Targeting PI3K and AMPKα Signaling Alone or in Combination to Enhance Radiosensitivity of Triple Negative Breast Cancer
Abstract
1. Introduction
2. Materials and Methods
2.1. Materials
2.2. Cell Culture
2.3. siRNA Transfection
2.4. Radiation Treatment
2.5. Analysis of Glycolytic Rate
2.6. Immunoblotting
2.7. Cell Counting Assay
2.8. Sulforhodamine B (SRB) Assay
2.9. Colony Formation Assay
2.10. Flow Cytometry
2.11. Immunohistochemistry
2.12. Statistical Analysis
3. Results
3.1. Analysis of AMPKα1 and AMPKα2 Expression in TNBC Patient Samples, Cell Lines and PDXs
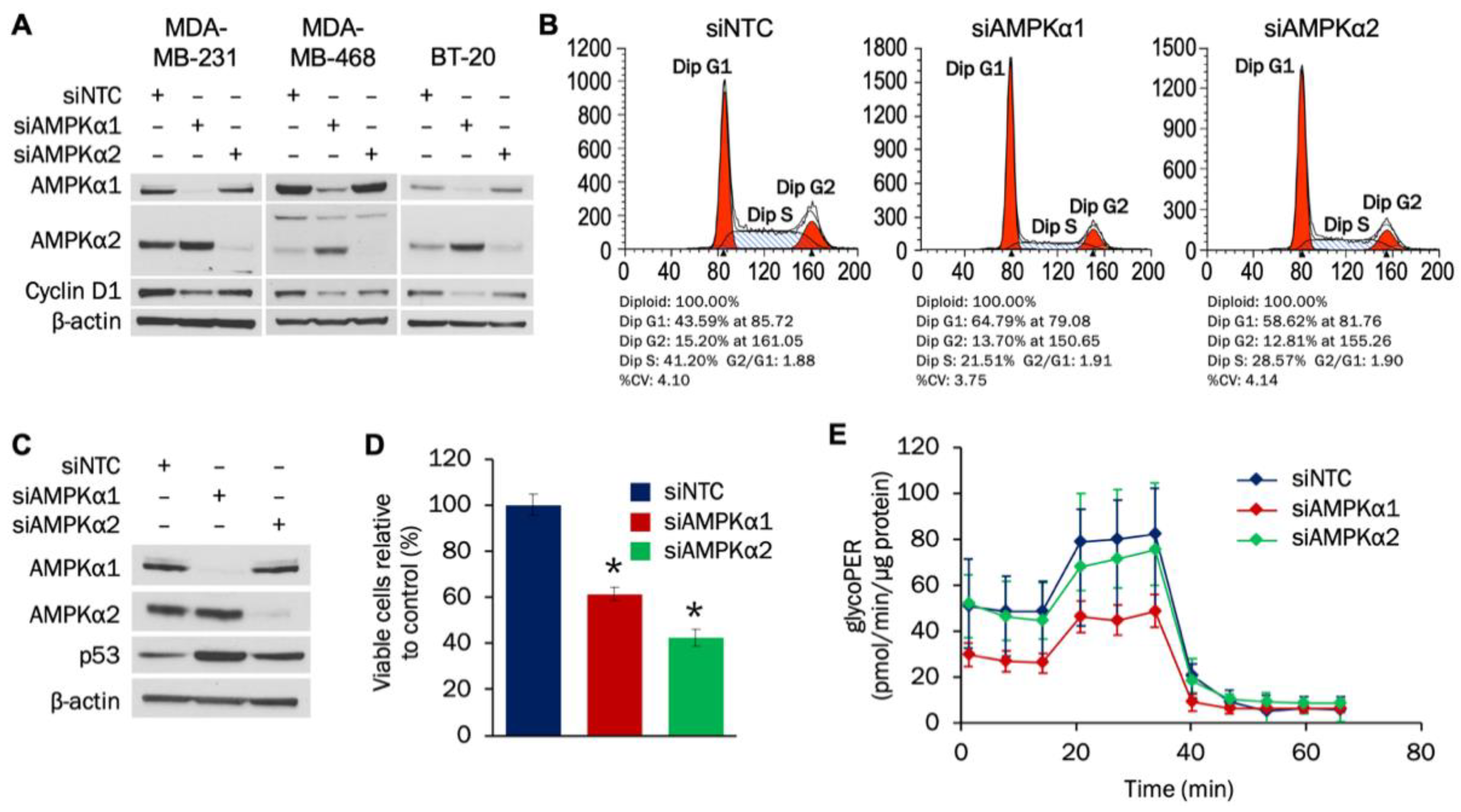
3.2. AMPKα1 and AMPKα2 Promote TNBC Proliferation and Cell Cycle Progression
3.3. Knockdown of Akt1 or p110α Induces Apoptosis and Reduces Proliferation in MDA-MB-231 Cells
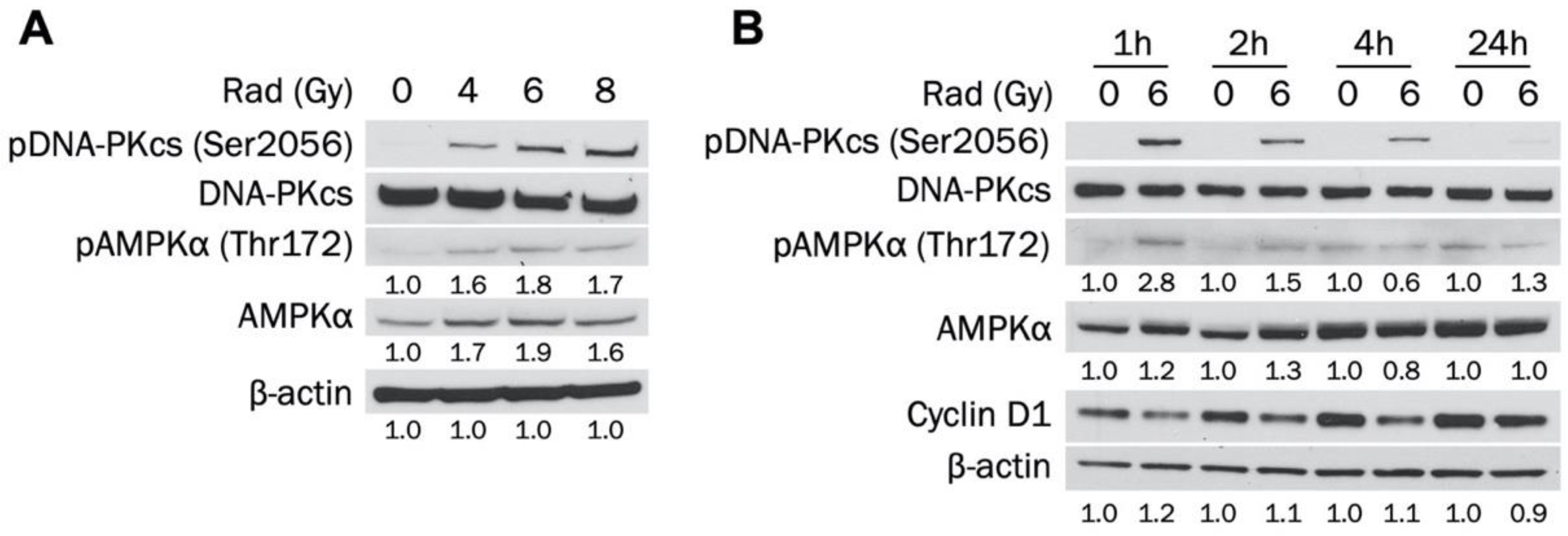
3.4. Radiotherapy Increases Activated and Total AMPKα in MDA-MB-231 Cells
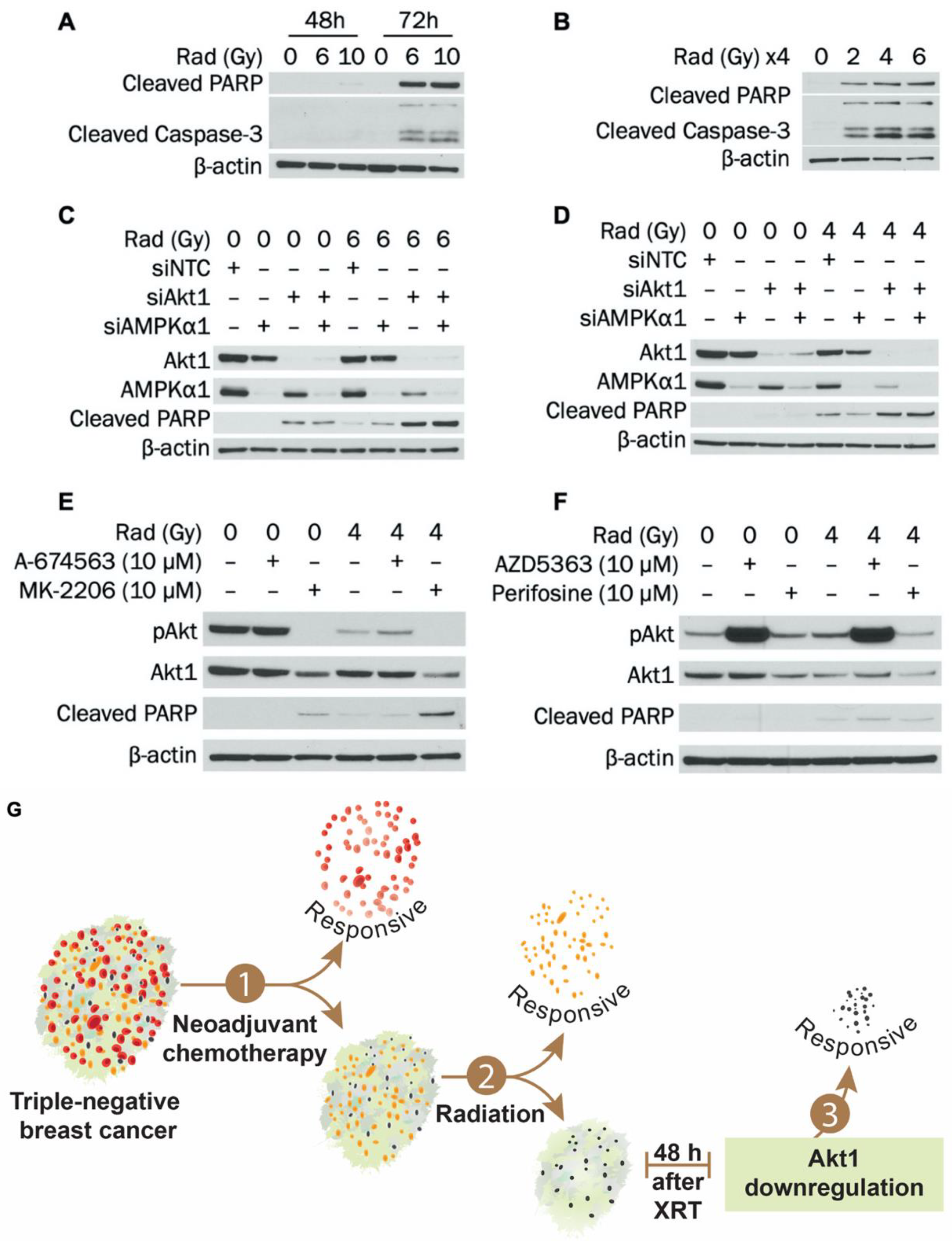
3.5. Akt1 Knockdown Potentiates Radiation-Induced Apoptosis and Further Suppresses Survival after Radiation in MDA-MB-231 Cells
3.6. MK-2206 Treatment Enhances Radiation-Induced Apoptosis in MDA-MB-231 Cells
4. Discussion
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Marotti, J.D.; De Abreu, F.B.; Wells, W.A.; Tsongalis, G.J. Triple-Negative Breast Cancer. Am. J. Pathol. 2017, 187, 2133–2138. [Google Scholar] [CrossRef] [PubMed]
- Andreopoulou, E.; Schweber, S.J.; Sparano, J.A.; McDaid, H.M. Therapies for triple negative breast cancer. Expert Opin. Pharmacother. 2015, 16, 983–998. [Google Scholar] [CrossRef]
- Elia, I.; Schmieder, R.; Christen, S.; Fendt, S.-M. Organ-Specific Cancer Metabolism and Its Potential for Therapy. Pharmacol. Ther. Cough 2015, 233, 321–353. [Google Scholar] [CrossRef]
- Lehmann, B.D.; Jovanović, B.; Chen, X.; Estrada, M.V.; Johnson, K.N.; Shyr, Y.; Moses, H.L.; Sanders, M.E.; Pietenpol, J.A. Refinement of Triple-Negative Breast Cancer Molecular Subtypes: Implications for Neoadjuvant Chemotherapy Selection. PLoS ONE 2016, 11, e0157368. [Google Scholar] [CrossRef] [PubMed]
