Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights
Abstract
:1. Introduction
2. Microbiota in Breast Cancer
3. Risk Factors Associated with Breast Cancer
3.1. Obesity, Breast Cancer and Microbiota
3.2. Aging, Breast Cancer and Microbiota
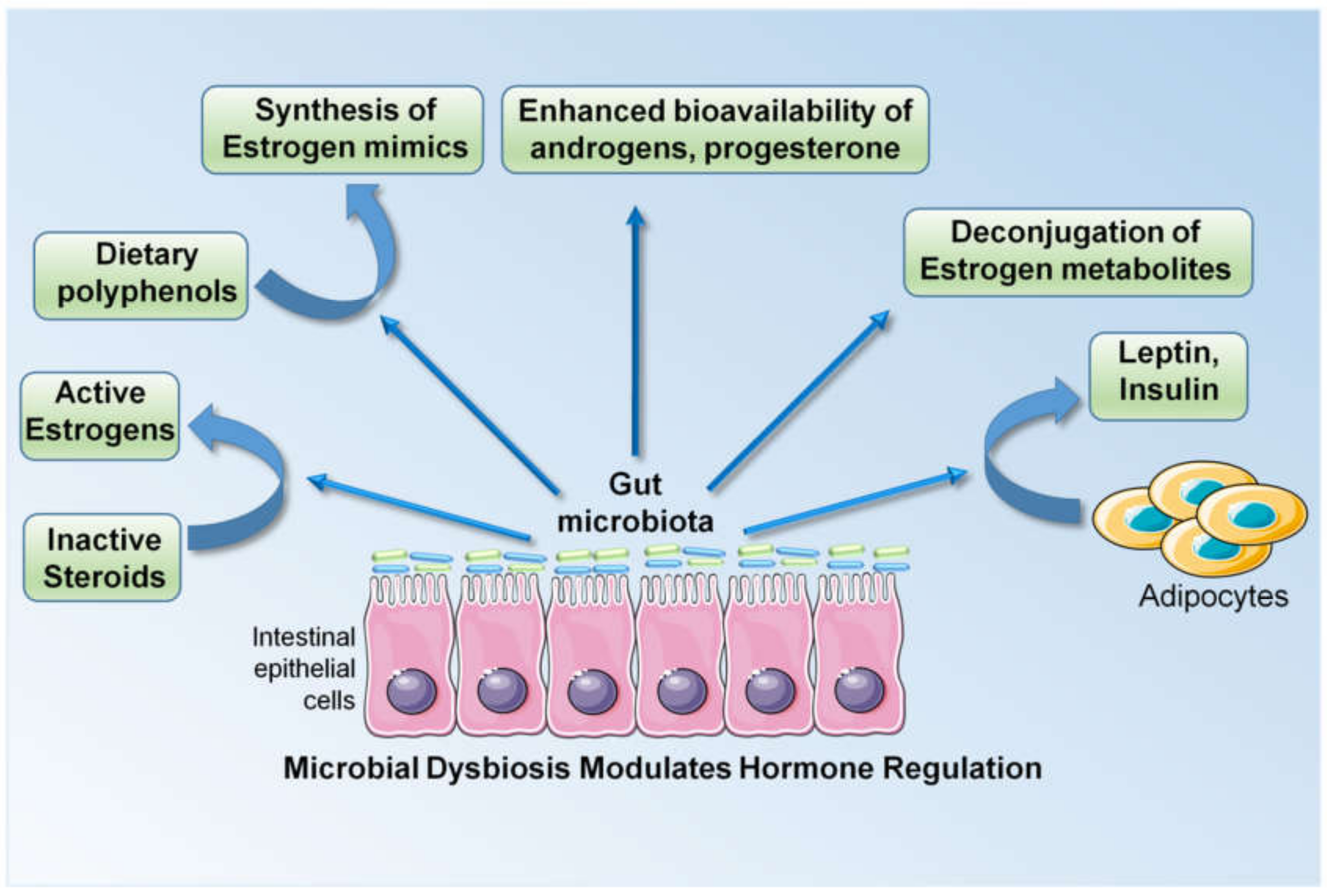
3.3. Higher Levels of Estrogens, Breast Cancer and Microbiota
3.4. Periodontal Disease, Breast Cancer and Microbiota
4. Mechanistic Insights into the Microbiota-Cancer Connection
4.1. Microbial Metabolites as Effector Molecules Influencing Breast Cancer
4.1.1. Short Chain Fatty Acids (SCFAs)
4.1.2. Amino Acid Metabolism (Tryptophan, Arginine, Lysine)
4.1.3. Secondary Bile Acids
4.1.4. Bacteriocins/Peptides and Antibiotics
4.2. Microbiota Modulates the Metabolism of Xenobiotics
4.3. Microbiota Induces Systemic Immune Modulation and Inflammatory Response in Breast Cancer
4.4. Microbial Dysbiosis May Modulate Response to Cancer Therapy
4.5. Beneficial Bugs as Drugs
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| IBD | Inflammatory bowel disease |
| BMI | Body mass index |
| TNBC | Triple-negative breast cancer |
| IBC | Inflammatory breast cancer |
| E coli | Escherichia coli |
| SCFA | Short chain fatty acids |
| IL | Interleukin |
| TNF | Tumor necrosis factor |
| CRP | C reactive protein |
| ERβ | Estrogen receptor beta |
| PD | Periodontal disease |
| NSAID | Non-steroidal anti-inflammatory drugs |
| HR | Hazard ratio |
| ERα | Estrogen receptor alpha |
| AR | Androgen receptor |
| PR | Progesterone receptor |
| Her2 | Human epidermal growth factor receptor 2 |
| GPCR | G-protein coupled receptor |
| HAT | Histone acetyltransferases |
| AhR | Aryl hydrocarbon receptor |
| GI | Gastrointestinal |
| SBA | Secondary bile acids |
| LCA | Lithocholic acid |
| TCA | Tricarboxylic acid |
| EMT | Epithelial to mesenchymal transition |
| VEGF | Vascular endothelial growth factor |
| COX-2 | Cyclooxygenase 2 |
| PGE2 | Prostaglandin E2 |
| TLR | Toll-like receptors |
| TIMER | Translocation, Immunomodulation, Metabolism, Enzymatic degradation and Reduced diversity |
References
- Fernández, M.F.; Reina-Pérez, I.; Astorga, J.M.; Rodríguez-Carrillo, A.; Plaza-Díaz, J.; Fontana, L. Breast Cancer and Its Relationship with the Microbiota. Int. J. Environ. Res. Public Health 2018, 15, 1747. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qin, J.; Li, R.; Raes, J.; Arumugam, M.; Burgdorf, K.S.; Manichanh, C.; Nielsen, T.; Pons, N.; Levenez, F.; Yamada, T.; et al. A human gut microbial gene catalogue established by metagenomic sequencing. Nature 2010, 464, 59–65. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, C.; Lagoudas, G.K.; Zhao, C.; Bullman, S.; Bhutkar, A.; Hu, B.; Ameh, S.; Sandel, D.; Liang, X.S.; Mazzilli, S.; et al. Commensal Microbiota Promote Lung Cancer Development via γδ T Cells. Cell 2019, 176, 998–1013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rose, U.D.; Vetizou, M.; Roy, S.; Edwards, B.; Peck, M.; Smith, L.; Robles, A.; Harris, C.; Difilippantonio, S.; Trinchieri, G. Role of the Microbiota in Primary Lung Cancer Initiation and Progression. J. Immunol. 2019, 202, 190–191. [Google Scholar]
- Gomes, S.; Cavadas, B.; Ferreira, J.; Marques, P.; Monteiro, C.; Sucena, M.; Sousa, C.; Vaz Rodrigues, L.; Teixeira, G.; Pinto, P.; et al. Microbiota profile of Non-small Cell Lung Cancer (NSCLC): The study of a large cohort. ERJ Open Res. 2019, 5, PP101. [Google Scholar]
- Riquelme, E.; Zhang, Y.; Zhang, L.; Montiel, M.; Zoltan, M.; Dong, W.; Quesada, P.; Sahin, I.; Chandra, V.; San Lucas, A.; et al. Tumor Microbiome Diversity and Composition Influence Pancreatic Cancer Outcomes. Cell 2019, 178, 795–806. [Google Scholar] [CrossRef]
- Tumor Microbiome Composition Influences Pancreatic Cancer Survival. Cancer Discov. 2019.
