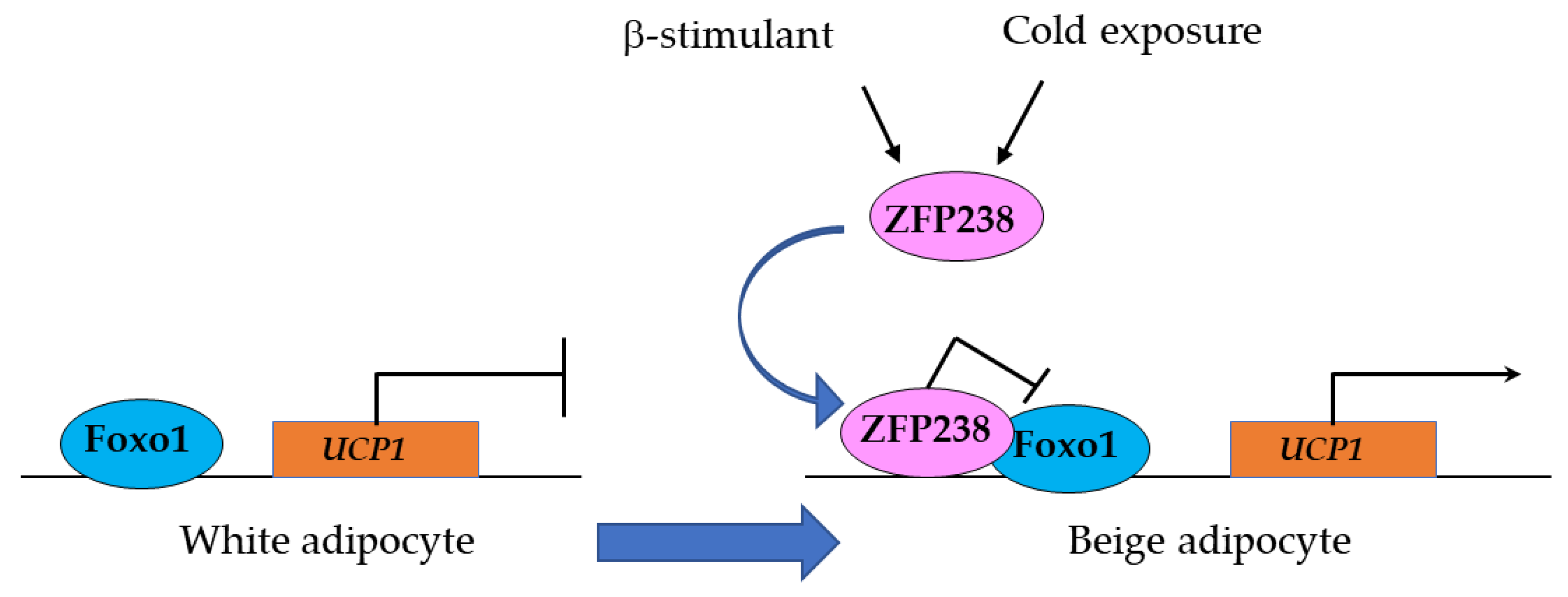
3.1.2. Zfp238 Regulates the Thermogenic Program in Cooperation with Foxo1
3.2. Liver
3.2.1. PPARα Binding to FOXO1 Suppresses apoC-III Expression in the Liver
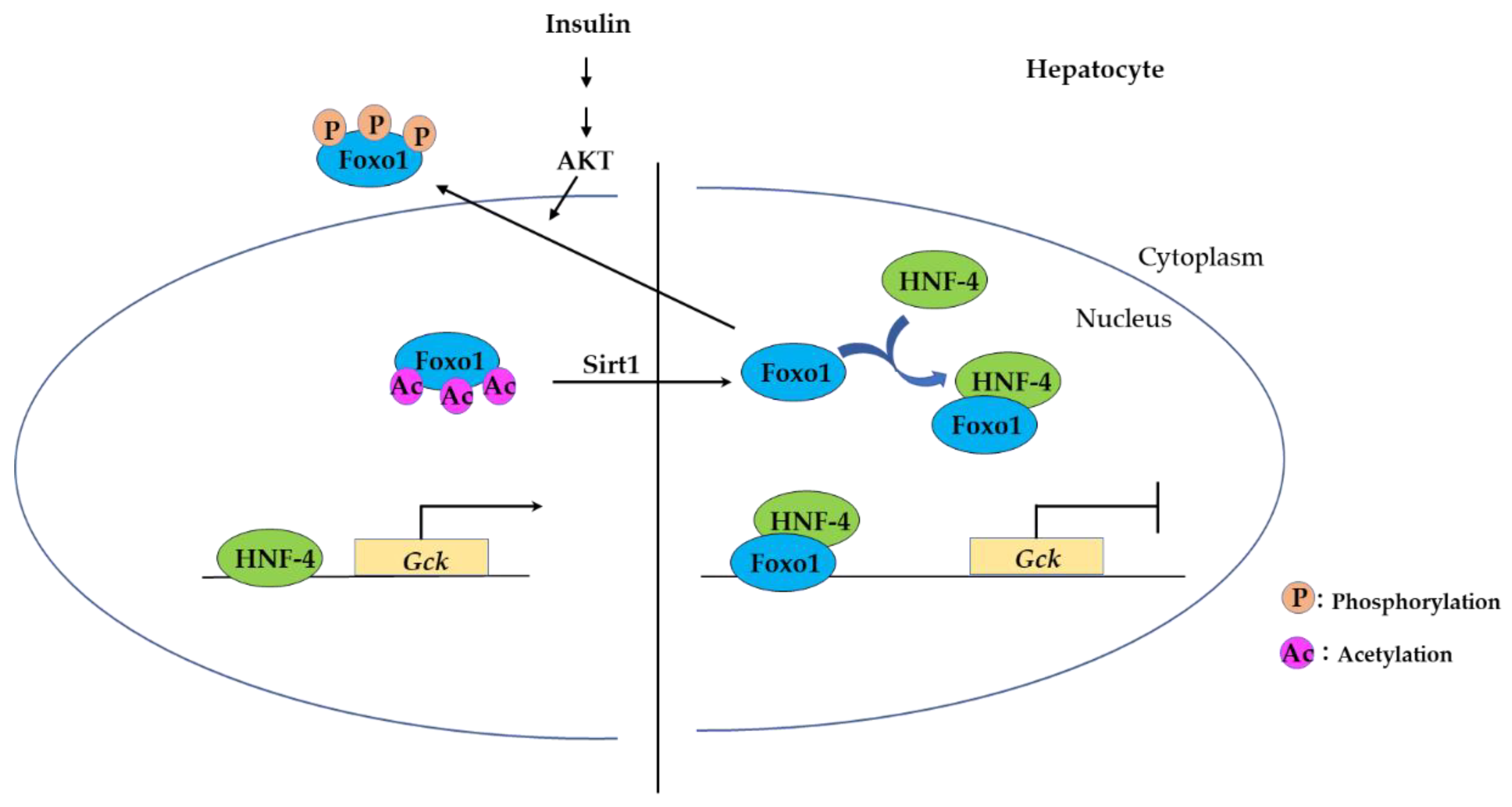
3.2.2. FOXO Binding to Hepatocyte Nuclear Factor-4 (HNF-4) Represses Expression of HNF-4 Target Genes
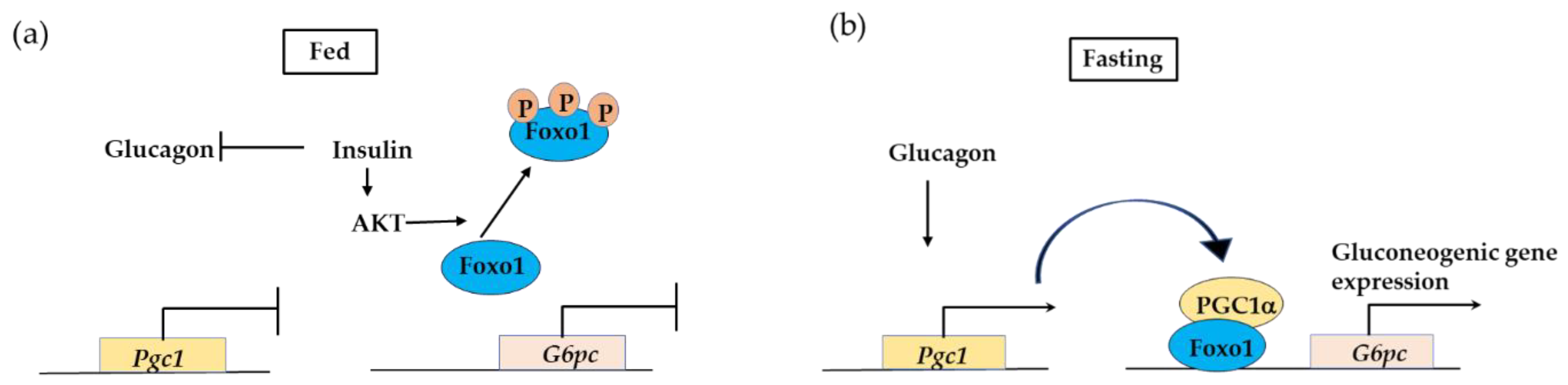
3.2.3. PPARγ Coactivator 1a (PGC1α) Interaction with FOXO1 Is Essential in Insulin-Regulated Hepatic Gluconeogenesis
3.2.4. FOXO1 Binding to C/EBPα Regulates Gluconeogenesis During Liver Development
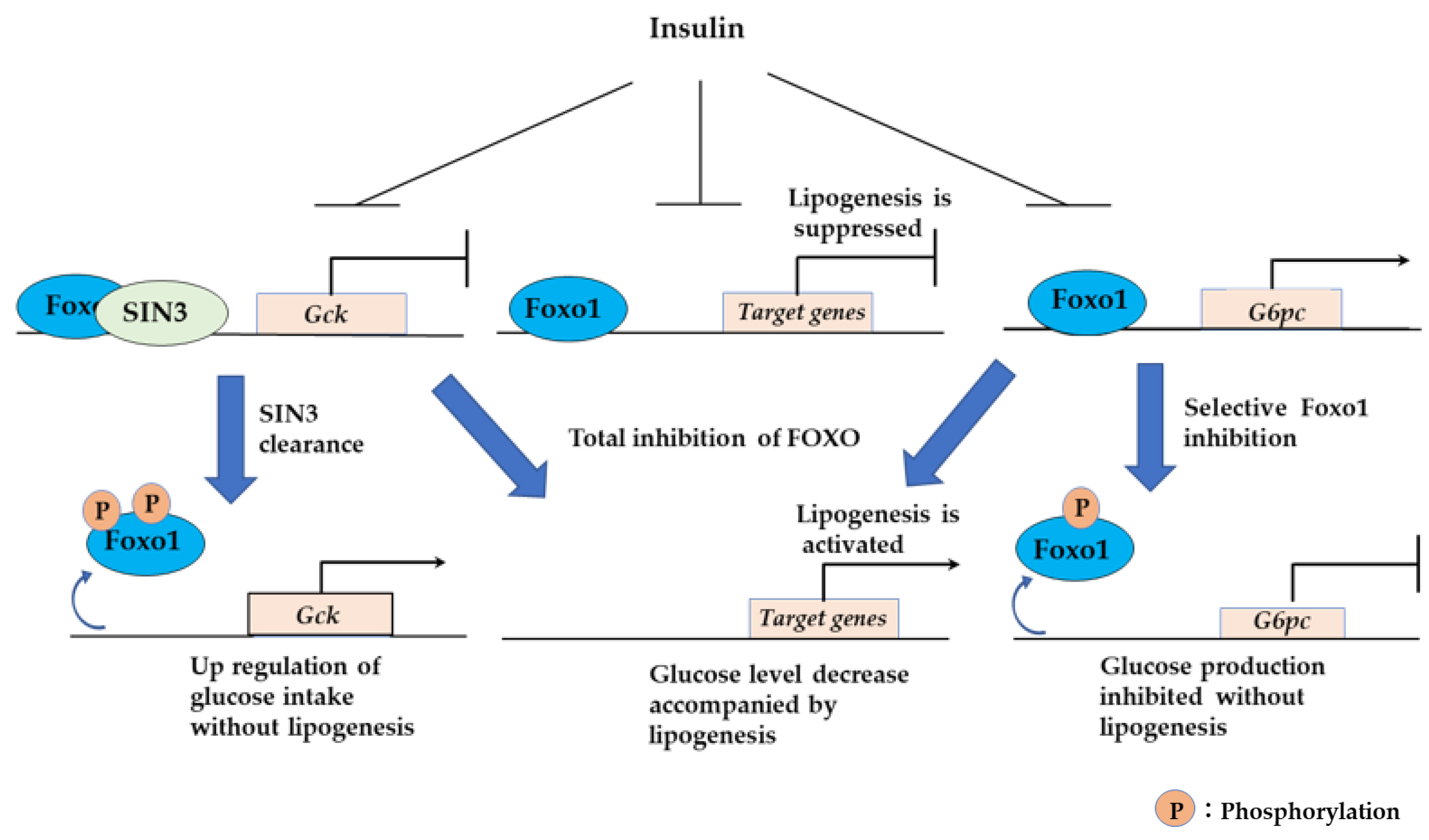
3.2.5. Sin3a Interaction with FOXO1 Confers Selective Regulation to Expression of G6pc and Gck in the Liver
3.3. Pancreas
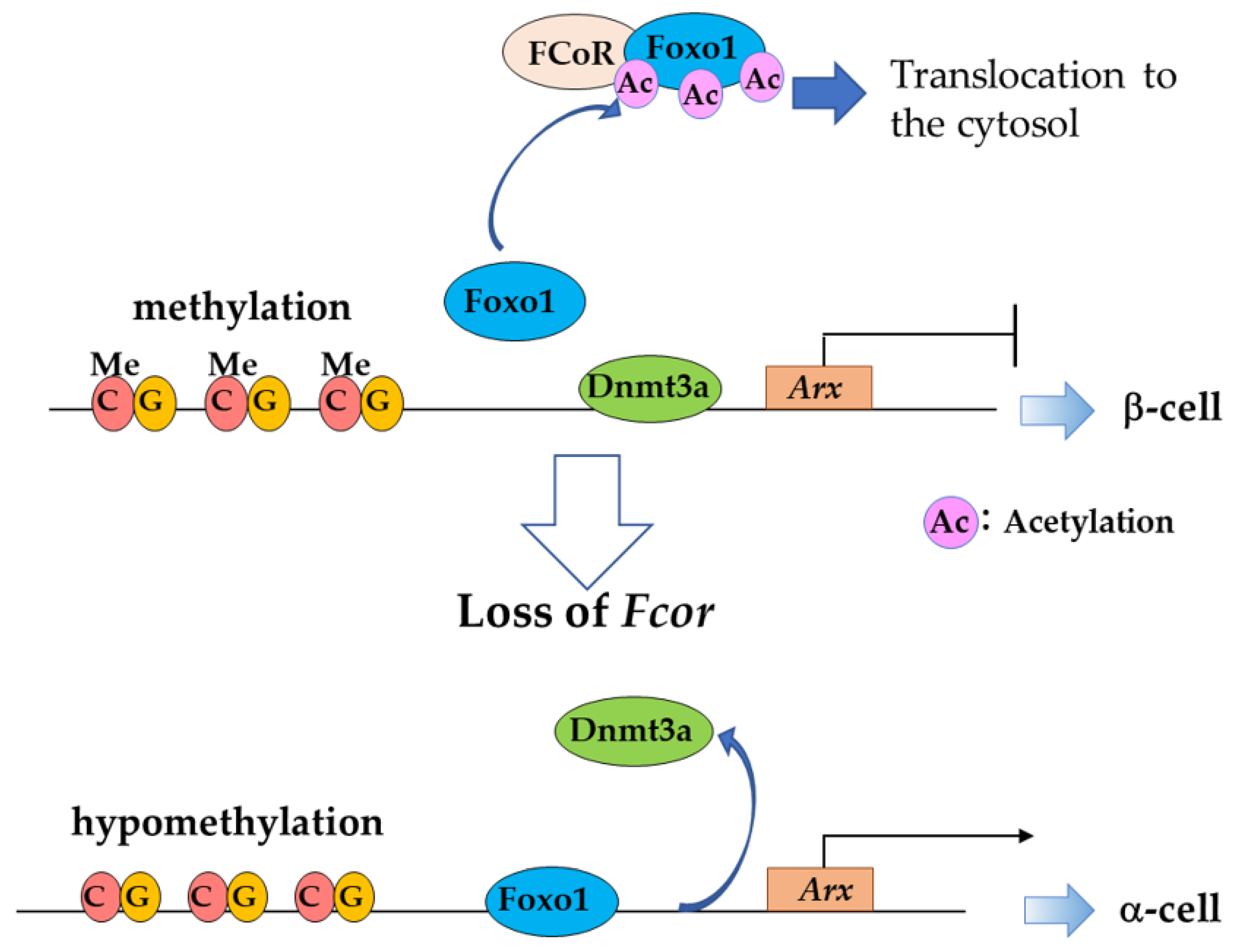
Foxo1 Corepressor (FCoR) Is a Main Regulator of Foxo1 Acetylation in Adipocytes and Pancreatic α- and β-cells
3.4. Smooth Muscle and Skeletal Muscle
3.4.1. Foxo4 Interacts with Myocardin and Represses Smooth Muscle Cell Differentiation
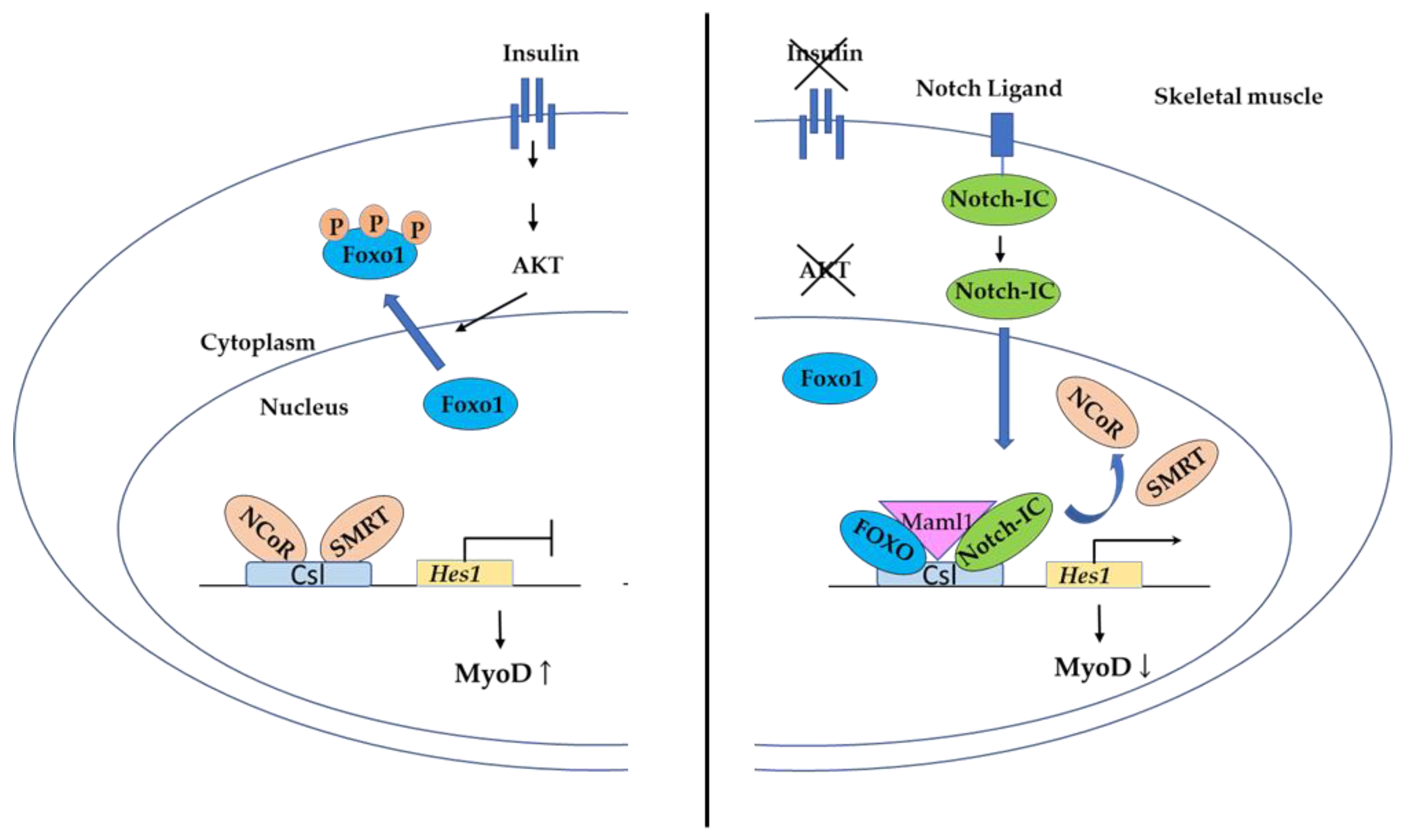
3.4.2. FOXO Binding to Csl in the Notch Pathway Controls Myogenic Differentiation and Fiber Type Specification in Skeletal Muscle
3.5. Cardiac Muscle
Sirt1–Foxo1 Interaction Activates Autophagy Flux Under Energy Deficiency in Cardiac Muscle
3.6. Hypothalamus
Sirt1 May Function as an Energy Sensor through FOXO1 Regulation in Hypothalamus
4. Conclusions and Future Perspectives
Author Contributions
Funding
Conflicts of Interest
References
- Accili, D.; Arden, K.C. FoxOs at the crossroads of cellular metabolism, differentiation, and transformation. Cell 2004, 117, 421–426. [Google Scholar] [CrossRef] [Green Version]
- Galili, N.; Davis, R.J.; Fredericks, W.J.; Mukhopadhyay, S.; Rauscher, F.J.; Emanuel, B.S.; Rovera, G.; Barr, F.G. Fusion of a fork head domain gene to PAX3 in the solid tumour alveolar rhabdomyosarcoma. Nat. Genet. 1993, 5, 230–235. [Google Scholar] [CrossRef] [PubMed]
- Hillion, J.; Le Coniat, M.; Jonveaux, P.; Berger, R.; Bernard, O.A. AF6q21, a novel partner of the MLL gene in t(6;11)(q21;q23), defines a forkhead transcriptional factor subfamily. Blood 1997, 90, 3714–3719. [Google Scholar] [CrossRef] [PubMed]
- Borkhardt, A.; Repp, R.; Haas, O.A.; Leis, T.; Harbott, J.; Kreuder, J.; Hammermann, J.; Henn, T.; Lampert, F. Cloning and characterization of AFX, the gene that fuses to MLL in acute leukemias with a t(X;11)(q13;q23). Oncogene 1997, 14, 195–202. [Google Scholar] [CrossRef] [Green Version]
- Corral, J.; Forster, A.; Thompson, S.; Lampert, F.; Kaneko, Y.; Slater, R.; Kroes, W.G.; van der Schoot, C.E.; Ludwig, W.D.; Karpas, A. Acute leukemias of different lineages have similar MLL gene fusions encoding related chimeric proteins resulting from chromosomal translocation. Proc. Natl. Acad. Sci. USA 1993, 90, 8538–8542. [Google Scholar] [CrossRef] [Green Version]
- Parry, P.; Wei, Y.; Evans, G. Cloning and characterization of the t(X;11) breakpoint from a leukemic cell line identify a new member of the forkhead gene family. Genes Chromosomes Cancer 1994, 11, 79–84. [Google Scholar] [CrossRef]
- Jacobs, F.M.; van der Heide, L.P.; Wijchers, P.J.; Burbach, J.P.; Hoekman, M.F.; Smidt, M.P. FoxO6, a novel member of the FoxO class of transcription factors with distinct shuttling dynamics. J. Biol. Chem. 2003, 278, 35959–35967. [Google Scholar] [CrossRef] [Green Version]
- Nakae, J.; Oki, M.; Cao, Y. The FoxO transcription factors and metabolic regulation. FEBS Lett. 2008, 582, 54–67. [Google Scholar] [CrossRef] [Green Version]