- Jerusalem, G.; Collignon, J.; Schroeder, H.; Lousberg, L. Triple-negative breast cancer: Treatment challenges and solutions. Breast Cancer Targets Ther. 2016, 8, 93–107. [Google Scholar] [CrossRef]
- Kindts, I.; Buelens, P.; Laenen, A.; Van Limbergen, E.; Janssen, H.; Wildiers, H.; Weltens, C. Omitting radiation therapy in women with triple-negative breast cancer leads to worse breast cancer-specific survival. Breast 2017, 32, 18–25. [Google Scholar] [CrossRef] [PubMed]
- Yao, Y.; Chu, Y.; Xu, B.; Hu, Q.; Song, Q. Radiotherapy after surgery has significant survival benefits for patients with triple-negative breast cancer. Cancer Med. 2019, 8, 554–563. [Google Scholar] [CrossRef]
- Tang, L.; Wei, F.; Wu, Y.; He, Y.; Shi, L.; Xiong, F.; Gong, Z.; Guo, C.; Li, X.; Deng, H.; et al. Role of metabolism in cancer cell radioresistance and radiosensitization methods. J. Exp. Clin. Cancer Res. 2018, 37, 87. [Google Scholar] [CrossRef]
- Baskar, R.; Dai, J.; Wenlong, N.; Yeo, R.; Yeoh, K.-W. Biological response of cancer cells to radiation treatment. Front. Mol. Biosci. 2014, 1. [Google Scholar] [CrossRef]
- Maier, P.; Hartmann, L.; Wenz, F.; Herskind, C. Cellular Pathways in Response to Ionizing Radiation and Their Targetability for Tumor Radiosensitization. Int. J. Mol. Sci. 2016, 17, 102. [Google Scholar] [CrossRef]
- Saha, M.; Kumar, S.; Bukhari, S.; Balaji, S.A.; Kumar, P.; Hindupur, S.K.; Rangarajan, A. AMPK-Akt Double-Negative Feedback Loop in Breast Cancer Cells Regulates Their Adaptation to Matrix Deprivation. Cancer Res. 2018, 78, 1497–1510. [Google Scholar] [CrossRef]
- Lage, R.; Diéguez, C.; Vidal-Puig, A.; López, M. AMPK: A metabolic gauge regulating whole-body energy homeostasis. Trends Mol. Med. 2008, 14, 539–549. [Google Scholar] [CrossRef] [PubMed]
- Luo, Z.; Zang, M.; Guo, W. AMPK as a metabolic tumor suppressor: Control of metabolism and cell growth. Future Oncol. 2010, 6, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Hardie, D.G.; Ross, F.A.; Hawley, S.A. AMP-activated protein kinase: A target for drugs both ancient and modern. Chem. Boil. 2012, 19, 1222–1236. [Google Scholar] [CrossRef]