- Archibugi, L.; Signoretti, M.; Capurso, G. The Microbiome and Pancreatic Cancer: An Evidence-based Association? J. Clin. Gastroenterol. 2018, 52, S82–S85. [Google Scholar] [CrossRef]
- Ma, X.; Chi, C.; Fan, L.; Dong, B.; Shao, X.; Xie, S.; Li, M.; Xue, W. The Microbiome of Prostate Fluid Is Associated With Prostate Cancer. Front. Microbiol. 2019, 10. [Google Scholar] [CrossRef] [Green Version]
- Banerjee, S.; Alwine, J.C.; Wei, Z.; Tian, T.; Shih, N.; Sperling, C.; Guzzo, T.; Feldman, M.D.; Robertson, E.S. Microbiome signatures in prostate cancer. Carcinogenesis 2019, 40, 749–764. [Google Scholar] [CrossRef] [Green Version]
- Massari, F.; Mollica, V.; Di Nunno, V.; Gatto, L.; Santoni, M.; Scarpelli, M.; Cimadamore, A.; Lopez-Beltran, A.; Cheng, L.; Battelli, N.; et al. The Human Microbiota and Prostate Cancer: Friend or Foe? Cancers 2019, 11, 459. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Smith, A.; Pierre, J.F.; Makowski, L.; Tolley, E.; Lyn-Cook, B.; Lu, L.; Vidal, G.; Starlard-Davenport, A. Distinct microbial communities that differ by race, stage, or breast-tumor subtype in breast tissues of non-Hispanic Black and non-Hispanic White women. Sci. Rep. 2019, 9, 11940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lakritz, J.R.; Poutahidis, T.; Levkovich, T.; Varian, B.J.; Ibrahim, Y.M.; Chatzigiagkos, A.; Mirabal, S.; Alm, E.J.; Erdman, S.E. Beneficial bacteria stimulate host immune cells to counteract dietary and genetic predisposition to mammary cancer in mice. J. Cancer 2014, 135, 529–540. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Viaud, S.; Daillère, R.; Boneca, I.G.; Lepage, P.; Langella, P.; Chamaillard, M.; Pittet, M.J.; Ghiringhelli, F.; Trinchieri, G.; Goldszmid, R.; et al. Gut microbiome and anticancer immune response: Really hot Sh*t! Cell Death Differ. 2015, 22, 199–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, C.; Han, M.; Heinrich, B.; Fu, Q.; Zhang, Q.; Sandhu, M.; Agdashian, D.; Terabe, M.; Berzofsky, J.A.; Fako, V.; et al. Gut microbiome–mediated bile acid metabolism regulates liver cancer via NKT cells. Science 2018, 360, eaan5931. [Google Scholar] [CrossRef] [Green Version]
- Spiljar, M.; Merkler, D.; Trajkovski, M. The Immune System Bridges the Gut Microbiota with Systemic Energy Homeostasis: Focus on TLRs, Mucosal Barrier, and SCFAs. Front. Immunol. 2017, 8, 1353. [Google Scholar] [CrossRef] [Green Version]
- Rao, V.P.; Poutahidis, T.; Ge, Z.; Nambiar, P.R.; Boussahmain, C.; Wang, Y.Y.; Horwitz, B.H.; Fox, J.G.; Erdman, S.E. Innate Immune Inflammatory Response against Enteric Bacteria Helicobacter hepaticus Induces Mammary Adenocarcinoma in Mice. Cancer Res. 2006, 66, 7395–7400. [Google Scholar] [CrossRef] [Green Version]
- Coussens, L.M.; Pollard, J.W. Leukocytes in mammary development and cancer. Cold Spring Harb. Perspect. Biol. 2011, 3, a003285. [Google Scholar] [CrossRef] [Green Version]
- Rutkowski MR, S.T.; Svoronos, N.; Allegrezza, M.J.; Tesone, A.J.; Perales-Puchalt, A.; Brencicova, E.; Escovar-Fadul, X.; Nguyen, J.M.; Cadungog, M.G.; Zhang, R.; et al. Microbially driven TLR5-dependent signaling governs distal malignant progression through tumor-promoting inflammation. Cancer Cell. 2015, 27, 27–40. [Google Scholar]
- Imani Fooladi, A.A.; Yazdi, M.H.; Pourmand, M.R.; Mirshafiey, A.; Hassan, Z.M.; Azizi, T.; Mahdavi, M.; Soltan Dallal, M.M. Th1 Cytokine Production Induced by Lactobacillus acidophilus in BALB/c Mice Bearing Transplanted Breast Tumor. Jundishapur J. Microbiol. 2015, 8, e17354. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Hamady, M.; Yatsunenko, T.; Cantarel, B.L.; Duncan, A.; Ley, R.E.; Sogin, M.L.; Jones, W.J.; Roe, B.A.; Affourtit, J.P.; et al. A core gut microbiome in obese and lean twins. Nature 2008, 457, 480. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Morrison, D.J.; Preston, T. Formation of short chain fatty acids by the gut microbiota and their impact on human metabolism. Gut Microbes 2016, 7, 189–200. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Regina Joice, K.Y.; Shafquat, A.; Xochitl, C.; Curtis Huttenhower, M. Determining microbial products and identifying molecular targets in the human microbiome. Cell Metab. 2014, 20, 731–741. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samuel, B.S.; Shaito, A.; Motoike, T.; Rey, F.E.; Backhed, F.; Manchester, J.K.; Hammer, R.E.; Williams, S.C.; Crowley, J.; Yanagisawa, M.; et al. Effects of the gut microbiota on host adiposity are modulated by the short-chain fatty-acid binding G protein-coupled receptor, Gpr41. Proc. Natl. Acad. Sci. USA 2008, 105, 16767–16772. [Google Scholar] [CrossRef] [Green Version]
- Wang, H.; Altemus, J.; Niazi, F.; Green, H.; Calhoun, B.C.; Sturgis, C.; Grobmyer, S.R.; Eng, C. Breast tissue, oral and urinary microbiomes in breast cancer. Oncotarget 2017, 8, 88122–88138. [Google Scholar] [CrossRef] [Green Version]
- Urbaniak, C.; Gloor, G.B.; Brackstone, M.; Scott, L.; Tangney, M.; Reid, G. The Microbiota of Breast Tissue and Its Association with Breast Cancer. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef] [Green Version]
- Ghazalpour, A.; Cespedes, I.; Bennett, B.J.; Allayee, H. Expanding role of gut microbiota in lipid metabolism. Curr. Opin. Lipidol. 2016, 27, 141–147. [Google Scholar] [CrossRef]
- Lee, I.O.; Kim, J.H.; Choi, Y.J.; Pillinger, M.H.; Kim, S.Y.; Blaser, M.J.; Lee, Y.C. Helicobacter pylori CagA phosphorylation status determines the gp130-activated SHP2/ERK and JAK/STAT signal transduction pathways in gastric epithelial cells. J. Biol Chem. 2010, 285, 16042–16050. [Google Scholar] [CrossRef] [Green Version]
- Ojanotko-Harri, A.; Nikkari, T.; Harrl, M.-P.; Paunio, K. Metabolism of progesterone and testosterone by Bacillus cereus strain Socransky 67 and Streptococcus mutans strain Ingbritt. Oral. Microbiol. Immunol. 1990, 5, 237–239. [Google Scholar] [CrossRef]
- Fulbright, L.E.; Ellermann, M.; Arthur, J.C. The microbiome and the hallmarks of cancer. PLoS Pathog. 2017, 13, e1006480. [Google Scholar] [CrossRef]
- Tikkanen, M.J.; Adlercreutz, H.; Pulkkinen, M.O. Effects of antibiotics on oestrogen metabolism. Br. Med. J. 1973, 2, 369. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Velicer, C.M.; Lampe, J.W.; Heckbert, S.R.; Potter, J.D.; Taplin, S.H. Hypothesis: Is antibiotic use associated with breast cancer? Cancer Causes Control.: CCC 2003, 14, 739–747. [Google Scholar] [CrossRef] [PubMed]
- Velicer, C.M.; Heckbert, S.R.; Lampe, J.W.; Potter, J.D.; Robertson, C.A.; Taplin, S.H. Antibiotic Use in Relation to the Risk of Breast Cancer. JAMA 2004, 291, 827–835. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Friedman, G.D.; Oestreicher, N.; Chan, J.; Quesenberry, C.P., Jr.; Udaltsova, N.; Habel, L.A. Antibiotics and risk of breast cancer: Up to 9 years of follow-up of 2.1 million women. Cancer Epidemiol. Biomark. Prev. 2006, 15, 2102–2106. [Google Scholar] [CrossRef] [Green Version]
- Velicer, C.M.; Heckbert, S.R.; Rutter, C.; Lampe, J.W.; Malone, K. Association between antibiotic use prior to breast cancer diagnosis and breast tumour characteristics (United States). Cancer Causes Control.: CCC 2006, 17, 307–313. [Google Scholar] [CrossRef] [PubMed]
- Tamim, H.M.; Hanley, J.A.; Hajeer, A.H.; Boivin, J.F.; Collet, J.P. Risk of breast cancer in relation to antibiotic use. Pharmacoepidemiol. Drug Saf. 2008, 17, 144–150. [Google Scholar] [CrossRef]
- Sergentanis, T.N.; Zagouri, F.; Zografos, G.C. Is antibiotic use a risk factor for breast cancer? A meta-analysis. Pharmacoepidemiol. Drug Saf. 2010, 19, 1101–1107. [Google Scholar] [CrossRef]
- Wirtz, H.S.; Buist, D.S.M.; Gralow, J.R.; Barlow, W.E.; Gray, S.; Chubak, J.; Yu, O.; Bowles, E.J.A.; Fujii, M.; Boudreau, D.M. Frequent antibiotic use and second breast cancer events. Cancer Epidemiol. Biomark. Prev. 2013, 22, 1588–1599. [Google Scholar] [CrossRef] [Green Version]
- Elkrief, A.; Derosa, L.; Kroemer, G.; Zitvogel, L.; Routy, B. The negative impact of antibiotics on outcomes in cancer patients treated with immunotherapy: A new independent prognostic factor? Ann. Oncol 2019, 30, 1572–1579. [Google Scholar] [CrossRef] [Green Version]