- Alessi, D.R.; Caudwell, F.B.; Andjelkovic, M.; Hemmings, B.A.; Cohen, P. Molecular basis for the substrate specificity of protein kinase B; comparison with MAPKAP kinase-1 and p70 S6 kinase. FEBS Lett. 1996, 399, 333–338. [Google Scholar] [CrossRef] [Green Version]
- Brunet, A.; Bonni, A.; Zigmond, M.J.; Lin, M.Z.; Juo, P.; Hu, L.S.; Anderson, M.J.; Arden, K.C.; Blenis, J.; Greenberg, M.E. Akt promotes cell survival by phosphorylating and inhibiting a Forkhead transcription factor. Cell 1999, 96, 857–868. [Google Scholar] [CrossRef] [Green Version]
- Biggs, W.H.; Meisenhelder, J.; Hunter, T.; Cavenee, W.K.; Arden, K.C. Protein kinase B/Akt-mediated phosphorylation promotes nuclear exclusion of the winged helix transcription factor FKHR1. Proc. Natl. Acad. Sci. USA 1999, 96, 7421–7426. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rena, G.; Guo, S.; Cichy, S.C.; Unterman, T.G.; Cohen, P. Phosphorylation of the transcription factor forkhead family member FKHR by protein kinase B. J. Biol. Chem. 1999, 274, 17179–17183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tang, E.D.; Nunez, G.; Barr, F.G.; Guan, K.L. Negative regulation of the forkhead transcription factor FKHR by Akt. J. Biol. Chem. 1999, 274, 16741–16746. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakae, J.; Park, B.C.; Accili, D. Insulin stimulates phosphorylation of the forkhead transcription factor FKHR on serine 253 through a Wortmannin-sensitive pathway. J. Biol. Chem. 1999, 274, 15982–15985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Furuyama, T.; Nakazawa, T.; Nakano, I.; Mori, N. Identification of the differential distribution patterns of mRNAs and consensus binding sequences for mouse DAF-16 homologues. Biochem. J. 2000, 349, 629–634. [Google Scholar] [CrossRef]
- Tzivion, G.; Avruch, J. 14-3-3 proteins: Active cofactors in cellular regulation by serine/threonine phosphorylation. J. Biol. Chem. 2002, 277, 3061–3064. [Google Scholar] [CrossRef] [Green Version]
- Tzivion, G.; Gupta, V.S.; Kaplun, L.; Balan, V. 14-3-3 proteins as potential oncogenes. Semin. Cancer Biol. 2006, 16, 203–213. [Google Scholar] [CrossRef]
- Morrison, D.K. The 14-3-3 proteins: Integrators of diverse signaling cues that impact cell fate and cancer development. Trends Cell Biol. 2009, 19, 16–23. [Google Scholar] [CrossRef] [Green Version]
- Brunet, A.; Kanai, F.; Stehn, J.; Xu, J.; Sarbassova, D.; Frangioni, J.V.; Dalal, S.N.; DeCaprio, J.A.; Greenberg, M.E.; Yaffe, M.B. 14-3-3 transits to the nucleus and participates in dynamic nucleocytoplasmic transport. J. Cell Biol. 2002, 156, 817–828. [Google Scholar] [CrossRef]
- Obsilova, V.; Vecer, J.; Herman, P.; Pabianova ASulc, M.; Teisinger, J.; Boura, E.; Obsil, T. 14-3-3 Protein interacts with nuclear localization sequence of forkhead transcription factor FoxO4. Biochemistry 2005, 30, 11608–11617. [Google Scholar] [CrossRef]
- Obsil, T.; Ghirlando, R.; Anderson, D.E.; Hickman, A.B.; Dyda, F. Two 14-3-3 binding motifs are required for stable association of Forkhead transcription factor FOXO4 with 14-3-3 proteins and inhibition of DNA binding. Biochemistry 2003, 42, 15264–15272. [Google Scholar] [CrossRef] [PubMed]
- Silhan, J.; Vacha, P.; Strnadova, P.; Vecer, J.; Herman, P.; Sulc, M.; Teisinger, J.; Obsilova, V.; Obsil, T. 14-3-3 protein masks the DNA binding interface of forkhead transcription factor FOXO4. J. Biol. Chem. 2009, 17, 19349–19360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boura, E.; Silhan, J.; Herman, P.; Vecer, J.; Sulc, M.; Teisinger, J.; Obsilova, V.; Obsil, T. Both the N-terminal loop and wing W2 of the forkhead domain of transcription factor Foxo4 are important for DNA binding. J. Biol. Chem. 2007, 282, 8265–8275. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Boura, E.; Rezabkova, L.; Brynda, J.; Obsilova, V.; Obsil, T. Structure of the human FOXO4-DBD-DNA complex at 1.9 Å resolution reveals new details of FOXO binding to the DNA. Acta Crystallogr. D Biol. Crystallogr. 2010, 66, 1351–1357. [Google Scholar] [CrossRef]