- Hardie, D.G. AMP-activated protein kinase: A key regulator of energy balance with many roles in human disease. J. Intern. Med. 2014, 276, 543–559. [Google Scholar] [CrossRef] [PubMed]
- Faubert, B.; Vincent, E.E.; Poffenberger, M.C.; Jones, R.G. The AMP-activated protein kinase (AMPK) and cancer: Many faces of a metabolic regulator. Cancer Lett. 2015, 356, 165–170. [Google Scholar] [CrossRef]
- Jeon, S.-M. Regulation and function of AMPK in physiology and diseases. Exp. Mol. Med. 2016, 48, e245. [Google Scholar] [CrossRef]
- Ross, F.A.; Mackintosh, C.; Hardie, D.G. AMP-activated protein kinase: A cellular energy sensor that comes in 12 flavours. FEBS J. 2016, 283, 2987–3001. [Google Scholar] [CrossRef]
- Garcia, D.; Shaw, R.J. AMPK: Mechanisms of Cellular Energy Sensing and Restoration of Metabolic Balance. Mol. Cell 2017, 66, 789–800. [Google Scholar] [CrossRef]
- Zannella, V.E.; Cojocari, D.; Hilgendorf, S.; Vellanki, R.N.; Chung, S.; Wouters, B.G.; Koritzinsky, M. AMPK regulates metabolism and survival in response to ionizing radiation. Radiother. Oncol. 2011, 99, 293–299. [Google Scholar] [CrossRef]
- Chen, Z.-T.; Zhao, W.; Qu, S.; Li, L.; Lu, X.-D.; Su, F.; Liang, Z.-G.; Guo, S.-Y.; Zhu, X.-D. PARP-1 promotes autophagy via the AMPK/mTOR pathway in CNE-2 human nasopharyngeal carcinoma cells following ionizing radiation, while inhibition of autophagy contributes to the radiation sensitization of CNE-2 cells. Mol. Med. Rep. 2015, 12, 1868–1876. [Google Scholar] [CrossRef] [PubMed]
- He, Q.; Li, J.; Dong, F.; Cai, C.; Zou, X. LKB1 promotes radioresistance in esophageal cancer cells exposed to radiation, by suppression of apoptosis and activation of autophagy via the AMPK pathway. Mol. Med. Rep. 2017, 16, 2205–2210. [Google Scholar] [CrossRef] [PubMed]
- Jin, H.; Gao, S.; Guo, H.; Ren, S.; Ji, F.; Liu, Z.; Chen, X. Re-sensitization of radiation resistant colorectal cancer cells to radiation through inhibition of AMPK pathway. Oncol. Lett. 2016, 11, 3197–3201. [Google Scholar] [CrossRef] [PubMed]