- Urbaniak, C.; Cummins, J.; Brackstone, M.; Macklaim, J.M.; Gloor, G.B.; Baban, C.K.; Scott, L.; O’Hanlon, D.M.; Burton, J.P.; Francis, K.P.; et al. Microbiota of Human Breast Tissue. Appl. Environ. Microbiol. 2014, 80, 3007–3014. [Google Scholar] [CrossRef] [Green Version]
- Chan, A.A.; Bashir, M.; Rivas, M.N.; Duvall, K.; Sieling, P.A.; Pieber, T.R.; Vaishampayan, P.A.; Love, S.M.; Lee, D.J. Characterization of the microbiome of nipple aspirate fluid of breast cancer survivors. Sci. Rep. 2016, 6, 28061. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hieken, T.J.; Chen, J.; Hoskin, T.L.; Walther-Antonio, M.; Johnson, S.; Ramaker, S.; Xiao, J.; Radisky, D.C.; Knutson, K.L.; Kalari, K.R.; et al. The Microbiome of Aseptically Collected Human Breast Tissue in Benign and Malignant Disease. Sci. Rep. 2016, 6, 30751. [Google Scholar] [CrossRef] [PubMed]
- Costantini, L.; Magno, S.; Albanese, D.; Donati, C.; Molinari, R.; Filippone, A.; Masetti, R.; Merendino, N. Characterization of human breast tissue microbiota from core needle biopsies through the analysis of multi hypervariable 16S-rRNA gene regions. Sci. Rep. 2018, 8, 16893. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Meng, S.; Chen, B.; Yang, J.; Wang, J.; Zhu, D.; Meng, Q.; Zhang, L. Study of Microbiomes in Aseptically Collected Samples of Human Breast Tissue Using Needle Biopsy and the Potential Role of in situ Tissue Microbiomes for Promoting Malignancy. Front. Oncol. 2018, 8, 318. [Google Scholar] [CrossRef] [Green Version]
- Xuan, C.; Shamonki, J.M.; Chung, A.; Dinome, M.L.; Chung, M.; Sieling, P.A.; Lee, D.J. Microbial dysbiosis is associated with human breast cancer. PLoS ONE 2014, 9, e83744. [Google Scholar] [CrossRef] [Green Version]
- Parida, S.; Sharma, D. The power of small changes: Comprehensive analyses of microbial dysbiosis in breast cancer. Biochim. Et Biophys. Acta. (BBA)—Rev. Cancer 2019, 1871, 392–405. [Google Scholar] [CrossRef]
- Henley, S.J.; Ward, E.M.; Scott, S.; Ma, J.; Anderson, R.N.; Firth, A.U.; Thomas, C.C.; Islami, F.; Weir, H.K.; Lewis, D.R.; et al. Annual Report to the Nation on the Status of Cancer, part I: National cancer statistics. Cancer 2020. [Google Scholar] [CrossRef]
- Henley, S.J.; Thomas, C.C.; Lewis, D.R.; Ward, E.M.; Islami, F.; Wu, M.; Weir, H.K.; Scott, S.; Sherman, R.L.; Ma, J.; et al. Annual Report to the Nation on the Status of Cancer, part II: Progress toward Healthy People 2020 objectives for 4 common cancers. Cancer 2020. [Google Scholar] [CrossRef]
- DeSantis, C.E.; Ma, J.; Gaudet, M.M.; Newman, L.A.; Miller, K.D.; Goding Sauer, A.; Jemal, A.; Siegel, R.L. Breast cancer statistics, 2019. CA Cancer J. Clin. 2019, 69, 438–451. [Google Scholar] [CrossRef]
- Gail, M.H.; Brinton, L.A.; Byar, D.P.; Corle, D.K.; Green, S.B.; Schairer, C.; Mulvihill, J.J. Projecting individualized probabilities of developing breast cancer for white females who are being examined annually. J. Natl. Cancer Inst. 1989, 81, 1879–1886. [Google Scholar] [CrossRef]
- Petracci, E.; Decarli, A.; Schairer, C.; Pfeiffer, R.M.; Pee, D.; Masala, G.; Palli, D.; Gail, M.H. Risk factor modification and projections of absolute breast cancer risk. J. Natl. Cancer Inst. 2011, 103, 1037–1048. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Decarli, A.; Calza, S.; Masala, G.; Specchia, C.; Palli, D.; Gail, M.H. Gail model for prediction of absolute risk of invasive breast cancer: Independent evaluation in the Florence-European Prospective Investigation Into Cancer and Nutrition cohort. J. Natl. Cancer Inst. 2006, 98, 1686–1693. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Banegas, M.P.; John, E.M.; Slattery, M.L.; Gomez, S.L.; Yu, M.; LaCroix, A.Z.; Pee, D.; Chlebowski, R.T.; Hines, L.M.; Thompson, C.A.; et al. Projecting Individualized Absolute Invasive Breast Cancer Risk in US Hispanic Women. J. Natl. Cancer Inst. 2017, 109. [Google Scholar] [CrossRef] [Green Version]
- Gail, M.H.; Costantino, J.P.; Pee, D.; Bondy, M.; Newman, L.; Selvan, M.; Anderson, G.L.; Malone, K.E.; Marchbanks, P.A.; McCaskill-Stevens, W.; et al. Projecting individualized absolute invasive breast cancer risk in African American women. J. Natl. Cancer Inst. 2007, 99, 1782–1792. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Costantino, J.P.; Gail, M.H.; Pee, D.; Anderson, S.; Redmond, C.K.; Benichou, J.; Wieand, H.S. Validation studies for models projecting the risk of invasive and total breast cancer incidence. J. Natl. Cancer Inst. 1999, 91, 1541–1548. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rockhill, B.; Spiegelman, D.; Byrne, C.; Hunter, D.J.; Colditz, G.A. Validation of the Gail et al. model of breast cancer risk prediction and implications for chemoprevention. J. Natl. Cancer Inst. 2001, 93, 358–366. [Google Scholar] [CrossRef]
- Picon-Ruiz, M.; Morata-Tarifa, C.; Valle-Goffin, J.J.; Friedman, E.R.; Slingerland, J.M. Obesity and adverse breast cancer risk and outcome: Mechanistic insights and strategies for intervention. CA Cancer J. Clin. 2017, 67, 378–397. [Google Scholar] [CrossRef]
- White, A.J.; Nichols, H.B.; Bradshaw, P.T.; Sandler, D.P. Overall and central adiposity and breast cancer risk in the Sister Study. Cancer 2015, 121, 3700–3708. [Google Scholar] [CrossRef] [Green Version]
- Cotterchio, M.; Kreiger, N.; Theis, B.; Sloan, M.; Bahl, S. Hormonal Factors and the Risk of Breast Cancer According to Estrogen- and Progesterone-Receptor Subgroup. Cancer Epidemiol. Prev. Biomark. 2003, 12, 1053–1060. [Google Scholar]
- Enger, S.M.; Ross, R.K.; Paganini-Hill, A.; Carpenter, C.L.; Bernstein, L. Body Size, Physical Activity, and Breast Cancer Hormone Receptor Status: Results from Two Case-Control Studies. Cancer Epidemiol. Prev. Biomark. 2000, 9, 681–687. [Google Scholar]
- Neuhouser, M.L.; Aragaki, A.K.; Prentice, R.L.; Manson, J.E.; Chlebowski, R.; Carty, C.L.; Ochs-Balcom, H.M.; Thomson, C.A.; Caan, B.J.; Tinker, L.F.; et al. Overweight, Obesity, and Postmenopausal Invasive Breast Cancer Risk: A Secondary Analysis of the Women’s Health Initiative Randomized Clinical Trials. JAMA Oncol. 2015, 1, 611–621. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, R.; Rylander-Rudqvist, T.; Ye, W.; Saji, S.; Wolk, A. Body weight and postmenopausal breast cancer risk defined by estrogen and progesterone receptor status among Swedish women: A prospective cohort study. Int. J. Cancer 2006, 119, 1683–1689. [Google Scholar] [CrossRef] [PubMed]
- Ahn, J.; Schatzkin, A.; Lacey, J.V., Jr.; Albanes, D.; Ballard-Barbash, R.; Adams, K.F.; Kipnis, V.; Mouw, T.; Hollenbeck, A.R.; Leitzmann, M.F. Adiposity, Adult Weight Change, and Postmenopausal Breast Cancer Risk. Arch. Intern. Med. 2007, 167, 2091–2102. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Setiawan, V.W.; Monroe, K.R.; Wilkens, L.R.; Kolonel, L.N.; Pike, M.C.; Henderson, B.E. Breast cancer risk factors defined by estrogen and progesterone receptor status: The multiethnic cohort study. Am. J. Epidemiol. 2009, 169, 1251–1259. [Google Scholar] [CrossRef] [Green Version]
- Rosenberg, L.U.; Einarsdóttir, K.; Friman, E.I.; Wedrén, S.; Dickman, P.W.; Hall, P.; Magnusson, C. Risk Factors for Hormone Receptor-Defined Breast Cancer in Postmenopausal Women. Cancer Epidemiol. Biomarkers Prev. 2006, 15, 2482–2488. [Google Scholar] [CrossRef] [Green Version]
- Phipps, A.I.; Chlebowski, R.T.; Prentice, R.; McTiernan, A.; Stefanick, M.L.; Wactawski-Wende, J.; Kuller, L.H.; Adams-Campbell, L.L.; Lane, D.; Vitolins, M.; et al. Body size, physical activity, and risk of triple-negative and estrogen receptor-positive breast cancer. Cancer Epidemiol. Biomark. Prev. 2011, 20, 454–463. [Google Scholar] [CrossRef] [Green Version]
- John, E.M.; Sangaramoorthy, M.; Hines, L.M.; Stern, M.C.; Baumgartner, K.B.; Giuliano, A.R.; Wolff, R.K.; Slattery, M.L. Body size throughout adult life influences postmenopausal breast cancer risk among hispanic women: The breast cancer health disparities study. Cancer Epidemiol. Biomark. Prev. 2015, 24, 128–137. [Google Scholar] [CrossRef] [Green Version]