- Greer, E.L.; Oskoui, P.R.; Banko, M.R.; Maniar, M.R.; Gygi, M.P.; Gygi, S.P.; Brunet, A. The Energy Sensor AMP-activated Protein Kinase Directly Regulates the Mammalian FOXO3 Transcription Factor. J. Biol. Chem. 2007, 282, 30107–30119. [Google Scholar] [CrossRef] [Green Version]
- Balaban, R.S.; Nemoto, S.; Finkel, T. Mitochondria, oxidants, and aging. Cell 2005, 120, 483–495. [Google Scholar] [CrossRef] [Green Version]
- Nemoto, S.; Takeda, K.; Yu, Z.X.; Ferrans, V.J.; Finkel, T. Role for mitochondrial oxidants as regulators of cellular metabolism. Mol. Cell Biol. 2000, 20, 7311–7318. [Google Scholar] [CrossRef] [Green Version]
- Lin, A. Activation of the JNK signaling pathway: Breaking the brake on apoptosis. Bioessays 2003, 25, 17–24. [Google Scholar] [CrossRef]
- Essers, M.A.; Weijzen, S.; de Vries-Smits, A.M.; Saarloos, I.; de Ruiter, N.D.; Bos, J.L.; Burgering, B.M. FOXO transcription factor activation by oxidative stress mediated by the small GTPase Ral and JNK. EMBO J. 2004, 23, 4802–4812. [Google Scholar] [CrossRef] [Green Version]
- Sunayama, J.; Tsuruta, F.; Masuyama, N.; Gotoh, Y. JNK antagonizes Akt-mediated survival signals by phosphorylating 14-3-3. J. Cell Biol. 2005, 170, 295–304. [Google Scholar] [CrossRef] [Green Version]
- Lehtinen, M.K.; Yuan, Z.; Boag, P.R.; Yang, Y.; Villen, J.; Becker, E.B.; DiBacco, S.; de la Iglesia, N.; Gygi, S.; Blackwell, T.K.; et al. A conserved MST-FOXO signaling pathway mediates oxidative-stress responses and extends life span. Cell 2006, 125, 987–1001. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, Z.; Lehtinen, M.K.; Merlo, P.; Villen, J.; Gygi, S.; Bonni, A. Regulation of neuronal cell death by MST1-FOXO1 signaling. J. Biol. Chem. 2009, 284, 11285–11292. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Graves, J.D.; Gotoh, Y.; Draves, K.E.; Ambrose, D.; Han, D.K.; Wright, M.; Chernoff, J.; Clark, E.A.; Krebs, E.G. Caspase-mediated activation and induction of apoptosis by the mammalian Ste20-like kinase Mst1. EMBO J. 1998, 17, 2224–2234. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sterner, D.E.; Berger, S.L. Acetylation of histones and transcription-related factors. Microbiol. Mol. Biol. Rev. 2000, 64, 435–459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daitoku, H.; Hatta, M.; Matsuzaki, H.; Aratani, S.; Ohshima, T.; Miyagishi, M.; Nakajima, T.; Fukamizu, A. Silent information regulator 2 potentiates Foxo1-mediated transcription through its deacetylase activity. Proc. Natl. Acad. Sci. USA 2004, 101, 10042–10047. [Google Scholar] [CrossRef] [Green Version]
- Matsuzaki, H.; Daitoku, H.; Hatta, M.; Aoyama, H.; Yoshimochi, K.; Fukamizu, A. Acetylation of Foxo1 alters its DNA-binding ability and sensitivity to phosphorylation. Proc. Natl. Acad. Sci. USA 2005, 102, 11278–11283. [Google Scholar] [CrossRef] [Green Version]
- Vogt, P.K.; Jiang, H.; Aoki, M. Triple layer control: Phosphorylation, acetylation and ubiquitination of FOXO proteins. Cell Cycle 2005, 4, 908–913. [Google Scholar] [CrossRef]
- Imai, S.; Armstrong, C.M.; Kaeberlein, M.; Guarente, L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature 2000, 403, 795–800. [Google Scholar] [CrossRef]
