Nitrogen Deficiency-Induced Decrease in Cytokinins Content Promotes Rice Seminal Root Growth by Promoting Root Meristem Cell Proliferation and Cell Elongation
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Growth Conditions
2.2. Chemicals and Treatments
2.3. CK Content Analysis
2.4. RNA Isolation and Quantitative Real-Time PCR (qRT-PCR) Analysis
2.5. Quantitative Analysis of Root Phenotypes
2.6. Statistical Analysis
2.7. Accession Numbers
3. Results
3.1. N Deficiency-Induced Growth of Rice Seminal Roots was Negated by Application of CK
3.2. N Deficiency Reduced CK Content in Rice Seminal Roots
3.3. N Deficiency Inhibited CK Biosynthesis and Promoted CK Degradation by Affecting the Transcription Levels of CK Metabolic Genes in Rice Seminal Roots
3.4. N Deficiency-Induced Decrease in CK Content Enhanced Root Meristem Cell Proliferation Rate and Promoted Root Cell Elongation
3.5. Decrease in CK Content Increased Root Meristem Cell Number by Affecting the Transcription Levels of Root Meristem Size-Related Genes
3.6. Decrease in CK Content Increased Root Cell Length by Up-Regulating Transcription of Root Cell Elongation-Related Genes
4. Discussion
4.1. Increasing Root Length is a Strategy for Responding to N Deficiency
4.2. N Deficiency Promotes Rice Seminal Root Growth by Reducing CK Content
4.3. N Deficiency Reduces CK Content by Inhibiting CK Biosynthesis and Promoting CK Degradation
4.4. N Deficiency-Induced Decrease in CK Content Promotes Rice Seminal Root Growth by Enhancing Meristem Cell Proliferation Rate through Increased Meristem Cell Number
4.5. Decrease in CK Content Promotes Rice Seminal Root Growth by Promoting Root Cell Elongation
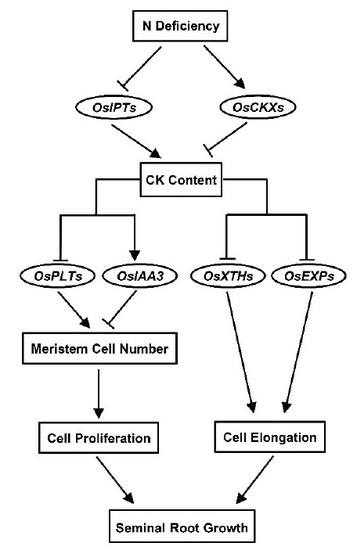
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| CK | Cytokinin |
| CKX | Cytokinin oxidase dehydrogenase |
| DZ | dihydrozeatin |
| EXP | Expansin |
| FW | Fresh weight |
| iP | Isopentenyladenine |
| N | Nitrogen |
| NS | Not significant |
| PLT | PLETHORA |
| XTH | Xyloglucan endotransglucosylase/hydrolase |
| Z | Zeatin |
References
- Fageria, N.K. Yield physiology of rice. J. Plant Nutr. 2007, 30, 843–879. [Google Scholar] [CrossRef]
- Peng, S.; Buresh, R.J.; Huang, J.; Zhong, X.; Zou, Y.; Yang, J.; Wang, G.; Liu, Y.; Hu, R.; Tang, Q.; et al. Improving nitrogen fertilization in rice by sitespecific N management. Agron. Sustain. Dev. 2010, 30, 649–656. [Google Scholar] [CrossRef]
- Ju, X.T.; Xing, G.X.; Chen, X.P.; Zhang, S.L.; Zhang, L.J.; Liu, X.J.; Cui, Z.L.; Yin, B.; Christie, P.; Zhu, Z.L.; et al. Reducing environmental risk by improving n management in intensive chinese agricultural systems. Proc. Natl. Acad. Sci. USA 2009, 106, 3041–3046. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Good, A.G.; Beatty, P.H. Fertilizing nature: A tragedy of excess in the commons. PLoS Biol. 2011, 9, e1001124. [Google Scholar] [CrossRef] [PubMed]
- Drew, M.C.; Saker, L.R.; Ashley, T.W. Nutrient supply and the growth of the seminal root system in barley. J. Exp. Bot. 1973, 24, 1189–1202. [Google Scholar] [CrossRef]
- Sun, H.W.; Tao, J.Y.; Liu, S.J.; Huang, S.J.; Chen, S.; Xie, X.N.; Yoneyama, K.; Zhang, Y.L.; Xu, G.H. Strigolactones are involved in phosphate- and nitrate-deficiency-induced root development and auxin transport in rice. J. Exp. Bot. 2014, 65, 6735–6746. [Google Scholar] [CrossRef]