- Michels, K.B.; Terry, K.L.; Willett, W.C. Longitudinal Study on the Role of Body Size in Premenopausal Breast Cancer. Arch. Intern. Med. 2006, 166, 2395–2402. [Google Scholar] [CrossRef] [Green Version]
- Berstad, P.; Coates, R.J.; Bernstein, L.; Folger, S.G.; Malone, K.E.; Marchbanks, P.A.; Weiss, L.K.; Liff, J.M.; McDonald, J.A.; Strom, B.L.; et al. A case-control study of body mass index and breast cancer risk in white and African-American women. Cancer Epidemiol. Biomark. Prev. 2010, 19, 1532–1544. [Google Scholar] [CrossRef] [Green Version]
- Harris, H.R.; Willett, W.C.; Terry, K.L.; Michels, K.B. Body fat distribution and risk of premenopausal breast cancer in the Nurses’ Health Study II. J. Natl. Cancer Inst. 2011, 103, 273–278. [Google Scholar] [CrossRef] [Green Version]
- Chang, S.; Buzdar, A.U.; Hursting, S.D. Inflammatory breast cancer and body mass index. J. Clin. Oncol. 1998, 16, 3731–3735. [Google Scholar] [CrossRef]
- Hooper, L.V.; Midtvedt, T.; Gordon, J. How host-microbial interactions shape the nutrient environment of the mammalian intestine. Annu Rev. Nutr 2002, 22, 283–307. [Google Scholar] [CrossRef] [Green Version]
- Brown, A.J.; Goldsworthy, S.M.; Barnest, A.A.; Eilert, M.M.; Tcheangt, L.; Daniels, D.; Muir, A.I.; Wigglesworth, M.J.; Kinghorn, I.; Fraser, N.J.; et al. The Orphan G Protein-coupled Receptors GPR41 and GPR43 Are Activated by Propionate and Other Short Chain Carboxylic Acids. J. Biol. Chem. 2003, 278, 11312–11319. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Le Roux, C.W.; Bloom, S.R. Peptide YY, appetite and food intake. Proc. Nutr. Soc. 2005, 64, 213–216. [Google Scholar] [CrossRef] [Green Version]
- Hullar, M.A.; Fu, B.C. Diet, the gut microbiome, and epigenetics. Cancer J. (SudburyMass) 2014, 20, 170. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Louis, P.; Flint, H.J. Diversity, metabolism and microbial ecology of butyrate-producing bacteria from the human large intestine. FEMS Microbiol Lett 2009, 294, 1–8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Million, M.; Lagier, J.C.; Yahav, D.; Paul, M. Gut bacterial microbiota and obesity. Clin. Microbiol. Infect. 2013, 19, 305–313. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Loscalzo, J. Lipid metabolism by gut microbes and atherosclerosis. Circ Res. 2011, 109, 127–129. [Google Scholar] [CrossRef] [Green Version]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.J.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef] [Green Version]
- Isaac, T.W.; Harley, C.L.K. Obesity and the gut microbiome: Striving for causality. Mol. Metab. 2012, 1, 21–31. [Google Scholar]
- Zheng, J.; Gänzle, M.G.; Lin, X.B.; Ruan, L.; Sun, M. Diversity and dynamics of bacteriocins from human microbiome. Environ Microbiol. 2015, 17, 2133–2143. [Google Scholar] [CrossRef] [PubMed]
- Gui, Y.; Pan, Q.; Chen, X.; Xu, S.; Luo, X.; Chen, L. The association between obesity related adipokines and risk of breast cancer: A meta-analysis. Oncotarget 2017, 8, 75389–75399. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cani, P.D.; Possemiers, S.; Van de Wiele, T.; Guiot, Y.; Everard, A.; Rottier, O.; Geurts, L.; Naslain, D.; Neyrinck, A.; Lambert, D.M.; et al. Changes in gut microbiota control inflammation in obese mice through a mechanism involving GLP-2-driven improvement of gut permeability. Gut 2009, 58, 1091–1103. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, S.; Pagovich, O.; Kriegel, M. Diet, microbiota and autoimmune diseases. Lupus 2014, 23, 518–526. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ridaura, V.K.; Faith, J.J.; Rey, F.E.; Knight, R.; Gordon, J.I. The Effect of Diet on the Human Gut Microbiome: A Metagenomic Analysis in Humanized Gnotobiotic Mice. Sci. Transl. Med. 2009, 1, ra14–ra16. [Google Scholar] [CrossRef] [Green Version]
- Paul, B.; Barnes, S.; Demark-Wahnefried, W.; Morrow, C.; Salvador, C.; Skibola, C.; Tollefsbol, T.O. Influences of diet and the gut microbiome on epigenetic modulation in cancer and other diseases. Clin. Epigenetics 2015, 7, 112. [Google Scholar] [CrossRef] [Green Version]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027–1031. [Google Scholar] [CrossRef]
- Ridlon, J.M.; Kang, D.J.; Hylemon, P.B.; Bajaj, J.S. Bile Acids and the Gut Microbiome. Curr. Opin. Gastroenterol. 2014, 30, 332–338. [Google Scholar]
- Takahashi, K.; Sugi, Y.; Nakano, K.; Tsuda, M.; Kurihara, K.; Hosono, A. Epigenetic control of the host gene by commensal bacteria in large intestinal epithelial cells. J. Biol. Chem. 2011, 286, 35755–35762. [Google Scholar] [CrossRef] [Green Version]
- Berni Canani, R.; Di Costanzo, M.; Leone, L. The epigenetic effects of butyrate: Potential therapeutic implications for clinical practice. Clin. Epigenetics 2012, 4, 4. [Google Scholar] [CrossRef]
- Pompei, A.; Cordisco, L.; Amaretti, A.; Zanoni, S.; Matteuzzi, D.; Rossi, M. Folate Production by Bifidobacteria as a Potential Probiotic Property. Appl. Environ. Microbiol. 2007, 73, 179–185. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Samsel, A.; Seneff, S. Glyphosate’s Suppression of Cytochrome P450 Enzymes and Amino Acid Biosynthesis by the Gut Microbiome: Pathways to Modern Diseases. Entropy 2013, 15, 1416–1463. [Google Scholar] [CrossRef] [Green Version]
- Shenderov, B.A. Gut indigenous microbiota and epigenetics. Microb. Ecol. Health Dis. 2012, 23. [Google Scholar] [CrossRef] [PubMed]
- Huda-Faujan, N.; Abdulamir, A.S.; Fatimah, A.B.; Anas, O.M.; Shuhaimi, M.; Yazid, A.M.; Loong, Y.Y. The Impact of the Level of the Intestinal Short Chain Fatty Acids in Inflammatory Bowel Disease Patients Versus Healthy Subjects. Open Biochem. J. 2010, 4, 53–58. [Google Scholar] [CrossRef]
- Ghanim, H.; Abuaysheh, S.; Sia, C.L.; Korzeniewski, K.; Chaudhuri, A.; Fernandez-Real, J.M.; Dandona, P. Increase in plasma endotoxin concentrations and the expression of Toll-like receptors and suppressor of cytokine signaling-3 in mononuclear cells after a high-fat, high-carbohydrate meal: Implications for insulin resistance. Diabetes Care. 2009, 32, 2281–2287. [Google Scholar]
- Takahashi, K. Influence of bacteria on epigenetic gene control. Cell Mol. Life Sci. 2014, 71, 1045–1054. [Google Scholar] [CrossRef]
- Davie, J.R. Inhibition of Histone Deacetylase Activity by Butyrate. J. Nutr. 2003, 133, 2485S–2493S. [Google Scholar] [CrossRef]
- Hýzd’alova, M.; Hofmanova, J.; Pachermk, J.; Vaculova, A.; Kozubik, A. The interaction of butyrate with TNF-a during differentiation and apoptosis of colon epithelial cells: Role of NF-k B activation. Cytokine 2008, 44, 33–43. [Google Scholar] [CrossRef]
- Koeth, R.A.; Wang, Z.; Levison, B.S.; Buffa, J.A.; Org, E.; Sheehy, B.T.; Britt, E.B.; Fu, X.; Wu, Y.; Li, L.; et al. Intestinal microbiota metabolism of l-carnitine, a nutrient in red meat, promotes atherosclerosis. Nat. Med. 2013, 19, 576. [Google Scholar] [CrossRef] [Green Version]
- Selkrig, J.; Wong, P.; Zhang, X.; Pettersson, S. Metabolic tinkering by the gut microbiome: Implications for brain development and function. Gut Microbes 2014, 5, 369–380. [Google Scholar] [CrossRef]
- Donohoe, D.R.; Garge, N.; Zhang, X.; Sun, W.; O’Connell, T.M.; Bunger, M.K. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab. 2011, 13, 517–526. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duggal, P.; Guo, X.; Haque, R.; Peterson, K.M.; Ricklefs, S.; Mondal, D.; Alam, F.; Noor, Z.; Verkerke, H.P.; Marie, C.; et al. A mutation in the leptin receptor is associated with Entamoeba histolytica infection in children. J. Clin. Investig. 2011, 121, 1191–1198. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Vieira, A.T.; Macia, L.; Galvão, I.; Martins, F.S.; Canesso, M.C.; Amaral, F.A. A role for gut microbiota and the metabolite-sensing receptor GPR43 in a murine model of gout. Arthritis Rheumatol. 2015, 67, 1646–1656. [Google Scholar] [CrossRef] [PubMed]
- Hague, A.; Manning, A.M.; Hanlon, K.A.; Hart, D.; Paraskeva, C.; Huschtscha, L.I. Sodium butyrate induces apoptosis in human colonic tumour cell lines in a p53-independent pathway: Implications for the possible role of dietary fibre in the prevention of large-bowel cancer. Int. J. Cancer 1993, 55, 498–505. [Google Scholar] [CrossRef] [PubMed]
