Citrate Mediates Crosstalk between Mitochondria and the Nucleus to Promote Human Mesenchymal Stem Cell In Vitro Osteogenesis
Abstract
1. Introduction
2. Materials and Methods
2.1. Cell Culture
2.2. Flow Cytometry
2.3. In Vitro Differentiation Protocol
2.4. Immunofluorescence Microscopy
2.5. Measurement of Mitochondrial Number, Volume, and Identification of Mitochondrion-Nucleus Contact Sites
2.6. Antibodies
2.7. XF Bioenergetic Analysis
2.8. Oil Red O Staining
2.9. Alizarin Red Staining
2.10. RNA Extraction and Quantitative Reverse Transcription Polymerase Chain Reaction (RT-PCR)
2.11. Cell Growth Assay
2.12. Statistical Analysis
3. Results
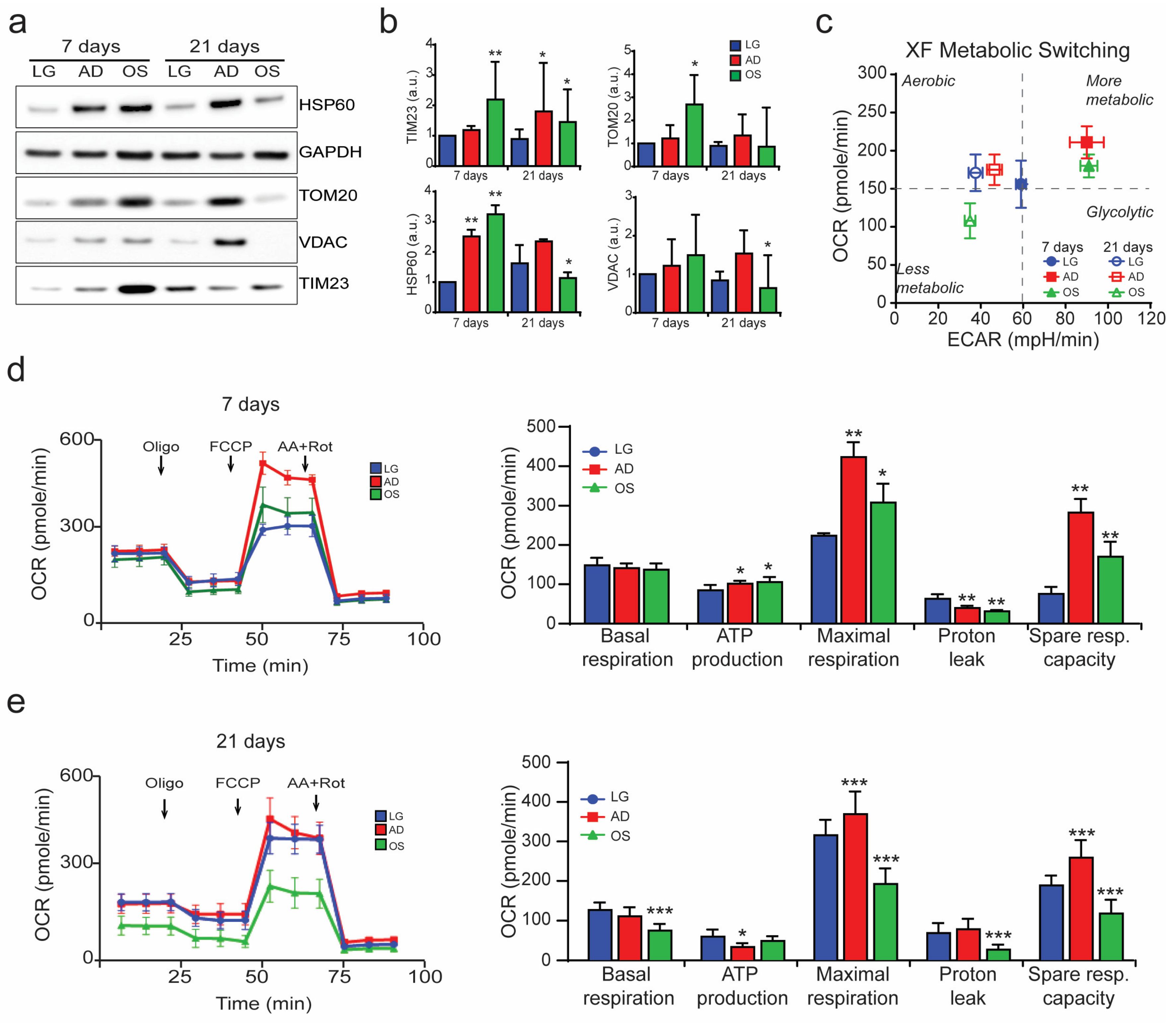
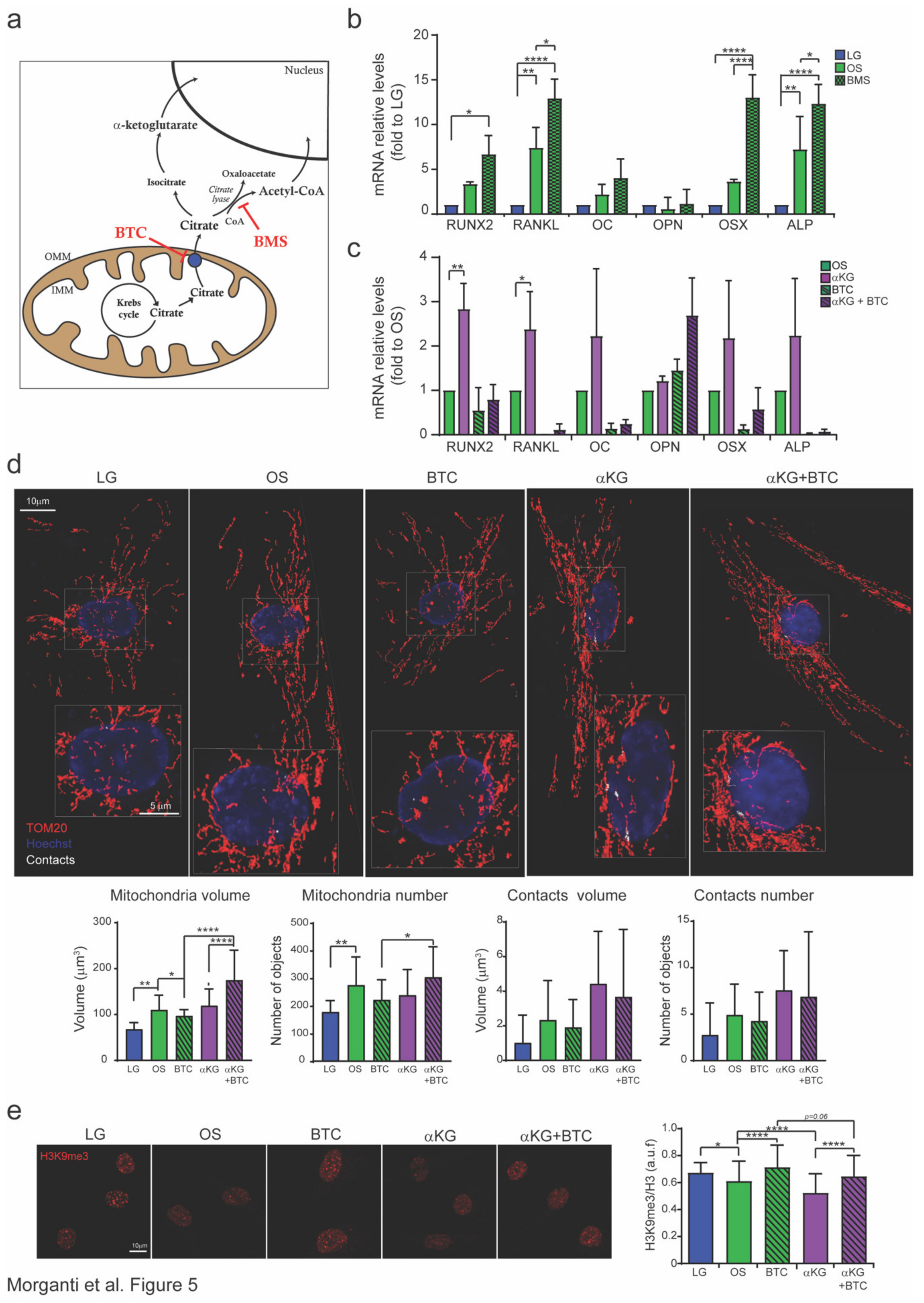
3.1. Mitochondrion-Nucleus Contact Sites Increase Following Differentiation Processes and Mitochondrial Modification
3.2. Inhibition of Citrate Transporters Impairs Osteogenesis
3.3. Citrate is Converted to αKG to Promote Osteogenesis
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Chen, Y.; Shao, J.-Z.; Xiang, L.-X.; Dong, X.-J.; Zhang, G.-R. Mesenchymal stem cells: A promising candidate in regenerative medicine. Int. J. Biochem. Cell Boil. 2008, 40, 815–820. [Google Scholar] [CrossRef] [PubMed]
- Parekkadan, B.; Milwid, J. Mesenchymal stem cells as therapeutics. Annu. Rev. Biomed. Eng. 2010, 12, 87–117. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Gao, Z.; Chen, Y.; Guan, M.-X. The role of mitochondria in osteogenic, adipogenic and chondrogenic differentiation of mesenchymal stem cells. Protein Cell 2017, 8, 439–445. [Google Scholar] [CrossRef] [PubMed]
- Cook, D.; Genever, P. Regulation of Mesenchymal Stem Cell Differentiation. Adv. Exp. Med. Biol. 2013, 786, 213–229. [Google Scholar] [CrossRef]
- Collu-Marchese, M.; Shuen, M.; Pauly, M.; Saleem, A.; Hood, D.A. The regulation of mitochondrial transcription factor A (Tfam) expression during skeletal muscle cell differentiation. Biosci. Rep. 2015, 35, 35. [Google Scholar] [CrossRef]
- Forni, M.F.; Peloggia, J.; Trudeau, K.; Shirihai, O.; Kowaltowski, A.J. Murine Mesenchymal Stem Cell Commitment to Differentiation Is Regulated by Mitochondrial Dynamics. Stem Cells 2015, 34, 743–755. [Google Scholar] [CrossRef]