- Yang, C.; Yang, L.; Yang, Y.; Ouyang, Z. Rice root growth and nutrient uptake as influenced by organic manure in continuously and alternately flooded paddy soils. Agr. Water Manage. 2004, 70, 67–81. [Google Scholar] [CrossRef]
- Forde, B.G. Nitrogen signalling pathways shaping root system architecture: An update. Curr. Opin. Plant Biol. 2014, 21, 30–36. [Google Scholar] [CrossRef]
- Trachse, S.; Kaeppler, S.M.; Brown, K.M.; Lynch, J.P. Maize root growth angles become steeper under low N conditions. Field Crop. Res. 2013, 140, 18–31. [Google Scholar] [CrossRef]
- Sakakibara, H. Nitrate-specific and cytokinin-mediated nitrogen signaling pathways in plants. J. Plant Res. 2003, 116, 253–257. [Google Scholar] [CrossRef]
- Sakakibara, H.; Takei, K.; Hirose, N. Interactions between nitrogen and cytokinin in the regulation of metabolism and development. Trends Plant Sci. 2006, 11, 440–448. [Google Scholar] [CrossRef] [PubMed]
- Petricka, J.J.; Winter, C.M.; Benfey, P.N. Control of Arabidopsis root development. Annu. Rev. Plant Biol. 2012, 63, 563–590. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jia, Z.; Giehl, R.F.H.; Meyer, R.C.; Altmann, T.; von Wirén, N. Natural variation of BSK3 tunes brassinosteroid signaling to regulate root foraging under low nitrogen. Nat. Commun. 2019, 10, 2378–2390. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Linkohr, B.I.; Williamson, L.C.; Fitter, A.H.; Leyser, H.M.O. Nitrate and phosphate availability and distribution have different effects on root system architecture of Arabidopsis. Plant J. 2002, 29, 751–760. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, Q.; Chen, F.; Zhang, F.; Mi, G. Possible involvement of cytokinin in nitrate-mediated root growth in maize. Plant Soil 2005, 277, 185–196. [Google Scholar] [CrossRef]
- Yu, P.; Li, X.; White, P.J.; Li, C. A large and deep root system underlies high nitrogen-use efficiency in maize production. PLoS ONE 2015, 10, e0126293. [Google Scholar] [CrossRef] [Green Version]
- Leskovar, D.; Othman, Y. Low nitrogen fertigation promotes root development and transplant quality in globe artichoke. Hortscience 2016, 51, 567–572. [Google Scholar] [CrossRef] [Green Version]
- Mu, X.; Chen, F.; Wu, Q.; Chen, Q.; Wang, J.; Yuan, L.; Mi, G. Genetic improvement of root growth increases maize yield via enhanced post-silking nitrogen uptake. Eur. J. Agron. 2015, 63, 55–61. [Google Scholar] [CrossRef]
- Zou, X.; Shao, J.; Wang, Q.; Chen, P.; Zhu, Y.; Yin, C. Supraoptimal cytokinin content inhibits rice seminal root growth by reducing root meristem size and cell length via increased ethylene content. Int. J. Mol. Sci. 2018, 19, 4051. [Google Scholar] [CrossRef] [Green Version]
- Werner, T.; Motyka, V.; Laucou, V.; Smets, R.; Onckelen, H.V.; Schmulling, T. Cytokinin-deficient transgenic Arabidopsis plants show multiple developmental alterations indicating opposite functions of cytokinins in the regulation of shoot and root meristem activity. Plant Cell 2003, 15, 2532–2550. [Google Scholar] [CrossRef] [Green Version]
- Miyawaki, K.; Tarkowski, P.; Matsumoto-Kitano, M.; Kato, T.; Sato, S.; Tarkowska, D.; Tabata, S.; Sandberg, G.; Kakimoto, T. Roles of Arabidopsis ATP/ADP isopentenyltransferases and tRNA isopentenyltransferases in cytokinin biosynthesis. Proc. Natl. Acad. Sci. USA 2006, 103, 16598–16603. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, R.G.; Hay, M.J.M. Shoot branching in response to nodal roots is mimicked by application of exogenous cytokinin in Trifolium repens. Funct. Plant Biol. 2015, 42, 115–125. [Google Scholar] [CrossRef]