- Candido, E.P.M.; Reeves, R.; Davie, J.R. Sodium butyrate inhibits histone deacetylation in cultured cells. Get Cell 1978, 14, 105–113. [Google Scholar] [CrossRef]
- Sharon, G.; Garg, N.; Debelius, J.; Knight, R.; Dorrestein, P.C.; Mazmanian, S.K. Specialized Metabolites from the Microbiome in Health and Disease. Cell Metab. 2014, 20, 719–730. [Google Scholar] [CrossRef] [Green Version]
- Hesson, L.B. Gut microbiota and obesity-related gastrointestinal cancer: A focus on epigenetics. Trans. Gastrointest Cancer 2013, 2, 204–210. [Google Scholar]
- Ley, R.E.; Turnbaugh, P.J.; S, K.; Gordon, J. Microbial ecology:human gut microbes associated with obesity. Nature 2006, 444, 1022–1023. [Google Scholar] [CrossRef]
- Ley, R.E. Obesity and the human microbiome. Curr. Opin. Gastroenterol. 2010, 26, 5–11. [Google Scholar] [CrossRef]
- Rosa, B.A.; Hallsworth-Pepin, K.; Martin, J.; Wollam, A.; Mitreva, M. Genome Sequence of Christensenella minuta DSM 22607T. Genome Announc 2017, 5, e01416–e01451. [Google Scholar] [CrossRef] [Green Version]
- Arumugam, M.; Raes, J.; Pelletier, E.; Le Paslier, D.; Yamada, T.; Mende, D.R.; Fernandes, G.R.; Tap, J.; Bruls, T.; Batto, J.M.; et al. Enterotypes of the human gut microbiome. Nature 2011, 473, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Mai, V.; McCrary, Q.M.; Sinha, R.; Glei, M. Associations between dietary habits and body mass index with gut microbiota composition and fecal water genotoxicity: An observational study in African American and Caucasian American volunteers. Nutr. J. 2009, 8, 49. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Million, M.; Maraninchi, M.; Henry, M.; Armougom, F.; Richet, H.; Carrieri, P.; Valero, R.; Raccah, D.; Vialettes, B.; Raoult, D. Obesity-associated gut microbiota is enriched in Lactobacillus reuteri and depleted in Bifidobacterium animalis and Methanobrevibacter smithii. Int. J. Obes. 2011, 36, 817. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jeffery, I.B.; O’Toole, P.W. Diet-microbiota interactions and their implications for healthy living. Nutrients 2013, 5, 234–252. [Google Scholar] [CrossRef] [PubMed]
- Hu, S.; Dong, T.S.; Dalal, S.R.; Wu, F.; Bissonnette, M.; Kwon, J.H.; Chang, E.B. The microbe-derived short chain fatty acid butyrate targets miRNA-dependent p21 gene expression in human colon cancer. PLoS ONE 2011, 6, e16221. [Google Scholar] [CrossRef] [PubMed]
- Xiong, Y.; Miyamoto, N.; Shibata, K.; Valasek, M.A.; Motoike, T.; Kedzierski, R.M.; Yanagisawa, M. Short-chain fatty acids stimulate leptin production in adipocytes through the G protein-coupled receptor GPR41. Proc. Natl. Acad. Sci. USA 2004, 101, 1045–1050. [Google Scholar] [CrossRef] [Green Version]
- Khan, M.J.; Gerasimidis, K.; Edwards, C.A.; Shaikh, M.G. Role of gut microbiota in the aetiology of obesity: Proposed mechanisms and review of the literature. J. Obes. 2016. [Google Scholar] [CrossRef] [Green Version]
- Bäckhed, F.; Ding, H.; Wang, T.; Hooper, L.V.; Koh, G.Y.; Nagy, A.; Semenkovich, C.F.; Gordon, J.I. The gut microbiota as an environmental factor that regulates fat storage. Proc. Natl. Acad. Sci. USA 2004, 101, 15718–15723. [Google Scholar] [CrossRef] [Green Version]
- Stappenbeck, T.S.; Hooper, L.V.; Gordon, J.I. Developmental regulation of intestinal angiogenesis by indigenous microbes via Paneth cells. Proc. Natl. Acad. Sci. USA 2002, 99, 15451–15455. [Google Scholar] [CrossRef] [Green Version]
- Hooper, L.V.; Wong, M.H.; Thelin, A.; Hansson, L.; Falk, P.G.; Gordon, J.I. Molecular Analysis of Commensal Host-Microbial Relationships in the Intestine. Science 2001, 291, 881–884. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Plaza-Díaz, J.; Álvarez-Mercado, A.I.; Ruiz-Marín, C.M.; Reina-Pérez, I.; Pérez-Alonso, A.J.; Sánchez-Andujar, M.B.; Torné, P.; Gallart-Aragón, T.; Sánchez-Barrón, M.T.; Reyes Lartategui, S.; et al. Association of breast and gut microbiota dysbiosis and the risk of breast cancer: A case-control clinical study. BMC Cancer 2019, 19, 495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradlow, H.L. Obesity and the gut microbiome: Pathophysiological aspects. Horm. Mol. Biol. Clin. Investig. 2014, 17, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Frugé, A.D.; Van der Pol, W.; Rogers, L.Q.; Morrow, C.D.; Tsuruta, Y.; Demark-Wahnefried, W. Fecal Akkermansia muciniphila Is Associated with Body Composition and Microbiota Diversity in Overweight and Obese Women with Breast Cancer Participating in a Presurgical Weight Loss Trial. J. Acad. Nutr. Diet. 2020, 120, 650–659. [Google Scholar] [CrossRef] [Green Version]
- Quong, J.; Eppenberger-Castori, S.; Moore, D., 3rd; Scott, G.K.; Birrer, M.J.; Kueng, W.; Eppenberger, U.; Benz, C.C. Age-dependent changes in breast cancer hormone receptors and oxidant stress markers. Breast Cancer Res. Treat. 2002, 76, 221–236. [Google Scholar] [CrossRef]
- Benz, C.C. Impact of aging on the biology of breast cancer. Crit. Rev. Oncol. Hematol. 2008, 66, 65–74. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lodi, M.; Scheer, L.; Reix, N.; Heitz, D.; Carin, A.J.; Thiebaut, N.; Neuberger, K.; Tomasetto, C.; Mathelin, C. Breast cancer in elderly women and altered clinico-pathological characteristics: A systematic review. Breast Cancer Res. Treat. 2017, 166, 657–668. [Google Scholar] [CrossRef]
- Thongaram, T.; Hoeflinger, J.L.; Chow, J.; Miller, M.J. Human milk oligosaccharide consumption by probiotic and human-associated bifidobacteria and lactobacilli. J. Dairy Sci. 2017, 100, 7825–7833. [Google Scholar] [CrossRef] [Green Version]
- Goldsmith, F.; O’Sullivan, A.; Smilowitz, J.T.; Freeman, S.L. Lactation and Intestinal Microbiota: How Early Diet Shapes the Infant Gut. J. Mammary Gland Biol. Neoplasia 2015, 20, 149–158. [Google Scholar] [CrossRef]
- Mariat, D.; Firmesse, O.; Levenez, F.; Guimarăes, V.; Sokol, H.; Doré, J.; Corthier, G.; Furet, J.P. The Firmicutes/Bacteroidetes ratio of the human microbiota changes with age. BMC Microbiol. 2009, 9, 123. [Google Scholar] [CrossRef]
- Reddy, B.S.; Rivenson, A. Inhibitory Effect of Bifidobacterium longum on Colon, Mammary, and Liver Carcinogenesis Induced by 2-Amino-3-methylimidazo[4,5-]quinoline, a Food Mutagen. Cancer Res. 1993, 53, 3914–3918. [Google Scholar]
- Biffi, A.; Coradini, D.; Larsen, R.; Riva, L.; Di Fronzo, G. Antiproliferative effect of fermented milk on the growth of a human breast cancer cell line. Nutr. Cancer 1997, 28, 93–99. [Google Scholar] [CrossRef] [PubMed]
- Orrhage, K.; Nord, C.E. Bifidobacteria and lactobacilli in human health. Drugs Exp Clin Res. 2000, 26, 95–111. [Google Scholar] [PubMed]
- Dinan, T.G.; Cryan, J.F. Gut instincts: Microbiota as a key regulator of brain development, ageing and neurodegeneration. J. Physiol. 2017, 595, 489–503. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J. Gut microbial metabolites in health and disease. Gut Microbes 2016, 7, 187–188. [Google Scholar] [CrossRef] [Green Version]
- Zapata, H.J.; Quagliarello, V.J. The microbiota and microbiome in aging: Potential implications in health and age-related diseases. J. Am. Geriatr. Soc. 2015, 63, 776–781. [Google Scholar] [CrossRef] [Green Version]
- Vaiserman, A.M.; Koliada, A.K.; Marotta, F. Gut microbiota: A player in aging and a target for anti-aging intervention. Ageing Res. Rev. 2017, 35, 36–45. [Google Scholar] [CrossRef]
- Landete, J.M.; Gaya, P.; Rodríguez, E.; Langa, S.; Peirotén, Á.; Medina, M.; Arqués, J.L. Probiotic Bacteria for Healthier Aging: Immunomodulation and Metabolism of Phytoestrogens. BioMed Res. Int. 2017, 2017, 5939818. [Google Scholar] [CrossRef]
- Key, T.; Appleby, P.; Barnes, I.; Reeves, G.; Endogenous, H.; Endogenous Hormones and Breast Cancer Collaborative Group. Endogenous sex hormones and breast cancer in postmenopausal women: Reanalysis of nine prospective studies. J. Natl. Cancer Inst. 2002, 94, 606–616. [Google Scholar]
- Breast cancer and hormone replacement therapy: Collaborative reanalysis of data from 51 epidemiological studies of 52,705 women with breast cancer and 108,411 women without breast cancer. Collaborative Group on Hormonal Factors in Breast Cancer. Lancet 1997, 350, 1047–1059.
