Large Animal Models of Inherited Retinal Degenerations: A Review
Abstract
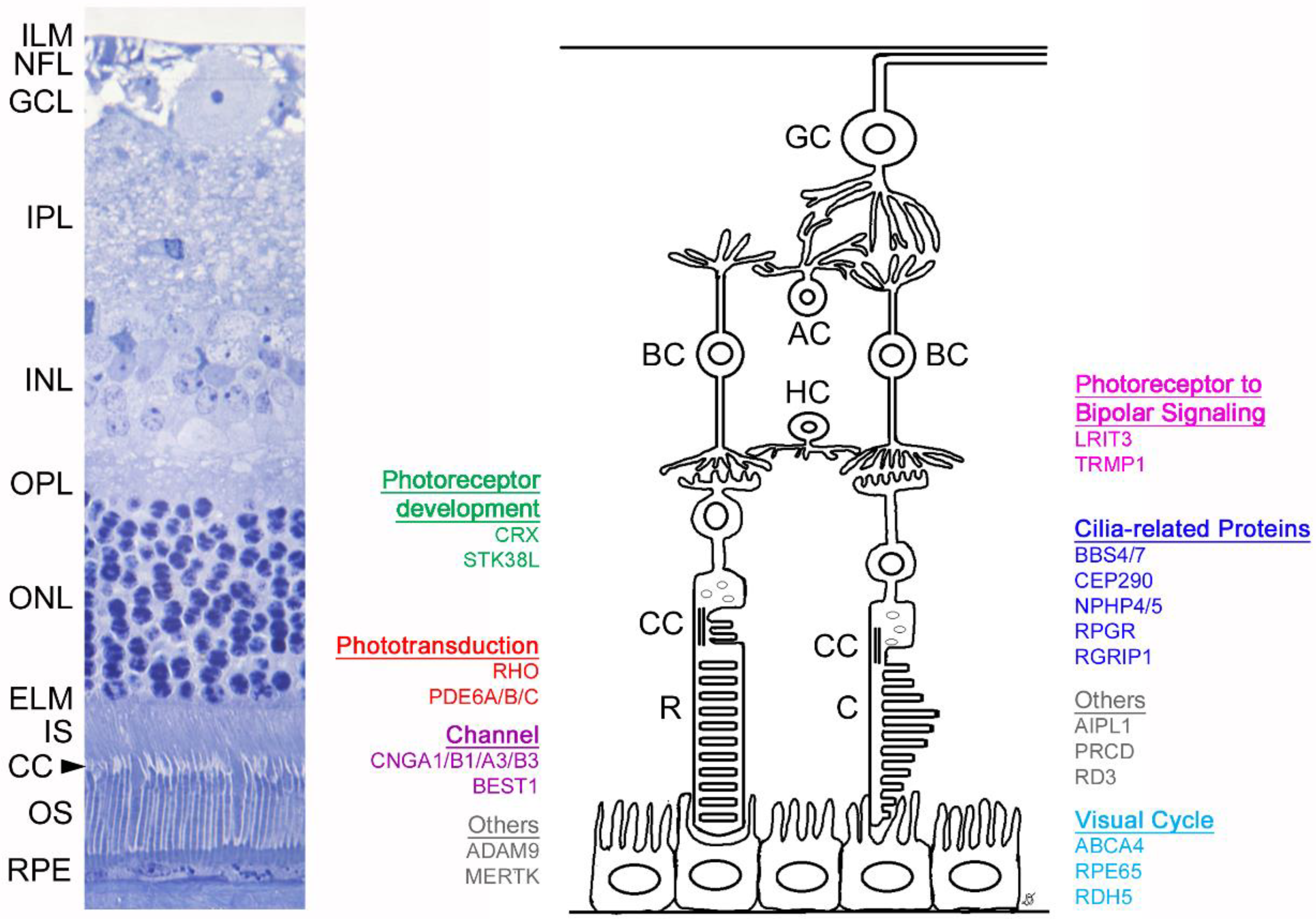
1. Introduction
2. Mutations in Phototransduction Genes
2.1. RHO
2.1.1. Dog Model
2.1.2. Pig Models
2.2. Phosphodiesterase 6 Genes
2.2.1. PDE6A
2.2.2. PDE6B
2.2.3. PDE6C
3. Visual Cycle
3.1. ABCA4
3.2. RPE65
3.3. RDH5
4. Channelopathies/Channel-Related Mutations
4.1. CNGA1
4.2. CNGB1
4.3. CNGA3
4.4. CNGB3
4.5. BEST1
5. Cilia-Related Proteins (Ciliopathies)
5.1. BBS4
5.2. BBS7
5.3. CEP290
5.4. NPHP4
5.5. NPHP5 (IQCB1)
5.6. RPGR
5.7. RPGRIP1
6. Photoreceptor Development
6.1. CRX
6.2. STK38L
7. Photoreceptor to Bipolar Cell Signaling
7.1. LRIT3
7.2. TRPM1
7.3. Whippet Dog Model of Incomplete CSNB with Retinal Degeneration
8. Structural/Other
8.1. ADAM9
8.2. AIPL1
8.3. MERTK
8.4. PRCD
8.5. RD3
9. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Fan, N.; Lai, L. Genetically Modified Pig Models for Human Diseases. J. Genet. Genom. 2013, 40, 67–73. [Google Scholar] [CrossRef] [PubMed]
- Petersen-Jones, S.M. Drug and gene therapy of hereditary retinal disease in dog and cat models. Drug Discov. Today Dis. Model. 2013, 10, e215–e223. [Google Scholar] [CrossRef]
- Peterson, S.M.; McGill, T.J.; Puthussery, T.; Stoddard, J.; Renner, L.; Lewis, A.D.; Colgin, L.M.; Gayet, J.; Wang, X.; Prongay, K.; et al. Bardet-Biedl Syndrome in rhesus macaques: A nonhuman primate model of retinitis pigmentosa. Exp. Eye Res. 2019, 189, 107825. [Google Scholar] [CrossRef]
- Moshiri, A.; Chen, R.; Kim, S.; Harris, R.A.; Li, Y.; Raveendran, M.; Davis, S.; Liang, Q.; Pomerantz, O.; Wang, J.; et al. A nonhuman primate model of inherited retinal disease. J. Clin. Investig. 2019, 129, 863–874. [Google Scholar] [CrossRef]
- Luo, X.; Li, M.; Su, B. Application of the genome editing tool CRISPR/Cas9 in non-human primates. Zool. Res. 2016, 37, 214–219. [Google Scholar]
- Kang, Y.; Chu, C.; Wang, F.; Niu, Y. CRISPR/Cas9-mediated genome editing in nonhuman primates. Dis. Model. Mech. 2019, 12, dmm039982. [Google Scholar] [CrossRef] [PubMed]
- Rahman, N.; Georgiou, M.; Khan, K.N.; Michaelides, M. Macular dystrophies: Clinical and imaging features, molecular genetics and therapeutic options. Br. J. Ophthalmol. 2019, 104, 451–460. [Google Scholar] [CrossRef]
- Michaelides, M.; Hunt, D.M.; Moore, A.T. The genetics of inherited macular dystrophies. J. Med. Genet. 2003, 40, 641–650. [Google Scholar] [CrossRef]
- Fritsche, L.G.; Igl, W.; Bailey, J.N.C.; Grassmann, F.; Sengupta, S.; Bragg-Gresham, J.L.; Burdon, K.P.; Hebbring, S.J.; Wen, C.; Gorski, M.; et al. A large genome-wide association study of age-related macular degeneration highlights contributions of rare and common variants. Nat. Genet. 2015, 48, 134–143. [Google Scholar] [CrossRef]
- Heesterbeek, T.J.; Lorés-Motta, L.; Hoyng, C.B.; Lechanteur, Y.T.E.; Hollander, A.I.D. Risk factors for progression of age-related macular degeneration. Ophthalmic Physiol. Opt. 2020, 40, 140–170. [Google Scholar] [CrossRef]
- Nishiguchi, K.; Yokoyama, Y.; Fujii, Y.; Furukawa, T.; Ono, F.; Shimozawa, N.; Togo, M.; Suzuki, M.; Nakazawa, T. Association between drusen and blood test results in a colony of 1174 monkeys. Acta Ophthalmol. 2015, 93, 93. [Google Scholar] [CrossRef]
- Pennesi, M.E.; Neuringer, M.; Courtney, R.J. Animal models of age related macular degeneration. Mol. Asp. Med. 2012, 33, 487–509. [Google Scholar] [CrossRef] [PubMed]
- Mowat, F.M.; Petersen-Jones, S.M.; Williamson, H.; Williams, D.L.; Luthert, P.J.; Ali, R.R.; Bainbridge, J.W. Topographical characterization of cone photoreceptors and the area centralis of the canine retina. Mol. Vis. 2008, 14, 2518–2527. [Google Scholar] [PubMed]
- Beltran, W.A.; Cideciyan, A.V.; Guziewicz, K.E.; Iwabe, S.; Swider, M.; Scott, E.M.; Savina, S.V.; Ruthel, G.; Stefano, F.; Zhang, L.; et al. Canine Retina Has a Primate Fovea-Like Bouquet of Cone Photoreceptors Which Is Affected by Inherited Macular Degenerations. PLoS ONE 2014, 9, e90390. [Google Scholar] [CrossRef]
- El-Aziz, M.M.A.; Barragan, I.; O’Driscoll, C.A.; Goodstadt, L.; Prigmore, E.; Borrego, S.; Mena, M.; Pieras, J.I.; El-Ashry, M.F.; Abu Safieh, L.; et al. EYS, encoding an ortholog of Drosophila spacemaker, is mutated in autosomal recessive retinitis pigmentosa. Nat. Genet. 2008, 40, 1285–1287. [Google Scholar] [CrossRef]
- Acland, G.M.; Aguirre, G.D.; Ray, J.; Zhang, Q.; Aleman, T.S.; Cideciyan, A.V.; Pearce-Kelling, S.E.; Anand, V.; Zeng, Y.; Maguire, A.M.; et al. Gene therapy restores vision in a canine model of childhood blindness. Nat. Genet. 2001, 28, 92–95. [Google Scholar] [CrossRef]
- Athanasiou, D.; Aguila, M.; Bellingham, J.; Li, W.; McCulley, C.; Reeves, P.J.; Cheetham, M.E. The molecular and cellular basis of rhodopsin retinitis pigmentosa reveals potential strategies for therapy. Prog. Retin. Eye Res. 2017, 62, 1–23. [Google Scholar] [CrossRef]
- Kijas, J.W.; Cideciyan, A.V.; Aleman, T.S.; Pianta, M.; Pearce-Kelling, S.E.; Miller, B.J.; Jacobson, S.G.; Aguirre, G.D.; Acland, G.M. Naturally occurring rhodopsin mutation in the dog causes retinal dysfunction and degeneration mimicking human dominant retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 2002, 99, 6328–6333. [Google Scholar] [CrossRef]
- Iwabe, S.; Ying, G.-S.; Aguirre, G.D.; Beltran, W.A. Assessment of visual function and retinal structure following acute light exposure in the light sensitive T4R rhodopsin mutant dog. Exp. Eye Res. 2016, 146, 341–353. [Google Scholar] [CrossRef]
- Born, L.I.V.D.; Van Schooneveld, M.J.; De Jong, L.A.M.S.; Riemslag, F.C.C.; DeJong, P.T.V.M.; Gal, A.; Bleeker-Wagemakers, E.M. Thr4Lys rhodopsin mutation is associated with autosomal dominant retinitis pigmentosa of the cone-rod type in a small Dutch family. Ophthalmic Genet. 1994, 15, 51–60. [Google Scholar] [CrossRef]
- Zhu, L.; Jang, G.-F.; Jastrzebska, B.; Filipek, S.; Pearce-Kelling, S.E.; Aguirre, G.D.; Stenkamp, R.E.; Acland, G.M.; Palczewski, K. A naturally occurring mutation of the opsin gene (T4R) in dogs affects glycosylation and stability of the G protein-coupled receptor. J. Boil. Chem. 2004, 279, 53828–53839. [Google Scholar] [CrossRef] [PubMed]
- Heckenlively, J.R.; Rodriguez, J.A.; Daiger, S.P. Autosomal Dominant Sectoral Retinitis Pigmentosa. Arch. Ophthalmol. 1991, 109, 84. [Google Scholar] [CrossRef] [PubMed]
- Orlans, H.O.; MacLaren, R.E. Comment on: ‘Sector retinitis pigmentosa caused by mutations of the RHO gene’. Eye 2019, 1–2. [Google Scholar] [CrossRef] [PubMed]