- Mochizuki, S.; Jikumaru, Y.; Nakamura, H.; Koiwai, H.; Sasaki, K.; Kamiya, Y.; Ichikawa, H.; Minami, E.; Nishizawa, Y. Ubiquitin ligase EL5 maintains the viability of root meristems by influencing cytokinin-mediated nitrogen effects in rice. J. Exp. Bot. 2014, 65, 2307–2318. [Google Scholar] [CrossRef] [PubMed]
- Ioio, R.D.; Linhares, F.S.; Scacchi, E.; Casamitjana-Martinez, E.; Heidstra, R.; Costantino, P.; Sabatini, S. Cytokinins determine Arabidopsis root-meristem size by controlling cell differentiation. Curr. Biol. 2007, 17, 678–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ioio, R.D.; Nakamura, K.; Moubayidin, L.; Perilli, S.; Taniguchi, M.; Morita, M.T.; Aoyama, T.; Costantino, P.; Sabatini, S. A genetic framework for the control of cell division and differentiation in the root meristem. Science 2008, 322, 1380–1384. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Takatsuka, H.; Umeda, M. Hormonal control of cell division and elongation along differentiation trajectories in roots. J. Exp. Bot. 2014, 65, 2633–2643. [Google Scholar] [CrossRef] [Green Version]
- Aida, M.; Beis, D.; Heidstra, R.; Willemsen, V.; Blilou, I.; Galinha, C.; Nussaume, L.; Noh, Y.; Amasino, R.; Scheres, B. The PLETHORA genes mediate patterning of the Arabidopsis root stem cell niche. Cell 2004, 119, 109–120. [Google Scholar] [CrossRef] [Green Version]
- Endo, M.; Nakayama, S.; Umeda-Hara, C.; Ohtsuki, N.; Saika, H.; Umeda, M.; Toki, S. CDKB2 is involved in mitosis and DNA damage response in rice. Plant J. 2012, 69, 967–977. [Google Scholar] [CrossRef] [Green Version]
- Galinha, C.; Hofhuis, H.; Luijten, M.; Willemsen, V.; Blilou, I.; Heidstra, R.; Scheres, B. PLETHORA proteins as dose-dependent master regulators of Arabidopsis root development. Nature 2007, 449, 1053–1057. [Google Scholar] [CrossRef] [Green Version]
- Nakamura, A.; Umemura, I.; Gomi, K.Y.; Kitano, H.; Sazuka, T.; Matsuoka, M. Production and characterization of auxin-insensitive rice by overexpression of a mutagenized rice IAA protein. Plant J. 2006, 46, 297–306. [Google Scholar] [CrossRef]
- Lee, Y.; Kende, H. Expression of alpha-expansin and expansin-like genes in deepwater rice. Plant Physiol. 2002, 130, 1396–1405. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yokoyama, R.; Rose, J.K.C.; Nishitani, K. A surprising diversity and abundance of xyloglucan endotransglucosylase/hydrolases in rice. Classification and expression analysis. Plant Physiol. 2004, 134, 1088–1099. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhu, X.F.; Shi, Y.Z.; Lei, G.J.; Fry, S.C.; Zhang, B.C.; Zhou, Y.H.; Braam, J.; Jiang, T.; Xu, X.Y.; Mao, C.Z.; et al. Xth31, encoding an in vitro XEH/XET-active enzyme, regulates aluminum sensitivity by modulating in vivo XET action, cell wall xyloglucan content, and aluminum binding capacity in Arabidopsis. Plant Cell 2012, 24, 4731–4747. [Google Scholar] [CrossRef] [Green Version]
- Zou, H.; Wenwen, Y.; Zang, G.; Kang, Z.; Zhang, Z.; Huang, J.; Wang, G. OsEXPB2, a beta-expansin gene, is involved in rice root system architecture. Mol. Breeding 2015, 35, 41–54. [Google Scholar] [CrossRef]
- Li, P.; Xue, H. Structural characterization and expression pattern analysis of the rice PLT gene family. Acta Biochim. Biophs. Sin. 2011, 43, 688–697. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yu, J.; Wang, J.; Lin, W.; Li, S.; Li, H.; Zhou, J.; Ni, P.; Dong, W.; Hu, S.; Zeng, C.; et al. The Genomes of Oryza sativa: A history of duplications. PLoS Biol. 2005, 3. [Google Scholar] [CrossRef] [Green Version]