- Hamajima, N.; Hirose, K.; Tajima, K.; Rohan, T.; Calle, E.E.; Heath, C.W., Jr.; Coates, R.J.; Liff, J.M.; Talamini, R.; Chantarakul, N.; et al. Alcohol, tobacco and breast cancer--collaborative reanalysis of individual data from 53 epidemiological studies, including 58,515 women with breast cancer and 95,067 women without the disease. Br. J. Cancer 2002, 87, 1234–1245. [Google Scholar]
- Dorgan, J.F.; Baer, D.J.; Albert, P.S.; Judd, J.T.; Brown, E.D.; Corle, D.K.; Campbell, W.S.; Hartman, T.J.; Tejpar, A.A.; Clevidence, B.A.; et al. Serum hormones and the alcohol-breast cancer association in postmenopausal women. J. Natl. Cancer Inst. 2001, 93, 710–715. [Google Scholar] [CrossRef] [Green Version]
- Vainio, H.; Kaaks, R.; Bianchini, F. Weight control and physical activity in cancer prevention: International evaluation of the evidence. Eur. J. Cancer Prev. 2002, 11, S94–S100. [Google Scholar]
- Stone, S.A.; Han, C.J.; Senn, T.; Korde, L.A.; Allott, K.; Reding, S.; Whittington, D.; Reding, K.W. Sex Hormones in Women With Elevated Breast Cancer Risk Undergoing Weight Loss. West. J. Nurs Res. 2019, 41, 1602–1622. [Google Scholar] [CrossRef]
- Plottel, C.S.; Blaser, M.J. Microbiome and Malignancy. Cell Host Microbe 2011, 10, 324–335. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Parida, S.; Sharma, D. The Microbiome-Estrogen Connection and Breast Cancer Risk. Cells 2019, 8, 1642. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Landete, J.M.; Arqués, J.; Medina, M.; Gaya, P.; de Las Rivas, B.; Muñoz, R. Bioactivation of Phytoestrogens: Intestinal Bacteria and Health. Crit. Rev. Food Sci. Nutr. 2016, 56, 1826–1843. [Google Scholar] [CrossRef]
- Fuhrman, B.J.; Feigelson, H.S.; Flores, R.; Gail, M.H.; Xu, X.; Ravel, J.; Goedert, J.J. Associations of the fecal microbiome with urinary estrogens and estrogen metabolites in postmenopausal women. J. Clin. Endocrinol. Metab 2014, 99, 4632–4640. [Google Scholar] [CrossRef] [PubMed]
- Fuhrman, B.J.; Schairer, C.; Gail, M.H.; Boyd-Morin, J.; Xu, X.; Sue, L.Y.; Buys, S.S.; Isaacs, C.; Keefer, L.K.; Veenstra, T.D.; et al. Estrogen metabolism and risk of breast cancer in postmenopausal women. J. Natl. Cancer Inst. 2012, 104, 326–339. [Google Scholar] [CrossRef] [PubMed]
- Flores, R.; Shi, J.; Fuhrman, B.; Xu, X.; Veenstra, T.D.; Gail, M.H.; Gajer, P.; Ravel, J.; Goedert, J.J. Fecal microbial determinants of fecal and systemic estrogens and estrogen metabolites: A cross-sectional study. J. Transl. Med. 2012, 10, 253. [Google Scholar] [CrossRef] [Green Version]
- Goedert, J.J.; Jones, G.; Hua, X.; Xu, X.; Yu, G.; Flores, R.; Falk, R.T.; Gail, M.H.; Shi, J.; Ravel, J.; et al. Investigation of the association between the fecal microbiota and breast cancer in postmenopausal women: A population-based case-control pilot study. J. Natl. Cancer Inst. 2015, 107, djv147. [Google Scholar] [CrossRef]
- Shao, J.; Wu, L.; Leng, W.-D.; Fang, C.; Zhu, Y.-J.; Jin, Y.-H.; Zeng, X.-T. Periodontal Disease and Breast Cancer: A Meta-Analysis of 1,73,162 Participants. Front. Oncol. 2018, 8, 601. [Google Scholar] [CrossRef] [Green Version]
- Freudenheim, J.L.; Genco, R.J.; LaMonte, M.J.; Millen, A.E.; Hovey, K.M.; Mai, X.; Nwizu, N.; Andrews, C.A.; Wactawski-Wende, J. Periodontal Disease and Breast Cancer: Prospective Cohort Study of Postmenopausal Women. Cancer Epidemiol. Biomark. Prev. 2016, 25, 43–50. [Google Scholar] [CrossRef] [Green Version]
- Mai, X.; LaMonte, M.J.; Hovey, K.M.; Freudenheim, J.L.; Andrews, C.A.; Genco, R.J.; Wactawski-Wende, J. Periodontal disease severity and cancer risk in postmenopausal women: The Buffalo OsteoPerio Study. Cancer Causes Control. 2016, 27, 217–228. [Google Scholar] [CrossRef] [Green Version]
- Nwizu, N.N.; Marshall, J.R.; Moysich, K.; Genco, R.J.; Hovey, K.M.; Mai, X.; LaMonte, M.J.; Freudenheim, J.L.; Wactawski-Wende, J. Periodontal Disease and Incident Cancer Risk among Postmenopausal Women: Results from the Women’s Health Initiative Observational Cohort. Cancer Epidemiol. Biomark. Prev. 2017, 26, 1255–1265. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Camila, S.; SfreddoDizdar, O.; Hayran, M.; Guven, D.C.; Yılmaz, T.B.; Taheri, S.; Akman, A.C.; Bilgin, E.; Hüseyin, B.; Berker, E. Increased cancer risk in patients with periodontitis. Curr. Med. Res. Opin. 2017, 33, 2195–2200. [Google Scholar]
- Sfreddo, C.S.; Maier, J.; De David, S.C.; Susin, C.; Moreira, C.H.C. Periodontitis and breast cancer: A case-control study. Community Dent. Oral Epidemiol. 2017, 45, 545–551. [Google Scholar] [CrossRef] [PubMed]
- Miller, M.B.; Bassler, B.L. Quorum Sensing in Bacteria. Annu. Rev. Microbiol. 2001, 55, 165–199. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Urbaniak, C.; Gloor, G.; Brackstone, M.; Tangney, M.; Reid, G. The microbiota of breast tissue and its association with tumours. Appl. Environ. Microbiol. 2016, 82, 5039–5048. [Google Scholar] [CrossRef] [Green Version]
- Cardona, F.; Andrés-Lacueva, C.; Tulipani, S.; Tinahones, F.J.; Queipo-Ortuño, M.I. Benefits of polyphenols on gut microbiota and implications in human health. J. Nutr. Biochem. 2013, 24, 1415–1422. [Google Scholar] [CrossRef] [Green Version]
- Kang, J.D.; Myers, C.J.; Harris, S.C.; Kakiyama, G.; Lee, I.-K.; Yun, B.-S.; Matsuzaki, K.; Furukawa, M.; Min, H.-K.; Bajaj, J.S.; et al. Bile Acid 7α-Dehydroxylating Gut Bacteria Secrete Antibiotics that Inhibit Clostridium difficile: Role of Secondary Bile Acids. Cell Chem. Biol. 2019, 26, 27–34.e24. [Google Scholar] [CrossRef] [Green Version]
- Ndeh, D.; Rogowski, A.; Cartmell, A.; Luis, A.S.; Baslé, A.; Gray, J.; Venditto, I.; Briggs, J.; Zhang, X.; Labourel, A.; et al. Complex pectin metabolism by gut bacteria reveals novel catalytic functions. Nature 2017, 544, 65–70. [Google Scholar] [CrossRef] [PubMed]
- Pan, W.-H.; Sommer, F.; Falk-Paulsen, M.; Ulas, T.; Best, P.; Fazio, A.; Kachroo, P.; Luzius, A.; Jentzsch, M.; Rehman, A.; et al. Exposure to the gut microbiota drives distinct methylome and transcriptome changes in intestinal epithelial cells during postnatal development. Genome Med. 2018, 10, 27. [Google Scholar] [CrossRef] [PubMed]
- Flint, H.J.; Scott, K.P.; Duncan, S.H.; Louis, P.; Forano, E. Microbial degradation of complex carbohydrates in the gut. Gut Microbes 2012, 3, 289–306. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hill, M.J. The role of colon anaerobes in the metabolism of bile acids and steroids, and its relation to colon cancer. Cancer 1975, 36, 2387–2400. [Google Scholar] [CrossRef]
- LeBlanc, J.G.; Milani, C.; de Giori, G.S.; Sesma, F.; van Sinderen, D.; Ventura, M. Bacteria as vitamin suppliers to their host: A gut microbiota perspective. Curr. Opin. Biotechnol. 2013, 24, 160–168. [Google Scholar] [CrossRef]
- Rowland, I.; Gibson, G.; Heinken, A.; Scott, K.; Swann, J.; Thiele, I.; Tuohy, K. Gut microbiota functions: Metabolism of nutrients and other food components. Eur. J. Nutr. 2018, 57, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Feng, X.; Li, Y.; Brobbey Oppong, M.; Qiu, F. Insights into the intestinal bacterial metabolism of flavonoids and the bioactivities of their microbe-derived ring cleavage metabolites. Drug Metab. Rev. 2018, 50, 343–356. [Google Scholar] [CrossRef]
- Tailford, L.E.; Crost, E.H.; Kavanaugh, D.; Juge, N. Mucin glycan foraging in the human gut microbiome. Front. Genet. 2015, 6, 81. [Google Scholar] [CrossRef] [Green Version]
- Geller, L.T.; Barzily-Rokni, M.; Danino, T.; Jonas, O.H.; Shental, N.; Nejman, D.; Gavert, N.; Zwang, Y.; Cooper, Z.A.; Shee, K.; et al. Potential role of intratumor bacteria in mediating tumor resistance to the chemotherapeutic drug gemcitabine. Science 2017, 357, 1156–1160. [Google Scholar] [CrossRef] [Green Version]
- Urdaneta, V.; Casadesús, J. Interactions between Bacteria and Bile Salts in the Gastrointestinal and Hepatobiliary Tracts. Front. Med. 2017, 4, 163. [Google Scholar] [CrossRef]
- Laparra, J.M.; Sanz, Y. Interactions of gut microbiota with functional food components and nutraceuticals. Pharmacol. Res. 2010, 61, 219–225. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Madsen, L.; Myrmel, L.S.; Fjære, E.; Liaset, B.; Kristiansen, K. Links between Dietary Protein Sources, the Gut Microbiota, and Obesity. Front. Physiol. 2017, 8, 1047. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Van Duynhoven, J.; Vaughan, E.E.; Jacobs, D.M.; Kemperman, A.R.; van Velzen, E.J.J.; Gross, G.; Roger, L.C.; Possemiers, S.; Smilde, A.K.; Doré, J.; et al. Metabolic fate of polyphenols in the human superorganism. Proc. Natl. Acad. Sci. USA 2011, 108, 4531–4538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zipperer, A.; Konnerth, M.C.; Laux, C.; Berscheid, A.; Janek, D.; Weidenmaier, C.; Burian, M.; Schilling, N.A.; Slavetinsky, C.; Marschal, M.; et al. Human commensals producing a novel antibiotic impair pathogen colonization. Nature 2016, 535, 511. [Google Scholar] [CrossRef] [PubMed]