- Tissenbaum, H.A.; Guarente, L. Increased dosage of a sir-2 gene extends lifespan in Caenorhabditis elegans. Nature 2001, 410, 227–230. [Google Scholar] [CrossRef]
- Wang, Y.; Tissenbaum, H.A. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing Dev. 2006, 127, 48–56. [Google Scholar] [CrossRef]
- Frye, R.A. Phylogenetic classification of prokaryotic and eukaryotic Sir2-like proteins. Biochem. Biophys. Res. Commun. 2000, 273, 793–798. [Google Scholar] [CrossRef] [PubMed]
- Nakae, J.; Cao, Y.; Daitoku, H.; Fukamizu, A.; Ogawa, W.; Yano, Y.; Hayashi, Y. The LXXLL motif of murine forkhead transcription factor FoxO1 mediates Sirt1-dependent transcriptional activity. J. Clin. Investig. 2006, 116, 2473–2483. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hariharan, N.; Maejima, Y.; Nakae, J.; Paik, J.; Depinho, R.A.; Sadoshima, J. Deacetylation of FoxO by Sirt1 Plays an Essential Role in Mediating Starvation-Induced Autophagy in Cardiac Myocytes. Circ. Res. 2010, 10, 1470–1482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brunet, A.; Sweeney, L.B.; Sturgill, J.F.; Chua, K.F.; Greer, P.L.; Lin, Y.; Tran, H.; Ross, S.E.; Mostoslavsky, R.; Cohen, H.Y.; et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science 2004, 303, 2011–2015. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- van der Horst, A.; Tertoolen, L.G.; de Vries-Smits, L.M.; Frye, R.A.; Medema, R.H.; Burgering, B.M. FOXO4 is acetylated upon peroxide stress and deacetylated by the longevity protein hSir2(SIRT1). J. Biol. Chem. 2004, 279, 28873–28879. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frescas, D.; Valenti, L.; Accili, D. Nuclear trapping of the forkhead transcription factor FoxO1 via Sirt-dependent deacetylation promotes expression of glucogenetic genes. J. Biol. Chem. 2005, 280, 20589–20595. [Google Scholar] [CrossRef] [Green Version]
- Banks, A.S.; Kim-Muller, J.Y.; Mastracci, T.L.; Kofler, N.M.; Qiang, L.; Haeusler, R.A.; Jurczak, M.J.; Laznik, D.; Heinrich, G.; Samuel, V.T.; et al. Dissociation of the glucose and lipid regulatory functions of FoxO1 by targeted knockin of acetylation-defective alleles in mice. Cell Metab. 2011, 14, 587–597. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Nguyen, M.; Qin, F.X.; Tong, Q. SIRT2 deacetylates FOXO3a in response to oxidative stress and caloric restriction. Aging Cell 2007, 6, 505–514. [Google Scholar] [CrossRef]
- Jing, E.; Gesta, S.; Kahn, C. SIRT2 regulates adipocyte differentiation through FoxO1 acetylation/deacetylation. Cell Metab. 2007, 6, 105–114. [Google Scholar] [CrossRef] [Green Version]
- Wang, F.; Tong, Q. SIRT2 suppresses adipocyte differentiation by deacetylating FOXO1 and enhancing FOXO1’s repressive interaction with PPARgamma. Mol. Biol. Cell 2009, 20, 801–808. [Google Scholar] [CrossRef] [Green Version]
- Dowell, P.; Otto, T.C.; Adi, S.; Lane, M.D. Convergence of peroxisome proliferator-activated receptor gamma and Foxo1 signaling pathways. J. Biol. Chem. 2003, 278, 45485–45491. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armoni, M.; Harel, C.; Karni, S.; Chen, H.; Bar-Yoseph, F.; Ver, M.R.; Quon, M.J.; Karnieli, E. FOXO1 represses peroxisome proliferator-activated receptor-gamma1 and gamma2 gene promoters in primary adipocytes. A novel paradigm to increase insulin sensitivity. J. Biol. Chem. 2006, 281, 19881–19891. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schwer, B.; North, B.J.; Frye, R.A.; Ott, M.; Verdin, E. The human silent information regulator (Sir) 2 homologue hSIRT3 is a mitochondrial nicotinamide adenine dinucleotide-dependent deacetylase. J. Cell Biol. 2002, 158, 647–657. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Scher, M.B.; Vaquero, A.; Reinberg, D. SirT3 is a nuclear NAD ± dependent histone deacetylase that translocates to the mitochondria upon cellular stress. Genes Dev. 2007, 21, 920–928. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shi, T.; Wang, F.; Stieren, E.; Tong, Q. SIRT3, a mitochondrial sirtuin deacetylase, regulates mitochondrial function and thermogenesis in brown adipocytes. J. Biol. Chem. 2005, 280, 13560–13567. [Google Scholar] [CrossRef] [Green Version]
- Ahn, B.H.; Kim, H.S.; Song, S.; Lee, I.H.; Liu, J.; Vassilopoulos, A.; Deng, C.X.; Finkel, T. A role for the mitochondrial deacetylase Sirt3 in regulating energy homeostasis. Proc. Natl. Acad. Sci. USA 2008, 105, 14447–14452. [Google Scholar] [CrossRef] [Green Version]
- Sundaresan, N.R.; Gupta, M.; Kim, G.; Rajamohan, S.B.; Isbatan, A.; Gupta, M.P. Sirt3 blocks the cardiac hypertrophic response by augmenting Foxo3a-dependent antioxidant defense mechanisms in mice. J. Clin. Investig. 2009, 119, 2758–2771. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Tindall, D.J. Regulation of FOXO protein stability via ubiquitination and proteasome degradation. Biochim. Biophys. Acta 2011, 1813, 1961–1964. [Google Scholar] [CrossRef] [Green Version]
- Huang, H.; Regan, K.M.; Wang, F.; Wang, D.; Smith, D.I.; van Deursen, J.M.; Tindall, D.J. Skp2 inhibits FOXO1 in tumor suppression through ubiquitin-mediated degradation. Proc. Natl. Acad. Sci. USA 2005, 102, 1649–1654. [Google Scholar] [CrossRef] [Green Version]
- Kikuchi, T.; Nakae, J.; Kawano, Y.; Watanabe, N.; Onodera, M.; Itoh, H. Foxo in T Cells Regulates Thermogenic Program through Ccr4/Ccl22 Axis. IScience 2019, 22, 81–96. [Google Scholar] [CrossRef] [Green Version]
- Cheng, Z.; White, M.F. Targeting Forkhead box O1 from the concept to metabolic diseases: Lessons from mouse models. Antioxid. Redox Signal. 2011, 14, 649–661. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Varga, T.; Czimmerer, Z.; Nagy, L. PPARs are a unique set of fatty acid regulated transcription factors controlling both lipid metabolism and inflammation. Biochim. Biophys. Acta 2011, 1812, 1007–1022. [Google Scholar] [CrossRef] [PubMed]
- Desvergne, B.; Wahli, W. Peroxisome proliferator-activated receptors: Nuclear control of metabolism. Endocr. Rev. 1999, 20, 649–688. [Google Scholar] [PubMed] [Green Version]
- Gilde, A.J.; Van Bilsen, M. Peroxisome proliferator-activated receptors (PPARS): Regulators of gene expression in heart and skeletal muscle. Acta Physiol. Scand 2003, 178, 425–434. [Google Scholar] [CrossRef] [PubMed]
- Fan, W.; Imamura, T.; Sonoda, N.; Sears, D.D.; Patsouris, D.; Kim, J.J.; Olefsky, J.M. FOXO1 transrepresses peroxisome proliferator-activated receptor gamma transactivation, coordinating an insulin-induced feed-forward response in adipocytes. J. Biol. Chem. 2009, 284, 12188–12197. [Google Scholar] [CrossRef] [Green Version]
- Nakae, J.; Kitamura, T.; Kitamura, Y.; Biggs, W.H.; Arden, K.C.; Accili, D. The forkhead transcription factor Foxo1 regulates adipocyte differentiation. Dev. Cell 2003, 4, 119–129. [Google Scholar] [CrossRef] [Green Version]
- Kim, J.J.; Li, P.; Huntley, J.; Chang, J.P.; Arden, K.C.; Olefsky, J.M. FoxO1 haploinsufficiency protects against high-fat diet-induced insulin resistance with enhanced peroxisome proliferator-activated receptor gamma activation in adipose tissue. Diabetes 2009, 58, 1275–1282. [Google Scholar] [CrossRef] [Green Version]