- Zhang, Y.; Marsboom, G.; Toth, P.T.; Rehman, J. Mitochondrial Respiration Regulates Adipogenic Differentiation of Human Mesenchymal Stem Cells. PLoS ONE 2013, 8, e77077. [Google Scholar] [CrossRef]
- Morganti, C.; Bonora, M.; Ito, K.; Ito, K. Electron transport chain complex II sustains high mitochondrial membrane potential in hematopoietic stem and progenitor cells. Stem Cell Res. 2019, 40, 101573. [Google Scholar] [CrossRef]
- Bonora, M.; Ito, K.; Morganti, C.; Pinton, P.; Ito, K. Membrane-potential compensation reveals mitochondrial volume expansion during HSC commitment. Exp. Hematol. 2018, 68, 30–37.e1. [Google Scholar] [CrossRef]
- Wanet, A.; Arnould, T.; Najimi, M.; Renard, P. Connecting Mitochondria, Metabolism, and Stem Cell Fate. Stem Cells Dev. 2015, 24, 1957–1971. [Google Scholar] [CrossRef]
- Chen, C.-T.; Shih, Y.-R.V.; Kuo, T.K.; Lee, O.K.; Wei, Y.-H. Coordinated Changes of Mitochondrial Biogenesis and Antioxidant Enzymes During Osteogenic Differentiation of Human Mesenchymal Stem Cells. Stem Cells 2008, 26, 960–968. [Google Scholar] [CrossRef] [PubMed]
- Sánchez-Aragó, M.; García-Bermúdez, J.; Martínez-Reyes, I.; Santacatterina, F.; Cuezva, J.M. Degradation of IF1 controls energy metabolism during osteogenic differentiation of stem cells. EMBO Rep. 2013, 14, 638–644. [Google Scholar] [CrossRef] [PubMed]
- Giorgi, C.; Marchi, S.; Pinton, P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat. Rev. Mol. Cell Boil. 2018, 19, 713–730. [Google Scholar] [CrossRef] [PubMed]
- Kaelin, W.G.; McKnight, S.L. Influence of metabolism on epigenetics and disease. Cell 2013, 153, 56–69. [Google Scholar] [CrossRef] [PubMed]
- Ferroni, L.; Gardin, C.; Bellin, G.; Vindigni, V.; Pavan, C.; Zavan, B. Effects of novel antidepressant drugs on mesenchymal stem cell physiology. Biomed. Pharmacother. 2019, 114, 108853. [Google Scholar] [CrossRef] [PubMed]
- Gardin, C.; Ferroni, L.; Bellin, G.; Rubini, G.; Barosio, S.; Zavan, B. Therapeutic Potential of Autologous Adipose-Derived Stem Cells for the Treatment of Liver Disease. Int. J. Mol. Sci. 2018, 19, 4064. [Google Scholar] [CrossRef]
- Youle, R.J.; Narendra, D.P. Mechanisms of mitophagy. Nat. Rev. Mol. Cell Boil. 2011, 12, 9–14. [Google Scholar] [CrossRef]