- Ross, J.W.; De Castro, J.P.F.; Zhao, J.; Samuel, M.; Walters, E.; Rios, C.; Bray-Ward, P.; Jones, B.W.; Marc, R.E.; Wang, W.; et al. Generation of an Inbred Miniature Pig Model of Retinitis Pigmentosa. Investig. Opthalmol. Vis. Sci. 2012, 53, 501–507. [Google Scholar] [CrossRef] [PubMed]
- Petters, R.M.; Alexander, C.A.; Wells, K.D.; Collins, E.B.; Sommer, J.; Blanton, M.R.; Rojas, G.; Hao, Y.; Flowers, W.L.; Banin, E.; et al. Genetically engineered large animal model for studying cone photoreceptor survival and degeneration in retinitis pigmentosa. Nat. Biotechnol. 1997, 15, 965–970. [Google Scholar] [CrossRef]
- Kraft, T.; Allen, D.; Petters, R.M.; Hao, Y.; Peng, Y.-W.; Wong, F. Altered light responses of single rod photoreceptors in transgenic pigs expressing P347L or P347S rhodopsin. Mol. Vis. 2005, 11, 1246–1256. [Google Scholar]
- Peng, Y.-W.; Hao, Y.; Petters, R.M.; Wong, F. Ectopic synaptogenesis in the mammalian retina caused by rod photoreceptor-specific mutations. Nat. Neurosci. 2000, 3, 1121–1127. [Google Scholar] [CrossRef]
- Banin, E.; Cideciyan, A.V.; Aleman, T.S.; Petters, R.M.; Wong, F.; Milam, A.H.; Jacobson, S.G. Retinal Rod Photoreceptor–Specific Gene Mutation Perturbs Cone Pathway Development. Neuron 1999, 23, 549–557. [Google Scholar] [CrossRef][Green Version]
- Shen, J.; Yang, X.; Dong, A.; Petters, R.M.; Peng, Y.-W.; Wong, F.; Campochiaro, P.A. Oxidative damage is a potential cause of cone cell death in retinitis pigmentosa. J. Cell. Physiol. 2005, 203, 457–464. [Google Scholar] [CrossRef]
- Sommer, J.R.; Wong, F.; Petters, R.M. Phenotypic stability of Pro347Leu rhodopsin transgenic pigs as indicated by photoreceptor cell degeneration. Transgenic Res. 2011, 20, 1391–1395. [Google Scholar] [CrossRef]
- Wang, W.; Lee, S.J.; Scott, P.A.; Lu, X.; Emery, D.; Liu, Y.; Ezashi, T.; Roberts, M.R.; Ross, J.W.; Kaplan, H.J.; et al. Two-Step Reactivation of Dormant Cones in Retinitis Pigmentosa. Cell Rep. 2016, 15, 372–385. [Google Scholar] [CrossRef] [PubMed]
- Daiger, S.P.; Bowne, S.J.; Sullivan, L.S. Perspective on Genes and Mutations Causing Retinitis Pigmentosa. Arch. Ophthalmol. 2007, 125, 151–158. [Google Scholar] [CrossRef] [PubMed]
- Baehr, W.; Devlin, M.J.; Applebury, M.L. Isolation and characterization of cGMP phosphodiesterase from bovine rod outer segments. J. Boil. Chem. 1979, 254, 11669–11677. [Google Scholar]
- Petersen-Jones, S.M.; Entz, D.D.; Sargan, D.R. cGMP phosphodiesterase-α mutation causes progressive retinal atrophy in the Cardigan Welsh corgi dog. Investig. Ophthalmol. Vis. Sci. 1999, 40, 1637–1644. [Google Scholar]
- Tuntivanich, N.; Pittler, S.J.; Fischer, A.J.; Omar, G.; Kiupel, M.; Weber, A.; Yao, S.; Steibel, J.P.; Khan, N.W.; Petersen-Jones, S.M. Characterization of a canine model of autosomal recessive retinitis pigmentosa due to a PDE6A mutation. Investig. Opthalmol. Vis. Sci. 2008, 50, 801–813. [Google Scholar] [CrossRef] [PubMed]
- Dryja, T.P.; Finn, J.T.; Peng, Y.W.; McGee, T.L.; Berson, E.L.; Yau, K.W. Mutations in the gene encoding the alpha subunit of the rod cGMP-gated channel in autosomal recessive retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 1995, 92, 10177–10181. [Google Scholar] [CrossRef]
- Huang, S.H.; Pittler, S.J.; Huang, X.; Oliveira, L.; Berson, E.L.; Dryja, T.P. Autosomal recessive retinitis pigmentosa caused by mutations in the α subunit of rod cGMP phosphodiesterase. Nat. Genet. 1995, 11, 468–471. [Google Scholar] [CrossRef]
- Suber, M.L.; Pittler, S.J.; Qin, N.; Wright, G.C.; Holcombe, V.; Lee, R.H.; Craft, C.M.; Lolley, R.N.; Baehr, W.; Hurwitz, R.L. Irish setter dogs affected with rod/cone dysplasia contain a nonsense mutation in the rod cGMP phosphodiesterase beta-subunit gene. Proc. Natl. Acad. Sci. USA 1993, 90, 3968–3972. [Google Scholar] [CrossRef]
- Hartong, D.T.; Berson, E.L.; Dryja, T.P. Retinitis pigmentosa. Lancet 2006, 368, 1795–1809. [Google Scholar] [CrossRef]
- McLaughlin, M.E.; Ehrhart, T.L.; Berson, E.L.; Dryja, T.P. Mutation spectrum of the gene encoding the beta subunit of rod phosphodiesterase among patients with autosomal recessive retinitis pigmentosa. Proc. Natl. Acad. Sci. USA 1995, 92, 3249–3253. [Google Scholar] [CrossRef]
- Occelli, L.M.; Schön, C.; Seeliger, M.W.; Biel, M.; Michalakis, S.; Petersen-Jones, S.M.; The RD-Cure Consortium. Gene Supplementation Rescues Rod Function and Preserves Photoreceptor and Retinal Morphology in Dogs, Leading the Way Toward Treating Human PDE6A-Retinitis Pigmentosa. Hum. Gene Ther. 2017, 28, 1189–1201. [Google Scholar] [CrossRef] [PubMed]
- Arango-Gonzalez, B.; Trifunović, D.; Sahaboglu, A.; Kranz, K.; Michalakis, S.; Farinelli, P.; Koch, S.; Koch, F.; Cottet, S.; Janssen-Bienhold, U.; et al. Identification of a Common Non-Apoptotic Cell Death Mechanism in Hereditary Retinal Degeneration. PLoS ONE 2014, 9, e112142. [Google Scholar] [CrossRef] [PubMed]
- Wensel, T.G.; Zhang, Z.; Anastassov, I.; Gilliam, J.C.; He, F.; Schmid, M.F.; Robichaux, M.A. Structural and molecular bases of rod photoreceptor morphogenesis and disease. Prog. Retin. Eye Res. 2016, 55, 32–51. [Google Scholar] [CrossRef] [PubMed]
- Mowat, F.M.; Occelli, L.M.; Bartoe, J.T.; Gervais, K.J.; Bruewer, A.R.; Querubin, J.; Dinculescu, A.; Boye, S.L.; Hauswirth, W.; Petersen-Jones, S.M. Gene Therapy in a Large Animal Model of PDE6A-Retinitis Pigmentosa. Front. Mol. Neurosci. 2017, 11, 342. [Google Scholar] [CrossRef]
- Clements, P.J.M.; Gregory, C.Y.; Petersen-Jones, S.M.; Sargan, D.R.; Bhattacharya, S.S. Confirmation of the rod cGMP phophodiesterase á-subunit (PDEá) nonsense mutation in affected rcd-1 Irish setters in the UK and development of a diagnostic test. Curr. Eye Res. 1993, 12, 861–866. [Google Scholar] [CrossRef]
- Farber, D.B.; Danciger, J.S.; Aguirre, G. The beta subunit of cyclic GMP phosphodiesterase mRNA is deficient in canine rod-cone dysplasia 1. Neuron 1992, 9, 349–356. [Google Scholar] [CrossRef]
- Aguirre, G.; Farber, D.; Lolley, R.; O’Brien, P.; Alligood, J.; Fletcher, R.T.; Chader, G. Retinal degenerations in the dog III abnormal cyclic nucleotide metabolism in rod-cone dysplasia. Exp. Eye Res. 1982, 35, 625–642. [Google Scholar] [CrossRef]
- Aguirre, G.D.; Rubin, L.F. Rod-cone dysplasia (progressive retinal atrophy) in Irish setters. J. Am. Veter Med. Assoc. 1975, 166, 157–164. [Google Scholar]
- Aquirre, G.; Farber, D.; Lolley, R.; Fletcher, R.; Chader, G. Rod-cone dysplasia in Irish setters: A defect in cyclic GMP metabolism in visual cells. Science 1978, 201, 1133–1134. [Google Scholar] [CrossRef]