- Mamo, G. Anaerobes as Sources of Bioactive Compounds and Health Promoting Tools. In Anaerobes in Biotechnology; Hatti-Kaul, R., Mamo, G., Mattiasson, B., Eds.; Springer International Publishing: Cham, Switzerland, 2016; pp. 433–464. [Google Scholar]
- Furusawa, Y.; Obata, Y.; Fukuda, S.; Endo, T.A.; Nakato, G.; Takahashi, D.; Nakanishi, Y.; Uetake, C.; Kato, K.; Kato, T.; et al. Commensal microbe-derived butyrate induces the differentiation of colonic regulatory T cells. Nature 2013, 504, 446. [Google Scholar] [CrossRef] [PubMed]
- Smith, P.M.; Howitt, M.R.; Panikov, N.; Michaud, M.; Gallini, C.A.; Bohlooly-Y, M.; Glickman, J.N.; Garrett, W.S. The Microbial Metabolites, Short-Chain Fatty Acids, Regulate Colonic Treg Cell Homeostasis. Science 2013, 341, 569–573. [Google Scholar] [CrossRef] [Green Version]
- Shibata, N.; Kunisawa, J.; Kiyono, H. Dietary and Microbial Metabolites in the Regulation of Host Immunity. Front. Microbiol. 2017, 8, 2171. [Google Scholar] [CrossRef]
- Levy, M.; Thaiss, C.A.; Elinav, E. Metabolites: Messengers between the microbiota and the immune system. Genes Dev. 2016, 30, 1589–1597. [Google Scholar] [CrossRef]
- Postler, T.S.; Ghosh, S. Understanding the Holobiont: How Microbial Metabolites Affect Human Health and Shape the Immune System. Cell Metab. 2017, 26, 110–130. [Google Scholar] [CrossRef] [Green Version]
- Thirunavukkarasan, M.; Wang, C.; Rao, A.; Hind, T.; Teo, Y.R.; Siddiquee, A.A.-M.; Goghari, M.A.I.; Kumar, A.P.; Herr, D.R. Short-chain fatty acid receptors inhibit invasive phenotypes in breast cancer cells. PLoS ONE 2017, 12, e0186334. [Google Scholar] [CrossRef]
- Elangovan, S.; Pathania, R.; Ramachandran, S.; Ananth, S.; Padia, R.N.; Lan, L.; Singh, N.; Martin, P.M.; Hawthorn, L.; Prasad, P.D.; et al. The Niacin/Butyrate Receptor GPR109A Suppresses Mammary Tumorigenesis by Inhibiting Cell Survival. Cancer Res. 2014, 74, 1166–1178. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schug, Z.T.; Vande Voorde, J.; Gottlieb, E. The metabolic fate of acetate in cancer. Nat. Rev. Cancer 2016, 16, 708. [Google Scholar] [CrossRef] [PubMed]
- Puccetti, P.; Fallarino, F.; Italiano, A.; Soubeyran, I.; MacGrogan, G.; Debled, M.; Velasco, V.; Bodet, D.; Eimer, S.; Veldhoen, M.; et al. Accumulation of an Endogenous Tryptophan-Derived Metabolite in Colorectal and Breast Cancers. PLoS ONE 2015, 10, e0122046. [Google Scholar] [CrossRef]
- Dewi, D.L.; Mohapatra, S.R.; Blanco Cabañes, S.; Adam, I.; Somarribas Patterson, L.F.; Berdel, B.; Kahloon, M.; Thürmann, L.; Loth, S.; Heilmann, K.; et al. Suppression of indoleamine-2,3-dioxygenase 1 expression by promoter hypermethylation in ER-positive breast cancer. OncoImmunology 2017, 6, e1274477. [Google Scholar] [CrossRef] [Green Version]
- Cao, Z.-G.; Qin, X.-B.; Liu, F.-F.; Zhou, L.-L. Tryptophan-induced pathogenesis of breast cancer. Afr. Health Sci. 2015, 15, 982–985. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bekki, K.; Vogel, H.; Li, W.; Ito, T.; Sweeney, C.; Haarmann-Stemmann, T.; Matsumura, F.; Vogel, C.F.A. The aryl hydrocarbon receptor (AhR) mediates resistance to apoptosis induced in breast cancer cells. Pestic. Biochem. Physiol. 2015, 120, 5–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Venkatesh, M.; Mukherjee, S.; Wang, H.; Li, H.; Sun, K.; Benechet, A.P.; Qiu, Z.; Maher, L.; Redinbo, M.R.; Phillips, R.S.; et al. Symbiotic bacterial metabolites regulate gastrointestinal barrier function via the xenobiotic sensor PXR and Toll-like receptor 4. Immunity 2014, 41, 296–310. [Google Scholar] [CrossRef] [Green Version]
- Verma, S.; Tabb, M.M.; Blumberg, B. Activation of the steroid and xenobiotic receptor, SXR, induces apoptosis in breast cancer cells. BMC Cancer 2009, 9, 3. [Google Scholar] [CrossRef] [Green Version]
- Pondugula, S.R.; Pavek, P.; Mani, S. Pregnane X Receptor and Cancer: Context-Specificity is Key. Nucl Recept. Res. 2016, 3. [Google Scholar] [CrossRef] [Green Version]
- Mikó, E.; Kovács, T.; Sebő, É.; Tóth, J.; Csonka, T.; Ujlaki, G.; Sipos, A.; Szabó, J.; Méhes, G.; Bai, P. Microbiome-Microbial Metabolome-Cancer Cell Interactions in Breast Cancer-Familiar, but Unexplored. Cells 2019, 8, 293. [Google Scholar] [CrossRef] [Green Version]
- Kovács, T.; Mikó, E.; Vida, A.; Sebő, É.; Toth, J.; Csonka, T.; Boratkó, A.; Ujlaki, G.; Lente, G.; Kovács, P.; et al. Cadaverine, a metabolite of the microbiome, reduces breast cancer aggressiveness through trace amino acid receptors. Sci. Rep. 2019, 9, 1300. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Mikó, E.; Vida, A.; Kovács, T.; Ujlaki, G.; Trencsényi, G.; Márton, J.; Sári, Z.; Kovács, P.; Boratkó, A.; Hujber, Z.; et al. Lithocholic acid, a bacterial metabolite reduces breast cancer cell proliferation and aggressiveness. Biochim. Et Biophys. Acta. (BBA)—Bioenerg. 2018, 1859, 958–974. [Google Scholar] [CrossRef] [PubMed]
- Kovács, P.; Csonka, T.; Kovács, T.; Sári, Z.; Ujlaki, G.; Sipos, A.; Karányi, Z.; Szeőcs, D.; Hegedűs, C.; Uray, K.; et al. Lithocholic Acid, a Metabolite of the Microbiome, Increases Oxidative Stress in Breast Cancer. Cancers 2019, 11, 1255. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Krishnamurthy, K.; Wang, G.; Rokhfeld, D.; Bieberich, E. Deoxycholate promotes survival of breast cancer cells by reducing the level of pro-apoptotic ceramide. Breast Cancer Res.: BCR 2008, 10, R106. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Koppel, N.; Maini Rekdal, V.; Balskus, E.P. Chemical transformation of xenobiotics by the human gut microbiota. Science 2017, 356, eaag2770. [Google Scholar] [CrossRef]
- Ren, D.; Li, L.; Schwabacher, A.W.; Young, J.W.; Beitz, D.C. Mechanism of cholesterol reduction to coprostanol by Eubacterium coprostanoligenes ATCC 51222. Steroids 1996, 61, 33–40. [Google Scholar] [CrossRef]
- Erdman, S.E.; Poutahidis, T. Gut bacteria and cancer. Biochim. Et Biophys. Acta. (BBA)—Rev. Cancer 2015, 1856, 86–90. [Google Scholar] [CrossRef] [Green Version]
- Moss, S.F.; Blaser, M.J. Mechanisms of Disease: Inflammation and the origins of cancer. Nat. Clin. Pract. Oncol. 2005, 2, 90–97. [Google Scholar] [CrossRef]
- Shapira, I.; Sultan, K.; Lee, A.; Taioli, E. Evolving concepts: How diet and the intestinal microbiome act as modulators of breast malignancy. ISRN Oncol. 2013, 2013, 693920. [Google Scholar] [CrossRef]
- Letran, S.E.; Le, S.J.; Atif, S.M.; Flores-Langarica, A.; Uematsu, S.; Akira, S.; Cunningham, A.F.; McSorley, S.J. TLR5-Deficient Mice Lack Basal Inflammatory and Metabolic Defects but Exhibit Impaired CD4 T Cell Responses to a Flagellated Pathogen. J. Immunol. 2011, 186, 5406–5412. [Google Scholar]
- Rao, V.P.; Poutahidis, T.; Fox, J.G.; Erdman, S.E. Breast Cancer: Should Gastrointestinal Bacteria Be on Our Radar Screen? Cancer Res. 2007, 67, 847–850. [Google Scholar] [CrossRef] [Green Version]
- Erdman SE, P.T.; Tomczak, M.; Rogers, A.B.; Cormier, K.; Plank, B.; Horwitz, B.H.; Fox, J.G. CD4+ CD25+ regulatory T lymphocytes inhibit microbially induced colon cancer in Rag2-deficient mice. Am. J. Pathol. 2003, 162, 691–702. [Google Scholar] [CrossRef] [Green Version]
- Jiang, X.; Shapiro, D.J. The immune system and inflammation in breast cancer. Mol. Cell. Endocrinol. 2014, 382, 673–682. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Margolis, K.L.; Rodabough, R.J.; Thomson, C.A.; Lopez, A.M.; McTiernan, A.; Group, W.s.H.I.R. Prospective Study of Leukocyte Count as a Predictor of Incident Breast, Colorectal, Endometrial, and Lung Cancer and Mortality in Postmenopausal Women. JAMA Intern. Med. 2007, 167, 1837–1844. [Google Scholar] [CrossRef]
- Azab, B.; Bhatt, V.R.; Phookan, J.; Murukutla, S.; Kohn, N.; Terjanian, T.; Widmann, W.D. Usefulness of the Neutrophil-to-Lymphocyte Ratio in Predicting Short- and Long-Term Mortality in Breast Cancer Patients. Ann. Surg. Oncol. 2012, 19, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Gritzapis, A.D.; Voutsas, I.F.; Lekka, E.; Tsavaris, N.; Missitzis, I.; Sotiropoulou, P.; Perez, S.; Papamichail, M.; Baxevanis, C.N. Identification of a Novel Immunogenic HLA-A*0201-Binding Epitope of HER-2/neu with Potent Antitumor Properties. J. Immunol. 2008, 181, 146–154. [Google Scholar] [CrossRef] [Green Version]