- Tanida, I.; Ueno, T.; Kominami, E. LC3 and Autophagy. Methods Mol. Biol. 2008, 445, 77–88. [Google Scholar] [CrossRef]
- Narendra, D.; Tanaka, A.; Suen, D.-F.; Youle, R.J. Parkin is recruited selectively to impaired mitochondria and promotes their autophagy. J. Cell Boil. 2008, 183, 795–803. [Google Scholar] [CrossRef]
- Matilainen, O.; Quiros, P.M.; Auwerx, J. Mitochondria and Epigenetics—Crosstalk in Homeostasis and Stress. Trends Cell Boil. 2017, 27, 453–463. [Google Scholar] [CrossRef]
- Menzies, K.J.; Zhang, H.; Katsyuba, E.; Auwerx, J. Protein acetylation in metabolism—Metabolites and cofactors. Nat. Rev. Endocrinol. 2015, 12, 43–60. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Menzies, K.J.; Auwerx, J. The role of mitochondria in stem cell fate and aging. Development 2018, 145, dev143420. [Google Scholar] [CrossRef] [PubMed]
- Palmieri, F. The mitochondrial transporter family (SLC25): Physiological and pathological implications. Pflugers Arch. 2004, 447, 689–709. [Google Scholar] [CrossRef] [PubMed]
- Aluvila, S.; Sun, J.; Harrison, D.H.T.; Walters, D.E.; Kaplan, R.S. Inhibitors of the Mitochondrial Citrate Transport Protein: Validation of the Role of Substrate Binding Residues and Discovery of the First Purely Competitive Inhibitor. Mol. Pharmacol. 2009, 77, 26–34. [Google Scholar] [CrossRef]
- Teven, C.M.; Liu, X.; Hu, N.; Tang, N.; Kim, S.H.; Huang, E.; Yang, K.; Li, M.; Gao, J.-L.; Liu, H.; et al. Epigenetic Regulation of Mesenchymal Stem Cells: A Focus on Osteogenic and Adipogenic Differentiation. Stem Cells Int. 2011, 2011, 1–18. [Google Scholar] [CrossRef]
- Eslaminejad, M.B.; Fani, N.; Shahhoseini, M. Epigenetic Regulation of Osteogenic and Chondrogenic Differentiation of Mesenchymal Stem Cells in Culture. Cell J. 2013, 15, 1–10. [Google Scholar] [PubMed]
- Brunet, A.; Rando, T.A. Interaction between epigenetic and metabolism in aging stem cells. Curr. Opin. Cell Boil. 2017, 45, 1–7. [Google Scholar] [CrossRef]
- Zheng, Y.; He, L.; Wan, Y.; Song, J. H3K9me-Enhanced DNA Hypermethylation of thep16INK4aGene: An Epigenetic Signature for Spontaneous Transformation of Rat Mesenchymal Stem Cells. Stem Cells Dev. 2013, 22, 256–267. [Google Scholar] [CrossRef]