- Fruman, D.; Chiu, H.; Hopkins, B.D.; Bagrodia, S.; Cantley, L.C.; Abraham, R.T. The PI3K Pathway in Human Disease. Cell 2017, 170, 605–635. [Google Scholar] [CrossRef]
- Martini, M.; De Santis, M.C.; Braccini, L.; Gulluni, F.; Hirsch, E. PI3K/AKT signaling pathway and cancer: An updated review. Ann. Med. 2014, 46, 372–383. [Google Scholar] [CrossRef]
- Jean, S.; Kiger, A.A. Classes of phosphoinositide 3-kinases at a glance. J. Cell Sci. 2014, 127, 923–928. [Google Scholar] [CrossRef]
- Cohen, M.M. The AKT genes and their roles in various disorders. Am. J. Med. Genet. Part A 2013, 161, 2931–2937. [Google Scholar] [CrossRef]
- Wang, Q.; Chen, X.; Hay, N. Akt as a target for cancer therapy: More is not always better (lessons from studies in mice). Br. J. Cancer 2017, 117, 159–163. [Google Scholar] [CrossRef]
- Kriegsmann, M.; Endris, V.; Wolf, T.; Pfarr, N.; Stenzinger, A.; Loibl, S.; Denkert, C.; Schneeweiss, A.; Budczies, J.; Sinn, H.-P.; et al. Mutational profiles in triple-negative breast cancer defined by ultradeep multigene sequencing show high rates of PI3K pathway alterations and clinically relevant entity subgroup specific differences. Oncotarget 2014, 5, 9952–9965. [Google Scholar] [CrossRef]
- Rocca, P.A.C.; Orrù, S.; Muroni, M.R.; Sanges, F.; Sotgiu, G.; Ena, S.; Pira, G.; Murgia, L.; Manca, A.; Uras, M.G.; et al. Analysis of PIK3CA Mutations and Activation Pathways in Triple Negative Breast Cancer. PLoS ONE 2015, 10, e0141763. [Google Scholar] [CrossRef]
- Söderlund, K.; Pérez-Tenorio, G.; Stål, O. Activation of the phosphatidylinositol 3-kinase/Akt pathway prevents radiation-induced apoptosis in breast cancer cells. Int. J. Oncol. 2005, 26, 25–32. [Google Scholar] [CrossRef] [PubMed]
- Albert, J.M.; Kim, K.W.; Cao, C.; Lu, B. Targeting the Akt/mammalian target of rapamycin pathway for radiosensitization of breast cancer. Mol. Cancer Ther. 2006, 5, 1183–1189. [Google Scholar] [CrossRef] [PubMed]