- Pichard, V.; Provost, N.; Mendes-Madeira, A.; Libeau, L.; Hulin, P.; Tshilenge, K.-T.; Biget, M.; Ameline, B.; Deschamps, J.-Y.; Weber, M.; et al. AAV-mediated Gene Therapy Halts Retinal Degeneration in PDE6β-deficient Dogs. Mol. Ther. 2016, 24, 867–876. [Google Scholar] [CrossRef]
- Dekomien, G.; Runte, M.; Gödde, R.; Epplen, J.T. Generalized progressive retinal atrophy of Sloughi dogs is due to an 8-bp insertion in exon 21 of the PDE6B gene. Cytogenet. Cell Genet. 2000, 90, 261–267. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, O.; Mezey, J.G.; Schweitzer, P.A.; Boyko, A.R.; Gao, C.; Bustamante, C.D.; Jordan, J.A.; Aguirre, G.D.; Acland, G.M. IQCB1 and PDE6B Mutations Cause Similar Early Onset Retinal Degenerations in Two Closely Related Terrier Dog Breeds. Investig. Opthalmol. Vis. Sci. 2013, 54, 7005–7019. [Google Scholar] [CrossRef] [PubMed]
- Quazi, F.; Lenevich, S.; Molday, R.S. ABCA4 is an N-retinylidene-phosphatidylethanolamine and phosphatidylethanolamine importer. Nat. Commun. 2012, 3, 925. [Google Scholar] [CrossRef] [PubMed]
- Mäkeläinen, S.; Gòdia, M.; Hellsand, M.; Viluma, A.; Hahn, D.; Makdoumi, K.; Zeiss, C.J.; Mellersh, C.; Ricketts, S.L.; Narfström, K.; et al. An ABCA4 loss-of-function mutation causes a canine form of Stargardt disease. PLoS Genet. 2019, 15, e1007873. [Google Scholar] [CrossRef] [PubMed]
- Maeda, A.; Maeda, T.; Golczak, M.; Palczewski, K. Retinopathy in mice induced by disrupted all-trans-retinal clearance. J. Boil. Chem. 2008, 283, 26684–26693. [Google Scholar] [CrossRef]
- Tsang, S.H.; Sharma, T. Stargardt Disease. In Atlas of Inherited Retinal Diseases; Tsang, S.H., Sharma, T., Eds.; Springer International Publishing: Cham, Switzerland, 2018; pp. 139–151. [Google Scholar] [CrossRef]
- Le Meur, G.; Stieger, K.; Smith, A.J.; Weber, M.; Deschamps, J.Y.; Nivard, D.; Mendes-Madeira, A.; Provost, N.; Péréon, Y.; Cherel, Y.; et al. Restoration of vision in RPE65-deficient Briard dogs using an AAV serotype 4 vector that specifically targets the retinal pigmented epithelium. Gene Ther. 2006, 14, 292–303. [Google Scholar] [CrossRef]
- Narfström, K.; Katz, M.L.; Bragadottir, R.; Seeliger, M.; Boulanger, A.; Redmond, T.M.; Caro, L.; Lai, C.-M.; Rakoczy, P.E. Functional and structural recovery of the retina after gene therapy in the RPE65 null mutation dog. Investig. Opthalmol. Vis. Sci. 2003, 44, 1663–1672. [Google Scholar] [CrossRef]
- Annear, M.J.; Bartoe, J.T.; Barker, S.E.; Smith, A.J.; Curran, P.G.; Bainbridge, J.W.; Ali, R.R.; Petersen-Jones, S.M. Gene therapy in the second eye of RPE65-deficient dogs improves retinal function. Gene Ther. 2010, 18, 53–61. [Google Scholar] [CrossRef]
- Annear, M.J.; Mowat, F.M.; Bartoe, J.T.; Querubin, J.; Azam, S.A.; Basche, M.; Curran, P.G.; Smith, A.; Bainbridge, J.W.; Ali, R.R.; et al. Successful Gene Therapy in Older Rpe65-Deficient Dogs Following Subretinal Injection of an Adeno-Associated Vector Expressing RPE65. Hum. Gene Ther. 2013, 24, 883–893. [Google Scholar] [CrossRef]
- Mowat, F.M.; Breuwer, A.R.; Bartoe, J.T.; Annear, M.J.; Zhang, Z.; Smith, A.J.; Bainbridge, J.W.; Petersen-Jones, S.M.; Ali, R.R. RPE65 gene therapy slows cone loss in Rpe65-deficient dogs. Gene Ther. 2012, 20, 545–555. [Google Scholar] [CrossRef]
- Mowat, F.M.; Gervais, K.J.; Occelli, L.M.; Annear, M.J.; Querubin, J.; Bainbridge, J.W.; Smith, A.J.; Ali, R.R.; Petersen-Jones, S.M. Early-Onset Progressive Degeneration of the Area Centralis in RPE65-Deficient Dogs. Investig. Opthalmol. Vis. Sci. 2017, 58, 3268. [Google Scholar] [CrossRef] [PubMed]
- Bainbridge, J.W.; Mehat, M.S.; Sundaram, V.; Robbie, S.J.; Barker, S.E.; Ripamonti, C.; Georgiadis, A.; Mowat, F.M.; Beattie, S.G.; Gardner, P.; et al. Long-Term Effect of Gene Therapy on Leber’s Congenital Amaurosis. N. Engl. J. Med. 2015, 372, 1887–1897. [Google Scholar] [CrossRef] [PubMed]
- Petersen-Jones, S.M.; Occelli, L.; Winkler, P.; Minella, A.; Sun, K.; Lyons, L.; Daruwalla, A.; Kiser, P.; Palczewski, K. New large animal model for RDH5-associated retinopathies. Investig. Ophthalmol. Vis. Sci. 2019, 60, 458. [Google Scholar]
- Gonzalez-Fernandez, F.; Kurz, D.; Bao, Y.; Newman, S.; Conway, B.P.; Young, J.E.; Han, D.P.; Khani, S.C. 11-cis retinol dehydrogenase mutations as a major cause of the congenital night-blindness disorder known as fundus albipunctatus. Mol. Vis. 1999, 5, 41. [Google Scholar] [PubMed]
- Hotta, K.; Nakamura, M.; Kondo, M.; Ito, S.; Terasaki, H.; Miyake, Y.; Hida, T. Macular dystrophy in a Japanese family with fundus albipunctatus. Am. J. Ophthalmol. 2003, 135, 917–919. [Google Scholar] [CrossRef]
- Nakamura, M.; Miyake, Y. Macular dystrophy in a 9-year-old boy with fundus albipunctatus. Am. J. Ophthalmol. 2002, 133, 278–280. [Google Scholar] [CrossRef]
- Nakamura, M.; Skalet, J.; Miyake, Y. RDH5 gene mutations and electroretinogram in fundus albipunctatus with or without macular dystrophy: RDH5 mutations and ERG in fundus albipunctatus. Doc. Ophthalmol. 2003, 107, 3–11. [Google Scholar] [CrossRef]
- Yamamoto, H.; Yakushijin, K.; Kusuhara, S.; Escaño, M.F.T.; Nagai, A.; Negi, A. A novel RDH5 gene mutation in a patient with fundus albipunctatus presenting with macular atrophy and fading white dots. Am. J. Ophthalmol. 2003, 136, 572–574. [Google Scholar] [CrossRef]
- Kuehlewein, L.; Nasser, F.; Gloeckle, N.; Kohl, S.; Zrenner, E. Fundus albipunctatus associated with cone dysfunction. Retin. Cases Brief Rep. 2017, 11, S73–S76. [Google Scholar] [CrossRef]
- Kim, T.S.; Maeda, A.; Maeda, T.; Heinlein, C.; Kedishvili, N.; Palczewski, K.; Nelson, P.S. Delayed dark adaptation in 11-cis-retinol dehydrogenase-deficient mice: A role of RDH11 in visual processes in vivo. J. Boil. Chem. 2005, 280, 8694–8704. [Google Scholar] [CrossRef]
- Kaupp, U.B.; Seifert, R. Cyclic Nucleotide-Gated Ion Channels. Physiol. Rev. 2002, 82, 769–824. [Google Scholar] [CrossRef] [PubMed]
- Zheng, J.; Trudeau, M.C.; Zagotta, W.N. Rod cyclic nucleotide-gated channels have a stoichiometry of three CNGA1 subunits and one CNGB1 subunit. Neuron 2002, 36, 891–896. [Google Scholar] [CrossRef]
- Zhong, H.; Molday, L.L.; Molday, R.S.; Yau, K.-W. The heteromeric cyclic nucleotide-gated channel adopts a 3A:1B stoichiometry. Nature 2002, 420, 193–198. [Google Scholar] [CrossRef] [PubMed]
- Johnson, A.A.; Guziewicz, K.E.; Lee, C.J.; Kalathur, R.C.; Pulido, J.S.; Marmorstein, L.Y.; Marmorstein, A.D. Bestrophin 1 and retinal disease. Prog. Retin. Eye Res. 2017, 58, 45–69. [Google Scholar] [CrossRef] [PubMed]
- Wiik, A.C.; Ropstad, E.O.; Ekesten, B.; Karlstam, L.; Wade, C.; Lingaas, F. Progressive retinal atrophy in Shetland sheepdog is associated with a mutation in theCNGA1gene. Anim. Genet. 2015, 46, 515–521. [Google Scholar] [CrossRef] [PubMed]