- Wang, S.J.; Ho, C.H.; Chen, H.W. Rice develop wavy seminal roots in response to light stimulus. Plant Cell Rep. 2011, 30, 1747–1758. [Google Scholar] [CrossRef]
- Zou, Z.; Zou, X.; Zhao, S.; Xia, C.; Qian, K.; Wang, P.; Yin, C. Fluridone induces leaf bleaching by inhibiting pigment biosynthesis via downregulated transcription levels of pigment biosynthetic genes in rice Oryza sativa L. J. Plant Growth Regul. 2018, 37, 1385–1395. [Google Scholar] [CrossRef]
- Liu, H.; Li, X.; Xiao, J.; Wang, S. A convenient method for simultaneous quantification of multiple phytohormones and metabolites: Application in study of rice-bacterium interaction. Plant Methods 2012, 8, 2–13. [Google Scholar] [CrossRef] [Green Version]
- Liu, L.; Xia, W.; Li, H.; Zeng, H.; Wei, B.; Han, S.; Yin, C. Salinity inhibits rice seed germination by reducing alpha-amylase activity via decreased bioactive gibberellin content. Front. Plant Sci. 2018, 9, 275–283. [Google Scholar] [CrossRef]
- Li, J.; Zhao, Y.; Chu, H.; Wang, L.; Fu, Y.; Liu, P.; Upadhyaya, N.; Chen, C.; Mou, T.; Feng, Y.; et al. SHOEBOX modulates root meristem size in rice through dose-dependent effects of gibberellins on cell elongation and proliferation. PLoS Genet. 2015, 11, e1005464. [Google Scholar] [CrossRef] [PubMed]
- Letham, D.S.; Palni, L.M. The biosynthesis and metabolism of cytokinins. Annu. Rev. Plant Physiol. 1983, 34, 163–197. [Google Scholar] [CrossRef]
- Kudo, T.; Makita, N.; Kojima, M.; Tokunaga, H.; Sakakibara, H. Cytokinin activity of cis-zeatin and phenotypic alterations induced by overexpression of putative cis-Zeatin-O-glucosyltransferase in rice. Plant Physiol. 2012, 160, 319–331. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frebort, I.; Kowalska, M.; Hluska, T.; Frebortova, J.; Galuszka, P. Evolution of cytokinin biosynthesis and degradation. J. Exp. Bot. 2011, 62, 2431–2452. [Google Scholar] [CrossRef] [PubMed]
- Hwang, I.; Sheen, J.; Müller, B. Cytokinin signaling networks. Annu. Rev. Plant Biol. 2012, 63, 353–380. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kakimoto, T. Identification of plant cytokinin biosynthetic enzymes as dimethylallyl diphosphate: ATP/ADP isopentenyltransferases. Plant Cell Physiol. 2001, 42, 677–685. [Google Scholar] [CrossRef]
- Sakamoto, T.; Sakakibara, H.; Kojima, M.; Yamamoto, Y.; Nagasaki, H.; Inukai, Y.; Sato, Y.; Matsuoka, M. Ectopic expression of KNOTTED1-like homeobox protein induces expression of cytokinin biosynthesis genes in rice. Plant Physiol. 2006, 142, 54–62. [Google Scholar] [CrossRef] [Green Version]
- Garnica, M.; Houdusse, F.; Zamarreño, A.M.; Garcia-Mina, J.M. The signal effect of nitrate supply enhances active forms of cytokinins and indole acetic content and reduces abscisic acid in wheat plants grown with ammonium. J. Plant Physiol. 2010, 167, 1264–1272. [Google Scholar] [CrossRef]
- Kellermeier, F.; Armengaud, P.; Seditas, T.J.; Danku, J.; Salt, D.E.; Amtmann, A. Analysis of the root system architecture of Arabidopsis provides a quantitative readout of crosstalk between nutritional signals. Plant Cell 2014, 26, 1480–1496. [Google Scholar] [CrossRef] [Green Version]
- Ma, W.; Li, J.; Qu, B.; He, X.; Zhao, X.; Li, B.; Fu, X.; Tong, Y. Auxin biosynthetic gene, TAR2 is involved in low nitrogen-mediated reprogramming of root architecture in Arabidopsis. Plant J. 2014, 78, 70–79. [Google Scholar] [CrossRef]