- Li, P.; Zhang, Q.; Torossian, A.; Li, Z.-B.; Xu, W.-C.; Lu, B.; Fu, S. Simultaneous Inhibition of EGFR and PI3K Enhances Radiosensitivity in Human Breast Cancer. Int. J. Radiat. Oncol. 2012, 83, e391–e397. [Google Scholar] [CrossRef] [PubMed]
- Kuger, S.; Cörek, E.; Polat, B.; Kämmerer, U.; Flentje, M.; Djuzenova, C.S. Novel PI3K and mTOR Inhibitor NVP-BEZ235 Radiosensitizes Breast Cancer Cell Lines under Normoxic and Hypoxic Conditions. Breast Cancer Basic Clin. Res. 2014, 8, 39–49. [Google Scholar] [CrossRef]
- Fatehi, D.; Soltani, A.; Ghatrehsamani, M. SRT1720, a potential sensitizer for radiotherapy and cytotoxicity effects of NVB-BEZ235 in metastatic breast cancer cells. Pathol. Res. Pr. 2018, 214, 889–895. [Google Scholar] [CrossRef]
- Steelman, L.S.; Navolanic, P.; Chappell, W.H.; Abrams, S.L.; Wong, E.W.; Martelli, A.M.; Cocco, L.; Stivala, F.; Libra, M.; Nicoletti, F.; et al. Involvement of Akt and mTOR in chemotherapeutic and hormonal-based drug resistance and response to radiation in breast cancer cells. Cell Cycle 2011, 10, 3003–3015. [Google Scholar] [CrossRef]
- Chen, Q.; Molloy, J.; Izumi, T.; Sterpin, E. Impact of backscatter material thickness on the depth dose of orthovoltage irradiators for radiobiology research. Phys. Med. Boil. 2019, 64, 055001. [Google Scholar] [CrossRef]
- Romero, N.; Swain, P.; Dranka, B. Characterization of Glycolysis with a Panel of Common Cellular Models Using Agilent Seahorse XF Technology; Agilent Technologies: Santa Clara, CA, USA, 2018. [Google Scholar]
- Bao, Y.; Mukai, K.; Hishiki, T.; Kubo, A.; Ohmura, M.; Sugiura, Y.; Matsuura, T.; Nagahata, Y.; Hayakawa, N.; Yamamoto, T.; et al. Energy Management by Enhanced Glycolysis in G1-phase in Human Colon Cancer Cells In Vitro and In Vivo. Mol. Cancer Res. 2013, 11, 973–985. [Google Scholar] [CrossRef]
- Laderoute, K.R.; Calaoagan, J.M.; Chao, W.-R.; Dinh, D.; Denko, N.; Duellman, S.; Kalra, J.; Liu, X.; Papandreou, I.; Sambucetti, L.; et al. 5′-AMP-activated Protein Kinase (AMPK) Supports the Growth of Aggressive Experimental Human Breast Cancer Tumors. J. Boil. Chem. 2014, 289, 22850–22864. [Google Scholar] [CrossRef]