- Winkler, P.A.; Ekenstedt, K.J.; Occelli, L.M.; Frattaroli, A.V.; Bartoe, J.T.; Venta, P.J.; Petersen-Jones, S.M. A Large Animal Model for CNGB1 Autosomal Recessive Retinitis Pigmentosa. PLoS ONE 2013, 8, 72229. [Google Scholar] [CrossRef]
- Petersen-Jones, S.M.; Occelli, L.M.; Winkler, P.A.; Lee, W.; Sparrow, J.R.; Tsukikawa, M.; Boye, S.L.; Chiodo, V.; Capasso, J.E.; Becirovic, E.; et al. Patients and animal models of CNGβ1-deficient retinitis pigmentosa support gene augmentation approach. J. Clin. Investig. 2017, 128, 190–206. [Google Scholar] [CrossRef]
- Hüttl, S.; Michalakis, S.; Seeliger, M.; Luo, N.-G.; Acar, N.; Geiger, H.; Hudl, K.; Mader, R.; Haverkamp, S.; Moser, M.; et al. Impaired channel targeting and retinal degeneration in mice lacking the cyclic nucleotide-gated channel subunit CNGB1. J. Neurosci. 2005, 25, 130–138. [Google Scholar] [CrossRef]
- Hull, S.; Attanasio, M.; Arno, G.; Carss, K.; Robson, A.; Thompson, D.; Plagnol, V.; Michaelides, M.; Holder, G.E.; Henderson, R.H.; et al. Clinical Characterization of CNGB1-Related Autosomal Recessive Retinitis Pigmentosa. JAMA Ophthalmol. 2017, 135, 137–144. [Google Scholar] [CrossRef]
- Wissinger, B.; Gamer, D.; Jägle, H.; Giorda, R.; Marx, T.; Mayer, S.; Tippmann, S.; Broghammer, M.; Jurklies, B.; Rosenberg, T.; et al. CNGA3 Mutations in Hereditary Cone Photoreceptor Disorders. Am. J. Hum. Genet. 2001, 69, 722–737. [Google Scholar] [CrossRef]
- Tanaka, N.; Dutrow, E.; Miyadera, K.; Delemotte, L.; MacDermaid, C.; Reinstein, S.L.; Crumley, W.R.; Dixon, C.J.; Casal, M.L.; Klein, M.L.; et al. Canine CNGA3 Gene Mutations Provide Novel Insights into Human Achromatopsia-Associated Channelopathies and Treatment. PLoS ONE 2015, 10, e0138943. [Google Scholar] [CrossRef] [PubMed]
- Reicher, S.; Seroussi, E.; Gootwine, E. A mutation in gene CNGA3 is associated with day blindness in sheep. Genomics 2010, 95, 101–104. [Google Scholar] [CrossRef] [PubMed]
- Gootwine, E.; Abu-Siam, M.; Obolensky, A.; Rosov, A.; Honig, H.; Nitzan, T.; Shirak, A.; Ezra-Elia, R.; Yamin, E.; Banin, E.; et al. Gene Augmentation Therapy for a Missense Substitution in the cGMP-Binding Domain of Ovine CNGA3 Gene Restores Vision in Day-Blind Sheep. Investig. Opthalmol. Vis. Sci. 2017, 58, 1577–1584. [Google Scholar] [CrossRef] [PubMed]
- Gootwine, E.; Ofri, R.; Banin, E.; Obolensky, A.; Averbukh, E.; Ezra-Elia, R.; Ross, M.; Honig, H.; Rosov, A.; Yamin, E.; et al. Safety and Efficacy Evaluation of rAAV2tYF-PR1.7-hCNGA3 Vector Delivered by Subretinal Injection in CNGA3 Mutant Achromatopsia Sheep. Hum. Gene Ther. Clin. Dev. 2017, 28, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Sidjanin, D.J.; Lowe, J.K.; McElwee, J.; Milne, B.S.; Phippen, T.M.; Sargan, D.R.; Aguirre, G.D.; Acland, G.M.; Ostrander, E.A. Canine CNGB3 mutations establish cone degeneration as orthologous to the human achromatopsia locus ACHM3. Hum. Mol. Genet. 2002, 11, 1823–1833. [Google Scholar] [CrossRef]
- Yeh, C.Y.; Goldstein, O.; Kukekova, A.; Holley, D.; Knollinger, A.M.; Huson, H.J.; Pearce-Kelling, S.E.; Acland, G.M.; Komáromy, A.M. Genomic deletion of CNGB3 is identical by descent in multiple canine breeds and causes achromatopsia. BMC Genet. 2013, 14, 27. [Google Scholar] [CrossRef]
- Kohl, S.; Varsányi, B.; Antunes, G.A.; Baumann, B.; Hoyng, C.B.; Jägle, H.; Rosenberg, T.; Kellner, U.; Lorenz, B.; Salati, R.; et al. CNGB3 mutations account for 50% of all cases with autosomal recessive achromatopsia. Eur. J. Hum. Genet. 2004, 13, 302–308. [Google Scholar] [CrossRef]
- Tanaka, N.; Delemotte, L.; Klein, M.L.; Komáromy, A.M.; Tanaka, J.C. A Cyclic Nucleotide-Gated Channel Mutation Associated with Canine Daylight Blindness Provides Insight into a Role for the S2 Segment Tri-Asp motif in Channel Biogenesis. PLoS ONE 2014, 9, e88768. [Google Scholar] [CrossRef]
- Ye, G.-J.; Komáromy, A.M.; Zeiss, C.; Calcedo, R.; Harman, C.D.; Koehl, K.L.; Stewart, G.A.; Iwabe, S.; Chiodo, V.A.; Hauswirth, W.; et al. Safety and Efficacy of AAV5 Vectors Expressing Human or Canine CNGB3 in CNGB3-Mutant Dogs. Hum. Gene Ther. Clin. Dev. 2017, 28, 197–207. [Google Scholar] [CrossRef]
- Guziewicz, K.E.; Zangerl, B.; Lindauer, S.J.; Mullins, R.F.; Sandmeyer, L.S.; Grahn, B.H.; Stone, E.M.; Acland, G.M.; Aguirre, G.D. Bestrophin gene mutations cause canine multifocal retinopathy: A novel animal model for best disease. Investig. Opthalmol. Vis. Sci. 2007, 48, 1959–1967. [Google Scholar] [CrossRef]
- Zangerl, B.; Wickström, K.; Slavik, J.; Lindauer, S.J.; Ahonen, S.; Schelling, C.; Lohi, H.; Guziewicz, K.E.; Aguirre, G.D. Assessment of canine BEST1 variations identifies new mutations and establishes an independent bestrophinopathy model (cmr3). Mol. Vis. 2010, 16, 2791–2804. [Google Scholar] [PubMed]
- Guziewicz, K.E.; Cideciyan, A.V.; Beltran, W.A.; Komaromy, A.M.; Dufour, V.L.; Swider, M.; Iwabe, S.; Sumaroka, A.; Kendrick, B.T.; Ruthel, G.; et al. BEST1 gene therapy corrects a diffuse retina-wide microdetachment modulated by light exposure. Proc. Natl. Acad. Sci. USA 2018, 115, E2839–E2848. [Google Scholar] [CrossRef] [PubMed]
- Guziewicz, K.E.; McTish, E.; Dufour, V.L.; Zorych, K.; Dhingra, A.; Boesze-Battaglia, K.; Aguirre, G.D. Underdeveloped RPE Apical Domain Underlies Lesion Formation in Canine Bestrophinopathies. Single Mol. Single Cell Seq. 2018, 1074, 309–315. [Google Scholar]
- Chew, T.; Haase, B.; Bathgate, R.; Willet, C.; Kaukonen, M.K.; Mascord, L.J.; Lohi, H.; Wade, C. A Coding Variant in the Gene Bardet-Biedl Syndrome 4 (BBS4) Is Associated with a Novel Form of Canine Progressive Retinal Atrophy. G3 Genes Genomes Genet. 2017, 7, 2327–2335. [Google Scholar] [CrossRef] [PubMed]
- Narfström, K. Progressive retinal atrophy in the Abyssinian cat. Clinical characteristics. Investig. Ophthalmol. Vis. Sci. 1985, 26, 193–200. [Google Scholar]
- Minella, A.L.; Occelli, L.M.; Narfström, K.; Petersen-Jones, S.M. Central retinal preservation in rdAc cats. Veter Ophthalmol. 2017, 21, 224–232. [Google Scholar] [CrossRef]
- Coppieters, F.; Lefever, S.; Leroy, B.P.; De Baere, E. CEP290, a gene with many faces: Mutation overview and presentation of CEP290base. Hum. Mutat. 2010, 31, 1097–1108. [Google Scholar] [CrossRef]
- Menotti-Raymond, M.; David, V.A.; Schäffer, A.A.; Stephens, R.; Wells, D.; Kumar-Singh, R.; O’Brien, S.; Narfström, K. Mutation in CEP290 Discovered for Cat Model of Human Retinal Degeneration. J. Hered. 2007, 98, 211–220. [Google Scholar] [CrossRef]