- Franchi, L.; Warner, N.; Viani, K.; Nuñez, G. Function of Nod-like receptors in microbial recognition and host defense. Immunol. Rev. 2009, 227, 106–128. [Google Scholar] [CrossRef] [Green Version]
- Roy, S.; Trinchieri, G. Microbiota: A key orchestrator of cancer therapy. Nat. Rev. Cancer 2017, 17, 271. [Google Scholar] [CrossRef]
- Maier, I.; Berry, D.M.; Schiestl, R.H. Intestinal Microbiota Reduces Genotoxic Endpoints Induced By High-Energy Protons. Radiat. Res. 2014, 181, 45–53. [Google Scholar] [CrossRef]
- Alexander, J.L.; Wilson, I.D.; Teare, J.; Marchesi, J.R.; Nicholson, J.K.; Kinross, J.M. Gut microbiota modulation of chemotherapy efficacy and toxicity. Nat. Rev. Gastroenterol. Hepatol. 2017, 14, 356. [Google Scholar] [CrossRef]
- Wallace, B.D.; Roberts, A.B.; Pollet, R.M.; Ingle, J.D.; Biernat, K.A.; Pellock, S.J.; Venkatesh, M.; Guthrie, L.; O’Neal, S.K.; Robinson, S.J.; et al. Structure and Inhibition of Microbiome β-Glucuronidases Essential to the Alleviation of Cancer Drug Toxicity. Chem. Biol. 2015, 22, 1238–1249. [Google Scholar]
- Chang, C.-W.; Liu, C.-Y.; Lee, H.-C.; Huang, Y.-H.; Li, L.-H.; Chiau, J.-S.C.; Wang, T.-E.; Chu, C.-H.; Shih, S.-C.; Tsai, T.-H.; et al. Lactobacillus casei Variety rhamnosus Probiotic Preventively Attenuates 5-Fluorouracil/Oxaliplatin-Induced Intestinal Injury in a Syngeneic Colorectal Cancer Model. Front. Microbiol. 2018, 9, 983. [Google Scholar] [CrossRef] [PubMed]
- Mu, Q.; Tavella, V.J.; Luo, X.M. Role of Lactobacillus reuteri in Human Health and Diseases. Front. Microbiol. 2018, 9, 757. [Google Scholar] [CrossRef] [PubMed]
- Motevaseli, E.; Dianatpour, A.; Ghafouri-Fard, S. The Role of Probiotics in Cancer Treatment: Emphasis on their In Vivo and In Vitro Anti-metastatic Effects. Int. J. Mol. Cell. Med. 2017, 6, 66–76. [Google Scholar] [PubMed]
- Ranjbar, S.; Seyednejad, S.A.; Azimi, H.; Rezaeizadeh, H.; Rahimi, R. Emerging Roles of Probiotics in Prevention and Treatment of Breast Cancer: A Comprehensive Review of Their Therapeutic Potential. Nutr. Cancer 2019, 71, 1–12. [Google Scholar] [CrossRef]
- Mendoza, L. Potential effect of probiotics in the treatment of breast cancer. Oncol. Rev. 2019, 13, 422. [Google Scholar] [CrossRef] [Green Version]
- Kevin, M.; Green, F.K.; Elnabawi, A.; Parveen, S.; Kwame Nyame, A.; Ishaque, A.B. Impact of Lactobacillus Reuteri DSM 20016 on breast cancer proliferation. In Proceedings of the Thirteenth International Symposium on Metal. Ions in Biology & Medicine Eleventh International Symposium on Recent Advances in Environmental Health Research, Jackson, MS, USA, 22 July 2014. [Google Scholar]
- Poutahidis, T.; Varian, B.J.; Levkovich, T.; Lakritz, J.R.; Mirabal, S.; Kwok, C.; Ibrahim, Y.M.; Kearney, S.M.; Chatzigiagkos, A.; Alm, E.J.; et al. Dietary microbes modulate transgenerational cancer risk. Cancer Res. 2015, 75, 1197–1204. [Google Scholar] [CrossRef] [Green Version]
- Aragón, F.; Carino, S.; Perdigón, G.; de Moreno de LeBlanc, A. Inhibition of Growth and Metastasis of Breast Cancer in Mice by Milk Fermented With Lactobacillus casei CRL 431. J. Immunother. 2015, 38, 185–196. [Google Scholar] [CrossRef]
- Hassan, Z.; Mustafa, S.; Rahim, R.A.; Isa, N.M. Anti-breast cancer effects of live, heat-killed and cytoplasmic fractions of Enterococcus faecalis and Staphylococcus hominis isolated from human breast milk. In Vitro Cell Dev. Biol. Anim. 2016, 52, 337–348. [Google Scholar] [CrossRef]
- Esfandiary, A.; Taherian-Esfahani, Z.; Abedin-Do, A.; Mirfakhraie, R.; Shirzad, M.; Ghafouri-Fard, S.; Motevaseli, E. Lactobacilli Modulate Hypoxia-Inducible Factor (HIF)-1 Regulatory Pathway in Triple Negative Breast Cancer Cell Line. Cell J. 2016, 18, 237–244. [Google Scholar]


| Biological Process | Microbial Species | Reference | ||
|---|---|---|---|---|
| Carbohydrate metabolism | ||||
| Parent carbohydrate | Metabolite | Most gut organisms Eubacterium hallii Roseburia Coprococcus catus Megasphera elsdenii Veillonella spp Ruminococcus obeum Bacteroidetes Eubacterium rectale Eubacterium hallii Roseburia spp Coprococcus spp Faecalibacterium prausnitzii Bifidobacterium spp Lactobacillus spp Methanobrevibacter smithii Desulfovibrio spp | [159,161,163,168] [22] | |
| Polysaccharides Oligosaccharides Resistant starch Mucins | Acetate Propionate Butyrate Lactate CH4 H2S | |||
| Fat metabolism | Eubacterium siraeum Roseburia faecis Roseburia intestinalis F. prausnitzii Bifidobacterium Propionibacterium Lactobacillus spp Bifidobacterium breve Propionibacterium freudenreichii Roseburia inulinivorans Roseburia hominis Butyrivibrio fibrisolvens Clostridia Peptostreptococci Peptococci | [27] | ||
Linoleic acid Conjugated lineloic acids | ||||
Stearic acid | ||||
| Fatty acids Short chain fatty acids(SCFA) Branched chain fatty acid(BCFA) (Isobutyrate from valine; 2-methylbutyrate from isoleucine; Isovalerate from leucine) | ||||
| Protein metabolism | Akkermansia Bifidobacterium | [166,172] | ||
| Bile metabolism | Cholic acid Chenodeoxycholic acid Deoxycholic acid Lithocholic acid Ursodeoxycholic acid | Bacteroides Bifidobacterium Clostridium Lactobacillus Listeria Clostridium Eubacterium Bacteroides Clostridium Egghertella Peptostreptococcus Ruminococcus Eubacterium | [170] | |
| Vitamin synthesis | Biotin Cobalamin Folate Nicotinic acid Panthothenic acid Pyridoxine Riboflavin Thiamine | Bacteroidetes (Most important) Fusobacteria Proteobacteria Firmicutes Actinobacteria | [165,166] | |
| Phytochemical metabolism | ||||
| Phytochemical Flavonoids Lignans Ferulic acid Isofllavones Caretenoids Glucosinolates Isothiocyanates Stilbens Organosulfur compounds | Process Deglycosylation Ring fission Dehydroxylation | Clostridium spp. Eubacterium ramulus Lactobacillus mucosae Enterococcus faecium Finegoldia magna Veillonella sp Slakia isoflavoniconvertens Slakia equolifaciens Adlercreutzia equolifaciens | [166,171,173] | |
| Antibiotics | Staphylococcus lugdunensis Pantoea agglomerans P. Vagans Clostridium sp. | [172,173] | ||
| Lugdunin Herbicolin I Andrimid Polykitide synthase | ||||
| Bacteriocins | Lactococcus lactis Butyrivibrio fibrisolvens Pedicoccus acidilactici Lactobacillus sakei Lactobacillus acidophillus Enterococcus faecium Lactobacillus helveticus Enterococcus sp. Lactobacillus plantarum | [85,174,175] | ||
| Nicin Lacticin 3147 Butyrivibriocin Pediocin Sakacin A Lactacin F Lactococcin G Enterolysin Helvecticin I Enterocin I F4-9 Glycocin F | ||||
| Bug | Animal Model | Mode of Action | Reference |
|---|---|---|---|
| Lactobacillus reuteri | FVB strain Her2 | Inhibits mammary tumorigenesis by CD4+CD25+ lymphocyte stimulation | [13] |
| Swiss mice | |||
| Lactobacillus casei CRL431 | 4T1 syngeneic breast cancer model in Balb/C mice | Inhibits tumor growth, vascularization and lung metastasis in mice by reducing infiltration of macrophages into the tumor and; enhances CD4+ and CD8+ antitumor immune response | [220] |
| Lactobacillus casei | 4T1 syngeneic breast cancer model in Balb/C mice | Reduces breast cancer incidence | [216] |
| Shirota | |||
| Lactobacillus acidophilus | 4T1 syngeneic breast cancer model in Balb/C mice | Improves survival of breast tumor bearing mice | [20] |
| Increased immune cell proliferation | |||
| Increased IFNγ production | |||
| Decreased IL4 production | |||
| Lactobacillus brevis | 4T1 syngeneic breast cancer model in Balb/C mice | Inhibits metastasis to vital organs | [215] |
| Improves NK cell activity | |||
| Increased IFN-γ | |||
| Increased IL-17 levels |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Parida, S.; Sharma, D. Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights. Cells 2020, 9, 1091. https://doi.org/10.3390/cells9051091
Parida S, Sharma D. Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights. Cells. 2020; 9(5):1091. https://doi.org/10.3390/cells9051091
Chicago/Turabian StyleParida, Sheetal, and Dipali Sharma. 2020. "Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights" Cells 9, no. 5: 1091. https://doi.org/10.3390/cells9051091
APA StyleParida, S., & Sharma, D. (2020). Microbial Alterations and Risk Factors of Breast Cancer: Connections and Mechanistic Insights. Cells, 9(5), 1091. https://doi.org/10.3390/cells9051091