- Cerutti, T.; Delatorre, C.A. Nitrogen and phosphorus interaction and cytokinin: Responses of the primary root of Arabidopsis thaliana and the pdr1 mutant. Plant Sci. 2013, 198, 91–97. [Google Scholar] [CrossRef] [PubMed]
- Kamada-Nobusada, T.; Makita, N.; Kojima, M.; Sakakibara, H. Nitrogen-dependent regulation of de novo cytokinin biosynthesis in rice: The role of glutamine metabolism as an additional signal. Plant Cell Physiol. 2013, 54, 1881–1893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sakakibara, H.; Suzuki, M.; Takei, K.; Deji, A.; Taniguchi, M.; Sugiyama, T. A response-regulator homologue possibly involved in nitrogen signal transduction mediated by cytokinin in maize. Plant J. 1998, 14, 337–344. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuelson, M.E.; Larsson, C.M. Nitrate regulation of zeation riboside levels in barley roots: Effects of inhibitors of N assimilation and comparison with ammonium. Plant Sci. 1993, 93, 77–84. [Google Scholar] [CrossRef]
- Werner, T.; Motyka, V.; Strnad, M.; Schmülling, T. Regulation of plant growth by cytokinin. Proc. Natl. Acad. Sci. USA 2001, 98, 10487–10492. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Werner, T.; Nehnevajova, E.; Kollmer, I.; Novak, O.; Strnad, M.; Kramer, U.; Schmulling, T. Root-specific reduction of cytokinin causes enhanced root growth, drought tolerance, and leaf mineral enrichment in Arabidopsis and tobacco. Plant Cell 2010, 22, 3905–3920. [Google Scholar] [CrossRef] [Green Version]
- Gao, S.; Fang, J.; Xu, F.; Wang, W.; Sun, X.; Chu, J.; Cai, B.; Feng, Y.; Chu, C. Cytokinin oxidase/dehydrogenase4 integrates cytokinin and auxin signaling to control rice crown root formation. Plant Physiol. 2014, 165, 1035–1046. [Google Scholar] [CrossRef] [Green Version]






| Treatments | Z (pmol g−1 FW) | DZ (pmol·g−1 FW) | iP (pmol·g−1 FW) |
|---|---|---|---|
| 1 N | 198.4 ± 11.1 a | 1.12 ± 0.06 a | 6.36 ± 0.16 a |
| 1/4 N | 102.3 ± 2.6 b | 1.09 ± 0.07 a | 6.32 ± 0.21 a |
| 1/16 N | 71.3 ± 2.8 c | 1.13 ± 0.06 a | 6.40 ± 0.17 a |
| 0 N | 68.2 ± 0.8 d | 1.09 ± 0.04 a | 6.30 ± 0.33 a |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.; Zhu, Y.; Zou, X.; Li, F.; Zhang, J.; Kang, Z.; Li, X.; Yin, C.; Lin, Y. Nitrogen Deficiency-Induced Decrease in Cytokinins Content Promotes Rice Seminal Root Growth by Promoting Root Meristem Cell Proliferation and Cell Elongation. Cells 2020, 9, 916. https://doi.org/10.3390/cells9040916
Wang Q, Zhu Y, Zou X, Li F, Zhang J, Kang Z, Li X, Yin C, Lin Y. Nitrogen Deficiency-Induced Decrease in Cytokinins Content Promotes Rice Seminal Root Growth by Promoting Root Meristem Cell Proliferation and Cell Elongation. Cells. 2020; 9(4):916. https://doi.org/10.3390/cells9040916
Chicago/Turabian StyleWang, Qi, Yanchun Zhu, Xiao Zou, Fengfeng Li, Jialiang Zhang, Ziyi Kang, Xuefei Li, Changxi Yin, and Yongjun Lin. 2020. "Nitrogen Deficiency-Induced Decrease in Cytokinins Content Promotes Rice Seminal Root Growth by Promoting Root Meristem Cell Proliferation and Cell Elongation" Cells 9, no. 4: 916. https://doi.org/10.3390/cells9040916
APA StyleWang, Q., Zhu, Y., Zou, X., Li, F., Zhang, J., Kang, Z., Li, X., Yin, C., & Lin, Y. (2020). Nitrogen Deficiency-Induced Decrease in Cytokinins Content Promotes Rice Seminal Root Growth by Promoting Root Meristem Cell Proliferation and Cell Elongation. Cells, 9(4), 916. https://doi.org/10.3390/cells9040916