- Wiik, A.C.; Wade, C.; Biagi, T.; Ropstad, E.-O.; Bjerkås, E.; Lindblad-Toh, K.; Lingaas, F. A deletion in nephronophthisis 4 (NPHP4) is associated with recessive cone-rod dystrophy in standard wire-haired dachshund. Genome Res. 2008, 18, 1415–1421. [Google Scholar] [CrossRef]
- Ropstad, E.O.; Narfström, K.; Lingaas, F.; Wiik, C.; Bruun, A.; Bjerkas, E. Functional and Structural Changes in the Retina of Wire-Haired Dachshunds with Early-Onset Cone-Rod Dystrophy. Investig. Opthalmol. Vis. Sci. 2008, 49, 1106–1115. [Google Scholar] [CrossRef]
- Ropstad, E.O.; Bjerkås, E.; Narfström, K. Electroretinographic findings in the Standard Wire Haired Dachshund with inherited early onset cone–rod dystrophy. Doc. Ophthalmol. 2006, 114, 27–36. [Google Scholar] [CrossRef] [PubMed]
- Ropstad, E.O.; Bjerkås, E.; Narfström, K. Clinical findings in early onset cone-rod dystrophy in the Standard Wire-haired Dachshund. Veter Ophthalmol. 2007, 10, 69–75. [Google Scholar] [CrossRef] [PubMed]
- Ronquillo, C.; Bernstein, P.S.; Baehr, W. Senior-Løken syndrome: A syndromic form of retinal dystrophy associated with nephronophthisis. Vis. Res. 2012, 75, 88–97. [Google Scholar] [CrossRef] [PubMed]
- Won, J.; De Evsikova, C.M.; Smith, R.S.; Hicks, W.L.; Edwards, M.M.; Longo-Guess, C.; Li, T.; Naggert, J.K.; Nishina, P.M. NPHP4 is necessary for normal photoreceptor ribbon synapse maintenance and outer segment formation, and for sperm development. Hum. Mol. Genet. 2010, 20, 482–496. [Google Scholar] [CrossRef]
- Downs, L.M.; Scott, E.M.; Cideciyan, A.V.; Iwabe, S.; Dufour, V.L.; Gardiner, K.L.; Genini, S.; Marinho, L.F.; Sumaroka, A.; Kosyk, M.S.; et al. Overlap of abnormal photoreceptor development and progressive degeneration in Leber congenital amaurosis caused by NPHP5 mutation. Hum. Mol. Genet. 2016, 25, 4211–4226. [Google Scholar] [CrossRef]
- Aguirre, G.D.; Cideciyan, A.V.; Boye, S.L.; Iwabe, S.; Dufour, V.; Swider, M.; Roszak, K.; Hauswirth, W.W.; Jacobson, S.G.; Beltran, W.A. Long-term preservation of photoreceptor function and structure following early-stage treatment by AAV-mediated gene augmentation in canine model of NPHP5 Leber congenital amaurosis. Investig. Ophthalmol. Vis. Sci. 2018, 59, 6006. [Google Scholar]
- Khanna, H. More Than Meets the Eye: Current Understanding of RPGR Function. In Advances in Experimental Medicine and Biology; Springer: Cham, Switzerland, 2018; pp. 521–538. [Google Scholar]
- Vervoort, R.; Lennon, A.; Bird, A.C.; Tulloch, B.; Axton, R.; Miano, M.G.; Meindl, A.; Meitinger, T.; Ciccodicola, A.; Wright, A.F. Mutational hot spot within a new RPGR exon in X-linked retinitis pigmentosa. Nat. Genet. 2000, 25, 462–466. [Google Scholar] [CrossRef]
- Kropatsch, R.; Akkad, D.A.; Frank, M.; Rosenhagen, C.; Altmüller, J.; Nürnberg, P.; Epplen, J.T.; Dekomien, G. A large deletion in RPGR causes XLPRA in Weimaraner dogs. Canine Genet. Epidemiol. 2016, 3, 7. [Google Scholar] [CrossRef]
- Zhang, Q.; Acland, G.M.; Wu, W.X.; Johnson, J.; Pearce-Kelling, S.; Tulloch, B.; Vervoort, R.; Wright, A.F.; Aguirre, G.D. Different RPGR exon ORF15 mutations in Canids provide insights into photoreceptor cell degeneration. Hum. Mol. Genet. 2002, 11, 993–1003. [Google Scholar] [CrossRef]
- Zeiss, C.J.; Acland, G.M.; Aguirre, G.D. Retinal pathology of canine X-linked progressive retinal atrophy, the locus homologue of RP3. Investig. Ophthalmol. Vis. Sci. 1999, 40, 3292–3304. [Google Scholar]
- Beltran, W.A.; Hammond, P.; Acland, G.M.; Aguirre, G.D. A Frameshift Mutation in RPGR Exon ORF15 Causes Photoreceptor Degeneration and Inner Retina Remodeling in a Model of X-Linked Retinitis Pigmentosa. Investig. Opthalmol. Vis. Sci. 2006, 47, 1669–1681. [Google Scholar] [CrossRef] [PubMed]
- Beltran, W.A.; Acland, G.M.; Aguirre, G.D. Age-dependent disease expression determines remodeling of the retinal mosaic in carriers of RPGR exon ORF15 mutations. Investig. Opthalmol. Vis. Sci. 2009, 50, 3985–3995. [Google Scholar] [CrossRef] [PubMed]
- Mellersh, C.; Boursnell, M.; Pettitt, L.; Ryder, E.; Holmes, N.; Grafham, D.; Forman, O.; Sampson, J.; Barnett, K.; Blanton, S.; et al. Canine RPGRIP1 mutation establishes cone–rod dystrophy in miniature longhaired dachshunds as a homologue of human Leber congenital amaurosis. Genomics 2006, 88, 293–301. [Google Scholar] [CrossRef] [PubMed]
- Lhériteau, E.; Petit, L.; Weber, M.; Le Meur, G.; Deschamps, J.-Y.; Libeau, L.; Mendes-Madeira, A.; Guihal, C.; François, A.; Guyon, R.; et al. Successful Gene Therapy in the RPGRIP1-deficient Dog: A Large Model of Cone–Rod Dystrophy. Mol. Ther. 2013, 22, 265–277. [Google Scholar] [CrossRef] [PubMed]
- Kuznetsova, T.; Iwabe, S.; Boesze-Battaglia, K.; Pearce-Kelling, S.; Chang-Min, Y.; McDaid, K.; Miyadera, K.; Komaromy, A.; Aguirre, G.D. Exclusion of RPGRIP1 ins44 from Primary Causal Association with Early-Onset Cone–Rod Dystrophy in Dogs. Investig. Opthalmol. Vis. Sci. 2012, 53, 5486–5501. [Google Scholar] [CrossRef]
- Forman, O.P.; Hitti, R.J.; Boursnell, M.; Miyadera, K.; Sargan, D.R.; Mellersh, C. Canine genome assembly correction facilitates identification of a MAP9 deletion as a potential age of onset modifier for RPGRIP1-associated canine retinal degeneration. Mamm. Genome 2016, 27, 237–245. [Google Scholar] [CrossRef]
- Miyadera, K.; Kato, K.; Boursnell, M.; Mellersh, C.; Sargan, D.R. Genome-wide association study in RPGRIP1−/− dogs identifies a modifier locus that determines the onset of retinal degeneration. Mamm. Genome 2011, 23, 212–223. [Google Scholar] [CrossRef]
- Miyadera, K.; Murgiano, L.; Spector, C.; Marinho, F.P.; Dufour, V.; Das, R.G.; Brooks, M.; Swaroop, A.; Aguirre, G.D. Isolated population helps tease out a third locus underlying a multigenic form of canine RPGRIP1 cone-rod dystrophy. Investig. Ophthalmol. Vis. Sci. 2018, 59, 1438. [Google Scholar]
- Chau, K.-Y.; Chen, S.; Zack, D.J.; Ono, S.J. Functional Domains of the Cone-Rod Homeobox (CRX) Transcription Factor. J. Boil. Chem. 2000, 275, 37264–37270. [Google Scholar] [CrossRef]
- Morrow, E.M.; Furukawa, T.; Raviola, E.; Cepko, C.L. Synaptogenesis and outer segment formation are perturbed in the neural retina of Crx mutant mice. BMC Neurosci. 2005, 6, 5. [Google Scholar] [CrossRef]
- Chen, S.; Wang, Q.-L.; Nie, Z.; Sun, H.; Lennon, G.; Copeland, N.; Gilbert, D.J.; Jenkins, N.A.; Zack, D.J. Crx, a novel Otx-like paired-homeodomain protein, binds to and transactivates photoreceptor cell-specific genes. Neuron 1997, 19, 1017–1030. [Google Scholar] [CrossRef]