- Liu, P.; Ye, F.; Xie, X.; Li, X.; Tang, H.; Li, S.; Huang, X.; Song, C.; Wei, W.; Xie, X. mir-101-3p is a key regulator of tumor metabolism in triple negative breast cancer targeting AMPK. Oncotarget 2016, 7, 35188–35198. [Google Scholar] [CrossRef]
- Salt, I.; Celler, J.W.; Hawley, S.A.; Prescott, A.; Woods, A.; Carling, D.; Hardie, D.G. AMP-activated protein kinase: Greater AMP dependence, and preferential nuclear localization, of complexes containing the α2 isoform. Biochem. J. 1998, 334, 177–187. [Google Scholar] [CrossRef] [PubMed]
- Fox, M.M.; Phoenix, K.N.; Kopsiaftis, S.G.; Claffey, K.P. AMP-Activated Protein Kinase α 2 Isoform Suppression in Primary Breast Cancer Alters AMPK Growth Control and Apoptotic Signaling. Genes Cancer 2013, 4, 3–14. [Google Scholar] [CrossRef] [PubMed]
- He, G.; Zhang, Y.-W.; Lee, J.-H.; Zeng, S.X.; Wang, Y.V.; Luo, Z.; Dong, X.C.; Viollet, B.; Wahl, G.M.; Lu, H. AMP-Activated Protein Kinase Induces p53 by Phosphorylating MDMX and Inhibiting Its Activity. Mol. Cell. Boil. 2013, 34, 148–157. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.-H.; Hsu, E.-C.; Guh, J.-H.; Yang, H.-C.; Wang, D.; Kulp, S.K.; Shapiro, C.L.; Chen, C.-S. Targeting Energy Metabolic and Oncogenic Signaling Pathways in Triple-negative Breast Cancer by a Novel Adenosine Monophosphate-activated Protein Kinase (AMPK) Activator. J. Boil. Chem. 2011, 286, 39247–39258. [Google Scholar] [CrossRef]
- Sanli, T.; Rashid, A.; Liu, C.; Harding, S.; Bristow, R.; Cutz, J.-C.; Singh, G.; Wright, J.; Tsakiridis, T. Ionizing Radiation Activates AMP-Activated Kinase (AMPK): A Target for Radiosensitization of Human Cancer Cells. Int. J. Radiat. Oncol. 2010, 78, 221–229. [Google Scholar] [CrossRef]
- Song, C.W.; Lee, H.; Dings, R.; Williams, B.; Powers, J.; Dos Santos, T.; Choi, B.-H.; Park, H.J. Metformin kills and radiosensitizes cancer cells and preferentially kills cancer stem cells. Sci. Rep. 2012, 2. [Google Scholar] [CrossRef]