- Fu, Y.; Zhang, P.; Ge, J.; Cheng, J.; Dong, W.; Yuan, H.; Du, Y.; Yang, M.; Sun, R.; Jiang, H. Histone deacetylase 8 suppresses osteogenic differentiation of bone marrow stromal cells by inhibiting histone H3K9 acetylation and RUNX2 activity. Int. J. Biochem. Cell Boil. 2014, 54, 68–77. [Google Scholar] [CrossRef]
- Klose, R.J.; Kallin, E.M.; Zhang, Y. JmjC-domain-containing proteins and histone demethylation. Nat. Rev. Genet. 2006, 7, 715–727. [Google Scholar] [CrossRef]
- Ye, L.; Fan, Z.; Yu, B.; Chang, J.; Al Hezaimi, K.; Zhou, X.; Park, N.-H.; Wang, C.-Y. Histone demethylases KDM4B and KDM6B promotes osteogenic differentiation of human MSCs. Cell Stem Cell 2012, 11, 50–61. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsov, A.; Margreiter, R. Heterogeneity of Mitochondria and Mitochondrial Function within Cells as Another Level of Mitochondrial Complexity. Int. J. Mol. Sci. 2009, 10, 1911–1929. [Google Scholar] [CrossRef] [PubMed]
- Folmes, C.D.; Dzeja, P.P.; Nelson, T.; Terzic, A. Metabolic Plasticity in Stem Cell Homeostasis and Differentiation. Cell Stem Cell 2012, 11, 596–606. [Google Scholar] [CrossRef] [PubMed]
- Parker, G.C.; Acsadi, G.; Brenner, C.A. Mitochondria: Determinants of Stem Cell Fate? Stem Cells Dev. 2009, 18, 803–806. [Google Scholar] [CrossRef]
- Pietilä, M.; Lehtonen, S.; Närhi, M.; Hassinen, I.E.; Leskelä, H.-V.; Aranko, K.; Nordström, K.; Vepsalainen, A.; Lehenkari, P. Mitochondrial Function Determines the Viability and Osteogenic Potency of Human Mesenchymal Stem Cells. Tissue Eng. Part C Methods 2010, 16, 435–445. [Google Scholar] [CrossRef]
- Lambertini, E.; Penolazzi, L.; Morganti, C.; Lisignoli, G.; Zini, N.; Angelozzi, M.; Bonora, M.; Ferroni, L.; Pinton, P.; Zavan, B.; et al. Osteogenic differentiation of human MSCs: Specific occupancy of the mitochondrial DNA by NFATc1 transcription factor. Int. J. Biochem. Cell Boil. 2015, 64, 212–219. [Google Scholar] [CrossRef]
- Wan, Y.; Zhuo, N.; Li, Y.; Zhao, W.; Jiang, D. Autophagy promotes osteogenic differentiation of human bone marrow mesenchymal stem cell derived from osteoporotic vertebrae. Biochem. Biophys. Res. Commun. 2017, 488, 46–52. [Google Scholar] [CrossRef]
- Qi, M.; Zhang, L.; Ma, Y.; Shuai, Y.; Li, L.; Luo, K.; Liu, W.; Jin, Y. Autophagy Maintains the Function of Bone Marrow Mesenchymal Stem Cells to Prevent Estrogen Deficiency-Induced Osteoporosis. Theranostics 2017, 7, 4498–4516. [Google Scholar] [CrossRef]