- Kajimura, S.; Spiegelman, B.M.; Seale, P. Brown and Beige Fat: Physiological Roles beyond Heat Generation. Cell Metab. 2015, 22, 546–559. [Google Scholar] [CrossRef] [Green Version]
- Wang, W.; Seale, P. Control of brown and beige fat development. Nat. Rev. Mol. Cell Biol. 2016, 17, 691–702. [Google Scholar] [CrossRef]
- Nakae, J.; Cao, Y.; Hakuno, F.; Takemori, H.; Kawano, Y.; Sekioka, R.; Abe, T.; Kiyonari, H.; Tanaka, T.; Sakai, J.; et al. Novel repressor regulates insulin sensitivity through interaction with Foxo1. EMBO J. 2012, 31, 2275–2295. [Google Scholar] [CrossRef]
- Kita, M.; Nakae, J.; Kawano, Y.; Asahara, H.; Takemori, H.; Okado, H.; Itoh, H. Zfp238 Regulates the Thermogenic Program in Cooperation with Foxo1. IScience 2019, 12, 87–101. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vamecq, J.; Latruffe, N. Medical significance of peroxisome proliferator-activated receptors. Lancet 1999, 354, 141–148. [Google Scholar] [CrossRef]
- Altomonte, J.; Cong, L.; Harbaran, S.; Richter, A.; Xu, J.; Meseck, M.; Dong, H.H. Foxo1 mediates insulin action on apoC-III and triglyceride metabolism. J. Clin. Investig. 2004, 114, 1493–1503. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qu, S.; Su, D.; Altomonte, J.; Kamagate, A.; He, J.; Perdomo, G.; Tse, T.; Jiang, Y.; Dong, H.H. PPARa mediates the hypolipidemic action of fibrates by antagonizing FoxO1. Am. J. Physiol. Endocrinol. Metab. 2007, 292, E421–E434. [Google Scholar] [CrossRef]
- Sladek, F.M.; Zhong, W.M.; Lai, E.; Darnell, J.E., Jr. Liver-enriched transcription factor HNF-4 is a novel member of the steroid hormone receptor superfamily. Genes Dev. 1990, 4, 2353–2365. [Google Scholar] [CrossRef] [Green Version]
- Drewes, T.; Senkel, S.; Holewa, B.; Ryffel, G.U. Human hepatocyte nuclear factor 4 isoforms are encoded by distinct and differentially expressed genes. Mol. Cell Biol. 1996, 16, 925–931. [Google Scholar] [CrossRef] [Green Version]
- Hayhurst, G.P.; Lee, Y.H.; Lambert, G.; Ward, J.M.; Gonzalez, F.J. Hepatocyte nuclear factor 4alpha (nuclear receptor 2A1) is essential for maintenance of hepatic gene expression and lipid homeostasis. Mol. Cell Biol. 2001, 21, 1393–1403. [Google Scholar] [CrossRef] [Green Version]
- Hirota, K.; Daitoku, H.; Matsuzaki, H.; Araya, N.; Yamagata, K.; Asada, S.; Sugaya, T.; Fukamizu, A. Hepatocyte nuclear factor-4 is a novel downstream target of insulin via FKHR as a signal-regulated transcriptional inhibitor. J. Biol. Chem. 2003, 278, 13056–13060. [Google Scholar] [CrossRef] [Green Version]
- Roth, U.; Curth, K.; Unterman, T.G.; Kietzmann, T. The transcription factors HIF-1 and HNF-4 and the coactivator p300 are involved in insulin-regulated glucokinase gene expression via the phosphatidylinositol 3-kinase/protein kinase B pathway. J. Biol. Chem. 2004, 279, 2623–2631. [Google Scholar] [CrossRef] [Green Version]
- Lin, J.; Handschin, C.; Spiegelman, B.M. Metabolic control through the PGC-1 family of transcription coactivators. Cell Metab. 2005, 1, 361–370. [Google Scholar] [CrossRef] [Green Version]
- Puigserver, P.; Rhee, J.; Donovan, J.; Walkey, C.J.; Yoon, J.C.; Oriente, F.; Kitamura, Y.; Altomonte, J.; Dong, H.; Accili, D.; et al. Insulin-regulated hepatic gluconeogenesis through FOXO1-PGC-1alpha interaction. Nature 2003, 423, 550–555. [Google Scholar] [CrossRef] [PubMed]
- Nerlov, C. C/EBPs: Recipients of extracellular signals through proteome modulation. Curr. Opin. Cell Biol. 2008, 20, 180–185. [Google Scholar] [CrossRef] [PubMed]