- Feng, T.; Li, L.; Ling, S.; Fan, N.; Fang, M.; Zhang, H.; Fang, X.; Lan, W.; Hou, Z.; Meng, Q.; et al. Metformin enhances radiation response of ECa109 cells through activation of ATM and AMPK. Biomed. Pharmacother. 2015, 69, 260–266. [Google Scholar] [CrossRef]
- Sanli, T.; Steinberg, G.R.; Singh, G.; Tsakiridis, T. AMP-activated protein kinase (AMPK) beyond metabolism: A novel genomic stress sensor participating in the DNA damage response pathway. Cancer Boil. Ther. 2013, 15, 156–169. [Google Scholar] [CrossRef]
- Green, B.D.; Jabbour, A.; Sandow, J.J.; Riffkin, C.D.; Masouras, D.; Daunt, C.P.; Salmanidis, M.; Brumatti, G.; Hemmings, B.A.; Guthridge, M.A.; et al. Akt1 is the principal Akt isoform regulating apoptosis in limiting cytokine concentrations. Cell Death Differ. 2013, 20, 1341–1349. [Google Scholar] [CrossRef]
- Hinz, N.; Jücker, M. Distinct functions of AKT isoforms in breast cancer: A comprehensive review. Cell Commun. Signal. 2019, 17, 154. [Google Scholar] [CrossRef]


© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Johnson, J.; Chow, Z.; Napier, D.; Lee, E.; Weiss, H.L.; Evers, B.M.; Rychahou, P. Targeting PI3K and AMPKα Signaling Alone or in Combination to Enhance Radiosensitivity of Triple Negative Breast Cancer. Cells 2020, 9, 1253. https://doi.org/10.3390/cells9051253
Johnson J, Chow Z, Napier D, Lee E, Weiss HL, Evers BM, Rychahou P. Targeting PI3K and AMPKα Signaling Alone or in Combination to Enhance Radiosensitivity of Triple Negative Breast Cancer. Cells. 2020; 9(5):1253. https://doi.org/10.3390/cells9051253
Chicago/Turabian StyleJohnson, Jeremy, Zeta Chow, Dana Napier, Eun Lee, Heidi L. Weiss, B. Mark Evers, and Piotr Rychahou. 2020. "Targeting PI3K and AMPKα Signaling Alone or in Combination to Enhance Radiosensitivity of Triple Negative Breast Cancer" Cells 9, no. 5: 1253. https://doi.org/10.3390/cells9051253
APA StyleJohnson, J., Chow, Z., Napier, D., Lee, E., Weiss, H. L., Evers, B. M., & Rychahou, P. (2020). Targeting PI3K and AMPKα Signaling Alone or in Combination to Enhance Radiosensitivity of Triple Negative Breast Cancer. Cells, 9(5), 1253. https://doi.org/10.3390/cells9051253