- Morganti, C.; Missiroli, S.; Lebiedzinska-Arciszewska, M.; Ferroni, L.; Morganti, L.; Perrone, M.; Ramaccini, D.; Occhionorelli, S.; Zavan, B.; Wieckowski, M.R.; et al. Regulation of PKCbeta levels and autophagy by PML is essential for high-glucose-dependent mesenchymal stem cell adipogenesis. Int. J. Obes. 2019, 43, 963–973. [Google Scholar] [CrossRef]
- Ito, K.; Turcotte, R.; Cui, J.; Zimmerman, S.E.; Pinho, S.; Mizoguchi, T.; Arai, F.; Runnels, J.M.; Alt, C.; Teruya-Feldstein, J.; et al. Self-renewal of a purified Tie2+ hematopoietic stem cell population relies on mitochondrial clearance. Science 2016, 354, 1156–1160. [Google Scholar] [CrossRef]
- Hofmann, A.D.; Beyer, M.; Krause, U.; Wobus, M.; Bornhäuser, M.; Rödel, G. OXPHOS Supercomplexes as a Hallmark of the Mitochondrial Phenotype of Adipogenic Differentiated Human MSCs. PLoS ONE 2012, 7, e35160. [Google Scholar] [CrossRef] [PubMed]
- Quinn, K.P.; Sridharan, G.V.; Hayden, R.S.; Kaplan, D.L.; Lee, K.; Georgakoudi, I. Quantitative metabolic imaging using endogenous fluorescence to detect stem cell differentiation. Sci. Rep. 2013, 3, 3432. [Google Scholar] [CrossRef] [PubMed]
- Gut, P.; Verdin, E. The nexus of chromatin regulation and intermediary metabolism. Nature 2013, 502, 489–498. [Google Scholar] [CrossRef] [PubMed]
- Lu, C.; Thompson, C.B. Metabolic regulation of epigenetics. Cell Metab. 2012, 16, 9–17. [Google Scholar] [CrossRef]
- Lisowski, P.; Kannan, P.; Mlody, B.; Prigione, A. Mitochondria and the dynamic control of stem cell homeostasis. EMBO Rep. 2018, 19, e45432. [Google Scholar] [CrossRef]
- Nagaraj, R.; Sharpley, M.S.; Chi, F.; Braas, D.; Zhou, Y.; Kim, R.; Clark, A.T.; Banerjee, U. Nuclear Localization of Mitochondrial TCA Cycle Enzymes as a Critical Step in Mammalian Zygotic Genome Activation. Cell 2017, 168, 210–223.e11. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B. A review of the important central role of altered citrate metabolism during the process of stem cell differentiation. J. Regen. Med. Tissue Eng. 2013, 2, 1. [Google Scholar] [CrossRef]