- Sekine, K.; Chen, Y.R.; Kojima, N.; Ogata, K.; Fukamizu, A.; Miyajima, A. Foxo1 links insulin signaling to C/EBPalpha and regulates gluconeogenesis during liver development. EMBO J. 2007, 26, 3607–3615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haeusler, R.A.; Hartil, K.; Vaitheesvaran, B.; Arrieta-Cruz, I.; Knight, C.M.; Cook, J.R.; Kammoun, H.L.; Febbraio, M.A.; Gutierrez-Juarez, R.; Kurland, I.J.; et al. Integrated control of hepatic lipogenesis versus glucose production requires FoxO transcription factors. Nat. Commun. 2014, 5, 5190. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakae, J.; Kitamura, T.; Silver, D.L.; Accili, D. The forkhead transcription factor Foxo1 (Fkhr) confers insulin sensitivity onto glucose-6-phosphatase expression. J. Clin. Investig. 2001, 108, 1359–1367. [Google Scholar] [CrossRef]
- Pajvani, U.B.; Accili, D. The new biology of diabetes. Diabetologia 2015, 58, 2459–2468. [Google Scholar] [CrossRef]
- Langlet, F.; Haeusler, R.A.; Lindén, D.; Ericson, E.; Norris, T.; Johansson, A.; Cook, J.R.; Aizawa, K.; Wang, L.; Buettner, C.; et al. Selective Inhibition of FOXO1 Activator/Repressor Balance Modulates Hepatic Glucose Handling. Cell 2017, 171, 824–835. [Google Scholar] [CrossRef]
- Kadamb, R.; Mittal, S.; Bansal, N.; Batra, H.; Saluja, D. Sin3: Insight into its transcription regulatory functions. Eur. J. Cell Biol. 2013, 92, 237–246. [Google Scholar] [CrossRef]
- Nakae, J.; Barr, V.; Accili, D. Differential regulation of gene expression by insulin and IGF-1 receptors correlates with phosphorylation of a single amino acid residue in the forkhead transcription factor FKHR. EMBO J. 2000, 19, 989–996. [Google Scholar] [CrossRef] [Green Version]
- Nakae, J.; Kitamura, T.; Ogawa, W.; Kasuga, M.; Accili, D. Insulin regulation of gene expression through the forkhead transcription factor Foxo1 (Fkhr) requires kinases distinct from Akt. Biochemistry 2001, 40, 11768–11776. [Google Scholar] [CrossRef]
- Kodani, N.; Nakae, J.; Kobayashi, M.; Kikuchi, O.; Kitamura, T.; Itoh, H. FCoR-Foxo1 Axis Regulates α-cell Mass through Repression of Arx Expression. IScience 2019, 23, 100798. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kitamura, Y.I.; Kitamura, T.; Kruse, J.P.; Raum, J.C.; Stein, R.; Gu, W.; Accili, D. FoxO1 protects against pancreatic beta cell failure through NeuroD and MafA induction. Cell Metab. 2005, 2, 153–163. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Owens, G.K.; Kumar, M.S.; Wamhoff, B.R. Molecular regulation of vascular smooth muscle cell differentiation in development and disease. Physiol. Rev. 2004, 84, 767–801. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.P.; Wang, Z.; Yanagisawa, H.; Olson, E.N. Phenotypic modulation of smooth muscle cells through interaction of Foxo4 and myocardin. Dev. Cell 2005, 9, 261–270. [Google Scholar] [CrossRef] [Green Version]
- Hribal, M.L.; Nakae, J.; Kitamura, T.; Shutter, J.R.; Accili, D. Regulation of insulin-like growth factor-dependent myoblast differentiation by Foxo forkhead transcription factors. J. Cell Biol. 2003, 162, 535–541. [Google Scholar] [CrossRef] [Green Version]
- Kitamura, T.; Kitamura, Y.I.; Funahashi, Y.; Shawber, C.J.; Castrillon, D.H.; Kollipara, R.; DePinho, R.A.; Kitajewski, J.; Accili, D. A Foxo/Notch pathway controls myogenic differentiation and fiber type specification. J. Clin. Investig. 2007, 117, 2477–2485. [Google Scholar] [CrossRef] [Green Version]
- Lai, E.C. Keeping a good pathway down: Transcriptional repression of Notch pathway target genes by CSL proteins. EMBO Rep. 2002, 3, 840–845. [Google Scholar] [CrossRef]