- Furukawa, T.; Morrow, E.M.; Cepko, C.L. Crx, a novel otx-like homeobox gene, shows photoreceptor-specific expression and regulates photoreceptor differentiation. Cell 1997, 91, 531–541. [Google Scholar] [CrossRef]
- Hennig, A.K.; Peng, G.-H.; Chen, S. Regulation of photoreceptor gene expression by Crx-associated transcription factor network. Brain Res. 2007, 1192, 114–133. [Google Scholar] [CrossRef] [PubMed]
- Peng, G.-H.; Chen, S. Crx activates opsin transcription by recruiting HAT-containing co-activators and promoting histone acetylation. Hum. Mol. Genet. 2007, 16, 2433–2452. [Google Scholar] [CrossRef] [PubMed]
- Menotti-Raymond, M.; Deckman, K.H.; David, V.; Myrkalo, J.; O’Brien, S.J.; Narfström, K. Mutation discovered in a feline model of human congenital retinal blinding disease. Investig. Opthalmol. Vis. Sci. 2010, 51, 2852–2859. [Google Scholar] [CrossRef] [PubMed]
- Occelli, L.M.; Tran, N.M.; Narfström, K.; Chen, S.; Petersen-Jones, S.M. CrxRdy Cat: A Large Animal Model for CRX-Associated Leber Congenital Amaurosis. Investig. Opthalmol. Vis. Sci. 2016, 57, 3780–3792. [Google Scholar] [CrossRef]
- Sohocki, M.M.; Sullivan, L.S.; Mintz-Hittner, H.A.; Birch, D.G.; Heckenlively, J.R.; Freund, C.L.; McInnes, R.R.; Daiger, S.P. A range of clinical phenotypes associated with mutations in CRX, a photoreceptor transcription-factor gene. Am. J. Hum. Genet. 1998, 63, 1307–1315. [Google Scholar] [CrossRef]
- Hull, S.; Arno, G.; Plagnol, V.; Chamney, S.; Russell-Eggitt, I.; Thompson, D.; Ramsden, S.C.; Black, G.C.; Robson, A.; Holder, G.E.; et al. The Phenotypic Variability of Retinal Dystrophies Associated With Mutations in CRX, With Report of a Novel Macular Dystrophy Phenotype. Investig. Opthalmol. Vis. Sci. 2014, 55, 6934–6944. [Google Scholar] [CrossRef]
- Huang, L.; Xiao, X.; Li, S.; Jia, X.; Wang, P.; Guo, X.; Zhang, Q. CRX variants in cone–rod dystrophy and mutation overview. Biochem. Biophys. Res. Commun. 2012, 426, 498–503. [Google Scholar] [CrossRef]
- Hanein, S.; Perrault, I.; Gerber, S.; Tanguy, G.; Barbet, F.; Ducroq, D.; Calvas, P.; Dollfus, H.; Hamel, C.; Löppönen, T.; et al. Leber congenital amaurosis: Comprehensive survey of the genetic heterogeneity, refinement of the clinical definition, and genotype-phenotype correlations as a strategy for molecular diagnosis. Hum. Mutat. 2004, 23, 306–317. [Google Scholar] [CrossRef]
- Kumaran, N.; Pennesi, M.E.; Yang, P.; Trzupek, K.M.; Schlechter, C.; Moore, A.T.; Weleber, R.G.; Michaelides, M. Leber Congenital Amaurosis/Early-Onset Severe Retinal Dystrophy Overview. In GeneReviews((R)); Adam, M.P., Ardinger, H.H., Pagon, R.A., Wallace, S.E., Bean, L.J.H., Stephens, K., Amemiya, A., Eds.; University of Washington: Seattle, WA, USA, 1993. [Google Scholar]
- Hollander, A.I.D.; Roepman, R.; Koenekoop, R.K.; Cremers, F.P. Leber congenital amaurosis: Genes, proteins and disease mechanisms. Prog. Retin. Eye Res. 2008, 27, 391–419. [Google Scholar] [CrossRef] [PubMed]
- Tran, N.M.; Chen, S. Mechanisms of blindness: Animal models provide insight into distinct CRX-associated retinopathies. Dev. Dyn. 2014, 243, 1153–1166. [Google Scholar] [CrossRef]
- Acland, G.M.; Aguirre, G.D. Retinal degenerations in the dog: IV. Early retinal degeneration (erd) in the Norwegian elkhound. Exp. Eye Res. 1987, 44, 491–521. [Google Scholar] [CrossRef]
- Goldstein, O.; Kukekova, A.V.; Aguirre, G.D.; Acland, G.M. Exonic SINE insertion in STK38L causes canine early retinal degeneration (erd). Genomics 2010, 96, 362–368. [Google Scholar] [CrossRef] [PubMed]
- Berta, Á.I.; Boesze-Battaglia, K.; Genini, S.; Goldstein, O.; O’Brien, P.J.; Szél, Á.; Acland, G.M.; Beltran, W.A.; Aguirre, G.D. Photoreceptor Cell Death, Proliferation and Formation of Hybrid Rod/S-Cone Photoreceptors in the Degenerating STK38L Mutant Retina. PLoS ONE 2011, 6, e24074. [Google Scholar] [CrossRef] [PubMed]
- Zeitz, C.; Jacobson, S.G.; Hamel, C.P.; Bujakowska, K.M.; Neuillé, M.; Orhan, E.; Zanlonghi, X.; Lancelot, M.-E.; Michiels, C.; Schwartz, S.B.; et al. Whole-Exome Sequencing Identifies LRIT3 Mutations as a Cause of Autosomal-Recessive Complete Congenital Stationary Night Blindness. Am. J. Hum. Genet. 2013, 92, 67–75. [Google Scholar] [CrossRef]
- Kondo, M.; Das, R.; Imai, R.; Santana, E.; Nakashita, T.; Imawaka, M.; Ueda, K.; Ohtsuka, H.; Sakai, K.; Aihara, T.; et al. A Naturally Occurring Canine Model of Autosomal Recessive Congenital Stationary Night Blindness. PLoS ONE 2015, 10, e0137072. [Google Scholar] [CrossRef]
- Das, R.; Becker, D.; Jagannathan, V.; Goldstein, O.; Santana, E.; Carlin, K.; Sudharsan, R.; Leeb, T.; Nishizawa, Y.; Kondo, M.; et al. Genome-wide association study and whole-genome sequencing identify a deletion in LRIT3 associated with canine congenital stationary night blindness. Sci. Rep. 2019, 9, 14166. [Google Scholar] [CrossRef]
- Oh, A.; Loew, E.R.; Foster, M.L.; Davidson, M.G.; English, R.V.; Gervais, K.J.; Herring, I.; Mowat, F.M. Phenotypic characterization of complete CSNB in the inbred research beagle: How common is CSNB in research and companion dogs? Doc. Ophthalmol. 2018, 137, 87–101. [Google Scholar] [CrossRef]
- Hasan, N.; Pangeni, G.; Cobb, C.A.; Ray, T.A.; Nettesheim, E.R.; Ertel, K.J.; Lipinski, D.M.; McCall, M.A.; Gregg, R.G. Presynaptic Expression of LRIT3 Transsynaptically Organizes the Postsynaptic Glutamate Signaling Complex Containing TRPM1. Cell Rep. 2019, 27, 3107–3116. [Google Scholar] [CrossRef]
- Morgans, C.; Zhang, J.; Jeffrey, B.G.; Nelson, S.M.; Burke, N.S.; Duvoisin, R.M.; Brown, R.L. TRPM1 is required for the depolarizing light response in retinal ON-bipolar cells. Proc. Natl. Acad. Sci. USA 2009, 106, 19174–19178. [Google Scholar] [CrossRef] [PubMed]
- Morgans, C.; Brown, R.L.; Duvoisin, R.M. TRPM1: The endpoint of the mGluR6 signal transduction cascade in retinal ON-bipolar cells. BioEssays 2010, 32, 609–614. [Google Scholar] [CrossRef] [PubMed]
- Witzel, D.A.; Smith, E.L.; Wilson, R.D.; Aguirre, G.D. Congenital stationary night blindness: An animal model. Investig. Ophthalmol. Vis. Sci. 1978, 17, 788–795. [Google Scholar]
- Sandmeyer, L.S.; Breaux, C.B.; Archer, S.; Grahn, B.H. Clinical and electroretinographic characteristics of congenital stationary night blindness in the Appaloosa and the association with the leopard complex. Veter Ophthalmol. 2007, 10, 368–375. [Google Scholar] [CrossRef] [PubMed]
- Bellone, R.; Brooks, S.A.; Sandmeyer, L.; Murphy, B.A.; Forsyth, G.; Archer, S.; Bailey, E.; Grahn, B. Differential Gene Expression of TRPM1, the Potential Cause of Congenital Stationary Night Blindness and Coat Spotting Patterns (LP) in the Appaloosa Horse (Equus caballus). Genetics 2008, 179, 1861–1870. [Google Scholar] [CrossRef] [PubMed]