- Hu, Y.-Y.; Rawal, A.; Schmidt-Rohr, K. Strongly bound citrate stabilizes the apatite nanocrystals in bone. Proc. Natl. Acad. Sci. USA 2010, 107, 22425–22429. [Google Scholar] [CrossRef]
- Costello, L.C.; Franklin, R.B.; Reynolds, M.A.; Chellaiah, M. The Important Role of Osteoblasts and Citrate Production in Bone Formation: “Osteoblast Citration” as a New Concept for an Old Relationship. Open Bone J. 2012, 4, 27–34. [Google Scholar] [CrossRef]
- Frezza, C. Mitochondrial metabolites: Undercover signalling molecules. Interface Focus 2017, 7, 20160100. [Google Scholar] [CrossRef]
- Yun, J.; Johnson, J.L.; Hanigan, C.L.; Locasale, J.W. Interactions between epigenetics and metabolism in cancers. Front. Oncol. 2012, 2, 163. [Google Scholar] [CrossRef] [PubMed]
- Carey, B.W.; Finley, L.W.S.; Cross, J.; Allis, C.D.; Thompson, C.B. Intracellular α-ketoglutarate maintains the pluripotency of embryonic stem cells. Nature 2014, 518, 413–416. [Google Scholar] [CrossRef] [PubMed]



© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Morganti, C.; Bonora, M.; Marchi, S.; Ferroni, L.; Gardin, C.; Wieckowski, M.R.; Giorgi, C.; Pinton, P.; Zavan, B. Citrate Mediates Crosstalk between Mitochondria and the Nucleus to Promote Human Mesenchymal Stem Cell In Vitro Osteogenesis. Cells 2020, 9, 1034. https://doi.org/10.3390/cells9041034
Morganti C, Bonora M, Marchi S, Ferroni L, Gardin C, Wieckowski MR, Giorgi C, Pinton P, Zavan B. Citrate Mediates Crosstalk between Mitochondria and the Nucleus to Promote Human Mesenchymal Stem Cell In Vitro Osteogenesis. Cells. 2020; 9(4):1034. https://doi.org/10.3390/cells9041034
Chicago/Turabian StyleMorganti, Claudia, Massimo Bonora, Saverio Marchi, Letizia Ferroni, Chiara Gardin, Mariusz R. Wieckowski, Carlotta Giorgi, Paolo Pinton, and Barbara Zavan. 2020. "Citrate Mediates Crosstalk between Mitochondria and the Nucleus to Promote Human Mesenchymal Stem Cell In Vitro Osteogenesis" Cells 9, no. 4: 1034. https://doi.org/10.3390/cells9041034
APA StyleMorganti, C., Bonora, M., Marchi, S., Ferroni, L., Gardin, C., Wieckowski, M. R., Giorgi, C., Pinton, P., & Zavan, B. (2020). Citrate Mediates Crosstalk between Mitochondria and the Nucleus to Promote Human Mesenchymal Stem Cell In Vitro Osteogenesis. Cells, 9(4), 1034. https://doi.org/10.3390/cells9041034