- Conboy, I.M.; Rando, T.A. The regulation of Notch signaling controls satellite cell activation and cell fate determination in postnatal myogenesis. Dev. Cell 2002, 3, 397–409. [Google Scholar] [CrossRef] [Green Version]
- Qi, Y.; Xu, Z.; Zhu, Q.; Thomas, C.; Kumar, R.; Feng, H.; Dostal, D.E.; White, M.F.; Baker, K.M.; Guo, S. Myocardial loss of IRS1 and IRS2 causes heart failure and is controlled by p38α MAPK during insulin resistance. Diabetes 2013, 62, 3887–3900. [Google Scholar] [CrossRef] [Green Version]
- Riehle, C.; Wende, A.R.; Zhu, Y.; Oliveira, K.J.; Pereira, R.O.; Jaishy, B.P.; Bevins, J.; Valdez, S.; Noh, J.; Kim, B.J.; et al. Insulin receptor substrates are essential for the bioenergetic and hypertrophic response of the heart to exercise training. Mol. Cell Biol. 2014, 34, 3450–3460. [Google Scholar] [CrossRef] [Green Version]
- Battiprolu, P.K.; Hojayev, B.; Jiang, N.; Wang, Z.V.; Luo, X.; Iglewski, M.; Shelton, J.M.; Gerard, R.D.; Rothermel, B.A.; Gillette, T.G.; et al. Metabolic stress-induced activation of FoxO1 triggers diabetic cardiomyopathy in mice. J. Clin. Investig. 2012, 122, 1109–1118. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, T.; Feng, Y.; Kitamura, Y.I.; Chua, S.C., Jr.; Xu, A.W.; Barsh, G.S.; Rossetti, L.; Accili, D. Forkhead protein FoxO1 mediates Agrp-dependent effects of leptin on food intake. Nat. Med. 2006, 12, 534–540. [Google Scholar] [CrossRef]
- Kim, M.S.; Pak, Y.K.; Jang, P.G.; Namkoong, C.; Choi, Y.S.; Won, J.C.; Kim, K.S.; Kim, S.W.; Kim, H.S.; Park, J.Y.; et al. Role of hypothalamic Foxo1 in the regulation of food intake and energy homeostasis. Nat. Neurosci. 2006, 9, 901–906. [Google Scholar] [CrossRef] [PubMed]
- Fukuda, M.; Jones, J.E.; Olson, D.; Hill, J.; Lee, C.E.; Gautron, L.; Choi, M.; Zigman, J.M.; Lowell, B.B.; Elmquist, J.K. Monitoring FoxO1 localization in chemically identified neurons. J. Neurosci. 2008, 28, 13640–13648. [Google Scholar] [CrossRef] [PubMed]
- Ramadori, G.; Lee, C.E.; Bookout, A.L.; Lee, S.; Williams, K.W.; Anderson, J.; Elmquist, J.K.; Coppari, R. Brain SIRT1: Anatomical distribution and regulation by energy availability. J. Neurosci. 2008, 28, 9989–9996. [Google Scholar] [CrossRef] [PubMed]
- Lafontaine-Lacasse, M.; Richard, D.; Picard, F. Effects of age and gender on Sirt 1 mRNA expressions in the hypothalamus of the mouse. Neurosci. Lett. 2010, 480, 1–3. [Google Scholar] [CrossRef]
- Sasaki, T.; Maier, B.; Bartke, A.; Scrable, H. Progressive loss of SIRT1 with cell cycle withdrawal. Aging Cell 2006, 5, 413–422. [Google Scholar] [CrossRef]
- Sasaki, T.; Kitamura, T. Roles of FoxO1 and Sirt1 in the central regulation of food intake. Endocr. J. 2010, 57, 939–946. [Google Scholar] [CrossRef] [Green Version]
- Sanese, P.; Forte, G.; Disciglio, V.; Grossi, V.; Simone, C. FOXO3 on the Road to Longevity: Lessons From SNPs and Chromatin Hubs. Comput. Struct. Biotechnol. J. 2019, 17, 737–745. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kodani, N.; Nakae, J. Tissue-Specific Metabolic Regulation of FOXO-Binding Protein: FOXO Does Not Act Alone. Cells 2020, 9, 702. https://doi.org/10.3390/cells9030702
Kodani N, Nakae J. Tissue-Specific Metabolic Regulation of FOXO-Binding Protein: FOXO Does Not Act Alone. Cells. 2020; 9(3):702. https://doi.org/10.3390/cells9030702
Chicago/Turabian StyleKodani, Noriko, and Jun Nakae. 2020. "Tissue-Specific Metabolic Regulation of FOXO-Binding Protein: FOXO Does Not Act Alone" Cells 9, no. 3: 702. https://doi.org/10.3390/cells9030702
APA StyleKodani, N., & Nakae, J. (2020). Tissue-Specific Metabolic Regulation of FOXO-Binding Protein: FOXO Does Not Act Alone. Cells, 9(3), 702. https://doi.org/10.3390/cells9030702