- Audo, I.; Kohl, S.; Leroy, B.P.; Munier, F.L.; Guillonneau, X.; Mohand-Saïd, S.; Bujakowska, K.M.; Nandrot, E.F.; Lorenz, B.; Preising, M.; et al. TRPM1 Is Mutated in Patients with Autosomal-Recessive Complete Congenital Stationary Night Blindness. Am. J. Hum. Genet. 2009, 85, 720–729. [Google Scholar] [CrossRef]
- Li, Z.; Sergouniotis, P.I.; Michaelides, M.; Mackay, D.; Wright, G.A.; Devery, S.; Moore, A.T.; Holder, G.E.; Robson, A.; Webster, A.R. Recessive Mutations of the Gene TRPM1 Abrogate ON Bipolar Cell Function and Cause Complete Congenital Stationary Night Blindness in Humans. Am. J. Hum. Genet. 2009, 85, 711–719. [Google Scholar] [CrossRef]
- Van Genderen, M.M.; Bijveld, M.M.; Claassen, Y.B.; Florijn, R.J.; Pearring, J.N.; Meire, F.M.; McCall, M.A.; Riemslag, F.C.; Gregg, R.G.; Bergen, A.A.; et al. Mutations in TRPM1 Are a Common Cause of Complete Congenital Stationary Night Blindness. Amer. J. Human Genet. 2009, 85, 730–736. [Google Scholar] [CrossRef]
- Bellone, R.; Holl, H.; Setaluri, V.; Devi, S.; Maddodi, N.; Archer, S.; Sandmeyer, L.; Ludwig, A.; Förster, D.; Pruvost, M.; et al. Evidence for a Retroviral Insertion in TRPM1 as the Cause of Congenital Stationary Night Blindness and Leopard Complex Spotting in the Horse. PLoS ONE 2013, 8, e78280. [Google Scholar] [CrossRef]
- Littink, K.W.; Van Genderen, M.M.; Collin, R.W.; Roosing, S.; De Brouwer, A.P.M.; Riemslag, F.C.C.; Venselaar, H.; Thiadens, A.A.H.J.; Hoyng, C.; Rohrschneider, K.; et al. A Novel Homozygous Nonsense Mutation inCABP4Causes Congenital Cone–Rod Synaptic Disorder. Investig. Opthalmol. Vis. Sci. 2009, 50, 2344–2350. [Google Scholar] [CrossRef]
- Marinho, L.L.P.; Occelli, L.M.; Pasmanter, N.; Somma, A.T.; Montiani-Ferreira, F.; Petersen-Jones, S.M. Autosomal recessive night blindness with progressive photoreceptor degeneration in a dog model. Investig. Ophthalmol. Vis. Sci. 2019, 60, 465. [Google Scholar]
- Somma, A.T.; Moreno, J.C.D.; Sato, M.T.; Rodrigues, B.D.; Occelli, L.M.; Bacellar-Galdino, M.; Petersen-Jones, S.M.; Montiani-Ferreira, F. Characterization of a novel form of progressive retinal atrophy in Whippet dogs: A clinical, electroretinographic, and breeding study. Veter Ophthalmol. 2016, 20, 450–459. [Google Scholar] [CrossRef] [PubMed]
- Kropatsch, R.; Petrasch-Parwez, E.; Seelow, D.; Schlichting, A.; Gerding, W.M.; Akkad, D.A.; Epplen, J.T.; Dekomien, G. Generalized progressive retinal atrophy in the Irish Glen of Imaal Terrier is associated with a deletion in the ADAM9 gene. Mol. Cell. Probes 2010, 24, 357–363. [Google Scholar] [CrossRef] [PubMed]
- Goldstein, O.; Mezey, J.G.; Boyko, A.R.; Gao, C.; Wang, W.; Bustamante, C.D.; Anguish, L.J.; Jordan, J.A.; Pearce-Kelling, S.E.; Aguirre, G.D.; et al. An ADAM9 mutation in canine cone-rod dystrophy 3 establishes homology with human cone-rod dystrophy 9. Mol. Vis. 2010, 16, 1549–1569. [Google Scholar] [PubMed]
- Sohocki, M.M.; Perrault, I.; Leroy, B.P.; Payne, A.; Dharmaraj, S.; Bhattacharya, S.S.; Kaplan, J.; Maumenee, I.H.; Koenekoop, R.; Meire, F.M.; et al. Prevalence of AIPL1 Mutations in Inherited Retinal Degenerative Disease. Mol. Genet. Metab. 2000, 70, 142–150. [Google Scholar] [CrossRef]
- Van Der Spuy, J.; Kim, J.H.; Yu, Y.S.; Szel, A.; Luthert, P.J.; Clark, B.J.; Cheetham, M.E. The expression of the Leber congenital amaurosis protein AIPL1 coincides with rod and cone photoreceptor development. Investig. Opthalmol. Vis. Sci. 2003, 44, 5396–5403. [Google Scholar] [CrossRef]
- Kumaran, N.; Moore, A.T.; Weleber, R.G.; Michaelides, M. Leber congenital amaurosis/early-onset severe retinal dystrophy: Clinical features, molecular genetics and therapeutic interventions. Br. J. Ophthalmol. 2017, 101, 1147–1154. [Google Scholar] [CrossRef]
- Sohocki, M.M.; Bowne, S.J.; Sullivan, L.S.; Blackshaw, S.; Cepko, C.L.; Payne, A.M.; Bhattacharya, S.S.; Khaliq, S.; Mehdi, S.Q.; Birch, D.G.; et al. Mutations in a new photoreceptor-pineal gene on 17p cause Leber congenital amaurosis. Nat. Genet. 2000, 24, 79–83. [Google Scholar] [CrossRef]
- Gopalakrishna, K.N.; Boyd, K.; Yadav, R.P.; Artemyev, N.O. Aryl Hydrocarbon Receptor-interacting Protein-like 1 Is an Obligate Chaperone of Phosphodiesterase 6 and Is Assisted by the γ-Subunit of Its Client. J. Boil. Chem. 2016, 291, 16282–16291. [Google Scholar] [CrossRef]
- Hidalgo-De-Quintana, J.; Evans, R.J.; Cheetham, M.E.; Van Der Spuy, J. The Leber congenital amaurosis protein AIPL1 functions as part of a chaperone heterocomplex. Investig. Opthalmol. Vis. Sci. 2008, 49, 2878–2887. [Google Scholar] [CrossRef]
- Ramamurthy, V.; Niemi, G.A.; Reh, T.A.; Hurley, J.B. Leber congenital amaurosis linked to AIPL1: A mouse model reveals destabilization of cGMP phosphodiesterase. Proc. Natl. Acad. Sci. USA 2004, 101, 13897–13902. [Google Scholar] [CrossRef]
- Yadav, R.P.; Artemyev, N.O. AIPL1: A specialized chaperone for the phototransduction effector. Cell. Signal. 2017, 40, 183–189. [Google Scholar] [CrossRef]
- Kolandaivelu, S.; Singh, R.K.; Ramamurthy, V. AIPL1, A protein linked to blindness, is essential for the stability of enzymes mediating cGMP metabolism in cone photoreceptor cells. Hum. Mol. Genet. 2013, 23, 1002–1012. [Google Scholar] [CrossRef]
- Rah, H.; Maggs, D.J.; Blankenship, T.N.; Narfström, K.; Lyons, L.A. Early-Onset, Autosomal Recessive, Progressive Retinal Atrophy in Persian Cats. Investig. Opthalmol. Vis. Sci. 2005, 46, 1742–1747. [Google Scholar] [CrossRef] [PubMed]
- Lyons, L.A.; Creighton, E.K.; Alhaddad, H.; Beale, H.; Grahn, R.; Rah, H.; Maggs, D.J.; Helps, C.; Gandolfi, B. Whole genome sequencing in cats, identifies new models for blindness in AIPL1 and somite segmentation in HES7. BMC Genom. 2016, 17, 265. [Google Scholar] [CrossRef] [PubMed]
- Ahonen, S.; Arumilli, M.; Seppälä, E.; Hakosalo, O.; Kaukonen, M.K.; Komaromy, A.M.; Lohi, H. Increased Expression of MERTK is Associated with a Unique Form of Canine Retinopathy. PLoS ONE 2014, 9, e114552. [Google Scholar] [CrossRef] [PubMed]
- Fanning, T.; Singer, M. LINE-1: A mammalian transposable element. Biochim. Biophys. Acta (BBA) Gene Struct. Expr. 1987, 910, 203–212. [Google Scholar] [CrossRef]
- Everson, R.; Pettitt, L.; Forman, O.P.; Dower-Tylee, O.; McLaughlin, B.; Ahonen, S.; Kaukonen, M.; Komaromy, A.M.; Lohi, H.; Mellersh, C.S.; et al. An intronic LINE-1 insertion in MERTK is strongly associated with retinopathy in Swedish Vallhund dogs. PLoS ONE 2017, 12, e0183021. [Google Scholar] [CrossRef] [PubMed]
- Zangerl, B.; Goldstein, O.; Philp, A.R.; Lindauer, S.J.; Pearce-Kelling, S.E.; Mullins, R.F.; Graphodatsky, A.; Ripoll, D.; Felix, J.S.; Stone, E.M.; et al. Identical mutation in a novel retinal gene causes progressive rod-cone degeneration in dogs and retinitis pigmentosa in humans. Genomics 2006, 88, 551–563. [Google Scholar] [CrossRef]
- Nevet, M.J.; Shalev, S.A.; Zlotogora, J.; Mazzawi, N.; Ben-Yosef, T. Identification of a prevalent founder mutation in an Israeli Muslim Arab village confirms the role of PRCD in the aetiology of retinitis pigmentosa in humans. J. Med. Genet. 2010, 47, 533–537. [Google Scholar] [CrossRef]
- Allon, G.; Mann, I.; Remez, L.; Sehn, E.; Rizel, L.; Nevet, M.J.; Perlman, I.; Wolfrum, U.; Ben-Yosef, T. PRCD is Concentrated at the Base of Photoreceptor Outer Segments and is Involved in Outer Segment Disc Formation. Hum. Mol. Genet. 2019, 28, 4078–4088. [Google Scholar] [CrossRef] [PubMed]
- Spencer, W.J.; Pearring, J.N.; Salinas, R.Y.; Loiselle, D.R.; Skiba, N.P.; Arshavsky, V.Y. Progressive Rod–Cone Degeneration (PRCD) Protein Requires N-Terminal S-Acylation and Rhodopsin Binding for Photoreceptor Outer Segment Localization and Maintaining Intracellular Stability. Biochemistry 2016, 55, 5028–5037. [Google Scholar] [CrossRef]
- Spencer, W.J.; Ding, J.-D.; Lewis, T.R.; Yu, C.; Phan, S.; Pearring, J.N.; Kim, K.-Y.; Thor, A.; Mathew, R.; Kalnitsky, J.; et al. PRCD is essential for high-fidelity photoreceptor disc formation. Proc. Natl. Acad. Sci. USA 2019, 116, 13087–13096. [Google Scholar] [CrossRef]
- Aguirre, G.D.; Alligood, J.; O’Brien, P.; Buyukmihci, N. Pathogenesis of progressive rod-cone degneration in miniature poodles. Investig. Ophthalmol. Vis. Sci. 1982, 23, 610–630. [Google Scholar]
- Friedman, J.S.; Chang, B.; Kannabiran, C.; Chakarova, C.; Singh, H.; Jalali, S.; Hawes, N.L.; Branham, K.; Othman, M.; Filippova, E.; et al. Premature Truncation of a Novel Protein, RD3, Exhibiting Subnuclear Localization Is Associated with Retinal Degeneration. Am. J. Hum. Genet. 2006, 79, 1059–1070. [Google Scholar] [CrossRef] [PubMed]
- Wolf, E.D.; Vainisi, S.J.; Santos-Anderson, R. Rod-cone dysplasia in the collie. J. Am. Veter Med Assoc. 1978, 173, 1331–1333. [Google Scholar]
- Woodford, B.; Liu, Y.; Fletcher, R.; Chader, G.; Farber, D.; Santos-Anderson, R.; Tso, M.O. Cyclic nucleotide metabolism in inherited retinopathy in collies: A biochemical and histochemical study. Exp. Eye Res. 1982, 34, 703–714. [Google Scholar] [CrossRef]
- Dizhoor, A.M.; Olshevskaya, E.V.; Peshenko, I.V. Retinal guanylyl cyclase activation by calcium sensor proteins mediates photoreceptor degeneration in an rd3 mouse model of congenital human blindness. J. Boil. Chem. 2019, 294, 13729–13739. [Google Scholar] [CrossRef]
- Kukekova, A.V.; Goldstein, O.; Johnson, J.; Richardson, M.A.; Pearce-Kelling, S.E.; Swaroop, A.; Friedman, J.S.; Aguirre, G.D.; Acland, G.M. Canine RD3 mutation establishes rod-cone dysplasia type 2 (rcd2) as ortholog of human and murine rd3. Mamm. Genome 2009, 20, 109–123. [Google Scholar] [CrossRef]
- Santos-Anderson, R.M.; Tso, M.O.; Wolf, E.D. An inherited retinopathy in collies. A light and electron microscopic study. Investig. Ophthalmol. Vis. Sci. 1980, 19, 1282–1294. [Google Scholar]
| Mechanism | Gene | Species | Mutation |
|---|---|---|---|
| Phototransduction | RHO* | dog pig pig pig | c.11C>G, p.Thr4Arg p.Pro23His c.1040C>T, p.Pro347Leu p.Pro347Ser |
| PDE6A* | dog | c.1939delA, p.Asn616ThrfsTer39 | |
| PDE6B* | dog | c.2420G>A, p.Trp807Ter; c.2449_2450insTGAAGTCC; p.Lys816Terfs817 c.2404_2406delAAC, p.Asn802del | |
| PDE6C* | NHP | c.1694G>A, p.Arg565Gln | |
| SAG | dog | c.1216T>C; p.Ter406extArg*25 | |
| Visual Cycle | ABCA4* | dog | c.4176insC, p.Phe1393LeufsTer3 |
| RPE65* | dog | c.487_490delAAGA; p.Lys154LeufsTer53 | |
| RDH5* | cat | unpublished | |
| Channelopathies/channel related | CNGA1* | dog | c.1752_1755delAACT, p.Thr584SerfsTer9 |
| CNGB1* | dog | c.2387delA;2389_2390insAGCTAC, p.Ser791ArgfsTer2 | |
| CNGA3* | dog dog sheep sheep | c.1270C>T; p.Arg424Trp c.1931_1933delTGG, p.Val644del p.Arg236Ter p.Gly540Ser | |
| CNGB3* | dog | c.784G>A; p.Asp262Asn CFA29:g.35,699,378-36,104,197del, c.0 | |
| BEST1* | dog | c.73C>T, p.Arg25Ter; c.482G>A, p.Gly161Asp; c.C1388del and c.1466G>T, p.Pro463fs and p.Gly489Val | |
| Ciliopathies | BBS4* | dog | c.58A>T, p.Lys20Ter |
| BBS7* | NHP | c.160delG, p.Ala54GlnfsTer18 | |
| c2orf71 | dog | c.3149_3150insC, p.Lys1051ValfsTer91 | |
| CCDC66 | dog | c.521_522insA, p.Asn174LysfsTer2 | |
| CEP290* | cat | c.6966+9T>G, p.Ile2323AlafsTer3 | |
| FAM161A | dog | c.1758-15_1758-16ins238, p.Ser588MetfsTer14 | |
| NPHP4* | dog | c.462_526del, p.Leu155LysfsTer2 | |
| NPHP5(IQCB1)* | dog cat | c.952-953insC, p.Ser319IlefsTer13 c.1282delCT, p.Leu428Ter | |
| RPGR* | dog | c.1084-1085delGA, c.1028-1032delGAGAA CFAX:g. 33106747+190-33102324del | |
| RGRIP1* | dog | CFA15:g.8228_8229insA29GGAAGCAACAGGATG | |
| TTC8 | dog | c.669delA, p.Lys223ArgfsTer15 | |
| Photoreceptor development | CRX* | cat | c.546delC, p.Pro185LysfsTer2 |
| STK38L* | dog | c.299_300ins [218;285_299]; p.Lys63_Glu103del | |
| Photoreceptor to Bipolar Cell | LRIT3* | dog | c.762_763delG, p.Lys246AsnfsTer5 |
| TRPM1* | horse | ECA1g.108,297,929_108,297,930ins1378 | |
| Whippet* | dog | unpublished | |
| Structural/Other | ADAM9* | dog | c.1592_1881del p.Lys531AsnfsTer3 |
| AIPL1* | cat | c.577C>T, p.Arg193Ter | |
| MERTK* | dog | CFA17:g.36338057_36338058ins[(6401);36338043-36338057] | |
| PRCD* | dog | c.5G>A, p.Cys2tyr | |
| RD3* | dog | c.418_419ins[22] | |
| NECAP1 | dog | c.544G>A, p.Gly182Arg | |
| PPT1 | dog | CFA15:g.[2,866,454_2,877,574dup; 2,874,661_2,875,048con2,877,563-2,877,607inv] | |
| SLC4A3 | dog | c.2601_2602insC, p.Glu868ArgfsTer104 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Winkler, P.A.; Occelli, L.M.; Petersen-Jones, S.M. Large Animal Models of Inherited Retinal Degenerations: A Review. Cells 2020, 9, 882. https://doi.org/10.3390/cells9040882
Winkler PA, Occelli LM, Petersen-Jones SM. Large Animal Models of Inherited Retinal Degenerations: A Review. Cells. 2020; 9(4):882. https://doi.org/10.3390/cells9040882
Chicago/Turabian StyleWinkler, Paige A., Laurence M. Occelli, and Simon M. Petersen-Jones. 2020. "Large Animal Models of Inherited Retinal Degenerations: A Review" Cells 9, no. 4: 882. https://doi.org/10.3390/cells9040882
APA StyleWinkler, P. A., Occelli, L. M., & Petersen-Jones, S. M. (2020). Large Animal Models of Inherited Retinal Degenerations: A Review. Cells, 9(4), 882. https://doi.org/10.3390/cells9040882




